Abstract
Listeria monocytogenes is the etiological factor of listeriosis. The main source of these organisms is food, including dairy products. The aim was to determine the multiple correlations between the drug susceptibility, virulence genes (VGs), and biofilm formation on silicone teat cups of milk-borne and human L. monocytogenes strains. The spread of L. monocytogenes via contaminated teat rubbers was assessed. The L. monocytogenes strains recovered from milk (18), human blood (10), and the reference strain ATCC®19111™ were used in the study. Penicillin resistance was the most prevalent resistance in the milk isolates (n=8; 44.4%), whereas among clinical strains erythromycin resistance was predominating – (n=6; 60%). The most frequent VGs among strains isolated from milk were hlyA (100%) and plcB (100%) whereas in strains isolated from blood – hlyA (100%) and prfA (90%). All tested VGs were present in 50% of blood isolates and 11% of milk-borne strains. The strains isolated from milk formed a significantly stronger biofilm. The strains with more numerous virulence genes were resistant to more antibiotics and formed a stronger biofilm. It was shown that contaminated teat cups might contribute to the transmission of L. monocytogenes in the herd. It seems reasonable to monitor the occurrence of L. monocytogenes biofilm in a dairy processing environment.
Key words: Listeria monocytogenes, biofilm formation, drug susceptibility, bacteria transmission, virulence genes frequency
Introduction
Listeria monocytogenes is Gram-positive, facultatively anaerobic, non-spore forming bacteria. L. monocytogenes causes listeriosis and is one of the most dangerous food-borne zoonotic pathogens (Schlech and Acheson 2000). Listeriosis mainly occurs in immunocompromised people, neonates, pregnant women, the elderly, and AIDS patients (Lawley et al. 2008; Jamali et al. 2013; Doijad et al. 2015). The major source of human infection is food contaminated with the pathogen, including raw milk and milk products. A recent trend to purchase fresh, unprocessed products from local suppliers has contributed to an increase in the consumption of unpasteurized milk and products thereof (Seremak-Bulge et al. 2013). Such products may be the source of pathogenic bacteria and lead to serious infections (Jamali and Radmehr 2013; Gould et al. 2014).
According to the European Food Safety Authority (EFSA) report in 2017 all tested milk samples included in the RTE (ready-to-eat) products category were in line with Food Safety Criteria (FSC). One of 85 samples (1.2%) from two member countries, in the category “Raw cow’s milk for direct consumption” in the retail trade, was positive for L. monocytogenes (EFSA 2018). In Poland and other European countries, the number of infections caused by this microorganism is constantly increasing. In 2017 2480 cases of listeriosis were recorded in the EU countries, while in 2013 only 1763 cases were found. The incidence rate per 100 000 inhabitants was 0.26 and 0.32, respectively, in 2016 and 2017 (EFSA 2018). In 2018 124 cases of listeriosis were reported in Poland that is a 20% increase when compared to 2015 (NIZP-PZH 2019). Listeriosis associated with raw milk consumption was reported in 2014 in two patients from California and Florida. The most likely source of Listeria spp. was raw chocolate milk (CDC 2016). In Los Angeles County (California) during an outbreak associated with the consumption of the contaminated cheese, the death of 48 out of 142 patients was recorded (Linnan et al. 1988). Another listeriosis outbreaks linked to the contaminated cheese took place in Germany in 2006–2007 (Koch et al. 2010), and in Spain in 2008–2009 (Jackson et al. 2011) where 189 and 8 cases were reported, respectively.
Despite the improvement in the hygiene of the production process, L. monocytogenes poses still a serious problem in the food processing plants, including dairies. Such environment favors biofilm formation and bacterial survival (Latorre et al. 2010). Since L. monocytogenes multiplies easily and quickly on improperly cleaned dairy appliances, biofilm formation starts within 20 minutes after bacterial contact with the surface (Hayes et al. 1986; Weiler et al. 2013). In dairy plants, L. monocytogenes colonizes processing surfaces, floors, equipment, and niches that are difficult to clean (e.g. hard to reach cavities), and becomes the potential source of milk and milk product contamination (Unnerstad et al. 1996; Latorre et al. 2010). Bacteria forming biofilm survive longer and are more resistant to disinfectants and mechanical washing (Frank and Koffi 1990; Walker et al. 1990; Meyer-Broseta et al. 2003). Therefore, it is of great importance to prevent biofilm formation on milking equipment to reduce the risk of milk contamination and, in consequence, human infections (Latorre et al. 2010).
Although most L. monocytogenes strains are susceptible to antimicrobial agents used in human and animal medicine, multidrug-resistant strains are increasingly frequently isolated. This is mainly due to the overuse of antibiotics in animal husbandry (Jamali et al. 2013). Also, several virulence factors enable L. monocytogenes to infect and spread in the host (Park et al. 2012). L. monocytogenes possesses many virulence genes responsible for the invasion of host cells (inlA, inlB, and iap), phagosomal escape (hlyA, plcA, and plcB) and cell to cell spread (actA) (Hamon et al. 2006).
This study aimed was to determine the multiple correlations between drug susceptibility, virulence genes and biofilm formation on silicone teat cups from milking machines of milk-borne and human L. monocytogenes strains. The spread of L. monocytogenes through contaminated teat rubbers was also assessed.
Experimental
Materials and Methods
Materials. Milk samples, obtained in Poland from cows without clinical signs of mastitis, were collected in 2015. For each cow, a sample of 100 ml from all the four teats was collected to one sterile container. Of 380 milk samples, 21 (5.5%) were positive for L. monocytogenes. Ten genetically different strains (a genetic similarity previously determined for the diagnostic reasons) isolated from the blood of patients by dr. A. Jurasz (University Hospital No. 1 in Bydgoszcz, Poland) were used to evaluate their drug susceptibility and ability to form biofilms. There was no epidemiological link between milk and blood isolate groups. The reference strain L. monocytogenes ATCC® 19111™ was included in the study. This strain is widely used as the reference strain in many studies, including biofilm formation assessment.
Isolation of L. monocytogenes from milk. Analysis of the intermediate and finished product samples was carried out following the ISO 11290-1 (ISO 2017). To isolate L. monocytogenes, 25 ml of milk was added to 225 ml of half-Fraser broth (Merck, Poland) and incubated for 24 h at 30°C. Then 0.1 ml of the culture was transferred into 10 ml of Fraser broth (Merck, Poland) and the secondary selective enrichment was performed at 37°C for 48 h. Finally, the culture was plated onto ALOA agar (ChromoCult Listeria Selective Agar®, Merck, Poland), and OXFORD Agar (Oxoid, United Kingdom), and incubated for 24 h at 37°C.
Identification of the strains isolated from milk. Initial species identification was performed based on morphological traits on the ALOA agar (Merck, Poland). The typical colony of L. monocytogenes is a turquoise and blue one surrounded with a turbidity zone.
Then, multiplex PCR was performed. For the genus Listeria identification primers (L1, L2) (Oligo.pl, Poland) based on the 16S rRNA sequence were used (Border et al. 1990) whereas primers (LM1, LM2) based on the sequence of the hly gene allowed the species identification (Bansal 1996).
DNA was isolated using the Genomic Mini AX Bacteria Spin kit (A&A Biotechnology, Poland), according to the manufacturer’s instructions. Amplification was performed in the mixture of 25 μl, containing: 1 × PCR buffer (Promega, United States), 2.0 mM MgCl2 (ABO, Poland), 1.25 mmol dNTPs (Promega, United States), 0.5 μM of each of the primers (Oligo.pl, Poland), 1.0 U Taq DNA polymerase (Promega, United States), ultrapure water (Merck, Poland) and 2.0 μl DNA. The amplification conditions and primer sequences are presented in Table I. L. monocytogenes strain ATCC 19111 was used as the reference strain. PCR products were separated on 1.5% agarose gel for 75 min at 80V and stained with a Midori Green dye (NIPPON Genetics EUROPE GmbH, Germany).
Table I.
Primers used for identification of L. monocytogenes.
| Primer Primer sequence (5' - 3') | Size of PCR product (bp) | Amplification conditions |
|---|---|---|
| L1 CAG CAG CCG CGG TAA TAC | 938 | Initial denaturation - 94°C/2 min 30 cycles: denaturation - 94°C/30 s annealing, - 50°C/30 s elongation - 72°C/1 min final elongation - 72°C/5 min |
| L2 CTC CAT AAA GGT GAC CCT | ||
| LM1 CCT AAG ACG CCA ATC GAA | 750 | |
| LM2 AAG CAC TTG CAA CTG CTC |
Determination of genetic relatedness of L. monocytogenes strains isolated from milk. All L. monocytogenes isolates were genotyped using Random Amplification of Polymorphic DNA (RAPD) with the nonspecific primer OPA-11 (5’-CA AT CG CC GT-3’) (Ozbey et al. 2006). Reactions were carried out in a mixture of 25 μl, containing: 1 × PCR buffer with 2.0 mM MgCl2 (Promega, United States), 1.5 mM MgCl2 (ABO, Poland), 200 μM dNTPs (Promega, United States), 1.0 μM primer OPA-11 (Oligo.pl, Poland), 1.25 U Taq DNA polymerase (Promega, United States), ultrapure water (Merck, Poland), and 3.0 μl DNA. The reaction consisted of six cycles of initial stage: denaturation (94°C/1 min), annealing (30°C/2 min) and elongation (72°C/1 min), followed by 35 cycles of: denaturation (94°C/15 s), annealing (37°C/1 min) and elongation (72°C/45 s), and the final elongation (72°C/10 min). PCR products were separated on 1.5% agarose gel for 150 min at a voltage of 80 V, and stained with a Midori Green dye (NIPPON Genetics EUROPE GmbH, Germany).
To determine the degree of genetic relationship between the isolates, the phylogenetic dendrogram was drawn in the program Phoretix 1D Pro (TotalLab). Data clustering was performed using the UPGMA hierarchic grouping technique with the Dice coefficient.
The frequency of the genes encoding virulence factors in milk and blood isolates. L. monocytogenes strains from milk (18) and blood (10) were examined for the multiplex PCR reactions. The study included the following virulence genes: actA (actin assembly-inducing protein), hlyA (listeriolysin O), iap (extracellular protein p60), inlA (internalin A), inlB (internalin B), plcA (phosphatidylinositol-specific phospholipase C), plcB (phosphatidylcholine-specific phospholipase C), and prfA (positive regulatory factor PrfA).
