Abstract
In vitro studies aimed at studying the mechanism of action of carvacrol and oregano as natural anti-bacterial agents to control multiple antibiotic-resistant avian pathogenic Escherichia coli (APEC) strain O23:H52 isolated from chicken were performed. Derivatives with increased minimum inhibitory concentrations (MIC) to the phytochemicals were selected after growing Escherichia coli (E. coli) strain O23:H52 at sub-lethal concentrations of carvacrol and oregano for a period of 60 days. Whole-genome sequencing (WGS) of two derivatives revealed a missense mutation in cadC and marR: the genes responsible for survival mechanisms and antibiotic resistance by efflux, respectively.
Key words: E. coli, APEC, phytochemicals, antibiotic resistance, WGS
Introduction
Phytochemicals are natural plant products produced as secondary metabolites of which some possess anti-microbial effects (Wink 2004), and therefore, may present a promising alternative strategy to antibiotics especially against antibiotic-resistant bacteria (Wong et al. 2006; Marcus et al. 2019) as well as pathogenic bacteria (Jayalakshmi et al. 2011; Rajamanickam et al. 2019). Several studies have investigated the antibacterial properties of a number of phytochemicals, but their mechanisms of action remain ill-defined. The interaction between phytochemicals and bacteria at the cellular level is due to the hydrophobic nature of phytochemicals which enables their entry into the lipid bilayer of the cytoplasmic membrane, to act as a membrane destabilizing agent that induces structural changes modifying the functionality of the lipid membrane and associated proteins (Sikkema et al. 1995; Luz et al. 2014; Yuan et al. 2019). Thus, the effects are pleiotropic and include: altering surface charge of the bacterial membrane (Cristani et al. 2007), ion (H+ and K+) transport (Ultee et al. 2002), stress responses (Richter et al. 2010; Di Pasqua et al. 2013), conjugation (Skalicka-Woźniak et al. 2018), motility and quorum sensing (Monte et al. 2014), amongst other effects.
Carvacrol and oregano exhibit anti-microbial activities against pathogenic microorganisms whether bacteria or fungi, irrespective of origin from the plant, animal or human sources (Baricevic and Bartol 2002; Mathlouthi et al. 2012). Given the focus of this study is on E. coli, previous studies have shown that oregano oil containing carvacrol and thymol is effective against E. coli in a dose-dependent manner (Friedman et al. 2002; Al-Mnaser 2019; Alvarez et al. 2019). Another study has shown that exposing E. coli to sub-lethal concentrations of carvacrol leads to changes in the ratio of unsaturated and saturated fatty acid components of the cell membrane (Di Pasqua et al. 2006) suggesting that E. coli develops an adaptive response upon exposure. To interrogate the many target sites of the E. coli cell, which could be affected by carvacrol and oregano, an approach used here was to grow and continuously expose E. coli cells for 60 days at sub-MIC level of phytochemicals (carvacrol and oregano) in growth medium, to generate derivatives that have reduced sensitivity (an increased resistance). This approach will select both temporary adaptations as well as mutational events. Focusing on the latter should identify the genes encoding cellular functions involved in response to the stress of carvacrol and oregano. Therefore, this work aimed at similar investigations into the anti-bacterial role of carvacrol and oregano at the genetic level.
Experimental
Materials and Methods
E. coli strain and growth conditions. One E. coli strain, designated C1, of poultry origin and previously characterized as harboring five virulence determinants (fim1, csgA, crl, astA, and hlyA) (Al-Mnaser 2019) and, therefore, potentially an APEC strain (Johnson et al. 2008), and being resistant to five antibiotics (cefotaxime, nalidixic acid, cefotaxime, ampicillin, and tetracycline) (Al-Mnaser 2019) was selected as a suitable poultry isolate for this study. Strain C1 was shown to have a MIC of 0.3 μg/ml against aqueous carvacrol and oregano. The E. coli strain was grown overnight in Luria-Bertani (LB) broth (inoculation was from a pure stock culture preserved in a cryotube at –80°C) at 37°C to yield approximately 109 CFU/ml (OD600 = 1.00) of which 100 μl was used to inoculate each of three sets of tubes (total volume of 10 ml) supplemented with 0.2 μg/ml aqueous carvacrol, with 0.2 μg/ml aqueous oregano, and without any supplement as a control. Re-inoculation by transfer of 100 μl to freshly prepared media was done every 48 h over a period of 60 days after which the bacteria were diluted and spread on non-selective LB agar to generate single well-defined colonies. Two derivative E. coli strains were chosen randomly and were designated as 22M and 26M; carvacrol-derivative strain and oregano derivative strain, respectively.
