Abstract
Elderly people living in nursing homes are a high-risk population for Staphylococcus aureus infection. Multiple comorbidities, a weakened immune system, inadequate hygienic conditions, and crowding might increase the prevalence rates of this opportunistic pathogen. However, the epidemiological aspects, genetic diversity, and transmission of S. aureus in nursing homes are still poorly understood, especially in Poland. This study aimed to determine the genetic relatedness and prevalence of colonization of S. aureus isolated from the anterior nares and the throat of residents and staff in a nursing home located in Lublin, Poland. The study showed a high S. aureus prevalence rate among participants (46.1%), yet there was a low frequency of MRSA strains among residents (1.7%) and staff (0%). The multiple-locus variable-number tandem-repeat fingerprinting (MLVF) analysis demonstrated a high degree of genetic diversity of S. aureus strains colonizing the anterior nares and the throat of the participants. The occurrence of simultaneous colonization with more than one unique S. aureus strain in any one individual as well as the incidence of colonization with the same genetic variant of S. aureus in different individuals was observed. These findings suggest that inter-participant S. aureus transmission might contribute to the development of cross-infections.
Key words: Staphylococcus aureus, colonization, elderly, genotyping, nursing home
Introduction
Staphylococcus aureus is an opportunistic human pathogen known for its increasing prevalence in both hospitals and the general community. An incremental rise in drug-resistance and high disease-causing capabilities has also been observed (Zhang et al. 2015; Zurita et al. 2016). The most common anatomical location for the commensal carriage of S. aureus is the anterior nares (Kluytmans et al. 1997; Pollitt et al. 2018), although recent epidemiological studies highlight the role of pharyngeal carriage in S. aureus transmission (Nilsson and Ripa 2006; Hamdan-Partida et al. 2010). The complexity of host-colonizer interactions has led researchers to investigate alternative contexts in pursuit of the mechanism for colonization. Long term care facilities (e.g. nursing homes) constitute communities in which crowding and inadequate hygienic conditions might contribute to higher MSSA (methicillin-sensitive S. aureus) and MRSA (methicillin-resistant S. aureus) prevalence rates. Chronic diseases, immune system disorders, and frequent hospitalization make elderly people vulnerable to S. aureus infection (Ledell et al. 2003; Chuang et al. 2015; Peters et al. 2017).
S. aureus is also one of the most genetically diverse bacteria and the mechanisms by which the predominant strains are selected within each biological niche depends on the geographic location as well as the studied human host (Zurita et al. 2016). MSSA strains are more prevalent than MRSA strains and have a higher genetic diversity, whereas MRSA outbreaks are usually caused by only a few different MRSA clones (Goering et al. 2008). Therefore, analyzing the genetic relatedness of colonizing MSSA strains in nursing homes might result in a better understanding of S. aureus epidemiology.
We aimed at determining the genetic diversity and relatedness of MSSA and MRSA strains colonizing the upper respiratory tract of nursing home residents and staff using the MLVF (multiple-locus variable-number tandem-repeat fingerprinting) method. We hypothesized that there would be the same genetic MLVF banding patterns of S. aureus retrieved from different individuals, and the presence of genetic homogeneity might indicate that bacterial transmission has occurred.
Experimental
Materials and Methods
Bacterial strains. Among 102 participants enrolled in our study, more than three-quarters were female and the mean age of the residents (81.5 years old) was almost twice as high as that of the staff members (43 years old) (Table I). Two anatomical locations (the anterior nares and the throat) were swabbed using sterile cotton swabs (Medlab, Poland) premoistened with sterile 0.9% saline solution (POCH, Poland). The samples were immediately transported to the laboratory and processed within two hours. The swabs were inoculated on 5% sheep blood agar plates (BioRad, USA) and mannitol salt agar plates (BioMaxima, Poland) at 35.5°C for 24 h. The β-hemolytic or/and mannitol-positive colonies were tested for the presence of coagulase (Biomed, Poland) and finally identified as S. aureus using the Vitek 2 Compact automated system and GP cards (Biomerieux, France).