Two multiplex-PCR reactions were optimized to detect these virulence genes. The first included the detection of iap, hlyA, inlB, and plcB, and the second one – actA, inlA, plcA,and prfA genes. The reaction mixture (25 μl) contained: 1 × PCR buffer with 2.0 mM MgCl2 (Promega, United States), 6.0 mM MgCl2(ABO, Poland), 1.0 mM dNTPs (Promega, United States), 1.0 μM of each primer (Oligo.pl, Poland) (Table II), 3.0 U Taq DNA polymerase (Promega, United States), ultrapure water, and 3.0 μl DNA. The course of PCR was as follows: the initial denaturation (95°C/2 min), 35 cycles of denaturation (95°C/15 s), annealing (60°C/30 s) and elongation (72°C/1,5 min) and the final elongation (72°C/10 min). The L. monocytogenes strain IW41 was used as the reference strain. PCR products were separated on 1.5% agarose gel and stained with a Midori Green dye (NIPPON Genetics EUROPE GmbH, Germany). The Perfect 100 bp DNA Ladder (EurX, Poland) was used.
Table II.
Primers used for the detection of virulence genes in Listeria monocytogenes strains (Franciosa et al. 2005; Rawool et al. 2007).
| Primer | Primer sequence (5’ - 3’) | Size of PCR product (bp) |
|---|---|---|
| actA - F | CGC CGC GGA AAT TAA AAA AAG A | 839 |
| actA - R | ACG AAG GAA CCG GGC TGC TAG | |
| hlyA - F | GCA GTT GCA AGC GCT TGG AGT GAA | 456 |
| hlyA - R | GCA ACG TAT CCT CCA GAG TGA TCG | |
| iap - F | ACA AGC TGC ACC TGT TGC AG | 131 |
| iap - R | TGA CAG CGT GTG TAG TAG CA | |
| inlA - F | CAG GCA GCT ACA ATT ACA CA | 2 341 |
| inlA - R | ATA TAG TCC GAA AAC CAC ATC T | |
| inlB - F | AGG AGA GGA TAG TGT GAA | 1 905 |
| inlB - R | TTA TTT CTG TGC CCT TAA | |
| plcA - F | CTG CTT GAG CGT TCA TGT CTC ATC CCC C | 1 484 |
| plcA - R | ATG GGT TTC ACT CTC CTT CTA C | |
| plcB - F | GCA AGT GTT CTA GTC TTT CCG G | 794 |
| plcB - R | ACC TGC CAA AGT TTG CTG TGA | |
| prfA - F | CAT GAA CGC TCA AGC AGA AG | 706 |
| prfA - R | AAT TTT CCC AAG TAG CAG GA |
Evaluation of drug susceptibility of L. monocytogenes strains. Drug susceptibility of 18 milk and ten blood isolates was determined using the disk diffusion method on the Mueller-Hinton agar with 5% defibrinated horse blood and β-NAD at a concentration of 20 mg/l (MH-F, bioMérieux, France). For each strain, a suspension of 0.5 McFarland’s scale (7.6 ×107 CFU/ml ±9.4×106 CFU/ml) in sterile saline was prepared. The disks with penicillin (1 U), ampicillin (2 μg), merope-nem (10 μg), erythromycin (15 μg) and cotrimoxazole (1.25–23.75 μg) (Becton Dickinson, United States) were used. Incubation of antibiograms was conducted in the atmosphere enriched in 5% CO2 at 35°C for 18 h. The results were interpreted according to the recommendations of EUCAST v. 8.0.
Evaluation of the biofilm formation ability of L. monocytogenes strains. Quantitative evaluation of biofilm formation by L. monocytogenes strains was performed on irradiated fragments of silicone teat cups from milking machines. The fragments of 1 × 1 cm were used in the study.
The intensity of biofilm formation was determined using a quantitative method by Kwiecińska-Piró (g et al. 2011) with some modifications. The study was conducted on one clinical strain, one strain isolated from milk, and the reference strain ATCC 19111. The sterile fragments of rubber (3 replications for each strain) were placed in tubes containing 3 ml of the bacterial suspensions in the Brain Heart Infusion Broth (BHI) (Merck, Poland) (0.5 of the McFarland scale). Incubation was conducted in the aerobic atmosphere at 37°C for 72 h, and the medium was replaced with a sterile one every 24 h. At each exchange of the medium, the rubber fragments were rinsed with sterile PBS (Phosphate Buffered Saline) (BTL, Poland). The fragments of rubbers incubated in the sterile BHI medium (Merck, Poland) were used as negative controls. After incubation, the samples were rinsed with PBS, placed in a tube containing 3 ml of PBS, and sonicated for 10 minutes (30 kHz, 150W) with the sonicator Ultrasonic DU-4 (Nickel-Electro Ltd.). Then, the samples were shaken for 10 minutes (400 rpm), and the serial 10-fold dilutions were prepared and inoculated on the Columbia Agar medium with 5% sheep blood (Becton Dickinson, United States). After 24-hours incubation at 37°C, the number of colonies per 1 cm2 of the fragment’s surface (CFU/cm-2) was calculated.
The intensity of biofilm formation by L. monocytogenes was observed under the confocal microscope. For this purpose, very thin rubber slides were prepared and the biofilms were grown, as described above. The biofilm-forming cells on the rubber slides were then stained with the LIVE/DEAD BacLight Bacterial Viability Kit (ThermoFisher Scientific, United States), according to the manufacturer’s instructions.
The spread of L. monocytogenes through contaminated teat rubbers. Transmission of L. monocytogenes on the skin of the udder and into the milk. The radiant sterilized, purified from fat, pieces of cow’s udder skin (1 × 1 cm) were used in the experiment. Biofilm formation on sterile teat rubbers was assessed as described above (section “Evaluation of biofilm formation ability of L. monocytogenes strains”). Two of the strongest and weakest biofilm-forming strains of L. monocytogenes derived from both milk and blood as well as the reference strain were selected for the study.
To evaluate the transmission of bacteria from the biofilm formed on the teat rubbers to the udder skin (according to our method), a piece of skin was rubbed with the contaminated rubber in two directions. This simulated the insertion and removal of the teat into the milking cup. For each strain, eight skin fragments were used in six replicates. After swabbing, each piece of skin (1–3 replicates) was placed in sterile PBS (BTL, Poland) and subjected to a 10-minute sonication. Subsequently, the serial 10-fold dilutions were made and plated onto Columbia Agar with 5% sheep blood (Becton Dickinson, United States). After 24 hours at 37°C, the number of bacteria in 1 cm2 of the udder skin was calculated. Also, to test the proliferation of L. monocytogenes on the skin, fragments (repeats 4–6) were left at 25°C for 12 hours. Subsequently, the samples were placed in sterile PBS, sonicated and plated onto Columbia Agar with 5% sheep blood (Becton Dickinson, United States).
To evaluate the transmission of L. monocytogenes from the biofilm formed on the teat rubbers to milk, each rubber was flushed with 100 ml of UHT milk. Then, 0.1 ml of milk was plated onto Columbia Agar medium with 5% sheep blood (Becton Dickinson, United States), incubated at 37°C for 24 hours, and the number of bacteria in 1 ml of milk was determined.
Transmission of L. monocytogenes in udder – teat rubber – udder model. Two of the strongest and weakest biofilm-forming strains of L. monocytogenes derived from milk and blood and the reference strain were selected for the study.
The bacterial suspensions in sterile PBS (BTL, Poland) (0.5 McF) were prepared and 50 μl of the suspension (six replicates) was poured on the sterile skin fragments. After drying at room temperature, the sterile teat rubber piece was rubbed in two directions with the contaminated skin. Then, the rubber pieces were placed (repeats 1–3) in sterile PBS (BTL, Poland) and sonicated. The number of bacteria per 1 cm2 of rubber was determined by plating the sample into Columbia Agar with 5% sheep blood (Becton Dickinson, United States), and incubation at 37°C for 24 hours. The remaining contaminated teat rubber fragments (repeats 4–6) were used to rub six sterile fragments of the udder skin. The number of bacteria transferred to them as well as proliferation on the udder skin was determined as described previously (“Transmission of L. monocytogenes on the skin of the udder and into the milk”).
Statistical analysis. The statistical analysis of the results was performed using the software Statistica 12 PL (StatSoft).
The frequency of the genes encoding virulence factors in L. monocytogenes strains isolated from milk and blood was established. Statistical analysis of the results was carried out using the chi-square test and a Fisher exact test, at the significance level α = 0.05. The virulence profiles of the strains tested from both groups were also determined.
The number of bacteria re-isolated from biofilm was averaged separately for both groups of strains and compared with each other and with the reference strain using the analysis of variance ANOVA and the post hoc Bonferroni test, at the significance level α = 0.05.
The multiple correlations between antibiotic resistance, virulence genes, and intensity of biofilm formation among clinical strains and milk isolates were tested. Single correlations between antibiotic resistance and prevalence of virulence genes, antibiotic resistance and intensity of biofilm formation, and incidence of virulence genes and the intensity biofilm formation were also evaluated. Correlation coefficients were evaluated according to Guilford’s scale.
Results
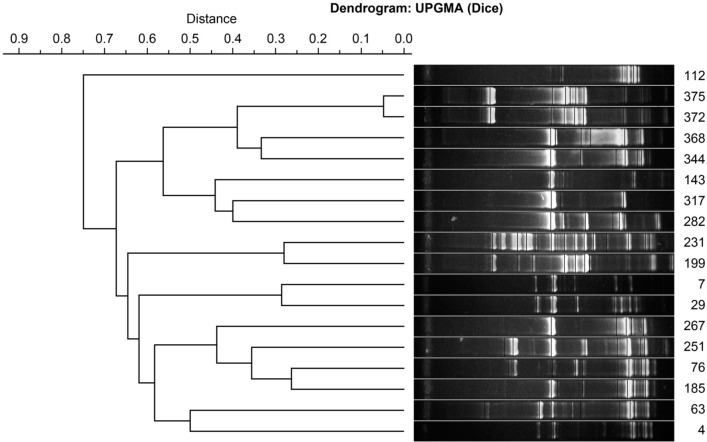
Of the 380 milk samples tested, 21 (5.5%) were positive for L. monocytogenes. Since four isolates were genetically identical, finally 18 unrelated genetically strains were subjected to evaluation of drug susceptibility and the ability to form a biofilm (Fig. 1).
Fig. 1.
Genetic similarity of tested Listeria monocytogenes strains.