Determination of MIC values of the derivative E. coli strains against aqueous phytochemicals. The two derivative E. coli strains (22M and 26M) were used to determine their MIC values against aqueous carvacrol and oregano using a quasi-microdilution method. 96-well plates with LB supplemented with a dilution series of oregano and carvacrol were inoculated with 22M and 26M and the OD600 was measured spectrophotometrically every 1 h for 24 h under aerobic conditions and at a temperature of 37°C (Fluostar Omega). The OD600 readings were used to plot the relationship between time and OD. Plots were used to calculate bacterial growth to determine the MIC value of carvacrol or oregano against the E. coli strains. The same procedure was done after two weeks of storage in cryotubes containing non-selective medium at –80°C, to ensure that the increase in MIC values was stable and not a result of an adaptative change.
WGS of the derivative E. coli strains. Strains 22M and 26M with their original wild-type strain C1 were sent to MicrobesNG at the University of Birmingham for WGS. In silico serotyping analysis using Serotype Finder 1.1 website (https://cge.cbs.dtu.dk/services/Serotype-Finder/) (Joensen et al. 2015) and MLST analysis using MLST 2.0 software (https://pubmlst.org/) (Sepehri et al. 2009) were performed prior to full genome analysis.
Results and Discussion
In this study, we have investigated the antibacterial properties of two phytochemicals; carvacrol (the active ingredient of oregano) and oregano using wild-type E. coli strain of poultry origin as a starter strain, which to our knowledge, this has not been done before. This initial in vitro study aimed at increasing our understanding of the mechanism of action of these phytochemicals to control APEC strain (the causative agent of colibacillosis disease in poultry) with multiple antibiotic-resistance, which will enable us to evaluate their anti-bacterial properties as possible feed additives in the poultry industry instead of antibiotics.
The continuous exposure of E. coli cells to sub-lethal concentrations of carvacrol and oregano resulted in an increased resistance (reduced sensitivity) to these phytochemicals, and this was demonstrated by increased MIC values from 0.3 μg/ml to 0.6 μg/ml to both carvacrol and oregano. This step was repeated twice in order to confirm that the elevated MIC was stable. After that, the identity of the derivative strains was confirmed by extracting data from the WGS to ensure that the derivative E. coli strains 22M and 26M were true derivatives of the E. coli strain C1. WGS data analysis revealed that the three strains shared the same in silico serotype and multi-locus sequence typing (MLST) profiles, O23:H52 and ST-373, respectively.
The next objective was to search for the genomic variations in the derivatives compared with the progenitor strain, as this might give us information on the evolution of these derivatives (Tenaillon et al. 2001; Bryant et al. 2012). WGS data analysis showed that there were missense mutations detected in two chromosomal genes; cadC which encodes for a transcriptional activator of the cad operon (Küper and Jung 2005) and marR which encodes for a repressor of mar operon (Cohen et al. 1993). These two mutations were found in the carvacrol-derivative strain (22M). However, the oregano-derivative strain (26M) contained only one missense mutation, which was in cadC.
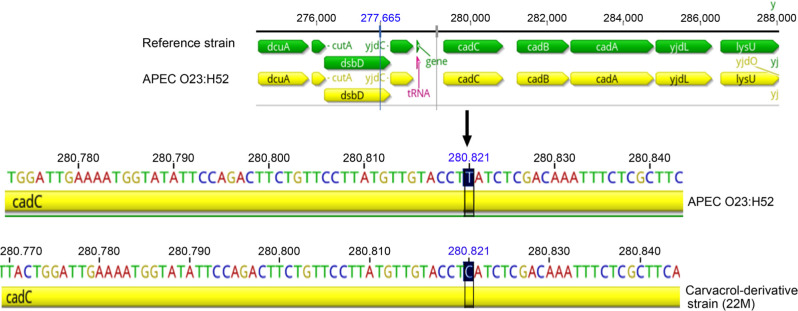
The cad operon is one of the survival mechanism systems in E. coli that is triggered in response to unfavorable acidic conditions (Tetsch et al. 2011). This system is composed of three genes; cadA (encodes a cytoplasmic CadA protein responsible for decarboxylation of lysine), cadB (encodes a transmembrane CadB protein responsible for excretion of the end products of lysine decarboxylation), and cadC (located upstream of the cadBA operon and encodes a transmembrane protein CadC) (Watson et al. 1992; Küper and Jung 2005). CadC has a dual function as a transcriptional activator of the cad operon in E. coli (Küper and Jung 2005) and as a sensor to external changes in pH in the environment (Tetsch et al. 2011). The missense mutation detected in cadC gene resulted in an amino acid substitution from tyrosine to histidine at position 504 of the CadC protein, caused by a transition substitution from T to C at the genome position 280821. The visualization of the mutation location in the carvacrol-derivative strain (22M), when compared with the wild-type strain (APEC O23:H52), is shown in Fig. 1. The increased phenotypic resistance to phytochemicals in the derivative strains 22M and 26M can be explained by two possible scenarios, assuming these mutations are not silent: 1) the substitution in cadC might effect on the expression of the Cad system, leading its over-expression of the CadC activator/sensor (Tetsch et al. 2008) to compensate for the constant presence of phytochemicals, 2) the substitution in cadC might affect the Cad system leading to over-expression of the speF-potE operon (another survival mechanism in E. coli) to replace the function of cadBA operon (Soksawatmaekhin et al. 2004). These options could be further investigated by Real-Time-PCR mRNA expression and/or complementation studies in order to get a clearer picture. These results suggest that carvacrol/oregano can trigger stress responses in multiple antibiotic-resistant APEC strain when used at sub-lethal concentration. Similar findings were documented when using sub-lethal concentrations of carvacrol which led to a missense mutation in soxR, which is another oxidative stress defence in E. coli (Chueca et al. 2018).
