Abstract
The recently discovered CRISPR-Cas gene editing system and its derivatives have found numerous applications in fundamental biology research and pharmaceutical sciences. The need for precise external control over the gene editing and regulatory events has driven the development of inducible CRISPR-Cas systems. While most of the light-controllable CRISPR-Cas systems are based on protein engineering, we developed an alternative synthetic approach based on modification of crRNA/tracrRNA duplex (guide RNA or gRNA) with photocaging groups, preventing the gRNA from recognizing its genome target sequence until its deprotection is induced within seconds of illumination. This approach relies on a straightforward solid-phase synthesis of the photocaged gRNAs, with simpler purification and characterization processes in comparison to engineering a light-responsive protein. We have demonstrated the feasibility of photocaging of gRNAs and light-mediated DNA cleavage upon brief exposure to light in vitro. We have achieved light-mediated spatiotemporally resolved gene editing as well as gene activation in cells, whereas photocaged gRNAs showed virtually no detectable gene editing or activation in the absence of light irradiation. Finally, we have applied this system to spatiotemporally control gene editing in zebrafish embryos in vivo, enabling the use of this strategy for developmental biology and tissue engineering applications.
Short abstract
We developed a simple approach for light-controlled CRISPR-Cas gene editing based on photocaging of the guide RNA and achieved spatiotemporally restricted gene regulation in living embryos.
Introduction
Genome sequencing has facilitated systematic studies of gene functions and regulation networks, thus providing the basis for understanding organogenesis and complexities of genetic diseases. Various biochemical tools aimed at altering gene expression have been essential for such discoveries. While several therapeutics based on temporal gene overexpression, suppression, or splicing correction have reached the market,1 the newer approach of permanent editing of the genomic DNA is being tested in the context of therapeutic intervention in cancer,2 viral infections,3 and monogenic hereditary disorders such as Duchenne muscular dystrophy4 or Huntington’s disease.5
Recent transfer of the microbial adaptive immune system CRISPR-Cas to mammalian cells has led to the development of programmable tools for gene editing based on RNA-guided DNA endonucleases (most commonly, Cas9 and its analogues).6−9 Their target specificity is determined by the presence of a nuclease-specific protospacer adjacent motif (PAM) sequence in the target DNA and by the guide RNA (gRNA), which binds through the Watson–Crick base pairing to the complementary sequence adjacent to the PAM. The main advantage of this system is that different genomic targets can be reached by simply altering the sequence of the gRNA. Multiple sites can be targeted simultaneously by using gRNAs with different sequences. Both gRNA and Cas9 can be chemically or genetically modified to improve stability10 or even add a specific functionality to the system.11−13 Thus, nuclease-deficient Cas9 protein (dCas9) can be fluorescently tagged for imaging purposes12,13 or fused with a transcription factor for specific regulation of target gene expression.13 Examples of gRNA modifications include appending aptamers for protein binding11 and chemical modifications of the backbone and nucleobases for improved stability and specificity.10
Given the permanent character of the gene editing, being able to target only a subset of cells at specific time would be a desirable improvement of the CRISPR-Cas systems. Conditional gene editing could potentially reduce off-target gene modifications due to restricted activity of the CRISPR-Cas in the cells.14 Studies of DNA repair pathways would immensely benefit from the higher spatiotemporal control over gene-editing events.15 Embryonic, tissue, or cancer development research is another area where precise spatiotemporal regulation of gene expression is of utmost need due to complex and dynamic gene expression patterns.16 This demand has triggered the development of several approaches for externally controlled gene editing and gene expression using small molecules,17 magnetic field,18 or light.19 Among these, light has the advantages of being relatively noninvasive, having minimal to no molecular footprint on treated cells, having fast on/off response, and having the highest spatial resolution.20
Several attempts have been made to render the CRISPR-Cas system light-responsive to enable precise temporal control over gene editing/expression.19,21−31 Examples of optogenetic approaches to control CRISPR-Cas include light-responsive Cas9 analogues,19,21,24−29 light-activatable transcription factors,30 photoswitchable calcium channel-mediated nuclear translocation of modified dCas9,31 gRNA-blocking oligonucleotides,22 and light-activatable transfection devices.23 These systems are designed to respond to far UV (365 nm),19,22 blue (450–470 nm),21,24−27,29,31 cyan (500 nm),28 red (650 nm),26 far red (730 nm),30 or near-infrared light (980 nm).23
The first demonstration of a light-controlled CRISPR-Cas-mediated gene editing in cancer cells was achieved by incorporation of UV light-sensitive photocaged unnatural amino acid in the structure of the Cas9 protein through expansion of the genetic code of the cells via an engineered pyrrolysyl tRNA/tRNA synthetase pair.19 As a more biocompatible alternative to in situ synthesis of photocaged Cas9, several systems based on fusion of Cas9 with photosensitive protein domains have been developed to achieve light-controlled gene editing or transcription.21,24−29 Such systems could be based on split-Cas925,27 or single-chain Cas9 analogues21,24,26−29 and be activated either through light-induced dimerization of cryptochrome-based CRY2/CIB1 pairs,21,24,26,27 pMag/nMag pairs,25,27 light oxygen voltage-based FKF1/GI pairs,26 phytochrome-based PHYB/PIF pairs,26 or dissociation of dimeric green fluorescent protein pdDronpa28 or a homodimeric light oxygen voltage domain.29
As an alternative to light-mediated assembly of photosensitive protein domains fused to Cas9 fragments, an approach based on light-induced transcription of CRISPR components has been developed.30 Upon irradiation with far-red light, bacteriophytochrome diguanylate cyclase induces formation of cyclic diguanylate monophosphate (c-di-GMP). This leads to dimerization of c-di-GMP–responsive hybrid transactivator and its binding to an orthogonal promoter, initiating expression of a MS2-transactivator fusion protein. The transactivator can further bind to gRNA fused to MS2 RNA aptamers and induce transcription of the target gene.30 Another approach to light-controlled regulation of transcription is based on light-mediated nuclear translocation of modified dCas9.31 The design of this system relies on a photoswitchable Ca2+ channel Opto-CRAC and a dCas9-VP64 transcription factor protein fused with Ca2+-responsive NFAT fragment. Upon blue light activation of the OptoCRAC, Ca2+ influx activates a Ca2+-dependent phosphatase calcineurin, which dephosphorylates NFAT and leads to NFAT-dCas9-VP64 nuclear translocation.31
Despite the variety of systems developed, several problems remain such as leakage of activity in the dark,21,25,27−30 incomplete recovery of Cas9 function,24−26,28,29 and the requirement for constant illumination during several hours,21,24−31 which could be partially explained by spontaneous self-assembly of split-CRISPR components,32 context-dependent structural folding,33 and low light sensitivity of the dimerizing domains,34 respectively.