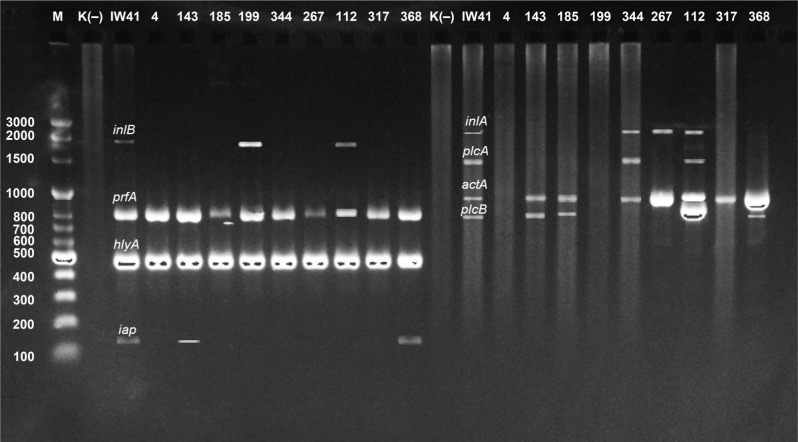
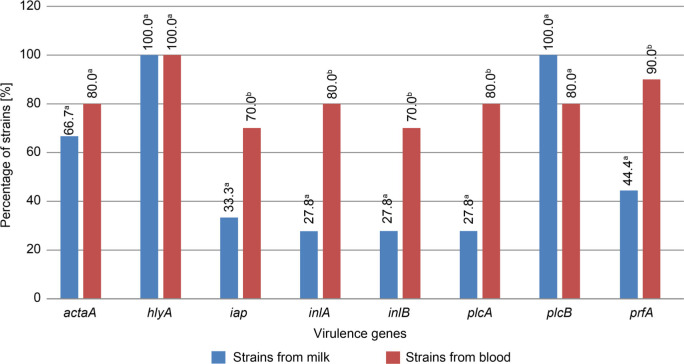
Assessment of the occurrence of virulence genes. The number of virulence genes varied in L. monocytogenes strains (Fig. 2). The most prevalent, found in all tested strains, was the hlyA gene. The plcB gene was found in all milk isolates, whereas the prfA gene was detected in 90.0% of blood strains (Fig. 3). The occurrence of the remaining genes ranged from 27.8% to 80% of the strains tested. The reference strain of L. monocytogenes ATCC 19111 had all virulence genes investigated. The occurrence of the genes iap, inlA, inlB, plcA, and prfA was significantly higher in the strains isolated from the blood than in milk strains (p ≤ 0.05).
Fig. 2.
Electrophoregram presenting the occurrence of virulence genes among the strains tested (M – marker, K+ – positive control (IW41), line 4, 143, 185, 199, 344, 267, 112, 317, 368 – L. monocytogenes strains from milk, K (−) – negative control).
Fig. 3.
Prevalence of virulence genes among L. monocytogenes strains isolated from milk and blood.
Among L. monocytogenes strains isolated from milk, 11 different profiles of virulence genes were detected. None of the profiles was represented by more than 2 strains (Table III). Six (33.3%) of L. monocytogenes strains from milk possessed three virulence genes. Two strains with all virulence genes investigated were identified. Among the L. monocytogenes isolates from the human blood six gene profiles were identified. The most frequent profile included all virulence genes tested (five strains, 50.0%) (Table III).
Table III.
The profiles of virulence genes in strains of L. monocytogenes isolated from milk and from the blood.
| Profile | Virulence genes | Number of strains | The strain identification number |
|---|---|---|---|
| Strains isolated from milk | |||
| I | hlyA, plcB | 2 | 4, 63 |
| II | hlyA, iap, plcB | 2 | 7, 29 |
| III | hlyA, inlB, plcB | 2 | 199, 231 |
| IV | actA, hlyA, plcB | 2 | 282, 317 |
| V | actA, hlyA, plcB, prfA | 2 | 76, 185 |
| VI | actA, hlyA, iap, plcB, prfA | 2 | 143,368 |
| VII | actA, hlyA, iap, inlA, inlB, plcA, plcB, prfA | 2 | 372, 375 |
| VIII | actA, hlyA, inlA, plcB | 1 | 267 |
| IX | actA, hlyA, inlA, plcA, plcB | 1 | 344 |
| X | actA, hlyA, plcA, plcB, prfA | 1 | 251 |
| XI | actA, hlyA, inlA, inlB, plcA, plcB, prfA | 1 | 112 |
| Strains isolated from the blood | |||
| I | actA, hlyA, iap, inlA, inlB, plcA, plcB, prfA | 5 | 1, 2, 5, 6, 9 |
| II | actA, hlyA, iap | 1 | 4 |
| III | actA, hlyA, iap, prfA | 1 | 10 |
| IV | hlyA, inlA, plcA, plcB, prfA | 1 | 3 |
| V | hlyA, inlA, inlB, plcA, plcB, prfA | 1 | 8 |
| VI | actA, hlyA, inlA, inlB, plcA, plcB, prfA | 1 | 7 |
Evaluation of drug susceptibility of the strains. It was found that strains derived from milk were the most frequently resistant to penicillin (8; 44.4%) and erythromycin (7; 38.9%). In turn, among strains isolated from blood, four (4, 40.0%) were resistant only to erythromycin, and another two strains (2, 20.0%) were resistant to erythromycin and meropenem (Table IV). Amongst all strains studied eight different profiles of drug susceptibility were found. Profile A was represented by 12 (42.86%) isolates susceptible to all the antibiotics tested, eight isolates (44.4%) from milk and four (40.0%) from the blood. We found three (16.7%) strains that were resistant only to penicillin (profile B), and five (17.86%) – resistant only to erythromycin (profile C). At the same time, two (11.1%) strains were resistant to two antibiotics; one of them to penicillin and erythromycin (profile D) and the other to erythromycin and cotrimoxazole (profile E). Further, one strain (5.6%) isolated from milk was susceptible only to erythromycin (profile F), and three (16.7%) strains isolated from milk were resistant to all antibiotics tested (profile G) (Table IV). In turn, among clinical strains of L. monocytogenes, the most prevalent was resistance to erythromycin (60.0%). We found also five isolates resistant only to erythromycin (profile C), one of them was acquired from milk and four were isolated from blood. Further, two (20.0%) strains isolated from blood were resistant to meropenem and erythromycin (profile H) (Table IV).
Table IV.
The profiles of drug resistance/susceptibility of strains of L. monocytogenes isolated from milk and the blood.
| Profile | Drug resistance/susceptibility | Number of strains | Total (n = 28) | |
|---|---|---|---|---|
| Strains from milk (n = 18) (the strain identification number) | Strains from the blood (n = 10) (the strain identification number) | |||
| A | R: - S: P, AM, MEM, E, SXT |
8 (44.44%) (7, 29, 63, 199, 231, 282, 317) |
4 (40.0%) (3, 4, 8, 10) |
12 (42.86%) |
| B | R: P S: AM, MEM, E, SXT |
3 (16.67%) (76, 185, 251) |
0 (0.0%) | 3 (10.71%) |
| C | R: E S: P, AM, MEM, SXT |
1 (5.56%) (267) |
4 (40.0%) (1, 2, 7, 9 ) |
5 (17.86%) |
| D | R: P, E S: AM, MEM, SXT |
1 (5.56%) (143) |
0 (0.0%) | 1 (3.57%) |
| E | R: E, SXT S: P, AM, MEM |
1 (5.56%) (344) |
0 (0.0%) | 1 (3.57%) |
| F | R: P, AM, MEM, SXT S: E |
1 (5.56%) (368) |
0 (0.0%) | 1 (3.57%) |
| G | R: P, AM, MEM, E, SXT S: - - - |
3 (16.67%) (112, 372, 375) |
0 (0.0%) | 3 (10.71%) |
| H | R: MEM, E S: P, AM, SXT |
0 (0. 0%) | 2 (20.0%) (5, 6) |
2 (7.14%) |
R - resistant
S - susceptible
P - penicillin, AM - ampicillin, MEM - meropenem, E - erythromycin, SXT - cotrimoxazole
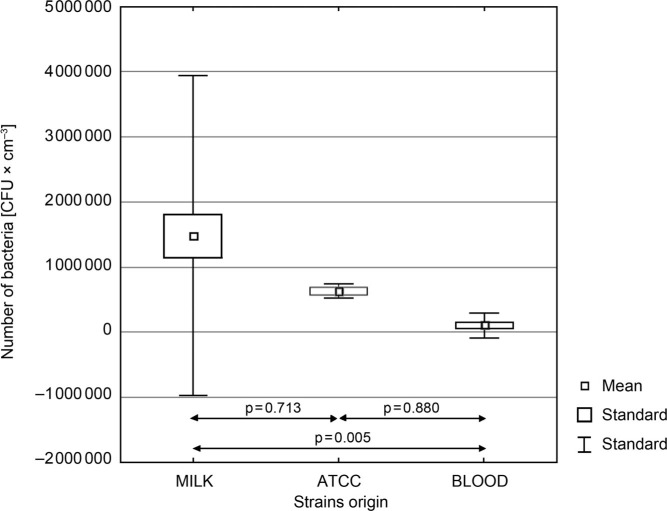
Evaluation of ability to form biofilm by L. monocytogenes strains. It was found that isolates derived from milk formed a biofilm with significantly higher efficiency than the clinical isolates from the blood (p = 0.005) (Fig. 4). The number of the cells recovered from biofilms formed on fragments of teat cup rubbers ranged from 4.64 to 6.87 log CFU/cm−2, and from 3.61 to 5.70 log CFU/cm−2 for the milk strains and the blood strains, respectively (Table V). Seven milk isolates developed more intense biofilms with a higher density of bacterial cell than the reference strain ATCC 19111 (5.80 log CFU/cm−2), whereas all clinical strains were weaker biofilm formers than the reference strain (Fig. 4).
Fig. 4.
Differences in biofilm formation among clinical L. monocytogenes strains and the strains isolated from milk.
Table V.
The intensity of biofilm formation by L. monocytogenes strains derived from cow’s milk and the blood.
| Strains origin | The strain identification number | Mean number of bacteria [log CFU/cm−2] | STD |
|---|---|---|---|
| Milk | 372 | 6.87 | ± 6.12* |
| Milk | 375 | 6.79 | ± 6.09 |
| Milk | 112 | 6.71 | ± 6.13 |
| Milk | 368 | 6.64 | ± 6.17 |
| Milk | 344 | 6.05 | ± 5.21 |
| Milk | 143 | 5.95 | ± 4.99 |
| Milk | 267 | 5.81 | ± 5.07 |
| Milk | 251 | 5.18 | ± 4.80 |
| Milk | 185 | 4.99 | ± 4.06 |
| Milk | 7m | 4.94 | ± 4.00 |
| Milk | 199 | 4.94 | ± 3.76 |
| Milk | 317 | 4.89 | ± 3.78 |
| Milk | 4m | 4.85 | ± 3.67 |
| Milk | 282 | 4.81 | ± 3.81 |
| Milk | 76 | 4.77 | ± 3.65 |
| Milk | 29 | 4.76 | ± 3.74 |
| Milk | 231 | 4.74 | ± 3.96 |
| Milk | 63 | 4.64 | ± 3.72 |
| ATCC | 19111 | 5.80 | ± 5.04 |
| Blood | 5 | 5.70 | ± 4.87 |
| Blood | 6 | 5.67 | ± 4.76 |
| Blood | 3 | 4.18 | ± 3.83 |
| Blood | 1 | 4.01 | ± 2.87 |
| Blood | 8 | 4.00 | ± 3.27 |
| Blood | 2 | 3.97 | ± 3.61 |
| Blood | 9 | 3.92 | ± 3.83 |
| Blood | 7 | 3.72 | ± 3.63 |
| Blood | 10 | 3.71 | ± 3.72 |
| Blood | 4 | 3.61 | ± 3.71 |
- Standard deviation
The biofilm formation intensity by L. monocytogenes isolates was confirmed under a confocal microscope (Fig. 5).