Table I.
Basic demographic characteristics of participants and the prevalence of S. aureus colonization.
| Total (n = 102) |
Residents (n = 60) |
Staff (n = 42) |
|
|---|---|---|---|
| Age (years) | |||
| Median (range) | 68 (26-97) | 81.5 (56-97) | 43 (26-67) |
| Gender n (%) | |||
| Female | 78 (76.5) | 43 (71.7) | 35 (83.3) |
| Male | 24 (23.5) | 17 (29.3) | 7 (16.7) |
| S. aureus colonization n (%) | 47 (46.1) | 28 (46.7) | 19 (45.2) |
| Nares | 17 (36.2) | 10 (35.7) | 7 (36.8) |
| Throat | 11 (23.4) | 6 (21.4) | 5 (26.4) |
| Nares and throat | 19 (40.4) | 12 (42.9) | 7 (36.8) |
Antimicrobial susceptibility testing was conducted using the Vitek 2 automated system and AST-P644 cards (BioMerieux, France) containing the following antibiotics: ceftaroline, ciprofloxacin, clindamycin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, oxacillin, rifampicin, teicoplanin, tetracycline, tigecycline, trimethoprim/sulfamethoxazole, and vancomycin. Additionally, resistance to cefoxitin (30 μg), tobramycin (10 μg), fusidic acid (10 μg) and mupirocin (200 μg) was determined using disc diffusion method. D-test was conducted to determine the mechanism of macrolide, lincosamide and streptogramin resistance (MLSB). All discs were obtained from the same manufacturer (Becton Dickinson, USA). Disc-diffusion tests were performed and interpreted according to EUCAST recommendations (EUCAST 2018). All S. aureus isolates have been stored in freezers (−70°C) for further analysis using a medium of tryptic soy broth (TSB; BTL, Poland) and glycerol (POCH, Poland) in equal proportions (1:1).
Multiple-locus variable-number tandem-repeat fingerprinting (MLVF). The MLVF genotyping was performed to analyze the genetic diversity and relatedness of all 66 identified S. aureus strains. The MLVF method involved a multiplex-PCR reaction targeting five variable number of tandem repeats (VNTRs) in the following loci: sspA, spa, sdr, clfA, and clfB. These five VNTRs are located within the genes that encode the following proteins: serine proteinase V8, protein A, sdrC, sdrD, sdrE fibrinogen binding proteins, clumping factor A, and clumping factor B (Sabat et al. 2003).
Genomic DNA was extracted from the 24-h TSB culture prepared from a single bacterial colony. DNA extraction and purification were conducted according to the manufacturer’s instruction (Genomic Mini, A&A Biotechnology, Poland), with the minor alteration of replacing lysozyme with an equal volume of lysostaphin (A&A Biotechnology, Poland). The quality and quantity of the DNA were assessed using BioPhotometer (Eppendorf, Germany) by measuring the absorbance at the wavelengths of 260 and 280 nm.
The PCR program consisted of predenaturation at 94°C for 5 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 60 s, followed by a final extension at 72°C for 10 min (Biometra, Germany) (Grzegorczyk and Malm 2014). The PCR products and DNA molecular size marker (100 bp DNA Ladder Plus, ThermoScientific, USA) were resolved in 2% agarose electrophoresis gels (Sigma-Aldrich, USA) stained with SimplySafe dye (EURx, Poland). The banding patterns were photographed, exported to BioGene (Vilber Lourmat, France), and analyzed. The dendrogram was constructed using a UPGMA algorithm (unweighted pair group method with arithmetic mean) by applying the Dice Similarity Ratio. A tolerance level of 1% was used in comparing the banding patterns.
Results and Discussion
Prevalence of S. aureus colonization. Among all participants, 46.1% (47/102) were colonized with S. aureus (Table I). The pattern of colonization was divided into three categories based on the anatomical location. The category that registered the highest number of individuals was the combination of the anterior nares and the throat (40.4% of colonized individuals), followed by the anterior nares only (36.2%), and the throat only (23.4%).
