Abstract
The aim of the study was mycological examination of ulcerated corneal tissues from an ophthalmic patient. Tissue fragments were analyzed on potato-glucose agar (PDA) and maltose (MA) (Difco) media using standard laboratory techniques. Cultures were identified using classical and molecular methods. Macro- and microscopic colony morphology was characteristic of fungi from the genus Aspergillus (restricted growth series), most probably Aspergillus penicillioides Speg. Molecular analysis of the following rDNA regions: ITS1, ITS2, 5.8S, 28S rDNA, LSU and β-tubulin were carried out for the isolates studied. A high level of similarity was found between sequences from certain rDNA regions, i.e. ITS1-5.8S-ITS2 and LSU, what confirmed the classification of the isolates to the species A. penicillioides. The classification of our isolates to A. penicillioides species was confirmed also by the phylogenetic analysis.
Key words: Aspergillus penicillioides, morphology, genetic characteristic, cornea
Introduction
Fungi from the genus Aspergillus are anamorphic stages of ascomycetes Ascomycota, class Eurotiomycetes, order Eurotiales and family Trichocomaceae (syn. Euro-tiaceae) (Kirk 2008). These are the fungi with a high morphological and genetic variability (Thom and Raper 1945; Peterson 2000, 2003).
They are most often saprotrophs or facultative parasites (Agrios 2005). They play a significant role in consumable and pharmaceutical industries. These fungi occur on plants, in soil, animal remnants, storage products or public utility places (Thom and Raper 1945; Ejdys 2007; Krijgsheld et al. 2013; Ogar et al. 2015). They are quite common in different climate zones, especially in subtropical and tropical regions (Thom and Raper 1945; Krijgsheld et al. 2013). These fungi are adapted to grow in high temperatures and a warm climate (Gock et al. 2003). They are the causative agents behind diseases such as aspergilloses (Bossche et al. 1988; Seyedmousavi et al. 2015). The aspergilloses of the human respiratory system, skin, ears, sinuses, especially in patients with immunodeficiency or cystic fibrosis has already been reported (Bossche et al. 1988; Sandhu et al. 1995; Hamilos 2010; Gupta et al. 2015; Walicka-Szyszko and Sands 2015).
Aspergilloses have been observed in bees, domestic birds and farm animals (Puerta et al. 1999; Lugauskas et al. 2004; Seyedmousavi et al. 2015). Many species produce secondary metabolites that are toxic to humans and warm-blooded animals. Aflatoxins and their derivatives (B1, B2, G1, G2, M1, M2) are Aspergillus spp. mycotoxins that are strongly poisonous to humans and animals. Aflatoxin B1 is considered the most carcinogenic substance (Agrios 2005). Ohratoxin A, sterygmatocistin and patulin are among other substances toxic to humans and animals (Bayman et al. 2002).
There is more and more information in the literature about ocular mycoses (Mendiratta et al. 2005; Mitani et al. 2009; Machowicz-Matejko and Zalewska 2015). Different taxa of fungi could be the cause of these infections i.e.: Candida, Colletotrichum, Fusarium, Penicilllium, Aspergillus and other (Sharma et al. 1993; Gunawardena et al. 1994; Thomas 2003; Chowdhury and Singh 2005; Nayak 2008). Aspergillus spp. are among the most frequently isolated fungi from the cornea (Nayak 2008).
Ocular mycoses are very difficult to eliminate, and it is essential to determine the aetiology of infection in order to select the proper treatment.
The aim of the study was the mycological examination of the affected corneal tissue from a patient with corneal ulceration.
Experimental
Materials and Methods
Material. The research material consisted of a fragment of the cornea with deep ulcer and inflammatory infiltration taken during the “on hot” surgery of the corneal transplant from the patient in the Department of Diagnostics and Microsurgery of Glaucoma at the Medical University in Lublin (Fig. 1).
Fig. 1.

The disease symptoms of the cornea from which A. penicillioides was isolated.
Photo E. Machowicz-Matejko.
Patient Characteristics. An 84-year-old man was treated for pneumonia a few months ago before his admission to the hospital. Moreover, he was also treated for diabetes, hyperthyroidism, atherosclerosis and kidney disorder. This patient was hospitalized because of severe keratitis of unknown aetiology. Cornel scrubs and cultures were taken for the microbiological analysis on different culture media. The negative results for bacteria and fungi were obtained after seven days of culture on the appropriate media. In spite of the empirical treatment with antibiotics and antifungal agents the corneal inflammation continued to be very severe. After nine days of local and systemic treatment we decided about corneal transplantation “on hot”. The tissue of the cornea was taken to the microbiological examination. After the seven days of the culture on PDA and MA culture media, using standard laboratory methods (FAO 2006) no colonies were visible on the media. Despite on this we decided to carry on our analysis for a longer time.
