Abstract
Probiotics are considered an alternative to antibiotics in the prevention and treatment of Salmonella diseases in poultry. However, to use probiotics as proposed above, it is necessary to evaluate their properties in detail and to select the most effective bacterial strains in the application targeted. In this study, probiotic properties of new Lactobacillus sp. strains were investigated and their antimicrobial activity against 125 environmental strains of Salmonella sp. was determined using the agar slab method. Furthermore, their survival in the presence of bile salts and at low pH, antibiotics susceptibility, aggregation and coaggregation ability, adherence to polystyrene and Caco-2 cells, and cytotoxicity were investigated. Each strain tested showed antagonistic activity against at least 96% of the environmental Salmonella sp. strains and thus representing a highly epidemiologically differentiated collection of poultry isolates. In addition, the probiotic properties of new Lactobacillus strains are promising. Therefore, all strains examined showed a high potential for use in poultry against salmonellosis.
Key words: Lactobacillus sp., Salmonella sp., antimicrobial activity, cytotoxicity, poultry, probiotic potential
Introduction
The well-being of farm animals is highly dependent on the prevention of disease outbreaks in husbandry. Numerous environmental stress factors favor the development of pathogenic bacteria that interfere with the composition of commensal microbes in the gastrointestinal tract and induce diseases (Gaggìa et al. 2010). Many infections can be easily transferred from animals to humans by contaminated food of animal origin. One of the most common zoonosis is salmonellosis, which can cause a wide range of illnesses such as fever, sepsis, infections of tissues and inflammation of the gastrointestinal tract. It is often transferred to humans from infected poultry meat (Ryan et al. 2017).
Salmonella is a genus of the family Enterobacteriaceae and contains two species: S. enterica and S. bongori. S. enterica is further subdivided into six subspecies (I, S. enterica subsp. enterica, II, S. enterica subsp. salamae, IIIa, S. enterica subsp. arizonae, IIIb, S. enterica subsp. diarizonae, IV, S. enterica subsp. houtenae, and VI, S. enterica subsp. indica) (Brenner et al. 2000; Ryan et al. 2017). In the European Union, salmonellosis is the second most common foodborne disease and S. enterica is the most frequently reported pathogen (Kirk et al. 2015; Bonardi et al. 2016; Shah et al. 2017; European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) 2018). The main source of Salmonella for humans is laying hens, followed by pigs, turkeys, and broilers (Kirk et al. 2015; Chousalkar and Gole 2016). Application of antibiotics to fight and prevent diseases is the most common practice but their overuse favors increasing number of antibiotic-resistant strains of bacteria (Davies and Davies 2010; Gao et al. 2017). Concern about the development of such pathogens and the transfer of antibiotic resistance genes to human microbiota have led to considerable interest in search of novel methods to fight pathogens.
Probiotic bacteria application appears a promising alternative to antibiotics (Musikasang et al. 2009; Carter et al. 2017). Probiotics have the potential of competitive exclusion of pathogens and growth-promoting effects by supporting the absorption of certain essential nutrients, thus increasing total body weight, feed intake, and feed conversion rate (Yeo et al. 2016; Angelakis 2017). Lactic acid bacteria (LAB) are the most common probiotic bacteria used in food fermentation, and in taste and texture enhancement in fermented food products. They are proved to be safe for animals and humans (Vankerckhoven et al. 2008; Mokoena 2017). To consider a strain as a potential probiotic, the strain has to possess certain health-promoting features such as the stimulation of host immune response or modulation of its microbiota (Romani Vestman et al. 2013). However, in the development of a probiotic product, also the safety, functional and technological aspects need to be considered (Hütt et al. 2006; Belicová et al. 2013). Probiotic has to withstand harsh gastrointestinal tract conditions (such as low pH, and presence of bile salts) and be able to adhere to intestinal epithelial cells (Kos et al. 2003; Kaushik et al. 2009; Tinrat et al. 2011).
The recent studies demonstrated that the application of probiotic cultures successfully reduced Salmonella sp. in farm animals. In 2003, La Ragione and Woodward described the controlling of Salmonella Enteritidis in young chickens by Bacillus subtilis, and in 2017 Carter et al. used a combination of Lactobacillus salivarius and Enterococcus faecium to treat the same pathogen also in poultry (La Ragione and Woodward 2003; Carter et al. 2017). Some other publications report the results of application of commercially available anti-Salmonella probiotic products like FloraMax-B11® containing Lactobacillus salivarius and Pediococcus parvulus (Prado-Rebolledo et al. 2017) or FM-B11™ containing 11 Lactobacillus strains (three of L. bulgaricus, three of L. fermentum, two of L. casei, two of L. cellobiosus, and one of L. helveticus) (Higgins et al. 2008; Vicente et al. 2008). However, there is still a need for novel strains of probiotics with a wide spectrum of activity that can be used as an antibacterial agent in farm animals (Reid 2017).
The aim of this research was to investigate the probiotic properties of new Lactobacillus sp. strains: L. rhamnosus LOCK 1131, L. casei LOCK 1132, and L. paracasei LOCK 1133, which are considered as a feed additive for prevention and treatment Salmonella in poultry. We investigated their survival in bile salts and low pH, antibiotic resistance, aggregation properties, and adherence to polystyrene and Caco-2 cells. The safety of the application of the strains was verified by the cytotoxicity test, and their effectivity was assessed by the competition assay and by the measurement of their antagonistic activity towards Salmonella strains.
Experimental
Materials and Methods
Bacterial strains. Three strains of Lactobacillus sp.: L. rhamnosus LOCK 1131, L. casei LOCK 1132, and L. paracasei LOCK 1133 were isolated. The first two strains were isolated from chicks ileal content, while the third strain derived from the domestic fermented cabbage. The strain were deposited in the collection of the Institute of Fermentation Technology and Microbiology (LOCK 105), Lodz University of Technology, Poland.
Additionally, 126 Salmonella sp. strains were used: three reference strains from American Type Culture Collection (ATCC, USA) and 123 environmental strains from the collection of Proteon Pharmaceuticals SA (Table III). These strains are the epidemiologically representative group of strains isolated from poultry in 2009–2016 from different geographical regions of Poland. Strains were confirmed as genetically different, as described by Zaczek et al. in 2015.
Table III.
Antagonistic activity of Lactobacillus sp. strains against Salmonella sp. (type of inhibition: +++ strong; ++ moderate; + weak; − no inhibition).