S. aureus colonizes multiple anatomical locations and the frequency of colonization in various niches might vary depending on the methodology and the context of the study. Our study demonstrated that the anterior nares are colonized more frequently than the throat, which is consistent with the findings of previous studies on the prevalence of both MSSA and MRSA occurring together (Roghmann et al. 2015; Zhang et al. 2015; Peters et al. 2017). Alternatively, some researchers have reported that colonization only in the throat is the most prevalent pattern (Nilsson and Ripa 2006; Hamdan-Partida et al. 2010) or have observed the high prevalence of extra-nasal colonization as well, i.e., in the groin and the perianal regions (Mody et al. 2008).
In the current study, 31.6% (6/19) of the participants colonized with two S. aureus strains (one in the anterior nares and one in the throat) registered dissimilar MLVF banding patterns, implying that the two S. aureus strains were unrelated. A similar observation has been made by (Mongkolrattanothai et al. 2011), who noted that simultaneously carriage of two or more unrelated S. aureus isolates was observed in 30.4% of positive swab samples collected from children. Colonization with more than one strain of bacteria entails a risk of heteroresistance, i.e., a situation in which an antibacterial agent is effective against one strain, but not against another strain of the same bacteria. Truly, in cases of co-colonization with MSSA and MRSA both together, heteroresistance might pose a therapeutic challenge.
Antimicrobial resistance profile of S. aureus. Table II shows the resistance profiles of S. aureus strains with the information about their location in the dendrogram. Among all S. aureus strains tested, 10.6% (7/66) were resistant to tetracycline; 6.1% (4/66) to macrolides, lincosamide, and streptogramin; 4.5% (3/66) to tobramycin and/or gentamicin, and 1.5% (1/66) to ciprofloxacin, levofloxacin, and daptomycin. We observed the presence of the MLSB-constitutive resistance only. The share of S. aureus isolates susceptible to all of the antimicrobial agents tested was high (77.3%, 51/66), although the presence of MDR (multi-drug resistant) MRSA strain (Table II) suggests the need of conducting longitudinal studies investigating its transmission and long-term colonization patterns (Sun et al. 2017). The antimicrobial susceptibility testing identified 98.5% (65/66) of S. aureus strains as MSSA, whereas only one isolate was identified as MRSA. Consequently, the MRSA prevalence rate in our studies was low when compared to similar studies: 1% among all participants and 1.7% specifically among the residents. The prevalence of MRSA colonization among residents of long term care facilities across the world varies within a wide range: from 0% in nine Swedish nursing homes (Andersson et al. 2012), 3.7% in nine nursing homes in Brasil (da Silveira et al. 2018), 7.2% in 19 long term care facilities in Luxemburg (Mossong et al. 2013), 20.4% in 36 residential care homes for the elderly in Hong Kong (Chuang et al. 2015), to 28% in 13 nursing homes in Michigan and Maryland (Roghmann et al. 2015).
Table II.
The antimicrobial resistance profiles of S. aureus strains isolated from residents and staff in a nursing home.
| Main cluster | Sub-cluster | S. aureus strain | Resistance profile |
|---|---|---|---|
| A | IV | R23T | TET |
| IV | S123T, S123N | TET | |
| IV | S133T, S133N | TET | |
| V | S116T | TOB, GEN | |
| B | VII | R38T | FOX, TOB, CIP, LEV, DAP |
| IX | R42T, R42N | TET | |
| IX | S139N | E, CC | |
| X | R2T | E, CC | |
| XI | R33T, R33N | E, CC | |
| XII | S134N | E, CC | |
| – | – | R41N | TET, TOB, GEN |
Note. S. aureus strains marked with the same number were retrieved from the anterior nares (N) and the throat (T) of one resident (R) or staff member (S) and had 100% genetic homology according to the MLVF analysis. Tetracycline (TET), tobramycin (TOB), gentamicin (GEN), cefoxitin (FOX), ciprofloxacin (CIP), levofloxacin (LEV), daptomycin (DAP), erythromycin (E), clindamycin (CC).