The research material was kept in a physiological solution of 0,9% NaCl at room temperature. The mycological analysis of cornea pieces was carried on PDA and MA culture media, using standard laboratory techniques.
Two isolation methods were used: the first (I) – 0.5 mm cornea fragments were placed on solidified culture media in Petri dishes – 4 pieces per one dish; and the second (II) – suspension of the fungus hyphae that passed from the cornea to the physiological solution was transferred to sterile Petri dishes in a volume of 0.1 ml and 0.5 ml, and poured over a chilled liquid PDA (own method). The samples were then incubated in a thermostat at a temperature of 25°C. The visible colonies of fungus were transferred to slants with PDA medium and subsequently single spore cultures were prepared and allowed to grow and develop. The cultures were identified using classical and molecular methods.
The species name and author’s initials were corrected in the current status of the Index Fungorum taxonomic species database in 2016 and according to Sklenář et al. (2017) elaboration.
Classical identification. Pure fungus cultures were placed as a small inoculum on agar media in Petri dishes, using three inocula per one Petri dish, which was then sealed with a parafilm to prevent drying. Five different culture media were used: Czapek-Dox, PDA, Sabouraud, YPD (Thom and Raper 1945; FAO 2006) and a poor medium (5 g – glucose, 20 g – agar, 1 dm3 – distilled water). The cultures of fungus were carried out at a temperature of 25°C for one month. Considering the very slow growth of fungi, the temperature was raised to 30°C and the period of incubation was extended.
Macroscopic and microscopic observations and photographic documentation were performed from the start of visible colony growth, i.e. from day 17 of culture. Photographs of the morphological structures using a Scanning Electron Microscope (SEM) (Tescan Vega/LMU) were taken at the Central Laboratory of Agro-Ecological (CLA), the University of Life Sciences in Lublin.
Molecular identification. The research material used in this study consisted of 3 different isolates, obtained from the human cornea (Nos 1–3). The colonies prepared for DNA isolation were grown on PDA medium. Identification of the isolates to the species level was carried out using universal primers for rDNA genes: the ITS1-5.8S-ITS2 – (ITS1 and ITS4) region, 28S rDNA region – LSU – (LR5 and LROR) and for the β-tubuline – gene (Bt2a and Bt2b) (Table I).
Table I.
Primers used in PCR reaction.
| Name | Primer | Primer sequence 5’-3’ | Melting temperature (Tm) °C | Primer orientation | Reference |
|---|---|---|---|---|---|
| ITS primers | ITS1 | TCCGTAGGTGAACCTGCGG | 59 | Forward | Ref. 28 |
| ITS4 | TCCTCCGCTTATTGATATGC | 59 | Reverse | Ref. 28 | |
| LSU primers | LROR | ACCCGCTGAACTTAAGC | 60 | Forward | Ref. 28 |
| LR5 | TCCTGAGGGAAACTTCG | 60 | Reverse | Ref. 29 | |
| β-tubulin primers | Bt2a | GGTAACCAAATCGGTGCTGCTTTC | 60 | Forward | Ref. 30 |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | 60 | Reverse | Ref. 30 |
DNA isolation. Genomic DNA isolation was performed using the CTAB method by Doyle and Doyle (1987) with some own modifications. Fungus isolates were grown at a temperature of 27°C for 45 days. The obtained mycelium was transferred to Eppendorf tubes of a 1.5 ml volume, and then frozen in liquid nitrogen. The frozen material was homogenized with a sterile pestle (Sigma-Aldrich) until mycelium homogeneity. Subsequently, 600 ml of CTAB lysis buffer was added to each tube and incubated at 65°C for 2 hours and then centrifuged (10 000 rpm for 10 minutes). In the next step, 1.0 ml phenol/chloroform/alcohol mixture in the volume ratio of 25:24:1 was added to the supernatant and centrifuged (10 000 rpm for 8 minutes). DNA was precipitated with 40 μl of sodium acetate (5 M) and 400 μl of isopropanol. The resulting precipitate was washed with 70% ethanol, and then centrifuged at 14 000 rpm for 15 minutes and dried. The supernatant was suspended in 50 μl of PCR water (Sigma-Aldrich), supplemented with RNase A, and then incubated for 25 hours at a temperature of 4°C, followed by one hour at 37°C, and 10 minutes at 65°C. DNA was run in 1.5% agarose gel electrophoresis and compared with GeneRuler 100 bp DNA Ladder Plus marker (Thermo Scientific). The DNA quantity was measured with a UV-Vis NanoDrop 2000c/2000 spectrophotometer (Thermo Scientific). Then, the DNA samples were diluted to a final concentration of 50 ng/μl and stored at 20°C for further analysis.