Fig. 5.
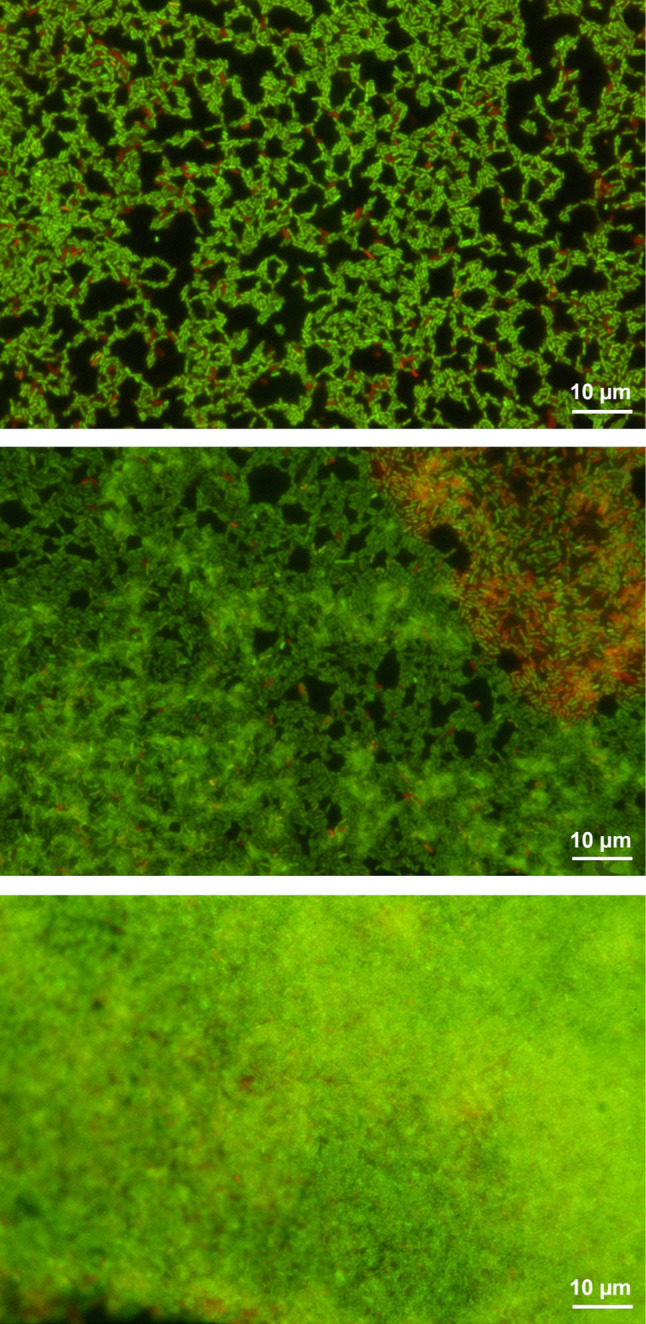
The intensity of biofilm formation by the selected L. monocytogenes strains.
A – weak biofilm, B – moderate biofilm, C – strong biofilm, green – live cells, red – death cells.
The correlation between antibiotic resistance, virulence genes, and biofilm formation ability. It was found that the strains with more virulence genes were resistant to a higher number of antibiotics, and their biofilms appeared to be more intense. There was a high positive correlation (r = 0.956) and positive correlation (r=0.896) between the efficiency of biofilm formation, the number of virulence genes and the antibiotic resistance level of isolates derived from milk (Fig. 6) as well as clinical strains (Fig. 7), respectively. For the clinical isolates, only the correlation between the number of virulence genes and the efficiency of biofilm formation was not significant.
Fig. 6.
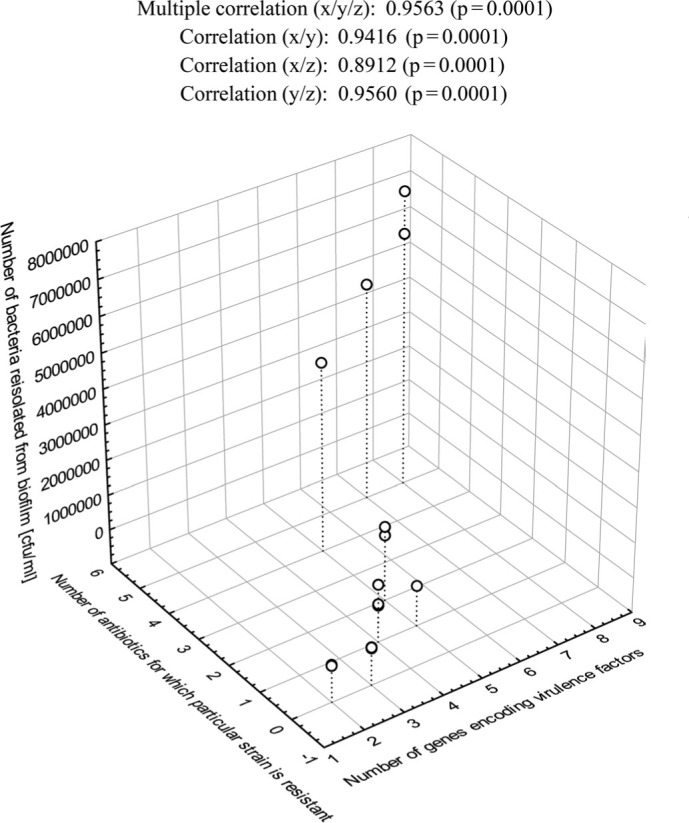
Multiple correlations between the intensity of biofilm formation, the number of virulence genes, and the drug resistance of L. monocytogenes strains isolated from milk.
Fig. 7.
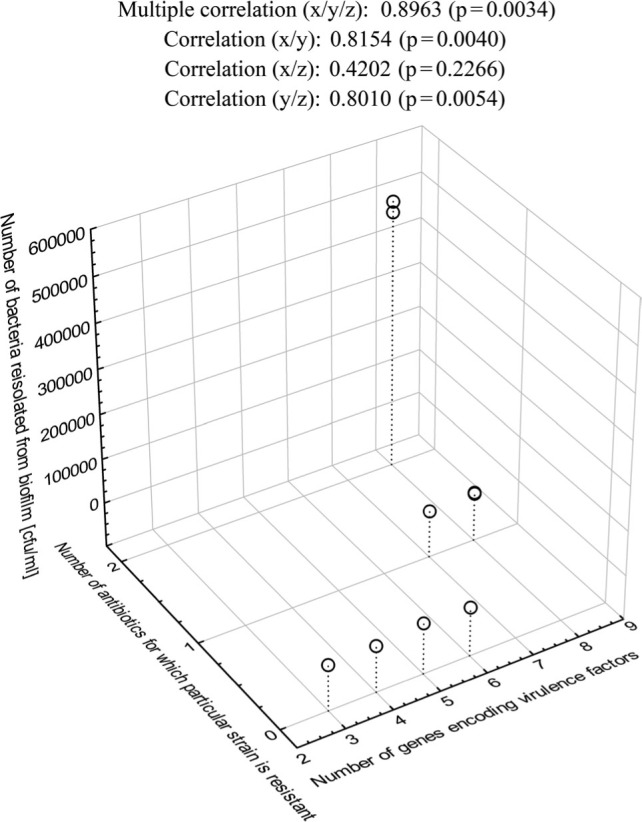
Multiple correlations between the intensity of biofilm formation, the number of virulence genes, and the drug resistance of the clinical L. monocytogenes strains.
The spread of L. monocytogenes through contaminated teat rubbers. Transmission of L. monocytogenes on the skin of the udder and into the milk. Bacteria were transferred to skin pieces by the contact with the contaminated teat rubber (Table VI). The number of bacterial cells from the strains isolated from cow’s milk that was detected in eight subsequent fragments of the udder skin in the number ranged from 1.08 log CFU/cm−2 (M231) to 6.87 log CFU/cm−2 (M372) (Table VI). Only the M63 strain was not isolated from the 8th fragment and its density was 1.59 log CFU/cm−2 on the 7th fragment (Table VI). Clinical strains (B5 and B6) that developed strong biofilms were isolated from seven consecutive fragments of the skin and the number of bacterial cells was 1.26 log CFU/cm−2 for the strain B6 and 1.43 log CFU/cm−2 for the strain B5. The clinical isolates that developed weak biofilms were detected in four consecutive fragments and the number of bacterial cells was 1.20 log CFU/cm−2 for the strain B4, and 1.49 log CFU/cm−2 for the strain B10 (Table VI). Strain ATCC 19111 was transferred from the teat rubber to seven consecutive fragments of the skin and the number of cells on the last one was 1.53 log CFU/cm−2 (Table VI). It was also shown that the contamination level of the first skin fragment was higher for the strains that formed a weaker biofilm (Table VI).
Table VI.