Multiple-locus variable-number tandem repeat fingerprinting of S. aureus. Using molecular typing methods to determine the genetic diversity of human pathogens is crucial in identifying the reservoirs and routes of transmission (Grzegorczyk and Malm 2014). Unlike other bacterial genotyping methods, such as PFGE (pulsed-field gel electrophoresis), MLST (multilocus sequence typing) or spa typing, MLVF has a comparable resolution, but is cheaper, less time- and labour-consuming (Luczak-Kadlubowska et al. 2008; Holmes et al. 2010; Kosecka-Strojek et al. 2016). The high discriminatory power of MLVF is its main advantage, enabling precise detection of single events of transmission between patients (Karynski et al. 2008).
The MLVF of the 66 S. aureus strains isolated from 47 individuals resulted in 51 banding patterns: 36 unique banding patterns were represented by only one strain and 15 banding patterns consisted of two identical S. aureus strains each. In accordance with our results, a high genetic diversity of MSSA has been reported in other studies (Emaneini et al. 2011; Karynski et al. 2008; Zurita et al. 2016). (Zurita et al. 2016) observed 69 MLVA (multiple-locus variable-number tandem-repeat analysis) genotypes among 70 tested MSSA strains from various populations.
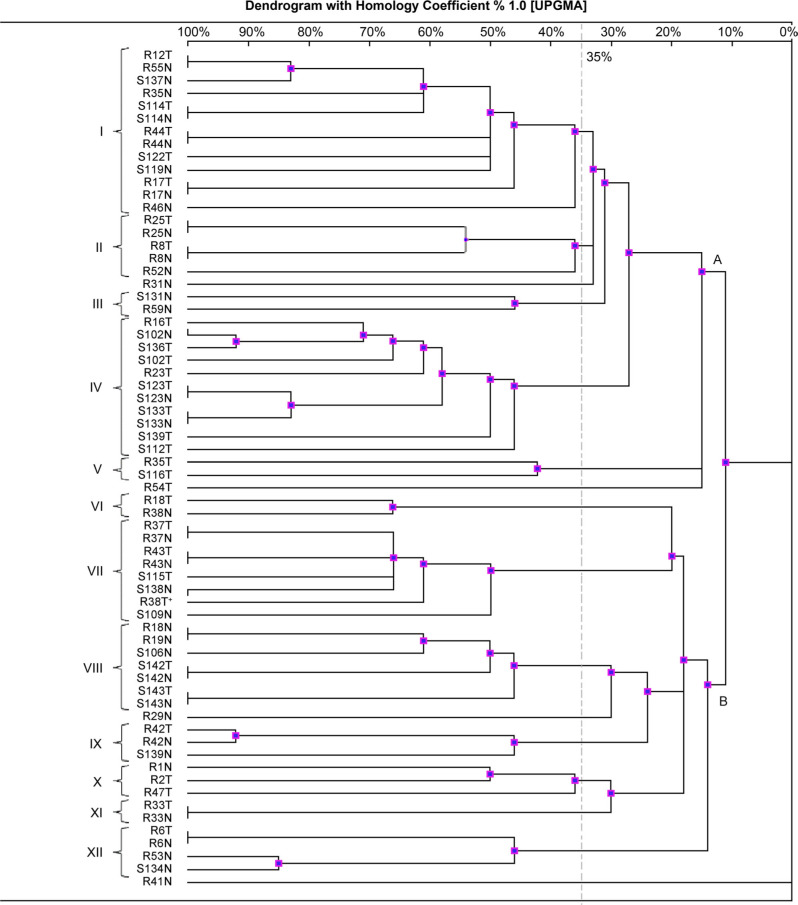
The MLVF typing allowed us to divide the 66 S. aureus isolates into two main clusters, herein designated as A and B (Fig. 1). One S. aureus strain (R41N) was not compatible with any other banding pattern. In the dendrogram, it represents a singleton with no association to the main clusters. The main cluster A was divided into five subclusters (herein designated I–V); the main cluster B was divided into seven subclusters (herein designated VI–XII). Altogether, the twelve subclusters included 94% (62/66) of all strains (cut-off >35%). Comparing the subclusters concerning the size, the two largest (I and IV) both included nine banding patterns, and 13 and 11 S. aureus strains, respectively. The six smallest subclusters (III, V, VI, IX, X, and XI) each included between one and three banding patterns and two or three S. aureus strains.