PCR amplification and sequence analysis. Three different primers were used for identification of Aspergillus spp. isolates: ITS1 and ITS4, LROR and LR5, and Bt2a and Bt2b (Table I). The amplification reaction volume was 25 μl and the reaction solution contained 20 ng/μl of genomic DNA, 1 × Buffer Tag (750 mM Tris HCl pH 8.8, 200 mM (NH4)2SO4, 0.1% Tween 20) (Thermo Scientific), 0.1 mM × MgCl2, dNTP mix 1 mM of each primer 0.2 μM and 1.0 U polymerase DNA (Fermentas). Amplification reaction was performed using a DNA Engine Dyad Thermal Cycler (Biorad). The PCR amplification reaction for ITS1 and ITS4 primer pair was carried out using the following program: initial denaturation for 5 min at 95°C, followed by 40 cycles consisting of 30 s at 95°C, 50 s at 57°C, 60 s at 72°C and final extension for 10 min at 72°C. PCR reaction programs for two primers pairs, the first, LROR and LR5, and the second, Bt2a and Bt2b, were subjected to minor changes, as follows: initial denaturation for 5 min at 94°C, then 36 cycles of annealing: 30 s at 94°C, 50 s at 56°C, 55 s at 72°C and final extension of the product at 72°C for 5 min. The obtained products were separated by electrophoresis using 1.5% agarose gel containing 7.5 μl of ethidium bromide in 1 × TBE buffer at 80 V for 1.5 hour in the presence of GeneRuler 100 bp Plus DNA Ladder (Thermo Scientific). The PCR products were further sequenced using Applied Biosystems BigDay®Terminator v. 3.1. kit (Life Technologies) and separated on a 3730xl DNA Analyser capillary sequencer. The nucleotide sequences obtained were compared with the nucleotide sequence deposited in GenBank at the National Centre for Biotechnology Information NCBI (http://www.ncbi.nlm.nih.gov/), using the Mega Blast program (Zhang et al. 2000; Morgulis et al. 2008). The “local sequence alignment” method was used as a comparison criterion. The highest degree of sequence identity was used to determine the species of the isolates. The phylogenetic analysis of ITS sequences of our three isolates of A. penicillioides studied was carried out in several steps using the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei 1993). Moreover, 13 other reference strains of four Aspergillus spp. i.e.: A. salinicola – strains EXF_10401, IBT_34266, KAS_6054, A. hordei – NRRL25826, NRRL25825, NRRL25830, A. clavato-phorus – DTO257-G5, NRRL_25874 and 5 strains of A. penicillioides (mentioned in Table II) were also included. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There was a total of 455 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016).
Table II.
The degree of identity and length of the covered sections of sequences as defined by the BLAST methods.
| Species | Strain/isolate | The analyzed sequences from GenBank accession No. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | LSU | β-tubulin | ||||||||
| No. GenBank | The degree of identity of sequence [%] = identity | Query cover nucleotides | No. GenBank | The degree of identity of sequence [%] = identity | Query cover nucleotides | No. GenBank | The degree of identity of sequence [%] = identity | Query cover nucleotides | ||
| Aspergillus penicillioides | isolate B1-8 | KF986414.1 | 100 | 569 | ||||||
| Aspergillus penicillioides | UFMGCB 6310 | KF373543.1 | 100 | 498 | ||||||
| Aspergillus sp. | DY115-21-7-M6 | KF411572.1 | 100 | 551 | ||||||
| Uncultured fungus | clone LX042233-122-012-B01 | GQ999240.1 | 99 | 574 | ||||||
| Aspergillus penicillioides | isolate KH00279 | GU017500.1 | 99 | 574 | GU017541.1 | 99 | 835 | |||
| Aspergillus penicillioides | isolate KH00251 | GU017494.1 | 99 | 574 | GU017535.1 | 100 | 833 | |||
| Uncultured fungus | clone PR-MAT-CV5-17 | FJ265955.1 | 99 | 574 | ||||||
| Aspergillus sp. | F55 | FJ755819.1 | 99 | 574 | ||||||
| Aspergillus penicillioides | strain WR1996 | KP997215.1 | 99 | 544 | ||||||
| Aspergillus penicillioides | Culture collection CCF<CZE> 3112 | FR727125.1 | 99 | 573 | ||||||
| Uncultured fungus | clone PR-MAT-CV5-19 | FJ265957.1 | 99 | 571 | ||||||
| Aspergillus penicillioides | isolate HNC15-78 | KT959298.1 | 99 | 530 | ||||||
| Uncultured fungus | clone xnh90 | KP063524.1 | 99 | 574 | ||||||
| Aspergillus sp. | 43m | KC834810.1 | 99 | 527 | ||||||
| Uncultured Aspergillus | clone CHiv38 | KP974191.1 | 99 | 574 | ||||||
| Uncultured fungus | clone xnh20 | KP063454.1 | 99 | 574 | ||||||
| Uncultured Aspergillus | clone Leof80 | KF225869.1 | 99 | 574 | ||||||
| Aspergillus penicillioides | clone KJ34-0.2-39 | KT390118.1 | 99 | 791 | ||||||
| Aspergillus penicillioides | U81265.1 APU81265 | 99 | 783 | |||||||
| Aspergillus sp. | CCF 3112 | FR775323.1 | 97 | 506 | ||||||
Results
Pathological changes in the eye involved the cornea. There were superficial epithelial changes and alterations in deep stromal layers. Corneal ulceration, 7 mm in diameter, was surrounded by a white infiltration and endothelial deposits were observed. Corneal tissue at the site of ulceration was completely opaque, while surrounding tissue was cloudy because of the inflammation (Fig. 1). Two fungal colonies were cultured from the examined tissues using method I and several colonies using method II of fungus isolation. The first colonies appeared only just after 15–17 days of incubation on PDA. There was no growth on MA medium regardless of the method of isolation.