| Salmonella sp. strain | Year of Salmonella acquisition | L. rhamnosus LOCK 1131 | L. casei LOCK 1132 | L. paracasei LOCK 1133 |
|---|---|---|---|---|
| S. Typhimurium ATCC 13311 | +++ | +++ | +++ | |
| S. Typhimurium ATCC 14028 | +++ | +++ | +++ | |
| S. Enteritidis ATCC 13076 | +++ | +++ | +++ | |
| S. Enteritidis 1067/S/09 K-2 | 2009 | + a | + a | +++ b |
| S. Enteritidis 848/S/09 | 2009 | ++ a | + b | ++ a |
| S. Enteritidis Ferma ‘K’ K-9 | 2010 | + a | + a | ++ b |
| S. Enteritidis Woź K-7 | 2010 | + a | ++ a | ++ b |
| S. Enteritidis Ferma ‘K’ K-10 | 2010 | ++ a | + b | ++ a |
| S. Enteritidis Ferma ‘K’ K-11 | 2010 | ++ a | + b | ++ a |
| S. Enteritidis Ferma ‘K’ K-12 | 2010 | + a | + b | + a |
| S. Enteritidis Ferma ‘K’ K-3 | 2010 | + a | ++ b | + a |
| S. Enteritidis Ferma ‘K’ K-4 | 2010 | + a | ++ b | + a |
| S. Enteritidis Ferma ‘K’ K-6 | 2010 | + a | + b | + a |
| S. Enteritidis Ferma ‘K’ K-14 | 2010 | + b | + a | ++ a |
| S. Enteritidis Ferma ‘K’ K-18 | 2010 | ++ a | + ab | + b |
| S. Enteritidis Pac K-piętro | 2010 | + ab | + a | + b |
| S. Enteritidis Woź K-6 | 2010 | + | + | + |
| S. Enteritidis Woź K-9 | 2010 | ++ a | + ab | + b |
| S. Enteritidis 64/S/10 | 2010 | + | + | + |
| S. Enteritidis 249 | 2010 | + | + | + |
| S. Enteritidis 517/S/09 | 2009 | ++ a | + ab | + b |
| S. Enteritidis 571/S/09 | 2009 | + a | + b | ++ c |
| S. Enteritidis 833/S/09 | 2009 | + | + | ++ |
| S. Enteritidis 838/S/09 | 2009 | ++ b | + a | + a |
| S. Enteritidis 838/S/09 B | 2009 | + | + | ++ |
| S. Enteritidis 847/S/09 | 2009 | + a | + a | − b |
| S. Enteritidis 865/S/09 | 2009 | + | + | + |
| S. Enteritidis 866/S/09 | 2009 | + a | − b | + a |
| S. Enteritidis 945/S/09 | 2009 | ++ b | + a | − a |
| S. Enteritidis 975/S/09 | 2009 | + | + | + |
| S. Enteritidis 1013/S/09 K-4 | 2009 | + a | ++ b | + a |
| S. Enteritidis 1014/S/09 K-1 | 2009 | + b | ++ a | + a |
| S. Enteritidis 1021/S/09 | 2009 | ++ b | + a | + a |
| S. Enteritidis 1022/S/09 K-9 | 2009 | ++ a | ++ a | + b |
| S. Enteritidis 1542/S/09 NWJ | 2009 | + ab | + a | + b |
| S. Enteritidis 1044/S/09 K-2 | 2009 | + a | − b | + a |
| S. Enteritidis 1445/S/09 K-46 | 2009 | + | + | + |
| S. Enteritidis 1446/S/09 K-31 | 2009 | ++ b | + a | + a |
| S. Enteritidis 1465/S/09 MEK | 2009 | + a | − b | + a |
| S. Enteritidis 1515/S/09 | 2009 | + a | + ab | + b |
| S. Enteritidis 1535/S/09 NWJ | 2009 | + b | + a | + a |
| S. Enteritidis 1714/09 | 2009 | + | + | + |
| S. Enteritidis 2050/S/09 | 2009 | + a | + ab | + b |
| S. Enteritidis 1085/S/09 MEK | 2009 | ++ a | ++ b | + c |
| S. Enteritidis 1231/S/09 | 2009 | + | + | + |
| S. Enteritidis 1206/S/09 MEK | 2009 | ++ a | + b | − c |
| S. Enteritidis 1171/S/09 | 2009 | − a | ++ b | + c |
| S. Enteritidis 1257/S/09 K7 | 2009 | ++ a | ++ b | + c |
| S. Enteritidis 1250/S/09 | 2009 | + a | ++ b | + a |
| S. Enteritidis 1143/S/09 | 2009 | + a | ++ b | ++ ab |
| S. Enteritidis 1422/S/09 MEK | 2009 | + a | ++ b | + c |
| S. Enteritidis 1106/S/09 K-1 | 2009 | + a | + a | ++ b |
| S. Enteritidis 1061/S/09/K-5 | 2009 | ++ b | ++ a | ++ a |
| S. Enteritidis 1067/S/09 K-3 | 2009 | + | ++ | + |
| S. Enteritidis 1573/S/09 NWJ | 2009 | ++ a | ++ b | ++ c |
| S. Enteritidis 1192/S/09 K-8 | 2009 | ++ ab | ++ a | + b |
| S. Enteritidis 1047/S/09 K-1 | 2009 | ++ a | ++ a | + b |
| S. Enteritidis 1545/S/09 MEK | 2009 | + ab | ++ a | + b |
| S. Enteritidis 1048/S/09 K-8 | 2009 | + | + | + |
| S. Enteritidis 1572/S/09 NWJ | 2009 | ++ ab | ++ a | ++ b |
| S. Enteritidis 002PP2014 | 2014 | + a | ++ ab | ++ b |
| S. Enteritidis 004PP2014 | 2014 | + a | ++ b | ++ b |
| S. Enteritidis 005PP2014 | 2014 | ++ a | + b | + c |
| S. Enteritidis 001PP2014 | 2014 | ++ | ++ | ++ |
| S. Enteritidis 006PP2014 | 2014 | + | + | + |
| S. Enteritidis 007PP2014 | 2014 | ++ a | + b | ++ a |
| S. Enteritidis 2149/09 | 2009 | ++ a | + b | ++ a |
| S. Enteritidis 2619/S/10 | 2010 | + b | + a | + a |
| S. Enteritidis 003PP2014 | 2014 | ++ a | + a | ++ b |
| S. Enteritidis 008PP2014 | 2014 | + b | ++ a | ++ a |
| S. Enteritidis 009PP2014 | 2014 | + a | + b | + a |
| S. Enteritidis 010PP2014 | 2014 | + a | + b | + a |
| S. Enteritidis 12617/2/S | 2014 | + a | + a | ++ b |
| S. Enteritidis 12633 | 2014 | + | + | + |
| S. Enteritidis 12961/1 | 2014 | + b | ++ a | ++ a |
| S. Enteritidis 13001 | 2014 | + a | ++ ab | ++ b |
| S. Enteritidis 011PP2014 | 2014 | ++ a | ++ b | ++ ab |
| S. Enteritidis 012PP2014 | 2014 | + a | + a | ++ b |
| S. Enteritidis 014PP2014 | 2014 | + a | + a | ++ b |
| S. Enteritidis 015PP2014 | 2014 | + a | + a | ++ b |
| S. Enteritidis 016PP2015 | 2015 | + a | + a | ++ b |
| S. Enteritidis 017PP2015 | 2015 | ++ a | + b | + ab |
| S. Enteritidis 018PP2015 | 2015 | ++ | ++ | ++ |
| S. Enteritidis 019PP2015 | 2015 | ++ a | + b | + c |
| S. Enteritidis 020PP2016 | 2016 | + | + | + |
| S. Enteritidis 021PP2016 | 2016 | ++ | ++ | ++ |
| S. Enteritidis 022PP2016 | 2016 | + a | + b | ++ a |
| S. Enteritidis 023PP2016 | 2016 | + | ++ | + |
| S. Enteritidis 024PP2016 | 2016 | + a | + b | ++ c |
| S. Enteritidis 025PP2016 | 2016 | + | + | + |
| S. Enteritidis 026PP2016 | 2016 | ++ a | ++ ab | + b |
| S. Enteritidis 027PP2016 | 2016 | ++ a | + b | + c |
| S. Enteritidis 028PP2016 | 2016 | + a | − b | + c |
| S. Enteritidis 029PP2016 | 2016 | + a | + b | + a |
| S. Enteritidis 030PP2016 | 2016 | + a | - b | + a |
| S. Enteritidis 031PP2016 | 2016 | + | + | + |
| S. Enteritidis 032PP2016 | 2016 | ++ a | ++ b | ++ c |
| S. Enteritidis 033PP2016 | 2016 | ++ | ++ | ++ |
| S. Enteritidis 034PP2016 | 2016 | + | + | + |
| S. Enteritidis 035PP2016 | 2016 | ++ a | ++ b | + c |
| S. Enteritidis 036PP2016 | 2016 | ++ | ++ | ++ |
| S. Enteritidis 037PP2016 | 2016 | + a | ++ b | + a |
| S. Enteritidis 038PP2016 | 2016 | ++ a | + b | ++ a |
| S. Enteritidis 039PP2016 | 2016 | ++ | ++ | + |
| S. Enteritidis 040PP2016 | 2016 | ++ a | ++ b | ++ ab |
| S. Enteritidis 041PP2016 | 2016 | ++ b | ++ a | ++ a |
| S. Enteritidis 042PP2016 | 2016 | ++ | ++ | ++ |
| S. Enteritidis 043PP2016 | 2016 | + a | + a | ++ b |
| S. Enteritidis 044PP2016 | 2016 | ++ a | +++ b | ++ a |
| S. Enteritidis 045PP2016 | 2016 | ++ | +++ | ++ |
| S. Enteritidis 046PP2016 | 2016 | + | ++ | + |
| S. Enteritidis 047PP2016 | 2016 | ++ a | ++ b | ++ ab |
| S. Enteritidis 048PP2016 | 2016 | + b | + a | + a |
| S. Enteritidis 049PP2016 | 2016 | ++ a | +++ b | + c |
| S. Enteritidis 050PP2016 | 2016 | ++ a | + a | + b |
| S. Enteritidis 051PP2016 | 2016 | +++ | +++ | +++ |
| S. Enteritidis 53A | 2013 | ++ a | + b | ++ a |
| S. Enteritidis Korczak 28A | 2013 | ++ a | + b | ++ a |
| S. Enteritidis Korczak 32A | 2013 | + | + | + |
| S. Enteritidis Korczak 40A | 2013 | ++ a | + b | + c |
| S. Enteritidis Korczak 41A | 2013 | ++ a | + b | ++ a |
| S. Enteritidis 4050/2014 D09 | 2014 | ++ a | ++ b | ++ ab |
| S. Enteritidis 6537/1/S/14 D09 | 2014 | + a | + b | + ab |
| S. Enteritidis 6670/1/S/14 D09 | 2014 | + a | + a | + b |
| S. Enteritidis 7660/1/D03 | 2014 | + a | + b | + c |
| S. Enteritidis 65/S/10 | 2010 | + a | + ab | + b |
| S. Enteritidis 1KSFA | 2013 | ++ a | + ab | + b |
Different superscripts in the same row denote statistically significant differences in antagonistic activity of different Lactobacillus strains (p < 0.05).