Fig. 1.
MLVF dendrogram of S. aureus strains isolated from the anterior nares and the throat of residents and staff in a nursing home.
Note. The name of S. aureus strain contains the following information: R – collected from resident, S – collected from staff member, number characteristic for each participant, N – nasal swab, T – throat swab. Two main clusters (A and B) and twelve subclusters (I–XII; cut-off > 35%) are marked in the dendrogram. MRSA strain (R38T) is marked with an asterisk.
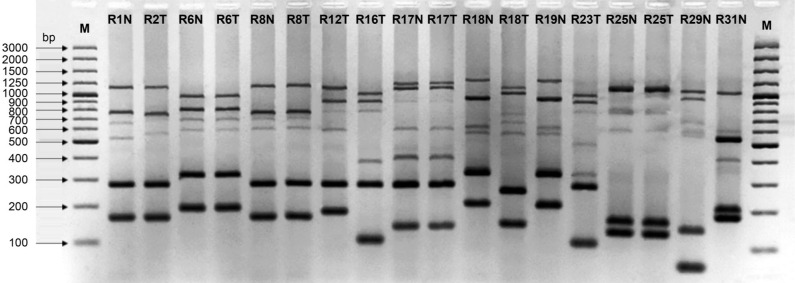
Exemplary MLVF banding patterns for S. aureus strains are shown in Fig. 2. The number of MLVF bands depends on the variable expression of genes in the sdr region of the bacterial DNA. The expression of between one and three genes (sdrC, sdrD and/or sdrE) translates into a range between five and seven MLVF bands (Sabat et al. 2003). In our study, the six-band and seven-band profile were the most numerous, accounting for 56.0% and 36.4% of all isolates, respectively. The less common five-band profile captured only 7.6% of all isolates.
Fig. 2.
MLVF of S. aureus strains isolated from the nose (N) and the throat (T) of thirteen residents living in a nursing home.
Note. Four residents were simultaneously colonized with the same genetic variant of S. aureus (strain R6N and R6T; strain R8N and R8T; strain R17N and R17T; strain R25N and R25T) and one resident was inhabited by two genetically different strains (R18N and R18T). M – molecular marker (100–3000 bp).
At the level of S. aureus isolates, we were able to group 26 isolates in even pairs based on perfect relatedness (one hundred percent homology). Without exception, each related pair of isolates was retrieved from the anterior nares and the throat of one individual, i.e., 13 individuals contributed one pair each. At the level of individuals, this meant that 27.6% (13/47) of the colonized individuals contributed a related pair of isolates. At the group level, i.e., the serendipitous detection of related isolates across the population, we were able to group four individuals in even couples, each couple sharing one bacterial strain. However, the anatomical location and pattern of co-colonization were inconsistent, rendering the similarities imperfect (for details, see subclusters I, VI and VIII in Fig. 1). These findings suggest that inter-participant transmission of S. aureus has occurred and might result in cross-infection – especially among immunocompromised individuals living in crowded conditions. (Andersen et al. 2002) have also reported that environmental factors, i.e., crowding and insufficient staffing, might increase the risk of S. aureus cross-infection.
Although our study provides valuable information about the genetic diversity, prevalence of colonization, and the occurrence of S. aureus transmission in a nursing home, it has several limitations. The MLVF method returns its most valid results when applied within a limited geographical area. As a result, making comparisons between studies from various countries or regions is difficult. Future research in the field of active microbial surveillance must include longitudinal aspects by subjecting the study populations to repeated sampling over extended periods. Understanding the temporal aspects of colonization might lead to new insights regarding the routes of bacterial transmission.