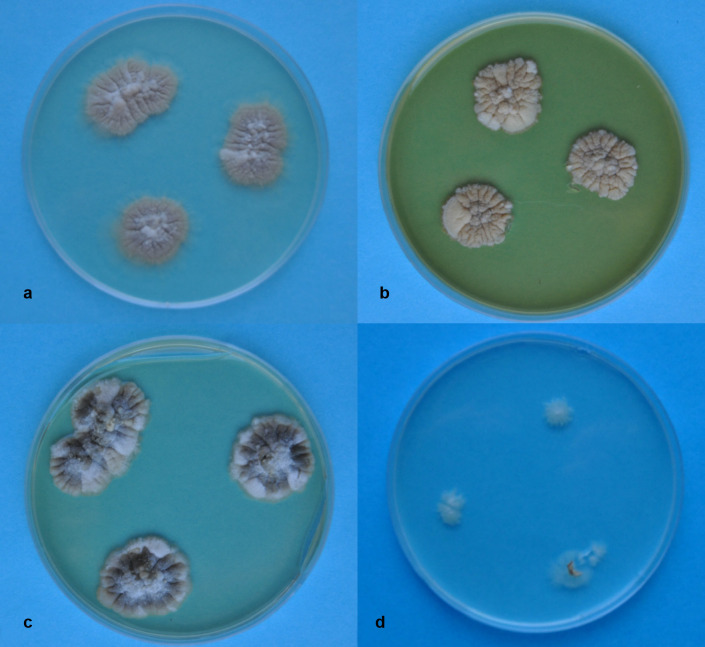
Morphological characteristics. A characteristic feature of the studied fungus was extremely limited growth of the colonies. After 1.5 months on Czapek-Dox medium, the diameter of the colony ranged from 0.8 to 1.0 cm, on PDA from 1.2 to 1.6 cm, and on YPD medium and Sabouraud from 1.2 to 1.4 (Fig. 2). On the poor medium, the growth of colony was hardly noticeable. Larger diameters of the colonies were observed after 2–2.5 months of culture.
Fig. 2.
Colonies of A. penicillioides after 8 weeks of growing on culture media.
a – PDA, b – YPD, c – Sabouraud, d – a poor medium (see M&M section). Photo E. Zalewska.
The colonies on Czapek-Dox, PDA, Sabouraud and YPD medium were light-beige in color, slightly darkening with age, while on poor medium they were white-grey (Fig. 2). The reverse of the colonies was bright to dark beige. An olive color of the colonies appeared only after two months of growth on Sabouraud medium.
Surface of the colonies was velvety, compact and strongly wrinkled with radial grooves, appearing after a long time of culture. Only on a poor medium, the colonies were flat (Fig. 2).
In microscopic slights of mycelium taken from Czapek-Dox, PDA, Sabourauda and YPD culture media, numerous conidiophores with smooth walls, growing from substrate mycelium or aerial mycelium, were observed (Fig. 3). Conidiophores contained oval or flask-shape vesicles (Figs. 4, 5). Four to several phialides grew from the upper part of vesicles in single row (Figs. 4, 5). On the other hand, single conidiophores with several long digitate phialides were observed on poor medium. Very sparse sporulation was a characteristic feature of the fungus. Short dense chains of conidia were produced only at a temperature of 30°C. Conidia with a smooth or slightly rough surface were cut at one or both ends (Figs. 4, 6). On Sabouraud medium, conidia were beige in mass but on other culture media, they were hyaline. Sizes on PDA were as follows: conidiophores from 50.2 to 205.0 µm, vesicles from 5.83 to 6.5 µm; phialides 8–10 × 2.4–3.2 µm and conidia 3.88–5.4 × 1.94–3.6 µm. No perithecia were found. The macro- and microscopic features of the colonies were characteristic of fungi of the genus Aspergillus, series Restrictus, and most likely A. penicillioides Speg. (Thom and Raper 1945; Sklenář 2017.)