Lactobacillus sp. strains were cultivated in De Man, Rogosa and Sharpe (MRS) broth (Merck-Millipore, Darmstadt, Germany). Salmonella sp. strains were cultivated in nutrient broth with glucose (Merck-Millipore, Darmstadt, Germany) or LB broth (BIOCORP, Warsaw, Poland). The strains were stored at –80°C in the broth supplemented with 25% glycerol. Strains were grown at 37°C. Stock cultures were stored for 24 h at 4–10°C.
Survival of lactobacilli in the presence of bile salts. As the concentration of bile salts in the small intestine ranges from 0.2 to 2% (Kristoffersen et al. 2007), the ability of LAB strains to survive in the presence of bile salts (1 and 2%) was evaluated during and after their exposure for 1–4 h (as the food passage through small intestine takes up to 5 h). Tubes containing 30 ml MRS broth were inoculated with bacterial stock cultures (3%, v/v) and incubated for 24 h at 37°C. After incubation, 1 ml aliquots were taken, serially diluted and plated on MRS agar in order to enumerate the initial bacterial population (at 0 h). Meanwhile, bacteria grown in MRS broth were centrifuged (3852 × g, 10 min, 4°C), washed with sterile phosphate buffer saline (PBS, pH 7.2, Sigma-Aldrich, St. Louis, MO, USA), divided into three portions, and finally suspended in distilled water containing 1% and 2% of bile salts (Sigma-Aldrich, St. Louis, MO, USA) or in distilled water without bile salts (control), respectively. The suspensions were then incubated aerobically at 37°C. After 1, 2, and 4 h, 1 ml aliquots were taken from each sample, serially diluted in sterile saline, and plated on MRS agar for the enumeration of total viable counts. Plates were incubated for 48 h at 37°C, colonies were counted and the bacterial population was calculated for each measurement point.
Survival of lactobacilli at low pH. The ability of LAB strains to survive at low pH (pH 2.0 and 3.0 – similar to pH of gastric juice, which is 1.5–3.5) was investigated during 1–4 h (as the food passage through the stomach is up to 5 h). Tubes containing 30 ml of MRS broth were inoculated with bacterial stock cultures (3%, v/v) and incubated for 24 h at 37°C. After incubation, 1 ml aliquots were taken, serially diluted, and plated on MRS agar in order to enumerate the initial bacterial population (at 0 h). Meanwhile, bacteria grown in MRS broth were centrifuged (3852 × g, 10 min, 4°C), washed with sterile PBS, divided into three portions and suspended in sterile saline, adjusted to pH 2.0 or 3.0 respectively (with 0.1 M HCl), and the sterile saline with unchanged pH that was included as a control. The suspensions were then incubated aerobically at 37°C. After 1, 2, and 4 h 1 ml aliquots were taken from each sample, serially diluted in sterile saline, and plated on MRS agar. After incubation of plates for 48 h at 37°C, the viable counts were determined.
Testing of susceptibility to antibiotics. The antibiotic resistance of LAB strains was screened by the agar disc diffusion method using discs impregnated with specified concentrations of antibiotics (Oxoid, Dardilly, France). A total of six antibiotics were tested: penicillin G (1 unit), kanamycin (30 μg), tetracycline (30 μg), erythromycin (30 μg), doxycycline (30 μg), and amoxicillin (25 μg). These were commercial antibiotics and their standard doses were commonly used for testing of probiotic bacteria. Antibiotics are mainly absorbed in the ileum and their therapeutic dose, which reaches the colon is lower than the initial dose (Moubareck et al. 2005). Briefly, 100 μl of the 24-hour culture of LAB was spread on MRS agar plates using a sterile L-shaped bacterial spreader and antibiotic discs were placed aseptically on the surface of the agar. Each antibiotic was tested in triplicate. Plates were incubated for 24 h at 37°C and the diameters of the inhibition zones were measured with a ruler. The results (average of three readings) were interpreted according to the standards defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2012).
Aggregation and coaggregation assays. Aggregation assay was performed as described by Kos et al. (2003) and Jankovic et al. (2012) with some modifications. LAB grown for 24 h in a liquid MRS were harvested by centrifugation (3852 × g, 10 min), washed once and re-suspended in PBS to give a final optical density of 1.0 at 600 nm, as measured by the spectrophotometer (Beckman DU 640, USA). For the autoaggregation assay, the cell suspension of each bacterial strain was vortexed for 10 s and incubated at room temperature without agitation. After 24 h, 100 μl of the suspension (the upper part) was transferred to another tube and the absorbance was measured at 600 nm. The autoaggregation percentage was determined using equation 1, as follows:
| (1) |
where At represents the absorbance at time t = 24 h, and A0 the absorbance at time t = 0 h.
The coaggregation was measured for LAB with reference to pathogenic strains of Salmonella sp. For the coaggregation assay, LAB and Salmonella sp. cell suspensions were prepared in the same way as that for the autoaggregation assay, but Salmonella sp. were cultivated in nutrient broth medium. Equal volumes of LAB and Salmonella sp. suspensions were mixed by vortexing for 10 s. As control tubes, samples from the autoaggregation assay containing aliquots of single bacterial suspension were used. The absorbance of suspensions was measured after 24 h, as described above. The percentage of coaggregation was calculated according to equation 2 (Handley et al. 1987), as follows:
| (2) |
where Ax and Ay represent absorbance of each of the two strains in the control tubes and Ax+y absorbance of their mixture. Each experiment was performed in triplicate.
Adherence to polystyrene. The adherence was determined according to the method described by Zeraik and Nitschke (2012) with some modifications (Aleksandrzak-Piekarczyk et al. 2016). Bacterial suspensions in PBS were prepared the same way as for the autoaggregation assay, then added into 96-well polystyrene plate (in eight repeats each), and incubated for 2 h at 37°C. Non-adherent bacteria were slightly washed away with water, and adherent bacteria were fixed with 80% methanol (15 min) and stained with 0.1% crystal violet (15 min, 120 rpm). Next, the wells were washed out with water and incubated with 33% glacial acetic acid (15 min, 120 rpm). The absorbance was measured at 630 nm with a microplate reader (Tristar2 LB 942, Berthold Technologies). To calculate the adhesion ratio of bacteria to polystyrene, the value of absorbance of bacteria was divided by the absorbance of the control sample (absorbance read in polystyrene wells only).
Adherence to human colon adenocarcinoma cells (Caco-2). The adherence of probiotic bacteria to colon cells of the gastrointestinal tract is a clue process for bacteria to survive and colonize this ecosystem as the adherence prevents probiotic cells from being washed out, and enables temporary colonization of the colon (Nowak and Motyl 2017). The adherence assay of Lactobacillus sp. strains to human colon adenocarcinoma cell line (Caco-2) was described previously (Nowak and Motyl 2017). Briefly, Caco-2 cells were seeded in 24-well tissue culture plates (Becton, Dickinson and Co., Franklin Lakes, NJ, USA) at a concentration of 2.5 × 105 cells per well to obtain confluent monolayers. Before the experiment, LAB strains were grown in MRS broth for 24 h at 37°C. After growth, bacteria were harvested by centrifugation (3852 × g, 10 min), washed with sterile PBS and re-suspended in fresh Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) without antibiotics and deposited on Caco-2 cells monolayer at the density of 2.8 × 108 CFU/ml. To provide each time the same density of the initial population of bacteria added to wells, the relationship between the number of bacteria of each LAB strain and its absorbance (λ = 540 nm) was drawn as a curve. Each strain was tested in triplicate. The plate was incubated for 2 h at 37°C in a 5% CO2 atmosphere. After incubation, the unbound bacteria were removed. They were aspirated and wells were slightly washed with sterile PBS. Then wells were treated with 1% trypsin-EDTA (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at 37°C to detach the cells and, additionally, the wells were scraped with a cell scraper. Subsequently, Caco-2 cells were harvested by centrifugation (3852 × g, 10 min), and treated with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min for lysis. The adherent bacterial cells were enumerated by the pour plating serial dilution method in MRS agar and the plates were incubated for 48 h at 37°C. The adherence (%) was expressed as a ratio of the number of adherent bacteria to the number of total bacteria added initially to each well (an adherence index).