Footnotes
Ethical approval
The study was approved by the Bioethics Committee at the Medical University of Lublin (KE 0254/59/2016). The samples were collected in January 2018 from residents and staff (n = 102) living and working in a nursing home (Lublin, Poland). Written informed consent was obtained from each individual before participation.
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Author contributions
M.K. and A.G. collected the samples and performed the experiments. All authors conceived, designed the experiments and wrote the paper.
ORCID
Martyna Kasela 0000-0002-9791-2932
Literature
- Andersen BM, Lindemann R, Bergh K, Nesheim BI, Syversen G, Solheim N, Laugerud F.. Spread of methicillin-resistant Staphylococcus aureus in a neonatal intensive unit associated with understaffing, overcrowding and mixing of patients. J Hosp Infect. 2002;50(1):18–24. 10.1053/jhin.2001.1128 [DOI] [PubMed] [Google Scholar]
- Andersson H, Lindholm C, Iversen A, Giske CG, Örtqvist Å, Kalin M, Fossum B.. Prevalence of antibiotic-resistant bacteria in residents of nursing homes in a Swedish municipality: healthcare staff knowledge of and adherence to principles of basic infection prevention. Scand J Infect Dis. 2012;44(9):641–649. 10.3109/00365548.2012.671956 [DOI] [PubMed] [Google Scholar]
- Chuang VWM, Tsang IHL, Keung JPY, Leung JYY, Yuk JMT, Wong DKW, Au S, Tam RKY, Lam WWY, Kwan MCT, et al.. Infection control intervention on meticillin resistant Staphylococcus aureus transmission in residential care homes for the elderly. J Infect Prev. 2015;16(2):58–66. 10.1177/1757177414556007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira M, da Cunha MLRS, de Souza CSM, Correa AAF, Fortaleza CMCB.. Nasal colonization with methicillin-resistant Staphylococcus aureus among elderly living in nursing homes in Brazil: risk factors and molecular epidemiology. Ann Clin Microbiol Antimicrob. 2018;17(1):18–22. 10.1186/s12941-018-0271-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaneini M, Jabalameli L, Eini HI, Aligholi M, Ghasemi A, Nakhjavani FA, Taherikalani M, Khoramian B, Asadollahi P, Jabalameli F.. Multiple-locus variable number of tandem repeats fingerprinting (MLVF) and virulence factor analysis of methicillin resistant Staphylococcus aureus SCCmec type III. Pol J Microbiol. 2011;60(4):303–307. [PubMed] [Google Scholar]
- EUCAST.. Breakpoints tables for interpretation of MICs and zones diameters Version 8.1 Basel (Switzerland): European Committee on Antimicrobial Susceptibility Testing; 2018. [cited 2019 Feb 6]. Available from http://www.eucast.org [Google Scholar]
- Goering RV, Shawar RM, Scangarella NE, O’Hara FP, Amrine-Madsen H, West JM, Dalessandro M, Becker JA, Walsh SL, Miller LA, et al.. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J Clin Microbiol. 2008;46(9):2842–2847. 10.1128/JCM.00521-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorczyk A, Malm A.. Genotyping of Staphylococcus aureus strains isolated from healthy persistent carriers. Folia Microbiol (Praha). 2014;59(4):349–353. 10.1007/s12223-013-0294-y [DOI] [PubMed] [Google Scholar]
- Hamdan-Partida A, Sainz-Espuñes T, Bustos-Martínez J.. Characterization and persistence of Staphylococcus aureus strains isolated from the anterior nares and throats of healthy carriers in a Mexican community. J Clin Microbiol. 2010;48(5):1701–1705. 10.1128/JCM.01929-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Edwards GF, Girvan EK, Hannant W, Danial J, Fitz-gerald JR, Templeton KE.. Comparison of two multilocus variable-number tandem-repeat methods and pulsed-field gel electrophoresis for differentiating highly clonal methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 2010;48(10):3600–3607. 10.1128/JCM.01039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karynski M, Sabat AJ, Empel J, Hryniewicz W.. Molecular surveillance of methicillin-resistant Staphylococcus aureus by multiple-locus variable number tandem repeat fingerprinting (formerly multiple-locus variable number tandem repeat analysis) and spa typing in a hierarchic approach. Diagn Microbiol Infect Dis. 2008;62(3): 255–262. 