Transmission of L. monocytogenes from a biofilm on silicone teat cups to the cow’s udder skin.
| Strain | Number of re-isolated L. monocytogenes strains [log CFU/cm2] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Silicone teat cup | Udder skin | |||||||||
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 | Regrowth on first negative or last positive sample (12 h/25°C) | ||
| M372 | 6.87* ± 6.12** | 4.33 ± 3.98 | 3.88 ± 3.77 | 3.71 ± 3.34 | 3.45 ± 2.88 | 2.92 ± 2.71 | 2.66 ± 2.35 | 2.09 ± 2.31 | 1.85 ± 1.84 | 2.92 ± 2.74 |
| M375 | 6.79 ± 6.09 | 4.54 ± 4.37 | 3.99 ± 3.82 | 3.65 ± 3.63 | 3.10 ± 3.45 | 2.83 ± 2.69 | 2.51 ± 1.96 | 1.92 ± 1.82 | 1.61 ± 1.31 | 2.79 ± 2.62 |
| M231 | 4.74 ± 3.96 | 3.87 ± 3.72 | 3.76 ± 3.79 | 3.47 ± 3.55 | 2.79 ± 2.71 | 2.52 ± 2.33 | 1.93 ± 1.20 | 1.67 ± 1.47 | 1.08 ± 1.54 | 1.99 ± 1.85 |
| M63 | 4.64 ± 3.72 | 3.96 ± 3.82 | 3.80 ± 3.33 | 3.34 ± 3.62 | 2.70 ± 2.58 | 2.33 ± 2.32 | 1.86 ± 1.78 | 1.59 ± 1.48 | n.d. | 1.87 ± 1.51 |
| ATCC 19111 | 5.80 ± 5.04 | 3.64 ± 3.67 | 3.02 ± 3.51 | 2.79 ± 2.39 | 2.62 ± 2.44 | 2.14 ± 2.48 | 1.79 ± 1.51 | 1.53 ± 1.63 | n.d. | 2.01 ± 2.31 |
| B5 | 5.70 ± 4.87 | 3.56 ± 3.71 | 2.85 ± 2.70 | 2.63 ± 2.20 | 2.48 ± 2.23 | 1.96 ± 1.86 | 1.76 ± 1.61 | 1.43 ± 1.01 | n.d. | 1.81 ± 1.20 |
| B6 | 5.67 ± 4.76 | 3.47 ± 3.17 | 2.91 ± 2.76 | 2.76 ± 1.99 | 2.39 ± 2.61 | 1.92 ± 1.65 | 1.64 ± 1.71 | 1.26 ± 1.33 | n.d. | 1.72 ± 1.80 |
| B10 | 3.71 ± 3.72 | 2.85 ± 2.58 | 2.57 ± 2.20 | 1.88 ± 1.57 | 1.49 ± 1.04 | n.d.*** | n.d. | n.d. | n.d. | 1.63 ± 1.41 |
| B4 | 3.61 ± 3.71 | 2.92 ± 2.88 | 2.73 ± 2.53 | 1.84 ± 161 | 1.20 ± 1.35 | n.d. | n.d. | n.d. | n.d. | 1.38 ± 1.50 |
- Mean
- Standard deviation
- Not detected
The obtained results showed that even if L. monocytogenes was not detected on the fragment of the udder skin (the number of bacteria was below the detection threshold), the bacteria were transferred from the teat rubber and their number increased to 1.38–2.92 log CFU/cm−2 along with incubation time (Table VI).
All L. monocytogenes strains that developed biofilms on teat rubbers also caused contamination of milk flowing through these rubbers. The number of bacteria recovered from milk ranged from 2.71 to 3.37 log CFU/ml for milk strains, and from 2.57 to 2.94 log CFU/ml for clinical strains (Table VII). The reference strain was grown in milk to the density of 2.97 log CFU/ml (Table VII).
Table VII.
Transmission of L. monocytogenes from a biofilm on silicone teat cups to milk.
| Strain | Number of re-isolated L. monocytogenes | |
|---|---|---|
| Silicone teat cup [log CFU/cm2] |
Milk [log CFU/ml] |
|
| M372 | 6.87* ± 6.12** | 3.37 ± 3.03 |
| M375 | 6.79 ± 6.09 | 3.29 ± 3.37 |
| M231 | 4.74 ± 3.96 | 2.75 ± 2.62 |
| M63 | 4.64 ± 3.72 | 2.71 ± 2.52 |
| ATCC 19111 | 5.80 ± 5.04 | 2.97 2.85 |
| B5 | 5.70 ± 4.87 | 2.94 ± 2.74 |
| B6 | 5.67 ± 4.76 | 2.84 ± 2.32 |
| B10 | 3.71 ± 3.72 | 2.63 ± 2.51 |
| B4 | 3.61 ± 3.71 | 2.57 ± 2.32 |
- Mean
- Standard deviation
Transmission of L. monocytogenes in udder – teat rubber – udder model. It was shown that L. monocytogenes found on the udder skin may be transferred to sterile teat rubber and then can cause the contamination of sterile udder skin (Table VIII). For all strains tested, the number of bacteria on the skin ranged from 5.13 to 5.91 log CFU/cm−2, irrespective of their origin (milk vs. human) (Table VIII). Bacteria from the skin fragments were transferred to the teat rubber and the number of the bacteria reisolated ranged from 3.58 to 3.81 log CFU/cm−2. In turn, the contact of the teat rubber with sic consecutive sterile pieces of udder skin resulted in the transmission of L. monocytogenes to from four (for strains No M63, B4, and B10) to six (for strains M372, M231, ATCC 19111, and B6) skin fragments (Table VIII). The number of bacteria isolated from the last skin fragment examined was from 1.11 to 1.92 log CFU/cm−2 (Table VIII). This may suggest the possibility of more than six consecutive udders contamination.
Table VIII.
Listeria spp. transmission system: udder skin – silicone teat cup – udder skin.
| Strain | Number of re-isolated L. monocytogenes [log CFU/cm2] | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Silicone teat cup | Udder skin | Udder skin | |||||||
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Recovery of bacteria (12 h/25°C) | |||
| M372 | 5.91* ± 5.81** | 3.81 ± 3.61 | 3.77 ± 3.73 | 3,. 0 ± 2.85 | 3.00 ± 2.85 | 2.81 ± 2.65 | 2.81 ± 2.65 | 1.92 ± 1.82 | 2.83 ± 2.53 |
| M375 | 5.84 ± 5.63 | 3.68 ± 3.45 | 3.61 ± 3.83 | 2.68 ± 2.50 | 2.68 ± 2.50 | 1.87 ± 1.93 | 1.87 ± 1.93 | n.d.*** | 1.66 ± 1.76 |
| M231 | 5.57 ± 5.60 | 3.71 ± 3.23 | 3.66 ± 3.54 | 2.68 ± 2.72 | 2.68 ± 2.72 | 2.00 ± 1.90 | 2.00 ± 1.90 | 1.11 ± 1.16 | 2.37 ± 2.48 |
| M63 | 5.52 ± 5.34 | 3.54 3.63 |
3.47 ± 3.55 | 1.47 ± 1.66 | 1.47 ± 1.66 | 1.00 ± 1.69 | 1.00 ± 1.69 | n.d | 1.26 ± 1.03 |
| ATCC 19111 | 5.66 ± 5.54 | 3.76 ± 3.57 | 3.72 ± 3.27 | 2.74 ± 1.99 | 2.74 ± 1.99 | 2.26 ± 1.98 | 2.26 ± 1.98 | 1.64 ± 1.47 | 2.57 ± 2.32 |
| B5 | 5.71 ± 5.61 | 3.62 ± 3.20 | 3.56 ± 3.47 | 2.57 ± 2.42 | 2.57 ± 2.42 | 1.84 ± 1.03 | 1.84 ± 1.03 | n.d. | 1.46 ± 1.58 |
| B6 | 5.69 ± 5.45 | 3.79 ± 3.86 | 3.73 ± 3.70 | 2.91 ± 2.78 | 2.91 ± 2.78 | 2.72 ± 2.64 | 2.72 ± 2.64 | 1.81 1.71 |
2.72 ± 2.63 |
| B10 | 5.32 ± 5.68 | 3.59 ± 3.02 | 3.53 ± 3.37 | 1.62 ± 1.48 | 1.62 ± 1.48 | 1.11 ± 1.46 | 1.11 ± 1.46 | n.d. | 1.20 ± 1.68 |
| B4 | 5.13 ± 5.40 | 3.58 ± 3.50 | 3.51 ± 3.58 | 1.52 ± 1.61 | 1.52 ± 1.61 | 0.90 ± 0.86 | 0.90 ± 0.86 | n.d. | 1.08 ± 1.71 |
- Mean
- Standard deviation
- Not detected
To confirm L. monocytogenes proliferation on skin fragments, the fragments were incubated 12 hours at 25°C. It appeared that bacteria were detected in both cases. Moreover, the increase in their numbers on the last tested fragments was observed (Table VIII).
Discussion
Since L. monocytogenes is widespread in the environment and survives in harsh environmental conditions, it may easily contaminate food (Wałecka 2011). L. monocytogenes is isolated from both unpasteurized and pasteurized cow’s milk, dairy products such as soft cheeses, and dairy farms (Fleming et al. 1985; Van Kessel et al. 2004; CDC 2007; Fox et al. 2009; Lomanco et al. 2009; Balandyte et al. 2011; Van Kessel et al. 2011). In recent years, such products have been linked to several outbreaks of listeriosis (Rocha et al. 2013; CDC 2014; CDC 2016). Although milk products have repeatedly proved to be the source of this pathogen, the characteristics of L. monocytogenes strains isolated from unpasteurized milk and dairy farms remain unclear.
In the present study, L. monocytogenes was isolated from 21 (5.5%) out of 380 milk samples. Similar results were obtained in the Czech Republic (3.2%) (Gelbíčová and Karpíšková 2012a), Ethiopia (4.0%) (Garedew et al. 2015), England, Wales, India (5.1%) (Siegman-Igra et al. 2002; Kalorey et al. 2008), Iran (5.4%) (Jamali and Radmehr 2013), and Spain (6.5%) (Vilar et al. 2007). In contrast, L. monocytogenes strains were isolated only three times out of 294 milk samples in Sweden (Waak et al. 2002). In another study carried out in India, the occurrence of L. monocytogenes in fresh milk was considerably higher and accounted for 21.73% (Sharma et al. 2012).
Our studies showed the presence of the hlyA gene in all strains tested, regardless of their origin (milk vs. human). This is in accordance with the study of (Indrawattana et al. 2011) and (Aurora et al. 2008). In the present study, we found the plcB gene in 100% of the strains isolated from milk and in 80% of clinical isolates. This gene was detected in all strains of L. monocytogenes in the studies of Gelbíčová and Karpíšková ( 2012b), (Mureddu et al. 2014), and Wieczorek and Osek ( 2017). Strains without the plcB gene divide but have reduced the ability to escape from phagosome (Bielecki 1994). We showed the presence of the iap gene in six (33.3%) strains cultured from milk and seven (70.0%) isolates from blood. In turn, Al- (Nabulsi et al. 2015) confirmed the presence of the iap gene only among 16.6% of L. monocytogenes strains isolated from processed meat whereas (Mureddu et al. 2014) demonstrated its presence in 97.1% strains of this species. Spontaneous mutants with reduced secretion of p60 protein do not lose their ability to grow intracellularly and the mutation is easily reversible. In contrast, insertional mutations in the region of the iap gene significantly reduce the hemolytic activity of L. monocytogenes (Rocourt et al. 2000).