Competition assay. The adherence assay to Caco-2 cells of Lactobacillus sp. strains in a competition test with pathogens was described previously (Nowak and Motyl 2017). In brief, the reference Salmonella sp. strains (108 CFU/ml) were added to the wells with Caco-2 cells monolayer as a single strains for individual adherence assay or in conjunction with probiotic strains (1:1) and then incubated at 37°C in 5% CO2 (Galaxy 48S, New Brunswick, United Kingdom) for 2 h. For each strain, the adherent bacteria were enumerated by plate counting with SS Agar medium (Merck-Millipore, Darmstadt, Germany) and were incubated in aerobic conditions at 37°C for 48 h. The inhibition rate [%] was calculated according to the equation 3, this represented a percentage reduction in adherent pathogens in the presence of a probiotic (as compared to the results for the pathogen alone):
| (3) |
Cytotoxicity. Cytotoxicity of the probiotics tested was measured by the neutral red uptake (NRU) assay as described by Repetto et al. (2008) and Borowiec et al. (2016) with some modifications. LAB supernatants were prepared from overnight culture by centrifugation (3852 × g, 10 min), and sterilization through a 0.22 μm filter. Caco-2 cells were seeded in 96-well plates in a concentration of 104 cells per well and exposed to LAB supernatants at different concentrations for 72 hours at 37°C in a humidified atmosphere of 5% CO2. The LAB supernatants were diluted as follows: 1, 1:2, 1:4, 1:8, 1:16, and the medium for cell culture was used as a control. After incubation, cells were washed with PBS and incubated with 50 g/ml neutral red in Hanks’ Balanced Salt Solution for 3 h, then washed with PBS again. Cell-bound dye was extracted with a solution containing 50% ethanol and 1% acetic acid by gentle shaking for 10 min. Absorbance at 550 nm was read at the Sunrise microplate reader (Tecan, Männedorf, Switzerland).
Antagonistic activity of lactobacilli towards reference and environmental strains of Salmonella sp. The inter-strain antagonism was investigated using the agar slab method (Strus 1998; Klewicka and Libudzisz 2004). To test the activity of LAB against Salmonella strains, strains of Lactobacillus sp. (at the density of 108 CFU/ml) were introduced into MRS agar medium and poured onto Petri dishes. Then, LAB strains were incubated at 37°C for 24 h. In the meantime Salmonella sp. stock cultures were prepared and 100 μl of each culture was spread on NB or LB agar plates using a sterile L-shaped bacterial spreader. Next, slabs around 9 mm in diameter (in three repeats) were cut out from solidified MRS medium overgrown with LAB and put on the prepared agar broth containing Salmonella sp. strains (106 CFU/ml). The dishes were incubated at 37°C for 18 h. Following the incubation, the diameter of the test strain growth inhibition zone was measured, the slab diameter was subtracted, and the result was presented in millimeters. To compare the antagonistic activity of the strains of LAB against the Salmonella cultures, the following criteria were adopted: the growth inhibition diameter above 11 mm – strong inhibition (+++), 6–10 mm – moderate inhibition (++), 1–5.9 mm – weak inhibition (+), 0 mm – no inhibition (–). The index of total antagonistic activity (IAA) was calculated as a sum of scores determined by the diameter of growth inhibition of bacteria (+++ 3 points; ++ 2 points; + 1 point; – 0 points), (Klewicka and Libudzisz 2004).
Statistical analysis. Statistical analysis was performed using Statistica (ver. 13.1) as the statistical program. 1-way ANOVA followed by Tukey’s post hoc test was performed for comparison of growth inhibition zone in the assay of antagonistic activity of Lactobacillus sp. strains against Salmonella sp. 1-way ANOVA followed by Dunnett’s post hoc test was performed for comparison of cytotoxic activity towards Caco-2 cells. Significant differences were declared at p < 0.05.
Results
The resistance of LAB to bile salts. The survival of the LAB strains examined in bile salts at concentrations of 1% and 2% is presented in Fig. 1. The highest decrease in the cell number was observed for L. rhamnosus LOCK 1131 after incubation in the presence of 1% of bile salt, with the highest drop occurring just after 1 h of incubation (–6 log). The increased bile salts tolerance was observed for 2% bile salts concentration for this bacteria. L. casei LOCK 1132 and L. paracasei LOCK 1133 demonstrated a relatively high survival rate even after 4 h incubation at 1% and 2% of bile salts (–3 log for L. casei and –3.9 log for L. paracasei). The decrease in the bacterial number at 1% and 2% of bile salts was statistically significant (p < 0.05) for all strains even after an hour incubation.
Fig. 1.
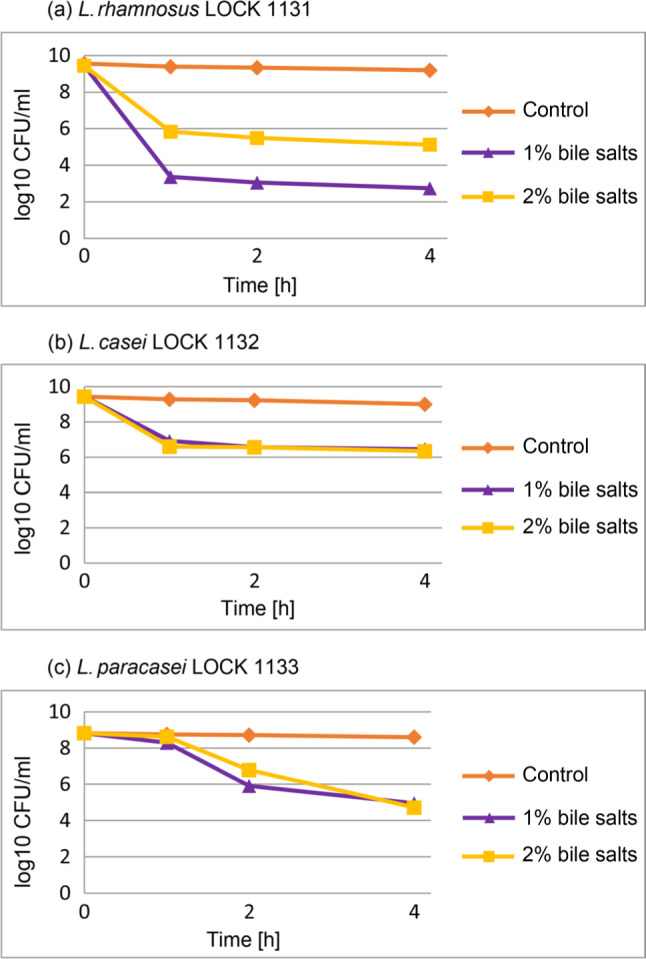
The survival of lactic acid bacteria at 1% and 2% bile salts.
Resistance of LAB to low pH. The resistance of lactic acid bacteria to pH 2.0 and pH 3.0 is shown in Fig. 2. L. rhamnosus LOCK 1131 and L. casei LOCK 1132 demonstrated very high survival at both pH values, as almost no decrease in viability was observed after 4 h of incubation, when compared to the control sample with physiological saline. However, the viability of L. paracasei LOCK 1133 at more acidic pH (pH 2.0) decreased significantly (p < 0.05) from 8.8 ± 0.04 log CFU/ml to approx. 2.9 ± 0.1 log CFU/ml after 4 h, while in less acidic environment (pH 3.0) the number of viable cells remained higher (4.4 ± 0.3 log CFU/ml).
Fig. 2.
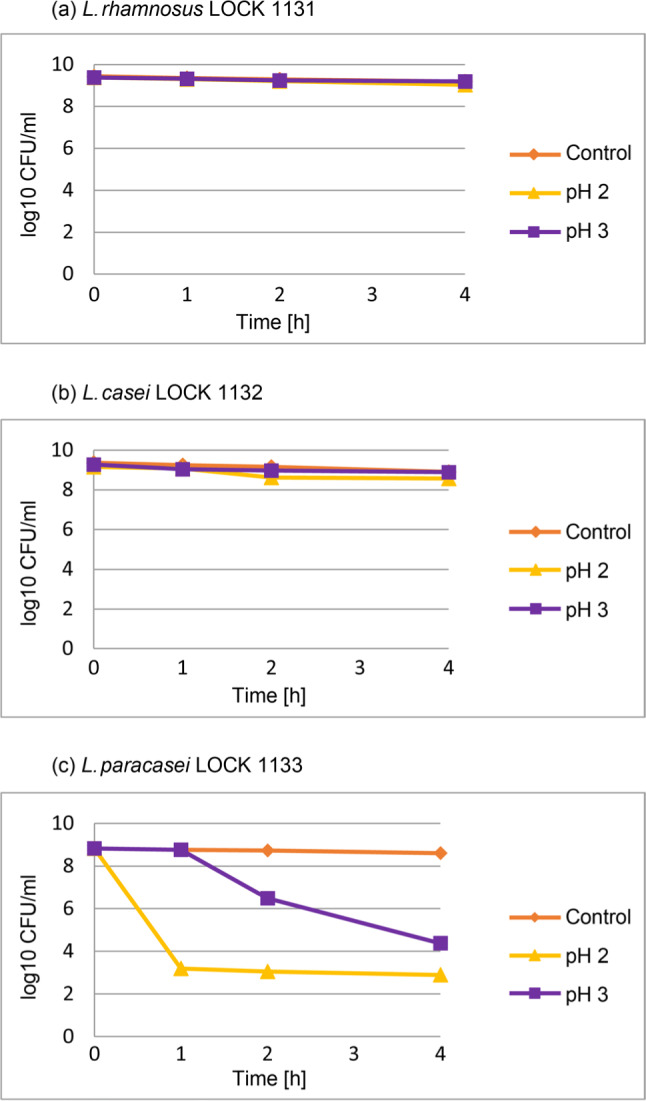
The resistance of LAB to low pH.