Fig. 3.

A. penicillioides cultured on PDA.
Photo E. Zalewska.
Fig. 4.
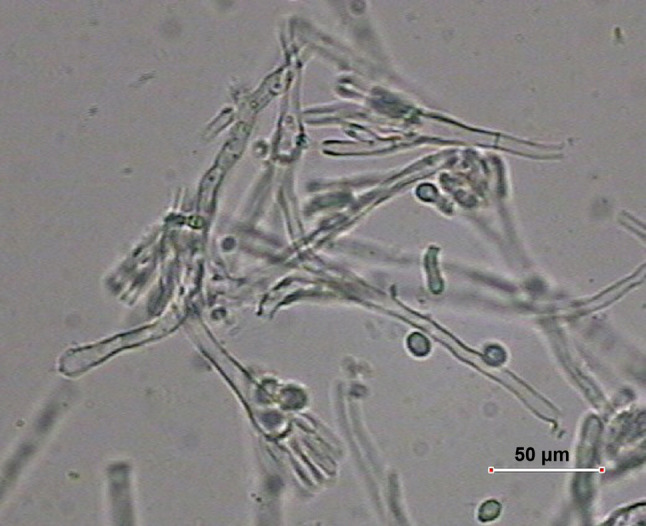
Conidiophores and conidia of A. penicillioides on PDA.
Photo E. Zalewska.
Fig. 5.

Conidiophores of A. penicillioides on PDA.
Photo M. Wróbel.
Fig. 6.

Conidia of A. penicillioides on PDA.
Photo M. Wróbel.
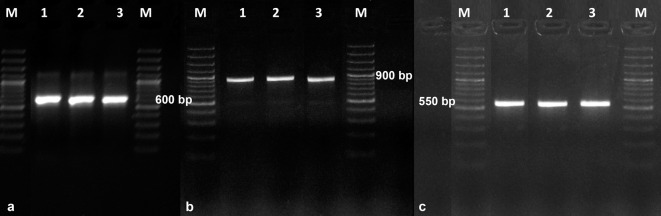
Molecular characterization. One specific product was obtained in the PCR reaction for each studied isolate (Fig. 7). The ITS1 and ITS2 primer pair generated a product with a length of approx. 600 bp, LROR and LR5 approx. 950 bp, and Bt2a and Bt2b approx. 550 bp.
Fig. 7.
Electrophoretic separation of PCR products using universal primers for regions of the genes.
a – rDNA (ITS1-5.8S-ITS4), b – LSU, c – β-tubulin. Photo A. Furmańczyk.
The ITS1-5.8S-ITS2 region (600 nt), a partial sequence of LSU (Large Submit RNA 25–28S, 959 nt) and part of the β-tubulin gene (550 nt) were sequenced. As a result of comparison of the sequences of ITS1-5.8S-ITS2 region, 56 sequences were selected from the GenBank database, but 43 of them (76.79%) belonged to the genus Aspergillus, with the degree of sequence identity ranging from 93 to 100%. Sixteen of the obtained sequences showed the highest similarity, i.e. 99–100% and seven of them belonged to the species Aspergillus penicillioides, five to undetermined species of the genus Aspergillus and four to unidentified species of fungi (Table II). The sequences of A. penicillioides strains: KF9864.1 (569 nt), KF373543.1 (498 nt) and Aspergillus sp. strain KF411572.1 (551 nt) showed 100% identity to the studied isolates. The degree of identity of other deposited sequences in GenBank was 99% for strains: GQ999240.1, GU17500.1, GU17494.1, FJ265955.1, FJ755819.1, KP063524.1, KP063454.1, KP974191.1 and KF22586.1 (574 nt), FR727125.1 (573 nt), FJ26597.1 (571 nt), KT959298.1 (530 nt) and for KC834810.1 (527 nt).
The comparison of 28S RNA (LSU regions) sequences of the isolates with the sequences deposited in GenBank resulted in the selection of 56 sequences. Their identity relative to the sequences tested ranged from 95 to 100% (Table II). A. penicillioides strain GU017535.1 was a species that showed the greatest sequence similarity (100%) over the length of about 833 nucleotides. Other three reference strains of this species showed 99% similarity; it was over 835, 791, and 783 nucleotides for the strains GU017541.1, KT390118.1 and U81265.1, respectively.
Moreover, the bioinformatic analysis of ITS1-5.8S-ITS2 and 28S RNA (LSU) regions showed a very strong similarity of our isolates to the reference strain K279 of A. penicillioides, the identity 99% for ITS1-5.8S-ITS2 and LSU, strain K251, ITS1-5.8S-ITS2 and LSU (100% sequence identity) (Table II). For the reference strain K279, the aligned sequences overlapped the sequences from nucleotide 6 to 590 in ITS1-5.8S-ITS2 regions, from nucleotide 1 to 871 in the 28S rRNA gene. For the strain K251 it was visible for the fragments from nucleotide 55 to 639 of ITS1-5.8S-ITS2 regions and from nucleotide 1 to 868 for the 28S rRNA gene.