To date, many studies evaluating the drug susceptibility of L. monocytogenes have been conducted (Morvan et al. 2010; Rahimi et al. 2010; Dalzini et al. 2016). Since the isolation of the first drug-resistant L. monocytogenes strains a systematic growth in resistance of these bacteria to antimicrobial agents has been observed (Poyart-Salmeron et al. 1990; Srinivasan et al. 2005; Morvan et al. 2010; Pesavento et al. 2010; Jamali et al. 2013). Therefore, it is important to monitor drug resistance of this pathogen. In the present study, eight (44.4%) isolates from milk and four (40.0%) isolates from blood were susceptible to all the antibiotics tested. On the contrary, most (98.2%) of Listeria spp. strains isolated from cow’s milk and products thereof showed resistance to at least one antibiotic in Iran (Rahimi et al. 2010). In our study, the greatest number of L. monocytogenes strains from milk was resistant to penicillin (44.4%), followed by erythromycin (33.3%), cotrimoxazole (27.7%), and ampicillin (22.2%). Similar results were obtained by (Srinivasan et al. 2005) and (Pesavento et al. 2010). A much higher percentage of the resistant strains was reported by (Jamali et al. 2013), who isolated L. monocytogenes strains from the milk of cows without signs of mastitis and from milk of cows with the clinical form of mastitis. In our study, four (40.0%) strains isolated from humans were resistant only to erythromycin, whereas two (20.0%) strains were resistant to meropenem and erythromycin. These results are similar to those obtained in other countries (Marco et al. 2000; Vitas et al. 2007; Prieto et al. 2016). In this study, we also found that almost half of the strains examined (eight, 44.4%) isolated from milk are resistant to penicillin, while no penicillin-resistant L. monocytogenes isolated from blood samples was observed. It may be due to the recommended empiric use of penicillins in the treatment of contagious disease, including mastitis, in ruminants (EVIRA 2018).
We found also three (16.7%) strains isolated from milk resistant to all of the antibiotics tested. The strain is described as MDR if is resistant to at least three various groups of antibiotic. According to this definition, we found four (22.2%) MDR strains among strains isolated from milk, and none MDR strain was isolated from blood samples. Similar results were shown by (Garedew et al. 2015) and (Pesavento et al. 2010), who isolated four (16.7%) and 11 (27.5%) multidrug-resistant strains from food, respectively. In contrast, in the study by (Jamali et al. 2013) as much as 71.4% of multidrug-resistant strains of L. monocytogenes from milk were detected. Charpentier and Courvalin ( 1999) have shown that the extensive use of antimicrobials in animal production, as well as clinical treatment of animals and humans, contributed to the continuous evolution of antimicrobial-resistant (AMR) bacteria with a diverse pool of genetically-transferred resistance determinants. Thereby, food of animal origin may be a source of AMR L. monocytogenes strains. In the food processing environment, L. monocytogenes may face many adverse conditions such as heat, high pressure, irradiation, acids, salts, and oxidants which induce the cross-protection against the same or other types of stresses (Wesche et al. 2009). It was shown, that the exposure of L. monocytogenes isolates derived from food to variable pH, cold, heat and salt stress, disinfectants, and low water activity increased their resistance to various antibiotics (Beuls et al. 2012; Al-Nabulsi et al. 2015; Faezi-Ghasemi and Kazemi 2015). Clinical strains usually are not subjected to such stresses. This may explain the difference in AMR between clinical and food isolates.
In Poland, among 471 L. monocytogenes strains isolated from different foods and food-related sources from 2004 to 2010, no resistance to ampicillin, amoxicillin, chloramphenicol, erythromycin, gentamicin, rifampicin, sulfamethoxazole, trimethoprim, and vancomycin was reported (Korsak et al. 2012). On the contrary, a Lebanese study assessing AMR in L. monocytogenes recovered from traditional dairy products showed that all isolates (n = 30) were resistant to at least one antimicrobial, including the resistance to ampicillin (60%), penicillin (90%), erythromycin (27%), gentamicin (7%), and SXT (17%) (Harakeh et al. 2009).
Monitoring of the occurrence of L. monocytogenes in food is a great challenge for food processing plants. The ability of these microorganisms to survive in a moist, cool environment and to form biofilm makes them difficult to eradicate. L. monocytogenes presence in milk and milk products may be caused by improper sanitization of surroundings and equipment used during milking, as well as ineffective disinfection in dairies (Tompkin et al. 2002). The present study evaluated the ability of the strains tested to form biofilm on silicone teat cups rubbers. Among L. monocytogenes strains derived from milk, seven isolates developed more intense biofilms than the reference ATCC® 19111™ strain. All clinical strains formed biofilm less intense than milk isolates (p = 0.005). (Latorre et al. 2010) isolated a considerable number of bacteria L. monocytogenes from milking machines. Using SEM micrography, they found that the bacteria were particularly visible in scratches on the inside plastic surface of the teat cups. In the study by (Doijad et al. 2015), only nine out of 98 strains isolated from different environments formed robust biofilms. It is worthy to note that all strains that developed intense biofilms were isolated from milk and dairy products. In previous studies, it has been proved that L. monocytogenes can form biofilm on many surfaces such as polystyrene, polypropylene, glass, stainless steel, quartz, marble, and granite (Silva et al. 2008). Though, studies by (Djordjevic et al. 2002) and (Harvey et al. 2007) indicated that L. monocytogenes formed only weak or moderate biofilm on various surfaces. In turn, the study by (Sinde et al. 2000) showed better adherence of L. monocytogenes to rubber when compared to stainless steel.
Bacteria within biofilms are much more resistant to antimicrobial agents than floating counterparts (Gong et al. 2013). To understand the relationship between biofilms and antimicrobial resistance in L. monocytogenes, we assessed biofilm formation ability of L. monocytogenes isolates together with their antimicrobial resistance. Our results showed very high positive correlation (r = 0.96) for isolates from milk and high positive correlation (r = 0.80) for clinical isolates. Such a relationship between biofilm production and antibiotic resistance was demonstrated for Salmonella Pullorum and uropathogenic E. coli (Adetunji et al. 2008; Gong et al. 2013). (Adetunji et al. 2008) showed that strains of L. monocytogenes producing intense biofilms were more virulent and drug-resistant. Intrinsic resistance of biofilm bacteria is related to the presence of persister cells or efflux pumps that remove antibiotics from the biofilm environment (Korsak et al. 2005; Sauvage et al. 2008). Bacterial cells from deeper layers of biofilm are protected against antimicrobials by the upper layers. The penicillin resistance among the strong-biofilm forming L. monocytogenes strains may be correlated with the presence of PBP5 that binds penicillin G. This protein is a DD-carboxypeptidase related with the membrane fraction and lateral wall growth of L. monocytogenes cell (Korsak et al. 2005; Sauvage et al. 2008).
Our study also revealed a positive correlation between the number of virulence genes, drug resistance, and biofilm formation ability. This is in agreement with the study by (Soni et al. 2013), which indicated that multi-drug resistant L. monocytogenes strains derived from clinical specimens, water, and milk possess a large number of virulence genes (inlA, inlC, plcA, prfA, actA, hlyA, and iap).
Our study showed that teat cups, contaminated during milking might contribute to the transmission of L. monocytogenes to cow’s udders and finally to the milk. It was found that this phenomenon might affect at least several successively milked animals. This supports the study of Benić et al. ( 2012), which demonstrated that bacteria might be transmitted by utensils used during milking, mainly by the teat cups. It was also confirmed by (Azevedo et al. 2016) studies, which found that improper hygiene of teat cups could cause transmission of Staphylococcus spp. both between the animals and to the milk tank.
A better understanding of the epidemiology of L. monocytogenes infections and the factors affecting their survival, spread and resistance is necessary to prevent contamination in the food industry and transmission of the pathogen. This may help in limiting listeriosis incidence in humans.
Conclusions
In the available literature, there are few data on the bacterial transmission via teat cups and the risks posed by this equipment. This study showed that L. monocytogenes isolates originating from cow’s milk are more resistant to antibiotics than the clinical strains. The most frequent virulence genes detected were hlyA and plcB among milk strains and hlyA and prfA in strains derived from the blood. The intensity of the biofilm formation was strain-dependent and was significantly higher in the milk strains. The association between biofilm formation the number of virulence genes, and antimicrobial resistance of L. monocytogenes strains was high positive for the isolates from milk (r = 0.96) as well as for the clinical isolates (r = 0.90). Our study showed also that teat cups, contaminated during the milking process, might play an important role in the transmission of L. monocytogenes to the milk, posing risk to the consumer health. For these reasons, it is reasonable to monitor incidence, susceptibility and biofilm formation by L. monocytogenes during milking and milk processing (especially unpasteurized milk).
Acknowledgments
We thank the owners of cattle farms for allowance of sampling of cow’s milk.
This research was financially supported by the Nicolaus Copernicus University with funds from the maintenance of the research potential of the Department of Microbiology (DS-UPB).
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Author’s contributions
Krzysztof Skowron – development of the concept of experiments and methodology of research, collecting the materials for research, participation in the performance of the laboratory part of experiments, collection, and analysis of results, participation in the writing of the manuscript, management of the research team and preparing the publication
Ewa Wałecka-Zacharska – development of the concept of experiments and methodology of research, translation of the manuscript text
Katarzyna Grudlewska – participation in the performance of the laboratory part of experiments and writing of the manuscript
Natalia Wiktorczyk – participation in the performance of the laboratory part of experiments and the collection of materials for research and references
Agnieszka Kaczmarek – participation in the performance of the laboratory part of experiments and the collection of references
Grzegorz Gryń – the collection and analysis of results
Joanna Kwiecińska-Piróg – participation in the performance of the laboratory part of experiments, collection of materials for research, adaptation of the manuscript to the editorial requirements
Klaudia Juszczuk – participation in the performance of the laboratory part of experiments Zbigniew Paluszak – obtaining funds for conducting research Katarzyna Kosek-Paszkowska – an adaptation of the manuscript to the editorial requirements
Eugenia Gospodarek-Komkowska – verification of the manuscript and final acceptance, obtaining funds for conducting research
All authors contributed to the draft of the manuscript and discussed results. All authors gave final approval for publication.