Antibiotic susceptibility. The results of antibiotic resistance testing of LAB strains are collected in Table I. All strains were resistant to amoxicillin (25 μg) and kanamycin (30 μg). Besides, all strains were susceptible to penicillin G (1 unit), tetracycline (30 μg), erythromycin (30 μg), and doxycycline (30 μg).
Table I.
Antibiotic resistance of Lactobacillus strains.
| Antibiotic inhibition zone diameter [mm ± SD] |
||||||
|---|---|---|---|---|---|---|
| PEN | KAN | TET | ERY | DOX | AMX | |
|
L. rhamnosus LOCK 1131 |
27 ± 2.5 (S) |
0 (R) |
34 ± 1.0 (S) |
33 ± 2.5 (S) |
32 ± 2.0 (S) |
21 ± 2.1 (R) |
|
L. casei LOCK 1132 |
28 ± 2.5 (S) |
0 (R) |
30 ± 0.6 (S) |
34 ± 1.5 (S) |
33 ± 1.0 (S) |
24 ± 0.6 (IR) |
|
L. paracasei LOCK 1133 |
27 ± 6.4 (S) |
0 (R) |
25 ± 1.5 (S) |
25 ± 0.0 (S) |
30 ± 0.6 (S) |
21 ± 1.0 (R) |
S: susceptible; R: resistant; IR: intermediate resistant
PEN: Penicillin G (1 unit); KAN: Kanamycin (30 μg); TET: Tetracycline (30 μg);
ERY: Erythromycin (30 μg); DOX: Doxycycline (30 μg); AMX: Amoxicillin (25 μg)
Aggregation and coaggregation with Salmonella sp. All LAB strains showed high autoaggregation ability with the autoaggregation rate (Table II) ranging from 93.5% ± 1.2 (for L. paracasei LOCK 1133) to 98.2% ± 3.6 (for L. casei LOCK 1132) observed after 24 h.
Table II.
Probiotic properties of Lactobacillus strains.
| Autoaggregation | Coaggregation | Coefficient of adherence to polystyrene | Adherence to Caco-2 cells | |||||
|---|---|---|---|---|---|---|---|---|
| S. Ent. 13076 | S. Typ. 13311 | S. Typ. 14028 | Init. No. | After 2 h | Adherence index | |||
| [log CFU/ml] | ||||||||
| L. rhamnosus LOCK 1131 | 96.1% ± 1.2% | 14.8% ± 0.01% | 84.2% ± 0.01% | 51.8% ± 0.03% | 1.18 ± 0.02 | 8.5 | 7.6 ± 1.1 | 89.8% |
| L. casei LOCK 1132 | 98.2% ± 3.6% | 26.1% ± 0.01% | 58.5% ± 0.01% | −11.1%± 0.01% | 1.22 ± 0.02 | 8.5 | 8.3 ± 0.5 | 98.0% |
| L. paracasei LOCK 1133 | 93.5% ± 0.9% | 41.8% ± 3.4% | 17.8% ± 3.3% | 43.7% ± 2.7% | 1.20 ± 0.07 | 8.0 | 6.3 ± 0.1 | 78.9% |
The coaggregation of LAB strains with three reference Salmonella sp. strains was also explored. L. rhamnosus LOCK 1131 exhibited the highest coaggregation with S. Typhimurium ATCC 13311 and S. Typhimurium ATCC 14028 (Table II), while L. casei LOCK 1132 with S. Typhimurium ATCC 13311. L. paracasei LOCK 1133 demonstrated moderate coaggregation with S. Enteritidis ATCC 13076 and S. Typhimurium ATCC 14028, but weak with S. Typhimurium ATCC 13311.
Adherence to polystyrene and Caco-2 cells. The adherence to polystyrene surface and Caco-2 cells was evaluated for all LAB strains. The results are collected in Table II. All strains demonstrated weak adherence properties with adherence ranging from 1.18 ± 0.02 to 1.22 ± 0.02. All strains strongly adhered to Caco-2 cells. The highest level of adherence was observed for L. casei LOCK 1132 (98.0%). However, L. rhamnosus LOCK 1131 also exhibited strong adherence to intestinal epithelial cells (89.8%). L. paracasei LOCK 1133 had a statistically significantly lower adherence index than other strains (78.9%).
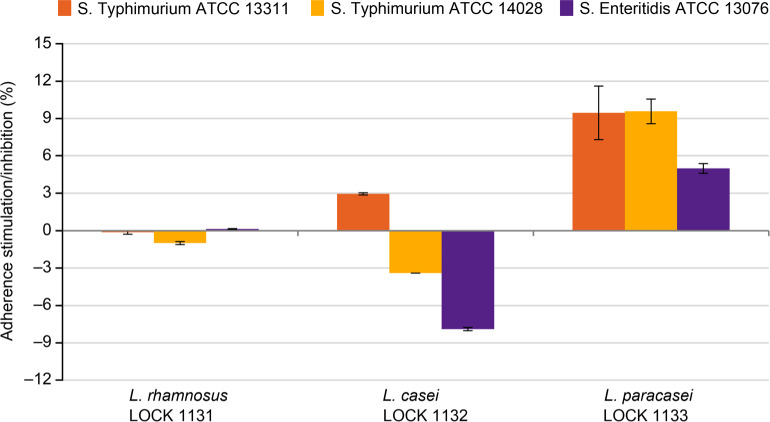
Antagonistic activity of LAB in competition assay – adherence to Caco-2 cells. In the competition test, the antagonistic activity of LAB on the adherence of Salmonella sp. ATCC strains to adenocarcinoma cells Caco-2 was tested. Only L. casei LOCK 1132 showed good efficiency in inhibition of two tested Salmonella strains: S. Enteritidis ATCC 13076 and S. Typhimurium ATCC 14028, up to 3.4% and 7.9%, respectively (Fig. 3). L. rhamnosus LOCK 1131 weakly reduced adherence only of S. Typhimurium ATCC 13311 (in 1%), while L. paracasei LOCK 1133 stimulated adherence of all tested Salmonella sp. strains (from approx. 5 to 10%).
Fig. 3.
Adherence of Salmonella sp. strains to adenocarcinoma cells Caco-2 in the presence of lactic acid bacteria presented as stimulation/inhibition rate (%).
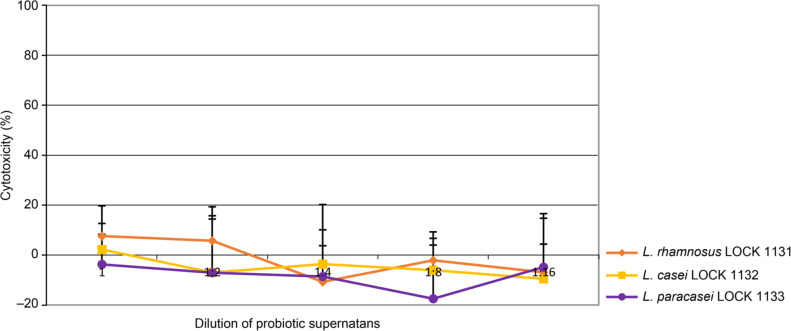
Cytotoxicity assay of LAB metabolic products. To assess cytotoxic effect of LAB strains, the effects of bacterial supernatants from LAB liquid cultures were examined in the comparison with the bacterial medium used for cell culture. The medium itself, regardless of the dilution factor, induced significant cytotoxic activity (p < 0.05 for dilutions between 1:1 and 1:8). There were no statistically significant differences between LAB supernatants and the medium itself for any strain or dilution. The results are presented as average from four experiments. Fig. 4 presents the cytotoxic effect of LAB supernatants. The obtained results indicate very low cytotoxicity towards Caco-2 cells of substances secreted into the medium by bacteria in the range of tested dilutions.
Fig. 4.
Cytotoxic effect of probiotics’ supernatants on Caco-2 cells in the range of tested dilutions presented as % of control.
Antagonistic activity of LAB against Salmonella sp. The antagonism of LAB was investigated against 126 Salmonella sp. strains, including three reference strains from ATCC and 123 environmental Salmonella Enteritidis. All strains of LAB demonstrated strong antagonistic activity towards all tested Salmonella sp. strains (Table III).
L. paracasei LOCK 1133 displayed the largest growth inhibition zone of all LAB strains tested for the highest number of Salmonella sp. strains; while, L. rhamnosus LOCK 1131 for the lowest number of strains. The calculated IAA values were similar for all three LAB strains with the highest value reported for L. rhamnosus LOCK 1131 (185 points) and lowest for L. casei LOCK 1132 (178 points) (Fig. 5).