Fig. 1.
A diagram showing the presence of T at 280821 in the genome of the C1 strain (APEC O23:H52) that is substituted by C at position 280821 in the genome of the carvacrol-derivative strain (22M). This graph was generated using the Geneious Prime 2019.1.1 (https://www.geneious.com).
The mar operon is responsible for chromosome-mediated multiple antibiotic resistance as a protective mechanism in response to environmental stresses such as the presence of antibiotics and oxidative stress (Ariza et al. 1994). This operon, which is short for multiple antibiotic resistance, consists of four genes; marA (encoding an activator protein of mar operon), marR (encoding a repressor protein of mar operon) (Cohen et al. 1993), marB and marC (with unknown function) (Alekshun and Levy 2004). The mar operon is responsible for the increased low level resistance in E. coli strains to different classes of antibiotics including tetracycline, chloramphenicol, β-lactams, and fluoroquinolones by efflux mechanisms (George and Levy 1983). Interestingly, the MarR repressor in E. coli found in the gut of animal hosts has another function which is detecting phenolic compounds of plant products (Sulavik et al. 1995), further supporting the role of MarR in carvacrol/oregano resistance. The missense mutation detected in marR gene was an amino acid substitution from arginine to histidine at position 94 of the protein MarR, caused by a transition substitution from C to T at the genome position 13346. The visualization of the mutation location in the carvacrol-derivative strain (22M), when compared with the wild-type strain (APEC O23:H52), is shown in Fig. 2. Given the increased resistance phenotype of the E. coli strains, this substitution is probably a non-silent mutation resulting in an increased activity of the mar efflux system due to the repressor failing to repress the mar operon, and therefore increased its resistance as was recently discovered (Chueca et al. 2018). Similar findings were demonstrated by previous work from our laboratory (AlKhandari 2017), which showed that thymol-derivative strain showed non-sense mutations in marR and acrR, genes encoding repressors involving in efflux pump systems were responsible for the reduced susceptibility to phytochemicals. These findings suggest that carvacrol can act as an efflux pump inhibitor when used at high concentrations as proposed in this study (Miladi et al. 2016).
Fig. 2.
A diagram showing the presence of C at 13346 in the genome of the C1 strain (APEC O23:H52) that is substituted by T at position 13346 in the genome of the carvacrol-derivative strain (22M). This graph was generated using the Geneious Prime 2019.1.1 (https://www.geneious.com).
This study might indicate the importance of giving carvacrol, oregano or thymol as a feed additive instead of antibiotics as a feed additive to chicken and what might happen after a long period of time use. However, this work was performed only once but the body of data from this current study supports the findings of previously mentioned studies which have suggested that exposure to phytochemicals (carvacrol, oregano, and thymol) select for mutants in different genes but each responsible for increased resistance phenotype. To confirm the effect of gene mutations on resistance, further studies could include: 1) gene complementation to show phenotype reversion and 2) use of mutant E. coli strains from the Keio library to study cadC and marR mutants in specific, and to study all the possible mechanisms of actions. In conclusion, the possible mechanisms of action of carvacrol/oregano against E. coli seem to be associated with missense mutations in the genes responsible for survival mechanisms under unfavorable conditions (cadC) and multiple antibiotic resistance (marR).