10.1016/j.diagmicrobio.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Kluytmans J, van Belkum A, Verbrugh H.. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. 10.1128/CMR.10.3.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosecka-Strojek M, Ilczyszyn WM, Buda A, Polakowska K, Murzyn K, Panz T, Bialecka A, Kasprowicz A, Jakubczak A, Krol J, et al.. Multiple-locus variable-number tandem repeat fingerprinting as a method for rapid and cost-effective typing of animal-associated Staphylococcus aureus strains from lineages other than sequence type 398. J Med Microbiol. 2016;65(12):1494–1504. 10.1099/jmm.0.000378 [DOI] [PubMed] [Google Scholar]
- Ledell K, Muto CA, Jarvis WR, Farr BM.. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and Enterococcus. Infect Control Hosp Epidemiol. 2003;24(9):639–641. 10.1086/502924 [DOI] [PubMed] [Google Scholar]
- Luczak-Kadlubowska A, Sabat A, Tambic-Andrasevic A, Payerl-Pal M, Krzyszton-Russjan J, Hryniewicz W.. Usefulness of Multiple-Locus VNTR Fingerprinting in detection of clonality of community- and hospital-acquired Staphylococcus aureus isolates. Antonie van Leeuwenhoek. 2008;94(4):543–553. 10.1007/s10482-008-9271-x [DOI] [PubMed] [Google Scholar]
- Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF.. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin Infect Dis. 2008;46(9):1368–1373. 10.1086/586751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolrattanothai K, Gray BM, Mankin P, Stanfill AB, Pearl RH, Wallace LJ, Vegunta RK.. Simultaneous carriage of multiple genotypes of Staphylococcus aureus in children. J Med Microbiol. 2011; 60(3):317–322. 10.1099/jmm.0.025841-0 [DOI] [PubMed] [Google Scholar]
- Mossong J, Gelhausen E, Decruyenaere F, Devaux A, Perrin M, Even J, Heisbourg E.. Prevalence, risk factors and molecular epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) colonization in residents of long-term care facilities in Luxembourg, 2010. Epidemiol Infect. 2013;141(6):1199–1206. 10.1017/S0950268812001999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P, Ripa T.. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J Clin Microbiol. 2006;44(9):3334–3339. 10.1128/JCM.00880-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Dulon M, Kleinmüller O, Nienhaus A, Schablon A.. MRSA prevalence and risk factors among health personnel and residents in nursing homes in Hamburg, Germany – A cross-sectional study. PLoS One. 2017;12(1):e0169425 10.1371/journal.pone.0169425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt EJG, Szkuta PT, Burns N, Foster SJ.. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018;14(6):e1007112 10.1371/journal.ppat.1007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghmann MC, Johnson JK, Sorkin JD, Langenberg P, Lydecker A, Sorace B, Levy L, Mody L.. Transmission of methicillin-resistant Staphylococcus aureus (MRSA) to healthcare worker gowns and gloves during care of nursing home residents. Infect Control Hosp Epidemiol. 2015;36(9):1050–1057. 10.1017/ice.2015.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat A, Krzyszton-Russjan J, Strzalka W, Filipek R, Kosowska K, Hryniewicz W, Travis J, Potempa J.. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J Clin Microbiol. 2003;41(4):1801–1804. 10.1128/JCM.41.4.1801-1804.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gu FF, Zhao SY, Xiao SZ, Wang YC, Guo XK, Ni YX, Han LZ.. Prevalence and molecular epidemiology of Staphylococcus aureus among residents of seven nursing homes in Shanghai. PLoS One. 2015;10(9):e0137593 10.1371/journal.pone.0137593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita J, Barba P, Ortega-Paredes D, Mora M, Rivadeneira S.. Local circulating clones of Staphylococcus aureus in Ecuador. Braz J Infect Dis. 2016;20(6):525–533. 10.1016/j.bjid.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]