Fig. 5.
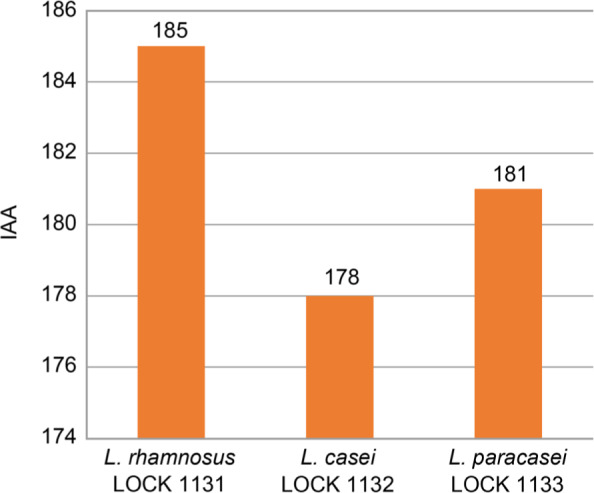
Total antagonistic activity index (IAA) of lactic acid bacteria.
Discussion
The increasing number of evidence demonstrates that probiotics are effective in the prevention and treatment of bacterial diseases in animals. Due to limitations in the application of antibiotics, probiotics may be a promising alternative. It holds especially in case of Salmonella, the second most widespread foodborne pathogen (European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) 2018), which is very difficult to control in poultry farming; thus, prevention and control strategies in commercial livestock are essential. When treatment is necessary it may occur that the causative strains can be resistant to most of the antibiotics.
Recent methods for Salmonella treatment, apart from antibiotics, include also application of probiotics (Patterson and Burkholder 2003), bacteriophages (Fiorentin et al. 2005; Toro et al. 2005; Kim et al. 2014), organic acids, plants extracts, and essential oils (Tellez et al. 2012). There is also evidence that ammonia at high pH is effective against Salmonella in poultry farming (Koziel et al. 2017). Promising results have been observed when treatments with bacteriophages and competitive exclusion products were applied simultaneously (Toro et al. 2005; Borie et al. 2009).
Therefore, lactic acid bacteria strains that reduce or even eliminate Salmonella in animal husbandry are highly desired. In this study, we examined three new Lactobacillus sp. strains selected from ileal content (L. rhamnosus LOCK 1131, L. casei LOCK 1132) and domestic fermented cabbage (L. paracasei LOCK 1133). To assess their potential as successful probiotics, certain features that allow them to withstand harsh gastrointestinal tract conditions and colonize the intestine were examined. Additionally, taking into account their potential application in controlling Salmonella sp. in poultry farming, the antagonistic activity towards environmental poultry isolates of Salmonella sp. bacteria and antibiotic susceptibility were evaluated.
The survival of bacteria in the gastrointestinal tract is highly dependent on bacteria’s resistance to bile salts and low pH (Verdenelli et al. 2009). The tolerance of probiotics strains to increased bile salts concentration is one of the crucial features responsible for their function and stability in the small intestine. Moreover, the high concentration of bile salt can be used as a strong tool for the selection of potential probiotic strains. In this study, all three strains survive under bile salts conditions higher than those observed in the intestinal tract (1% and 2%) even after 4 h of incubation. Moreover, L. rhamnosus LOCK 1131 strain showed better tolerance to the higher bile salt concentration. These results are consistent with those of Duary et al. (2012) and Wang et al. (2010) who also reported that the potential probiotic strains were able to survive at bile salt concentration higher than physiological levels. Our finding suggests that the selected Lactobacillus strains have the potential to survive in the harsh condition of the gastrointestinal tract.
In the case of pH tolerance, probiotic’s passage through media with low pH is strain-dependent (Belicová et al. 2013). The results of our study also confirmed this observation. L. rhamnosus LOCK 1131 and L. casei LOCK 1132 demonstrated very high survival during the incubation in the media with pH of 2.0 and 3.0 whereas L. paracasei LOCK 1133 cell viability decreased significantly at pH 2.0.
To colonize GIT, probiotic bacteria need to adhere to mucosal surfaces (Tropcheva et al. 2011). It is a multistep process based on non-specific physical interactions between cells and bacteria, which then enable specific interactions between complementary receptors (Kos et al. 2003). Aggregation is the process of reversible gathering of bacterial cells belonging to the same bacterial strain (autoaggregation) or two different bacterial strains (coaggregation). Adhesion is dependent on autoaggregation properties of bacteria, while coaggregation provides close interaction with pathogenic bacteria, in which lactobacilli could release antimicrobial substances in very close proximity. These features also allow probiotics to form a barrier preventing pathogens from colonization (Kos et al. 2003; Janković et al. 2012). All LAB strains tested showed a high autoaggregation rate above 93% observed after 24 h. The coaggregation rate with Salmonella sp. was strain-dependent and each lactobacilli had different coaggregation rates depending on the Salmonella strain examined. The highest coaggregation was observed for a pair of L. rhamnosus LOCK 1131 and S. Typhimurium ATCC 13311 (84%), while the lowest for a pair of L. casei LOCK 1132 and S. Typhimurium ATCC 14028 (–11%).
One of the most essential features of probiotics is their ability to adhere to intestinal cells and colonize the gastrointestinal tract of the host. Colonization is related to many beneficial effects such as the stimulation of immune system and the prevention of pathogen colonization either by probiotics antagonistic activity towards pathogens or by their competition for a limited number of receptors on the surface of GIT (Cukrowska et al. 2009; Ostad et al. 2009; Verdenelli et al. 2009). Thus, a high rate of adherence to the biotic surface is the desired feature, whereas adherence to the abiotic surface can be problematic in the production of a probiotic formulation. Adherence properties of lactic acid bacteria depend on many factors – especially on the cell wall structure and the individual strain. In this study, the ability of bacteria to form a biofilm on an abiotic surface was examined by measuring the adherence to a polystyrene surface. All strains revealed weak adherence properties with the adherence values ranging from 1.18 ± 0.02 to 1.22 ± 0.02 at 630 nm. Adherence to epithelial cells was assessed by measuring the adherence index of LAB strains to Caco-2 cells. The highest level of adherence was observed for L. casei LOCK 1132 (98.0%) and the weakest adherence of all tested strains (78.9%) had L. paracasei LOCK 1133. However, all strains strongly adhered to Caco-2 cells which was confirmed by microscopic observations (results not shown).
Another important method, used in this study, was a competition test conducted to observe the stimulation or inhibition of Salmonella sp. adherence to Caco-2 cells. By adhering to the biotic surface, the probiotics may prevent the colonization of pathogenic bacteria. It was found that L. casei LOCK 1132 inhibited S. Enteritidis ATCC 13076 and S. Typhimurium ATCC 14028, up to 3.4% and 7.9%, respectively. L. rhamnosus LOCK 1131 weakly reduced adherence of S. Typhimurium ATCC 13311 (1%), while L. paracasei LOCK 1133 stimulated adherence of all tested Salmonella sp. strains (5 to 10%). The inhibition of Salmonella sp. adherence determined in this study is not very high when compared to the results obtained by others (Jankowska et al. 2008). However, to gain deeper insight into this phenomenon one can consider that the adherence of the pathogens in vivo can be interrupted not only by the competition for the cell receptors but also by the secretion of antimicrobial compounds by the probiotic strains. Further tests are necessary to confirm these findings.
To assess the cytotoxicity of probiotic’s metabolic products and, thus, to confirm their safety for the application, the NRU assay was performed. No significant cytotoxic effect was detected when compared to control samples. The undiluted supernatant from L. rhamnosus LOCK 1131 culture had the highest cytotoxic effect (8%), whereas eight-times diluted supernatant from L. paracasei LOCK 1133 culture had lower cytotoxic effect than control sample (–18%).
As the target application of the described LAB strains for using in poultry farming their susceptibility to antibiotics (penicillin G, tetracycline, erythromycin, and doxycycline) was evaluated. All three strains were found to be resistant to amoxicillin and kanamycin, which is not a rare phenomenon among probiotics as they are naturally resistant to certain antibiotics. Several studies reported lactobacilli intrinsic resistance to kanamycin and amoxicillin (Varankovich et al. 2015; Imperial and Ibana 2016; Wang et al. 2018). In some cases, antibiotic resistance of probiotics could be beneficial due to the accompanying administration of various antimicrobials during animal farming or the application of antibiotic-probiotic combination treatment (Wright et al. 2015). What is more important, the probiotic strains identified in our study belong to species included in the EFSA QPS list.
Finally, the broad antagonistic spectrum against Salmonella sp. was examined as the most desired feature of tested LAB strains. The antagonistic activity of probiotics towards pathogenic bacteria, could prevent their attachment to the gastrointestinal tract and stimulate their removal from the intestine (Gaggìa et al. 2010; Varankovich et al. 2015; Mokoena 2017).