The comparative analysis of the β-tubulin gene sequence showed higher variation than in the two other loci, and the identity of sequences ranged from 78 to 97%. The highest sequence identity was 97% at the same coverage percentage as the sequence deposited in NCBI. This identity was observed for 506 nucleotides and it was the sequence of CCCF 3112 strain of Aspergillus.
Phylogenetic analysis of ITS sequences of three isolates of A. penicillioides and other reference strains of Aspergillus indicated segregation of all isolates into two main clusters (Fig. 8). The first main cluster contained the reference strains of A. penicillioides and all our isolates of this species (Fig. 8). The second main cluster included the reference strains of A. hordei and A. salinicola (Fig. 8).
Fig. 8.
Phylogenetic tree of three native A. penicillioides isolates and the reference strains of Aspergillus spp. generated using the Maximum Likelihood method.
Discussion
The results obtained indicated that A. penicillioides could be a fungus species involved in corneal inflammation. This species was described by Spegazzini et al. (1896) and it was included into Aspergillus series of limited growth in a monograph of Thom and Raper (1945). A. penicillioides firstly was isolated from mouldy sugar cane in Argentina, cured fish in Australia and desiccated chilli in New Gwinea (Spegazzini et al. 1896; Tamura et al. 2000). The first report of disseminated aspergillosis caused by A. penicilioides in humans was described by Gupta et al. (2015). This species was isolated during an autopsy of an infant with cystic fibrosis (Gupta et al. 2015). A. penicillioides, as a xerophilous species, occurs in closed spaces, dust, on dry products, clothes, mattresses and paper products (Gallo 1993; Ejdys 2007; Visagie et al. 2014; Sklenář et al. 2017). The results of aeromycological studies confirmed a very high occurrence of Aspergillus spp. in the air (Bayman et al. 2002; Pusz et al. 2014). It seems that the human eye is naturally exposed to A. penicillioides, especially in poor hygienic conditions. Conidia of thermophilic A. penicillioides find very advantageous thermal and humidity conditions in the eye for the development and metabolic activity (Gock et al. 2003). That is why symptoms of aspergillosis are dangerous and more frequently observed than other mycoses. The weakened immunity due to illnesses of the patient in this study could be a factor favoring colonization of the cornea, which may confirm the opportunistic nature of the parasitic fungus identified. The detailed analyses are required to identify Aspergillus spp. in human tissues and organs. Our results demonstrated that the isolation of A. penicillioides was possible, instead of extremely slow development and sporulation. The temperature of 25°C, suitable for the development and growth of many fugal species (Agrios 2005), appeared to be to low, regardless of culture duration. Very slow sporulation at temperature below 30°C caused serious difficulties in the culture optimization. This observation is consistent with other authors’ opinion (Gock et al. 2003). The latter authors showed A. penicillioides sporulation at a temperature of 33°C. Among the culture media suitable for the isolation and culture of A. penicillioides, PDA and Sabouraud proved to be the best for the isolation and culture at a temperature of about 30°C, while above 30°C for stimulation and sporulation (Gock et al. 2003).
The presence of ambiguous microscopic features of A. penicillioides, for example, the overgrowth of conidiophores from aerial mycelium or substrate mycelium, various shapes and sizes of vesicle, smoothness or roughness of the surface of the conidia indicated that the identification of this species by the classical method was insufficient.
Molecular studies are required to confirm the phylogeny of A. penicillioides. Operons of rDNA belong to the most commonly used regions in molecular identification, as well as certain regions of mitochondrial DNA (mtDNA). Ribosomal (rDNA) regions contain both conservative regions and sequences with a very high genetic variability, such as those that separate the non-coding regions of ITS. Both are used in the diagnostics and genotyping of pathogenic fungi, and they constitute a standard method in molecular genotyping of fungi (Vilgalys and Hester 1990; Kuba 2008).
The very high degree of similarity of the studied ITS1-5.8S-ITS2 and LSU sequences indicated that the isolates analyzed belonged to A. penicillioides. The present study confirmed that, among the analyzed loci, ITS1-5.8S-ITS2 and LSU regions are useful for the identification of A. penicillioides.