ORCID
Krzysztof Skowron 0000-0003-0868-864X
Literature
- Adetunji VO, Adegoke GO.. Formation of biofilm by strains of Listeria monocytogenes isolated from soft cheese ‘wara’ and its processing environment. Afr J Biotechnol. 2008;7(16):2893–2897. [Google Scholar]
- Al-Nabulsi AA, Osaili TM, Awad AA, Olaimat AN, Shaker RR, Holley RA.. Occurrence and antibiotic susceptibility of Listeria monocytogenes isolated from raw and processed meat products in Amman, Jordan. J Food. 2015;13:346–352. [Google Scholar]
- Aurora R, Prakash A, Prakash S, Rawool DB, Barbuddhe SB.. Comparison of PI-PLC based assays and PCR along with in vivo pathogenicity tests for rapid detection of pathogenic Listeria monocytogenes. Food Control. 2008;19(7):641–647. 10.1016/j.foodcont.2007.07.002 [DOI] [Google Scholar]
- Azevedo C, Pacheco D, Soares L, Romão R, Moitoso M, Maldonado J, Guix R, Simões J.. Prevalence of contagious and environmental mastitis-causing bacteria in bulk tank milk and its relationships with milking practices of dairy cattle herds in São Miguel Island (Azores). Trop Anim Health Prod. 2016;48(2):451–459. 10.1007/s11250-015-0973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandyté L, Brodard I, Frey J, Oevermann A, Abril C.. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to a multilocus variable-number tandem-repeat analysis complex. Appl Environ Microbiol. 2011;77(23):8325–8335. 10.1128/AEM.06507-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal NS.. Development of a polymerase chain reaction assay for the detection of Listeria monocytogenes in foods. Lett Appl Microbiol. 1996;22(5):353–356. 10.1111/j.1472-765X.1996.tb01177.x [DOI] [PubMed] [Google Scholar]
- Benić M, Habrun B, Kompes G.. Clinical and epidemiological aspects of cow mastitis caused by Staphylococcus aureus and its methicillin-resistant strains. Rad Medical Sciences. 2012;37:113–122. [Google Scholar]
- Beuls E, Modrie P, Deserranno C, Mahillon J.. High-salt stress conditions increase the pAW63 transfer frequency in Bacillus thuringiensis. Appl Environ Microbiol. 2012;78(19):7128–7131. 10.1128/AEM.01105-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki J.. Molekularne podstawy mechanizmów patogenezy Listeria monocytogenes. Postepy Mikrobiol. 1994;23:85–105. [Google Scholar]
- Border PM, Howard JJ, Plastow GS, Siggens KW.. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett Appl Microbiol. 1990;11(3):158–162. 10.1111/j.1472-765X.1990.tb00149.x [DOI] [PubMed] [Google Scholar]
- CDC.. Multistate outbreak of listeriosis linked to raw milk produced by miller’s organic farm in pennsylvania (Final Update) Atlanta (USA): Centers for Disease Control and Prevention; 2016. [Google Scholar]
- CDC.. Multistate outbreak of listeriosis linked to roos foods dairy products (Final Update) Atlanta (USA): Centers for Disease Control and Prevention; 2014. 4. [Google Scholar]
- CDC.. Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy – Massachusetts, 2007. MMWR Morb Mortal Wkly Rep. 2008;57(40):1097–1100. [PubMed] [Google Scholar]
- Charpentier E, Courvalin P.. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother. 1999;43(9):2103–2108. 10.1128/AAC.43.9.2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalzini E, Bernini V, Bertasi B, Daminelli P, Losio MN, Varisco G.. Survey of prevalence and seasonal variability of Listeria monocytogenes in raw cow milk from Northern Italy. Food Control. 2016;60: 466–470. 10.1016/j.foodcont.2015.08.019 [DOI] [Google Scholar]
- Djordjevic D, Wiedmann M, McLandsborough LA.. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol. 2002;68(6):2950–2958. 10.1128/AEM.68.6.2950-2958.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doijad SP, Barbuddhe SB, Garg S, Poharkar KV, Kalorey DR, Kurkure NV, Rawool DB, Chakraborty T.. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS One. 2015;10(9):e0137046 10.1371/journal.pone.0137046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA.. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16(12):5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST.. Breakpoints tables for interpretation of MICs and zones diameters Version 8.0. Basel (Switzerland): European Committee on Antimicrobial Susceptibility Testing; 2018 [cited 2019 Jan 21]. Available from http://www.eucast.org
- EVIRA ; The Faculty of Veterinary Medicine at the University of Helsinki Recommendations for the use of antimicrobials in the treatment of the most significant infectious and contagious diseases in animals. Helsinki (Finland): The Finnish Food Safety Authority; 2018. [Google Scholar]
- Faezi-Ghasemi M, Kazemi S.. Effect of sub-lethal environmental stresses on the cell survival viability and antibacterial susceptibility of Listeria monocytogenes PTCC1297 (serotype 4a). Zahedan J Res Med Sci. 2015;17(1):1–6. [Google Scholar]
- Fleming DW, Cochi SL, MacDonald KL, Brondum J, Hayes PS, Plikaytis BD, Holmes MB, Audurier A, Broome CV, Reingold AL.. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N Engl J Med. 1985;312(7):404–407. 10.1056/NEJM198502143120704 [DOI] [PubMed] [Google Scholar]
- Fox E, O’Mahony, Clancy M, Dempsey R, O’Brien M, Jordan K.. Listeria monocytogenes in the Irish dairy farm environment. J Food Prot. 2009;72(7):1450–1456. 10.4315/0362-028X-72.7.1450 [DOI] [PubMed] [Google Scholar]
- Franciosa G, Maugliani A, Floridi F, Aureli P.. Molecular and experimental virulence of Listeria monocytogenes strains isolated from cases with invasive listeriosis and febrile gastroenteritis. FEMS Immunol Med Microbiol. 2005;43(3):431–439. 10.1016/j.femsim.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Frank JF, Koffi RA.. Surface-adherent growth of Listeria monocytogenes is associated with increased resistence to surfactant sanitizers and heat. J Food Prot. 1990;53(7):550–554. 10.4315/0362-028X-53.7.550 [DOI] [PubMed] [Google Scholar]
- Garedew L, Taddese A, Biru T, Nigatu S, Kebede E, Ejo M, Fikru A, Birhanu T.. Prevalence and antimicrobial susceptibility profile of listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 2015;15(1):100 10.1186/s12866-015-0434-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbícová T, Karpísková R.. [Occurrence and typing of Listeria monocytogenes isolated from raw cow’s milk collected on farms and from vending machines] (in Czech). Klin Mikrobiol Infekc Lek. 2012a;18(2):38–42. [PubMed] [Google Scholar]
- Gelbíčová T, Karpíšková R.. Outdoor environment as a source of Listeria monocytogenes in food chain. Czech J Food Sci. 2012b; 30(1):83–88. 10.17221/7/2011-CJFS [DOI] [Google Scholar]
- Gong J, Xu M, Zhu C, Miao J, Liu X, Xu B, Zhang J, Yu Y, Jia X.. Antimicrobial resistance, presence of integrons and biofilm formation of Salmonella Pullorum isolates from eastern China (1962– 2010). Avian Pathol. 2013;42(3):290–294. 10.1080/03079457.2013.788129 [DOI] [PubMed] [Google Scholar]
- Gould LH, Mungai E, Barton Behravesh C.. Outbreaks attributed to cheese: differences between outbreaks caused by unpasteurized and pasteurized dairy products, United States, 1998-2011. Foodborne Pathog Dis. 2014;11(7):545–551. 10.1089/fpd.2013.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P.. Listeria monocytogenes: a multi-faceted model. Nat Rev Microbiol. 2006;4(6):423–434. 10.1038/nrmicro1413 [DOI] [PubMed] [Google Scholar]
- Harakeh S, Saleh I, Zouhairi O, Baydoun E, Barbour E, Alwan N.. Antimicrobial resistance of Listeria monocytogenes isolated from dairy-based food products. Sci Total Environ. 2009;407(13): 4022–4027. 10.1016/j.scitotenv.2009.04.010 [DOI] [PubMed] [Google Scholar]
- Harvey J, Keenan KP, Gilmour A.. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 2007;24(4):380–392. 10.1016/j.fm.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Hayes PS, Feeley JC, Graves LM, Ajello GW, Fleming DW.. Isolation of Listeria monocytogenes from raw milk. Appl Environ Microbiol. 1986;51(2):438–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrawattana N, Nidabbhasobon T, Sookrung N, ChongsaNguan M, Tungtrongchitr A, Makino S, Tungyong W, Chaicumpa W.. Prevalence of Listeria monocytogenes in raw meats marketed in Bangkok and characterization of the isolates by phenotypic and molecular methods. J Health Popul Nutr. 2011;29(1):26–38. 10.3329/jhpn.v29i1.7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO 11290-1.. Microbiology of food and animal feeding stuffs – horizontal method for the detection and enumeration of Listeria monocytogenes – Part 1: Detection method. 2017.