All tested LAB bacteria revealed the ability to inhibit the growth of most of the environmental strains of Salmonella sp. and all reference strains. L. rhamnosus LOCK 1131 inhibited growth of 125 out of 126 tested strains (99.2% coverage), L. casei LOCK 1132 had 98% coverage and L. paracasei LOCK 1133 had 96% coverage.
While acting in the gastrointestinal tract, probiotic formulations affect an entire body of the animal. It is known that lactic acid bacteria compete for food and the place of adhesion to the intestinal epithelium with pathogens; however, the mechanisms of this actions are still not fully understood (Trzeciak et al. 2016). Probiotic mixtures seem to have a better effect than single bacteria preparation because they can address a wide spectrum of pathogens and may have synergistic adhesion effects (Verdenelli et al. 2009; Chapman et al. 2011). The synergistic effect on adhesion and antagonistic activity of presented bacteria still need to be evaluated. There is also the requirement for further research that will investigate other properties of studied bacteria such as the presence of antibiotic resistance plasmids, the nature of the antimicrobial agents produced by the bacteria, and the ability to withstand conditions of simulated gastrointestinal tract.
Our results showed that L. rhamnosus LOCK 1131, L. casei LOCK 1132 and L. paracasei LOCK 1133 possess a number of interesting properties and can be considered as a component of functional feed. They can withstand the gut conditions, interact with the host system and their safety was proved in the cytotoxicity test. However, what is most important, they possess antagonistic activity towards a wide spectrum of environmental Salmonella species. The S. Enteritidis collection used in this study is an epidemiologically representative group of strains isolated from poultry in 2009–2016 that were confirmed as genetically different (Zaczek et al. 2015). Results of antagonistic activity obtained on such representative collection of Salmonella sp. allow the conclusion that the probiotics tested in this study have a high potential for use in commercial poultry farming.
Acknowledgments
Agnieszka Chlebicz, Anna Paliwoda, Monika Terenowicz and Marta Zygmunt for assistance in conducting the assays; dr Aurelia Walczak-Drzewiecka for conducting cytotoxicity test; dr Ewelina Wójcik for revising the manuscript.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
ORCID
Justyna D. Kowalska https://orcid.org/0000-0003-1102-9389
Funding
This research was funded by project “Innovative bacteriophage preparation to ensure the microbiological safety of poultry farming” co-financed by the European Regional Development Fund under the Smart Growth Operational Programme 2014–2020 (POIR.01.01.02-00-0052/16).
Literature
- Aleksandrzak-Piekarczyk T, Koryszewska-Bagińska A, Grynberg M, Nowak A, Cukrowska B, Kozakova H, Bardowski J.. Genomic and functional characterization of the unusual pLOCK 0919 plasmid harboring the spaCBA pili cluster in Lactobacillus casei LOCK 0919. Genome Biol Evol. 2016;8(1):202–217. 10.1093/gbe/evv247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelakis E. 2017. Weight gain by gut microbiota manipulation in productive animals. Microb Pathog. 106:162–170. 10.1016/j.micpath.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Belicová A, Mikulášová M, Dušinský R.. Probiotic potential and safety properties of Lactobacillus plantarum from Slovak Bryndza cheese. BioMed Res Int. 2013;2013:1–8. 10.1155/2013/760298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi S, Alpigiani I, Bruini I, Barilli E, Brindani F, Morganti M, Cavallini P, Bolzoni L, Pongolini S.. Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Int J Food Microbiol. 2016;218:44–50. 10.1016/j.ijfoodmicro.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Borie C, Sánchez ML, Navarro C, Ramírez S, Morales MA, Retamales J, Robeson J.. Aerosol spray treatment with bacteriophages and competitive exclusion reduces Salmonella enteritidis infection in chickens. Avian Dis. 2009. Jun;53(2):250–254. 10.1637/8406-071008-Reg.1 [DOI] [PubMed] [Google Scholar]
- Borowiec M, Gorzkiewicz M, Grzesik J, Walczak-Drzewiecka A, Salkowska A, Rodakowska E, Steczkiewicz K, Rychlewski L, Dastych J, Ginalski K.. Towards engineering novel PE-based immunotoxins by targeting them to the nucleus. Toxins (Basel). 2016. Nov 10;8(11):321 10.3390/toxins8110321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B.. Salmonella nomenclature. J Clin Microbiol. 2000. Jul;38(7):2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A, Adams M, La Ragione RM, Woodward MJ.. Colonisation of poultry by Salmonella Enteritidis S1400 is reduced by combined administration of Lactobacillus salivarius 59 and Enterococcus faecium PXN-33. Vet Microbiol. 2017;199:100–107. 10.1016/j.vetmic.2016.12.029 [DOI] [PubMed] [Google Scholar]
- Chapman CMC, Gibson GR, Rowland I.. Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr. 2011. Feb;50(1):1–17. 10.1007/s00394-010-0166-z [DOI] [PubMed] [Google Scholar]
- Chousalkar K, Gole VC.. Salmonellosis acquired from poultry. Curr Opin Infect Dis. 2016. Oct;29(5):514–519. 10.1097/QCO.0000000000000296 [DOI] [PubMed] [Google Scholar]
- Cukrowska B, Motyl I, Kozáková H, Schwarzer M, Górecki RK, Klewicka E, Śliżewska K, Libudzisz Z.. Probiotic Lactobacillus strains: in vitro and in vivo studies. Folia Microbiol (Praha). 2009. Nov;54(6):533–537. 10.1007/s12223-009-0077-7 [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D.. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010. Sep 01;74(3):417–433. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duary RK, Batish VK, Grover S.. Relative gene expression of bile salt hydrolase and surface proteins in two putative indigenous Lactobacillus plantarum strains under in vitro gut conditions. Mol Biol Rep. 2012. Mar;39(3):2541–2552. 10.1007/s11033-011-1006-9 [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA), European Centre for Disease Prevention and Control (ECDC) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16(12). 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentin L, Vieira ND, Barioni W Jr.. Oral treatment with bacteriophages reduces the concentration of Salmonella Enteritidis PT4 in caecal contents of broilers. Avian Pathol. 2005. Jun;34(3):258–263. 10.1080/01445340500112157 [DOI] [PubMed] [Google Scholar]
- Gaggìa F, Mattarelli P, Biavati B.. Probiotics and prebiotics in animal feeding for safe food production. Int J Food Microbiol. 2010; 141(SUPPL.):S15–S28. 10.1016/j.ijfoodmicro.2010.02.031 [DOI] [PubMed] [Google Scholar]
- Gao P, Ma C, Sun Z, Wang L, Huang S, Su X, Xu J, Zhang H.. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome. 2017. Dec; 5(1):91 10.1186/s40168-017-0315-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley PS, Harty DWS, Wyatt JE, Brown CR, Doran JP, Gibbs ACC.. A comparison of the adhesion, coaggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J Gen Microbiol. 1987. Nov;133(11): 3207–3217. 10.1099/00221287-133-11-3207 [DOI] [PubMed] [Google Scholar]
- Higgins SE, Higgins JP, Wolfenden AD, Henderson SN, Torres-Rodriguez A, Tellez G, Hargis B.. Evaluation of a Lactobacillus-based probiotic culture for the reduction of Salmonella enteritidis in neonatal broiler chicks. Poult Sci. 2008. Jan 01;87(1):27–31. 10.3382/ps.2007-00210 [DOI] [PubMed] [Google Scholar]
- Hütt P, Shchepetova J, Lõivukene K, Kullisaar T, Mikelsaar M.. Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero- and uropathogens. J Appl Microbiol. 2006. Jun;100(6): 1324–1332. 10.1111/j.1365-2672.2006.02857.x [DOI] [PubMed] [Google Scholar]
- Imperial ICVJ, Ibana JA.. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front Microbiol. 2016. Dec 15;07:1983 10.3389/fmicb.2016.01983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janković T, Frece J, Abram M, Gobin I.. Aggregation ability of potential probiotic Lactobacillus plantarum strains. Int J Sanit Eng Res. 2012;6(1):19–24. [Google Scholar]