The variable region of 28S rDNA (LSU) was detected in the genome of different species of pathogenic fungi by Sandhu et al. in 1995. The analysis of the amplified region formed the basis for the differentiation of species. Single strains of Scopulariopsis, i.e. S. brevicaulis and S. brumptii that cause skin and nail fungal infections were identified in this manner (Sandhu et al. 1995). However, ITS barcodes are often considered insufficient to distinguish between species (Skouboe et al. 1999; Peterson 2000, 2003; Samson et al. 2011). Therefore, multilocus sequence analyses of the genus are used in the case of species identification as well as polygenetic analyses for Septoria (Quaedvlieg et al. 2013; Verkley et al. 2013), Aspergillus, Penicillium and Talaromyces (Visagie et al. 2014). The phylogenetic analysis of the isolates studied confirmed their identification as A. penicillioides.
The above results provide the first information about the colonization of the human cornea by A. penicillioides.
When determining the phylogeny of the studied Aspergillus isolates using bioinformatic methods, the strains obtained from cornea and other human tissues were not found in the available database. The compared sequences, deposited in GenBank, belonged to strains obtained from water bodies (https://www.ncbi.nlm.nih.gov/nucleotide/984294657?report=genbank&log$=nucltop&blastrank=3&RID=MSTGJ4SC01R), sea grass (https://www.ncbi.nlm.nih.gov/nucleotide/294471398?report=genbank&log$=nucltop&blast_rank=5&RID=MSFXJGHB015, microbiological materials (https://www.ncbi.nlm.nih.gov/nucleotide/221047144?report=genbank&log$=nucltop&blastrank=7&RID=MSFXJGHB015 as well as from wall paint (https://www.ncbi.nlm.nih.gov/nucleotide/951503016?report=genbank&log$=nucltop&blastrank=12&RID=MSFXJGHB015), (https://www.ncbi.nlm.nih.gov/nucleotide/927092372?report=genbank&log$=nucltop&blast_rank=15&RID=MSFXJGHB015) and personal clothing (https://www.ncbi.nlm.nih.gov/nucleotide/313574881?report=genbank&log$=nucltop&blastrank=10&RID=MSFXJGHB015.
Footnotes
Conflict of interest
Authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Agrios GN. 2005. Plant Pathology Fifth Edition Amsterdam (Netherlands): Elsevier Academic Press; 922 p. [Google Scholar]
- Bayman P, Baker JL, Doster MA, Michailides TJ, Mahoney NE. 2002. Ochratoxin production by the Aspergillus ochraceus group and Aspergillus alliaceus. Appl Environ Microbiol. 68(5):2326–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bossche H, Mackenzie DWR, Cauwenbergh G. 1988. Aspergillus and aspergillosis New York (USA): Plenum Press. [Google Scholar]
- Chowdhury A, Singh K. 2005. Spectrum of fungal keratitis in North India. Cornea. 24:8–15. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Ejdys E. 2007. Fungi isolated in school buildings. Acta Mycol. 42(2):245–254. [Google Scholar]
- FAO 2006. Handbook of mycological methods. Project GCP/INT/743/CFC [Internet] Rome (Italy): Food and Agriculture Organization of the United Nations; [cited 7 Jul 2016]. Available from: http://www.fao.org/fileadmin/user_upload/agns/pdf/coffee/Annex-F.2.pdf [Google Scholar]
- Gallo F. 1993. Aerobiological research and problems in libraries. Aerobiologia 9:117–130. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl Environ Microb. 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gock MA, Hocking AD, Pitt JI, Poulos PG. 2003. Influence of temperature, water activity and pH on growth of some xerophilic fungi. Int J Food Microbiol. 81(1):11–19. [DOI] [PubMed] [Google Scholar]
- Gunawardena SA, Ranasinghe KP, Arseculeratne SN, Seimon CR, Ajello L. 1994. Survey of mycotic and bacterial keratitis in Sri Lanca. Mycopathologia. 127:77–81. [DOI] [PubMed] [Google Scholar]
- Gupta S, Radhakrishnan A, Raharja-Liu P, Lin G, Steinmetz LM, Gagneur J, Shina H. 2015. Temporal expression profiling identifies pathways mediating effect of causal variant on phenotype. PLoS Gent. 11(6):e1005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilos DL. 2010. Allergic fungal rhinitis and rhinosinusitis. Proc Am Thorac Soc. 7:245–252. [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Ainsworth and Bisby’s dictionary of the fungi Wallingford (UK): CAB International; 771 p. [Google Scholar]
- Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Müller WH, Dijksterhuis J, Wösten HAB. 2013. Development in Aspergillus. Stud Mycol. 74(1):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K. 2008. Molecular and immunological methods applied in diagnosis of mycoses. Wiad Parazytol. 54(3):187–197. [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugauskas A, Krikštaponis A, Šveistyle L. 2004. Airborne fungi in industrial environments – potential agents of respiratory diseases. Ann Agric Environ Med. 11:19–25. [PubMed] [Google Scholar]