- Jackson KA, Biggerstaff M, Tobin-D’Angelo M, Sweat D, Klos R, Nosari J, Garrison O, Boothe E, Saathoff-Huber L, Hainstock L, et al.. Multistate outbreak of Listeria monocytogenes associated with Mexican-style cheese made from pasteurized milk among pregnant, Hispanic women. J Food Prot. 2011;74(6):949–953. 10.4315/0362-028X.JFP-10-536 [DOI] [PubMed] [Google Scholar]
- Jamali H, Radmehr B, Thong KL.. Prevalence, characterization, and antimicrobial resistance of Listeria species and Listeria monocytogenes isolates from raw milk in farm bulk tanks. Food Control. 2013;34(1):121–125. 10.1016/j.foodcont.2013.04.023 [DOI] [Google Scholar]
- Jamali H, Radmehr B.. Frequency, virulence genes and antimicrobial resistance of Listeria spp. isolated from bovine clinical mastitis. Vet J. 2013;198(2):541–542. 10.1016/j.tvjl.2013.06.012 [DOI] [PubMed] [Google Scholar]
- Kalorey DR, Warke SR, Kurkure NV, Rawool DB, Barbuddhe SB.. Listeria species in bovine raw milk: A large survey of Central India. Food Control. 2008;19(2):109–112. 10.1016/j.foodcont.2007.02.006 [DOI] [Google Scholar]
- Koch J, Dworak R, Prager R, Becker B, Brockmann S, Wicke A, Wichmann-Schauer H, Hof H, Werber D, Stark K.. Large listeriosis outbreak linked to cheese made from pasteurized milk, Germany, 2006-2007. Foodborne Pathog Dis. 2010;7(12):1581–1584. 10.1089/fpd.2010.0631 [DOI] [PubMed] [Google Scholar]
- Korsak D, Borek A, Daniluk S, Grabowska A, Pappelbaum K.. Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int J Food Microbiol. 2012;158(3):203–208. 10.1016/j.ijfoodmicro.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Korsak D, Vollmer W, Markiewicz Z.. Listeria monocytogenes EGD lacking penicillin-binding protein 5 (PBP5) produces a thicker cell wall. FEMS Microbiol Lett. 2005;251(2):281–288. 10.1016/j.femsle.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Kwiecińska-Piróg J, Bogiel T, Gospodarek E.. [Evaluation of biofilm formation by Proteus mirabilis strains on the surface of different biomaterials by two methods] (in Polish). Med Dosw Mikrobiol. 2011;63(2):131–138. [PubMed] [Google Scholar]
- Latorre AA, Van Kessel JS, Karns JS, Zurakowski MJ, Pradhan AK, Boor KJ, Jayarao BM, Houser BA, Daugherty CS, Schukken YH.. Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with Listeria monocytogenes. J Dairy Sci. 2010;93(6):2792–2802. 10.3168/jds.2009-2717 [DOI] [PubMed] [Google Scholar]
- Lawley R, Curtis L, Davis J.. The Food Safety Hazard Guide Book Food Safety Information. London (UK): RSC Publishing; 2008. p. 47–49. [Google Scholar]
- Linnan MJ, Mascola L, Lou XD, Goulet V, May S, Salminen C, Hird DW, Yonekura ML, Hayes P, Weaver R, et al. . Epidemic listeriosis associated with Mexican-style cheese. N Engl J Med. 1988; 319(13):823–828. 10.1056/NEJM198809293191303 [DOI] [PubMed] [Google Scholar]
- Lomonaco S, Decastelli L, Nucera D, Gallina S, Manila Bianchi D, Civera T.. Listeria monocytogenes in Gorgonzola: Subtypes, diversity and persistence over time. Int J Food Microbiol. 2009;128(3): 516–520. 10.1016/j.ijfoodmicro.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Marco F, Almela M, Nolla-Salas J, Coll P, Gasser I, Ferrer MD, de Simon M; The Collaborative Study Group of Listeriosis of Barcelona.. In vitro activities of 22 antimicrobial agents against Listeria monocytogenes strains isolated in Barcelona, Spain. Diagn Microbiol Infect Dis. 2000;38(4):259–261. 10.1016/S0732-8893(00)00208-X [DOI] [PubMed] [Google Scholar]
- Meyer-Broseta S, Diot A, Bastian S, Rivière J, Cerf O.. Estimation of low bacterial concentration: listeria monocytogenes in raw milk. Int J Food Microbiol. 2003;80(1):1–15. 10.1016/S0168-1605(02)00117-4 [DOI] [PubMed] [Google Scholar]
- Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, Courvalin P, Le Monnier A.. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother. 2010;54(6):2728–2731. 10.1128/AAC.01557-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mureddu A, Mazza R, Fois F, Meloni D, Bacciu R, Piras F, Mazzette R.. Listeria monocytogenes persistence in ready-to-eat sausages and in processing plants. Ital J Food Saf. 2014;3(1):1697 10.4081/ijfs.2014.1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIZP-PZH.. Choroby zakaźne i zatrucia w Polsce. Warszawa (Poland): Narodowy Instytut Zdrowia Publicznego – Państwowy Zakład Higieny; 2019. [Google Scholar]
- Ozbey G, Ertas HB, Kok F.. Prevalence of Listeria species in camel sausages from retail markets in Aydin province in Turkey and RAPD analysis of Listeria monocytogenes isolates. Ir Vet J. 2006;59(6): 342–344. 10.1186/2046-0481-59-6-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Jung J, Choi S, Oh Y, Lee J, Chae H, Ryu S, Jung H, Park G, Choi S, et al.. Molecular Characterization of Listeria monocytogenes Based on the PFGE and RAPD in Korea. Adv Microbiol. 2012; 02(04):605–616. 10.4236/aim.2012.24079 [DOI] [Google Scholar]
- Pesavento G, Ducci B, Nieri D, Comodo N, Lo Nostro A.. Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control. 2010;21(5):708–713. 10.1016/j.foodcont.2009.10.012 [DOI] [Google Scholar]
- Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu AL, Courvalin P.. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet. 1990;335(8703):1422–1426. 10.1016/0140-6736(90)91447-I [DOI] [PubMed] [Google Scholar]
- Prieto M, Martínez C, Aguerre L, Rocca MF, Cipolla L, Callejo R.. Antibiotic susceptibility of Listeria monocytogenes in Argentina. Enferm Infecc Microbiol Clin. 2016;34(2):91–95. 10.1016/j.eimc.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Rahimi E, Ameri M, Momtaz H.. Prevalence and antimicrobial resistance of Listeria species isolated from milk and dairy products in Iran. Food Control. 2010;21(11):1448–1452. 10.1016/j.foodcont.2010.03.014 [DOI] [Google Scholar]
- Rawool DB, Malik SVS, Barbuddhe SB, Shakuntala I, Aurora R.. A multiplex PCR for detection of virulence associated genes in Listeria monocytogenes. Internet J Food Saf. 2007;9:56–62. [DOI] [PubMed] [Google Scholar]
- Rocha PRDA, Lomonaco S, Bottero MT, Dalmasso A, Dondo A, Grattarola C, Zuccon F, Iulini B, Knabel SJ, Capucchio MT, et al.. Ruminant rhombencephalitis-associated Listeria monocytogenes strains constitute a genetically homogeneous group related to human outbreak strains. Appl Environ Microbiol. 2013;79(9):3059–3066. 10.1128/AEM.00219-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocourt J, Jacquet C, Reilly A.. Epidemiology of human listeriosis and seafoods. Int J Food Microbiol. 2000;62(3):197–209. 10.1016/S0168-1605(00)00336-6 [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P.. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32(2):234–258. 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- Schlech WF 3, Acheson D. Foodborne Listeriosis. Clin Infect Dis. 2000;31(3):770–775. 10.1086/314008 [DOI] [PubMed] [Google Scholar]
- Seremak-Bulge J, Świetlik K, Mieczkowski M, Szajner P, Zdziarska T.. Rynek mleka Stan i perspektywy, nr 45. Analizy Rynkowe, Warszawa (Poland): IERiGŻ-PIB, ARR, MRiRW; 2013. [Google Scholar]
- Sharma D, Sharma PK, Saharan BS, Malik A.. Isolation, identification and antibiotic susceptibility profiling of antimicrobial resistant Listeria monocytogenes from dairy milk. Int J Microbial Res Technol. 2012;1:1–4. [Google Scholar]
- Siegman-Igra Y, Levin R, Weinberger M, Golan Y, Schwartz D, Samra Z, Konigsberger H, Yinnon A, Rahav G, Keller N, et al.. Listeria monocytogenes infection in Israel and review of cases worldwide. Emerg Infect Dis. 2002;8(3):305–310. 10.3201/eid0803.010195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Teixeira P, Oliveira R, Azeredo J.. Adhesion to and viability of Listeria monocytogenes on food contact surfaces. J Food Prot. 2008;71(7):1379–1385. 10.4315/0362-028X-71.7.1379 [DOI] [PubMed] [Google Scholar]
- Sinde E, Carballo J.. Attachment of Salmonella spp. and Listeria monocytogenes to stainless steel, rubber and polytetrafluorethylene: the influence of free energy and the effect of commercial sanitizers. Food Microbiol. 2000;17(4):439–447. 10.1006/fmic.2000.0339 [DOI] [Google Scholar]
- Soni DK, Singh RK, Singh DV, Dubey SK.. Characterization of Listeria monocytogenes isolated from Ganges water, human clinical and milk samples at Varanasi, India. Infect Genet Evol. 2013;14:83–91. 10.1016/j.meegid.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Nam HM, Nguyen LT, Tamilselvam B, Murinda SE, Oliver SP.. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog Dis. 2005;2(3):201–211. 10.1089/fpd.2005.2.201 [DOI] [PubMed] [Google Scholar]
- Tompkin RB.. Control of Listeria monocytogenes in the food-processing environment. J Food Prot. 2002;65(4):709–725. 10.4315/0362-028X-65.4.709 [DOI] [PubMed] [Google Scholar]
- Unnerstad H, Bannerman E, Bille J, Danielsson-Tham M-L, Waak E, Tham W.. Prolonged contamination of a dairy with Listeria monocytogenes. Neth Milk Dairy J. 1996;50:493–499. [Google Scholar]
- Van Kessel JAS, Karns JS, Lombard J, Kopral CA.. Prevalence of Salmonella enterica, Listeria monocytogenes, and Escherichia coli virulence factors in bulk tank milk and in-line filters from U.S. dairies. J Food Prot. 2011;74(5):759–768. 10.4315/0362-028X.JFP-10-423 [DOI] [PubMed] [Google Scholar]
- Van Kessel JS, Karns JS, Gorski L, McCluskey BJ, Perdue ML.. Prevalence of Salmonellae, Listeria monocytogenes, and fecal coliforms in bulk tank milk on US dairies. J Dairy Sci. 2004;87(9):2822–2830. 10.3168/jds.S0022-0302(04)73410-4 [DOI] [PubMed] [Google Scholar]
- Vilar MJ, Yus E, Sanjuán ML, Diéguez FJ, Rodríguez-Otero JL.. Prevalence of and risk factors for Listeria species on dairy farms. J Dairy Sci. 2007;90(11):5083–5088. 10.3168/jds.2007-0213 [DOI] [PubMed] [Google Scholar]
- Vitas AI, María Sánchez R, Aguado V, García-Jalón I.. Antimicrobial susceptibility of Listeria monocytogenes isolated from food and clinical cases in Navarra, Spain. J Food Prot. 2007;70(10):2402–2406. 10.4315/0362-028X-70.10.2402 [DOI] [PubMed] [Google Scholar]
- Waak E, Tham W, Danielsson-Tham ML.. Prevalence and finger-printing of Listeria monocytogenes strains isolated from raw whole milk in farm bulk tanks and in dairy plant receiving tanks. Appl Environ Microbiol. 2002;68(7):3366–3370. 10.1128/AEM.68.7.3366-3370.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wałecka E.. Badanie wpływu stresu środowiskowego na wirulencję Listeria monocytogenes [dissertation]. Wroclaw Medical University; 2011.
- Walker SJ, Archer P, Banks JG.. Growth of Listeria monocytogenes at refrigeration temperatures. J Appl Bacteriol. 1990;68(2):157–162. 10.1111/j.1365-2672.1990.tb02561.x [DOI] [PubMed] [Google Scholar]
- Weiler C, Ifland A, Naumann A, Kleta S, Noll M. Incorporation of Listeria monocytogenes strains in raw milk biofilms. Int J Food Microbiol. 2013;161(2):61–68. 10.1016/j.ijfoodmicro.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Wesche AM, Gurtler JB, Marks BP, Ryser ET.. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J Food Prot. 2009;72(5):1121–1138. 10.4315/0362-028X-72.5.1121 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Osek J.. Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 2017;64:164–171. 10.1016/j.fm.2016.12.022 [DOI] [PubMed] [Google Scholar]