Acknowledgments
We are grateful to Fatemah Al-Khandari for providing us with the E. coli strain and to Geoffrey Woodward of the NHS, Southmead Hospital, Genetic Analysis Department for carrying out some of the bioinformatics analyses.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
ORCID
Afnan A. Al-Mnaser https://orcid.org/0000-0003-1792-3170
Literature
- Alekshun MN, Levy SB.. The Escherichia coli mar locus-antibiotic resistance and more. ASM News. 2004;70:451–456. [Google Scholar]
- Alkhandari F. Characterization of Escherichia coli in poultry and their interaction with phytochemicals [dissertation]. University of Reading; 2017. [Google Scholar]
- Al-Mnaser A. Phytochemicals (carvacrol and oregano extract) as possible alternatives to antibiotics in poultry feed [dissertation] University of Reading; 2019. [Google Scholar]
- Alvarez MV, Ortega-Ramirez LA, Silva-Espinoza BA, Gonzalez-Aguilar GA, Ayala-Zavala JF.. Antimicrobial, antioxidant, and sensorial impacts of oregano and rosemary essential oils over broccoli florets. J Food Process Preserv. 2019. Mar;43(3):e13889 10.1111/jfpp.13889 [DOI] [Google Scholar]
- Ariza RR, Cohen SP, Bachhawat N, Levy SB, Demple B.. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994. Jan;176(1):143–148. 10.1128/jb.176.1.143-148.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricevic D, Bartol T.. The biological/pharmacological activity of the Origanum genus. Oregano, the genera Origanum and Lippia. New York (USA): Taylor & Francis Inc.; 2002. p. 177–201. [Google Scholar]
- Bryant J, Chewapreecha C, Bentley SD.. Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol. 2012. Nov;7(11):1283–1296. 10.2217/fmb.12.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueca B, Renzoni A, Berdejo D, Pagán R, Kelley WL, García-Gonzalo D.. Whole-genome sequencing and genetic analysis reveals novel stress responses against individual constituents of essential oils in Escherichia coli. Appl Environ Microbiol. 2018. Mar 19;84(7):e02538-17. 10.1128/AEM.02538-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, Hächler H, Levy SB.. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993. Mar;175(5):1484–1492. 10.1128/jb.175.5.1484-1492.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristani M, D’Arrigo M, Mandalari G, Castelli F, Sarpietro MG, Micieli D, Venuti V, Bisignano G, Saija A, Trombetta D.. Interaction of four monoterpenes contained in essential oils with model membranes: implications for their antibacterial activity. J Agric Food Chem. 2007. Jul;55(15):6300–6308. 10.1021/jf070094x [DOI] [PubMed] [Google Scholar]
- Di Pasqua R, Hoskins N, Betts G, Mauriello G.. Changes in membrane fatty acids composition of microbial cells induced by addiction of thymol, carvacrol, limonene, cinnamaldehyde, and eugenol in the growing media. J Agric Food Chem. 2006. Apr;54(7): 2745–2749. 10.1021/jf052722l [DOI] [PubMed] [Google Scholar]
- Di Pasqua R, Mauriello G, Mamone G, Ercolini D.. Expression of DnaK, HtpG, GroEL and Tf chaperones and the corresponding encoding genes during growth of Salmonella Thompson in presence of thymol alone or in combination with salt and cold stress. Food Res Int. 2013. Jun;52(1):153–159. 10.1016/j.foodres.2013.02.050 [DOI] [Google Scholar]
- Friedman M, Henika PR, Mandrell R.. Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J Food Prot. 2002. Oct;65(10):1545–1560. 10.4315/0362-028X-65.10.1545 [DOI] [PubMed] [Google Scholar]
- George AM, Levy SB.. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983. Aug;155(2):531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayalakshmi B, Raveesha K, Amruthesh K.. Phytochemical investigations and antibacterial activity of some medicinal plants against pathogenic bacteria. J Appl Pharm Sci. 2011;1:124. [Google Scholar]
- Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F.. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J Clin Microbiol. 2015. Aug;53(8): 2410-26. 10.1128/JCM.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK.. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol. 2008. Dec 01;46(12):3987–3996. 10.1128/JCM.00816-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küper C, Jung K.. CadC-mediated activation of the cadBA promoter in Escherichia coli. J Mol Microbiol Biotechnol. 2005;10(1):26–39. 10.1159/000090346 [DOI] [PubMed] [Google Scholar]
- Luz IS, de Melo ANF, Bezerra TKA, Madruga MS, Magnani M, de Souza EL.. Sublethal amounts of Origanum vulgare L. essential oil and carvacrol cause injury and changes in membrane fatty acid of Salmonella Typhimurium cultivated in a meat broth. Foodborne Pathog Dis. 2014. May;11(5):357–361. 10.1089/fpd.2013.1695 [DOI] [PubMed] [Google Scholar]