- Machowicz-Matejko E, Zalewska ED. 2015. Pharmacological substances in vitro in limiting growth and development of fungi Colletotrichum genera. J Ocul Pharmacol and Ther. 31(5):303–309. [DOI] [PubMed] [Google Scholar]
- Mendiratta DK, Thamke D, Shukla AK, Narang P. 2005. Keratitis due to Colletotrichum dematium – A case report. Indian J Med Microbiol [Internet]. [cited 7 Jul 2016]; 23:56–58. Available from: http://www.ijmm.org/text.asp?2005/23/1/56/13876 [DOI] [PubMed] [Google Scholar]
- Mitani A, Shiraishi A, Uno T, Miyamoto H, Hara Y, Yamaguchi M, Ohashi Y. 2009. In vivo and in vitro investigations of fungal keratitis caused by Colletotrichum gloeosporioides. J Ocul Pharmacol Ther. 25:563–565. [DOI] [PubMed] [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. 2008. Database indexing for production MegaBLAST searches. Bioinformatics. 15:1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak N. 2008. Fungal infections of the eye – laboratory diagnosis and treatment. Nepal Med Coll J. 10(1):48–63. [PubMed] [Google Scholar]
- Ogar A, Tylko G, Turnau K. 2015. Antifungal properties of silver nanoparticles against indoor mould growth. Sci Total Environ. 521–522:305–314. [DOI] [PubMed] [Google Scholar]
- Peterson SW. 2000. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam (Netherlands): Harwood Acad. Publ. 323–355 p. [Google Scholar]
- Peterson SW. 2000. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam (Netherlands): Harwood Acad. Publ. 163–178 p. [Google Scholar]
- Puerta F, Flores JM, Ruiz JA, Ruz JM, Campano F. 1999. Fungal diseases of the honeybee (Apis mellifera L.) In: Colin ME, Ball BV, Kilani M, editors. Bee disease diagnosis. Zaragoza (Spain): CIHEAM (Options Méditerranéennes: Série B. Etudes et Recherché; n. 25) 61–68 p. Available from: http://om.ciheam.org/om/pdf/b25/99600236.pdf [Google Scholar]
- Pusz W, Kita W, Weber R. 2014. Microhabitat influences the occurrence of airborne fungi in copper mine in Poland. J Cave Karst Stud. 76(1):14–19. [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin H-D, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW. 2013. Sizing up Septoria. Stud Mycol. 75:307–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 70:159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu GS, Kline B, Stockman L, Roberts G. 1995. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 33: 2913–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S, Guillot J, Arné P, de Hoog GS, Mouton JW, Melchers WJ, Verweij PE. 2015. Aspergillus and aspergilloses in wild and domestic animals: a global health concern with parallels to human disease. Medicinal Mycol. 53:765–797. [DOI] [PubMed] [Google Scholar]
- Sharma S, Srinivasan M, George C. 1993. The current status of Fusarium species in mycotic keratitis in South India. Indian J Med Microbiol. 11:140–147. [Google Scholar]
- Sklenář F, Jurjević Ž, Zalar P, Frisvad JC, Visagie CM, Kolařík M, Houbraken J, Chen AJ, Yilmaz N, Seifert KA, Coton M, et al. . 2017. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Stud Mycol. 88:161–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouboe P, Frisvad JC, Taylor JW, Lauritsen D, Boysen M, Rossen L. 1999. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol Res. 103(7):873–881. [Google Scholar]
- Spegazzini C. 1896. Hongos de la Caña de Azúcar. Revista de la Facultad de Agronomía Universidade Nacional de La Plata. 2(19): 227–258. [Google Scholar]
- Tamura M, Kawahara K, Sugiyama J. 2000. Molecular phylogeny of Aspergillus and associated teleomorphs in the Trichocomaceae In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam (Netherlands): Harwood Acad. Publ. 369 p. [Google Scholar]
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526. [DOI] [PubMed] [Google Scholar]
- Thom Ch, Raper KB. 1945. A manual of the Aspergilli Baltimore (USA): The Williams & Wilkins Company. [Google Scholar]
- Thomas PA. 2003. Current perspectives on ophthalmic mycoses. Clin Microbiol Rev. 16(4):730–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walicka-Szyszko K, Sands D. 2015. The clinical presentations of pulmonary aspergillosis in children with cystic fibrosis-preliminary report. Dev. Period Med. 19(1):66–79. [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a guide to methods and applications. New York (USA): Academic Press Inc. 315–322 p. [Google Scholar]
- Verkley GJM, Quaedvlieg W, Shin HD, Crous PW. 2013. A new approach to species delimitation in Septoria. Stud Mycol. 75:213–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172:4239–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Hirooko Y, Tanney JB, Whitfield E, Mwange K, Meijer M, Amend AS, Seifert KA, Samson RA. 2014. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol 78:63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7(1–2):203–214. [DOI] [PubMed] [Google Scholar]