- Jankowska A, Laubitz D, Antushevich H, Zabielski R, Grzesiuk E.. Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco-2 cells. J Biomed Biotechnol. 2008(1). 10.1155/2008/357964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik JK, Kumar A, Duary RK, Mohanty AK, Grover S, Batish VK.. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One. 2009. Dec 1;4(12): e8099 10.1371/journal.pone.0008099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Ingale SL, Kim JS, Lee SH, Lee JH, Kwon IK, Chae BJ.. Bacteriophage and probiotics both enhance the performance of growing pigs but bacteriophage are more effective. Anim Feed Sci Technol. 2014;196:88–95. 10.1016/j.anifeedsci.2014.06.012 [DOI] [Google Scholar]
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, et al. World Health Organization Estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;12(12):1–21. 10.1371/journal.pmed.1001920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klewicka E, Libudzisz Z.. Antagonistic activity of Lactobacillus acidophilus bacteria towards selected food-contaminating bacteria. Pol J Food Nutr Sci. 2004;13/54(2):169–174. [Google Scholar]
- Kos B, Šušković J, Vuković S, Šimpraga M, Frece J, Matošić S.. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003. Jun;94(6):981–987. 10.1046/j.1365-2672.2003.01915.x [DOI] [PubMed] [Google Scholar]
- Koziel JA, Frana TS, Ahn H, Glanville TD, Nguyen LT, van Leeuwen JH.. Efficacy of NH3 as a secondary barrier treatment for inactivation of Salmonella Typhimurium and methicillin-resistant Staphylococcus aureus in digestate of animal carcasses: proof-of-concept. PLoS One. 2017. May 5;12(5):e0176825 10.1371/journal.pone.0176825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen SM, Ravnum S, Tourasse NJ, Økstad OA, Kolstø AB, Davies W.. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579 [corrected]. J Bacteriol. 2007. Jul 15;189(14):5302–5313. 10.1128/JB.00239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ragione R, Woodward MJ.. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet Microbiol. 2003. Jul 17;94(3):245–256. 10.1016/S0378-1135(03)00077-4 [DOI] [PubMed] [Google Scholar]
- Mokoena MP. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules. 2017. Jul 26;22(8):1255 10.3390/molecules22081255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubareck C, Gavini F, Vaugien L, Butel MJ, Doucet-Populaire F.. Antimicrobial susceptibility of bifidobacteria. J Antimicrob Chemother. 2005. Jan 01;55(1):38–44. 10.1093/jac/dkh495 [DOI] [PubMed] [Google Scholar]
- Musikasang H, Tani A, H-kittikun A, Maneerat S.. Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World J Microbiol Biotechnol. 2009. Aug; 25(8):1337–1345. 10.1007/s11274-009-0020-8 [DOI] [Google Scholar]
- Nowak A, Motyl I.. In vitro anti-adherence effect of probiotic Lactobacillus strains on human enteropathogens. Biotechnol Food Sci. 2017;81(2):103–112. [Google Scholar]
- Ostad SN, Salarian AA, Ghahramani MH, Fazeli MR, Samadi N, Jamalifar H.. Live and heat-inactivated lactobacilli from feces inhibit Salmonella typhi and Escherichia coli adherence to Caco-2 cells. Folia Microbiol (Praha). 2009. Mar;54(2):157–160. 10.1007/s12223-009-0024-7 [DOI] [PubMed] [Google Scholar]
- Patterson J, Burkholder K.. Application of prebiotics and probiotics in poultry production. Poult Sci. 2003. Apr 01;82(4):627–631. 10.1093/ps/82.4.627 [DOI] [PubMed] [Google Scholar]
- Prado-Rebolledo OF, Delgado-Machuca JJ, Macedo-Barragan RJ, Garcia-Márquez LJ, Morales-Barrera JE, Latorre JD, Hernandez-Velasco X, Tellez G.. Evaluation of a selected lactic acid bacteria-based probiotic on Salmonella enterica serovar Enteritidis colonization and intestinal permeability in broiler chickens. Avian Pathol. 2017. Jan 02;46(1):90–94. 10.1080/03079457.2016.1222808 [DOI] [PubMed] [Google Scholar]
- Reid G. Probiotic use in an infectious disease setting. Expert Rev Anti Infect Ther. 2017;15(5):449–455. 10.1080/14787210.2017.1300061 [DOI] [PubMed] [Google Scholar]
- Repetto G, del Peso A, Zurita JL.. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc. 2008. Jul;3(7): 1125–1131. 10.1038/nprot.2008.75 [DOI] [PubMed] [Google Scholar]
- Romani Vestman N, Timby N, Holgerson P, Kressirer CA, Claesson R, Domellöf M, Öhman C, Tanner ACR, Hernell O, Johansson I.. Characterization and in vitro properties of oral lactobacilli in breastfed infants. BMC Microbiol. 2013;13(1):193 10.1186/1471-2180-13-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, O’Dwyer J, Adley CC.. Evaluation of the complex nomenclature of the clinically and veterinary significant pathogen Salmonella. BioMed Res Int. 2017;2017:1–6. 10.1155/2017/3782182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DH, Paul NC, Sischo WC, Crespo R, Guard J.. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci. 2017. Mar 1; 96(3): 687–702. 10.3382/ps/pew342 [DOI] [PubMed] [Google Scholar]
- Strus M. A new method for testing antagonistic activity of lactic acid bacteria (LAB) on selected pathogenic indicator bacteria (in Polish). Med Dosw Mikrobiol. 1998;50:123–130 [PubMed] [Google Scholar]
- Tellez G, Pixley C, Wolfenden RE, Layton SL, Hargis BM.. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int. 2012;45(2):628–633. 10.1016/j.foodres.2011.03.047 [DOI] [Google Scholar]
- Tinrat S, Saraya S, Traidej Chomnawang M.. Isolation and characterization of Lactobacillus salivarius MTC 1026 as a potential probiotic. J Gen Appl Microbiol. 2011;57(6):365–378. 10.2323/jgam.57.365 [DOI] [PubMed] [Google Scholar]
- Toro H, Price SB, McKee S, Hoerr FJ, Krehling J, Perdue M, Bauermeister L.. Use of bacteriophages in combination with competitive exclusion to reduce Salmonella from infected chickens. Avian Dis. 2005. Mar;49(1):118–124. 10.1637/7286-100404R [DOI] [PubMed] [Google Scholar]
- Tropcheva R, Georgieva R, Danova S.. Adhesion ability of Lactobacillus plantarum AC131. Biotechnol Biotechnol Equip. 2011. Jan;25(sup1) SUPPL. 4:121–124. 10.5504/BBEQ.2011.0123 [DOI] [Google Scholar]
- Trzeciak M, Wolna-Murawska A, Kosicka D.. Wpływ preparatów probiotycznych na stan zdrowia i cechy hodowlane zwierząt. Kosmos (Stockh). 2016;65(1):57–67. [Google Scholar]
- Vankerckhoven V, Huys G, Vancanneyt M, Vael C, Klare I, Romond MB, Entenza JM, Moreillon P, Wind RD, Knol J, et al. Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci Technol. 2008. Feb;19(2):102–114. 10.1016/j.tifs.2007.07.013 [DOI] [Google Scholar]
- Varankovich NV, Nickerson MT, Korber DR.. Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front Microbiol. 2015. Jul 14;6(JUN):685 10.3389/fmicb.2015.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdenelli MC, Ghelfi F, Silvi S, Orpianesi C, Cecchini C, Cresci A.. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur J Nutr. 2009. Sep;48(6):355–363. 10.1007/s00394-009-0021-2 [DOI] [PubMed] [Google Scholar]
- Vicente JL, Torres-Rodriguez A, Higgins SE, Pixley C, Tellez G, Donoghue AM, Hargis BM.. Effect of a selected Lactobacillus spp. based probiotic on Salmonella enterica serovar enteritidis-infected broiler chicks. Avian Dis. 2008. Mar;52(1):143–146. 10.1637/7847-011107-ResNote [DOI] [PubMed] [Google Scholar]
- Wang CY, Lin PR, Ng CC, Shyu YT.. Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe. 2010;16(6):578–585. 10.1016/j.anaerobe.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Wang J, Guo H, Cao C, Zhao W, Kwok LY, Zhang H, Zhang W.. Characterization of the adaptive amoxicillin resistance of Lactobacillus casei Zhang by proteomic analysis. Front Microbiol. 2018. Feb 20;9(FEB):292 10.3389/fmicb.2018.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K, Wright H, Murray M.. Probiotic treatment for the prevention of antibiotic-associated diarrhoea in geriatric patients: A multicentre randomised controlled pilot study. Australas J Ageing. 2015. Mar;34(1):38–42. 10.1111/ajag.12116 [DOI] [PubMed] [Google Scholar]
- Yeo S, Lee S, Park H, Shin H, Holzapfel W, Huh CS.. Development of putative probiotics as feed additives: validation in a porcine-specific gastrointestinal tract model. Appl Microbiol Biotechnol. 2016; 100(23):10043–10054. 10.1007/s00253-016-7812-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaczek A, Wojtasik A, Izdebski R, Gorecka E, Wojcik EA, Nowak T, Kwiecinski P, Dziadek J.. PCR melting profile as a tool for outbreak studies of Salmonella enterica in chickens. BMC Vet Res. 2015. Dec;11(1):137 10.1186/s12917-015-0451-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeraik AE, Nitschke M.. Influence of growth media and temperature on bacterial adhesion to polystyrene surfaces. Braz Arch Biol Technol. 2012. Aug;55(4):569–576. 10.1590/S1516-89132012000400012 [DOI] [Google Scholar]