- Marcus A, Edori O, Maduagu M.. Phytochemical and antimicrobial screening of Phyllantus fratenus and Taraxacuim officinale leaves. Biochem Anal Biochem. 2019;8(1):376 10.35248/2161-1009.19.8.376 [DOI] [Google Scholar]
- Mathlouthi N, Bouzaienne T, Oueslati I, Recoquillay F, Hamdi M, Urdaci M, Bergaoui R.. Use of rosemary, oregano, and a commercial blend of essential oils in broiler chickens: in vitro antimicrobial activities and effects on growth performance1. J Anim Sci. 2012. Mar 01;90(3):813–823. 10.2527/jas.2010-3646 [DOI] [PubMed] [Google Scholar]
- Miladi H, Zmantar T, Chaabouni Y, Fedhila K, Bakhrouf A, Mahdouani K, Chaieb K.. Antibacterial and efflux pump inhibitors of thymol and carvacrol against food-borne pathogens. Microb Pathog. 2016. Oct;99:95–100. 10.1016/j.micpath.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Monte J, Abreu A, Borges A, Simões L, Simões M.. Antimicrobial activity of selected phytochemicals against Escherichia coli and Staphylococcus aureus and their biofilms. Pathogens. 2014. Jun 18; 3(2):473–498. 10.3390/pathogens3020473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamanickam K, Yang J, Sakharkar MK.. Phytochemicals as alternatives to antibiotics against major pathogens involved in bovine respiratory disease (BRD) and bovine mastitis (BM). Bioinformation. 2019. Jan 31;15(1):32–35. 10.6026/97320630015032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J.. The heat shock response: life on the verge of death. Mol Cell. 2010. Oct;40(2):253–266. 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Sepehri S, Kotlowski R, Bernstein CN, Krause DO.. Phylogenetic analysis of inflammatory bowel disease associated Escherichia coli and the fimH virulence determinant. Inflamm Bowel Dis. 2009. Nov;15(11):1737–1745. 10.1002/ibd.20966 [DOI] [PubMed] [Google Scholar]
- Sikkema J, de Bont JA, Poolman B.. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995. Jun;59(2):201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicka-Woźniak K, Walasek M, Aljarba TM, Stapleton P, Gibbons S, Xiao J, Łuszczki JJ.. The anticonvulsant and anti-plasmid conjugation potential of Thymus vulgaris chemistry: an in vivo murine and in vitro study. Food Chem Toxicol. 2018. Oct;120: 472–478. 10.1016/j.fct.2018.07.045 [DOI] [PubMed] [Google Scholar]
- Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K.. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol. 2004. Mar; 51(5): 1401–1412. 10.1046/j.1365-2958.2003.03913.x [DOI] [PubMed] [Google Scholar]
- Sulavik MC, Gambino LF, Miller PF.. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995. May;1(4): 436–446. 10.1007/BF03401581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Taddei F, Radman M, Matic I.. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res Microbiol. 2001. Jan; 152(1):11–16. 10.1016/S0923-2508(00)01163-3 [DOI] [PubMed] [Google Scholar]
- Tetsch L, Koller C, Dönhöfer A, Jung K.. Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 2011;11(1):74 10.1186/1471-2180-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsch L, Koller C, Haneburger I, Jung K.. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol Microbiol. 2008. Feb;67(3):570–583. 10.1111/j.1365-2958.2007.06070.x [DOI] [PubMed] [Google Scholar]
- Ultee A, Bennik MHJ, Moezelaar R.. The phenolic hydroxyl group of carvacrol is essential for action against the food-borne pathogen Bacillus cereus. Appl Environ Microbiol. 2002. Apr 01;68(4):1561–1568. 10.1128/AEM.68.4.1561-1568.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N, Dunyak DS, Rosey EL, Slonczewski JL, Olson ER.. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992. Jan;174(2):530–540. 10.1128/jb.174.2.530-540.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M. Phytochemical diversity of secondary metabolites In: Goodman RM, editor. Encyclopedia of plant and crop science. New ork (USA): Marcel Dekker, Inc.; 2004. [Google Scholar]
- Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA.. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006. Mar;40(3):235–243. 10.1097/00004836-200603000-00015 [DOI] [PubMed] [Google Scholar]
- Yuan W, Teo CHM, Yuk HG.. Combined antibacterial activities of essential oil compounds against Escherichia coli O157:H7 and their application potential on fresh-cut lettuce. Food Control. 2019. Feb;96:112–118. 10.1016/j.foodcont.2018.09.005 [DOI] [Google Scholar]