An alternative approach to light-activatable protein engineering is to block the gRNA activity by hybridization with a complementary photolyzable DNA oligonucleotide.22 While achieving fast response to the UV light irradiation, this system suffers from unsatisfactory leakage of activity in the dark (7.4% vs 16.7% of indel in GFP gene before and after UV exposure, respectively), possibly due to complex dissociation inside cells.22 Another drawback is the relatively high UV dose of 4.0 J/cm2 required to degrade the protectors bearing three photolyzable linkages and to restore gene editing.22
Another example of light-activatable CRISPR-Cas system is based on upconversion nanoparticles, to which Cas9 protein is attached via a photocleavable linker.23 To assist with the endosomal escape of these particles, a coating with a positively charged polymer PEI was added. Despite complex multistep synthetic process, this delivery system was capable of NIR-mediated gene editing in vitro and in vivo upon intratumoral injection; however, these particles failed to fully exert their effect on tumor tissues due to clearance via mononuclear phagocyte system upon intravenous administration.23
In this project, we used a simpler synthetic strategy to directly modify the gRNA of the CRISPR-Cas system with photocaging groups, thus preventing gRNA from recognizing the target genomic DNA sequence until its light-deprotection is induced upon brief illumination. This approach relies on a straightforward solid-phase synthesis of the photocaged gRNA, with simpler purification and characterization processes in comparison to engineering a light-responsive protein or formation of a heteroduplex or engineered nanoparticles. In addition, chemical modification of the gRNA allows for tighter and faster control over Cas9 function, enabling the use of this strategy for developmental biology and tissue engineering applications.
Results and Discussion
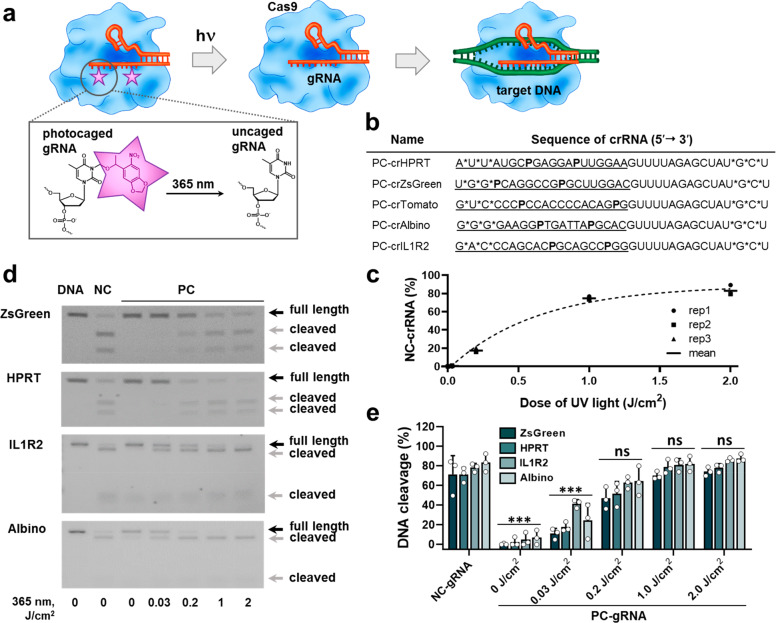
Our platform for light-controlled CRISPR-Cas gene editing and regulation is based on chemical modification of the gRNAs with photocaging groups (Figure 1a). Due to the higher yields and lower costs of shorter RNA molecules obtained via the solid-phase synthesis,35 we employed the crRNA/tracrRNA format for the gRNAs (36 and 67 bases long RNAs, respectively) rather than using the single chain gRNAs (100 bases long). The constant part of the duplex (Alt-R tracrRNA) was chemically stabilized using proprietary modifications (Integrated DNA Technologies, Inc.). For the variable part of the duplex (crRNAs), we introduced three phosphorothioate linkages at each terminus to improve their intracellular stability (Figure 1b). We positioned the photocaging groups in the targeting region of the crRNA so that the Watson–Crick base pairing with the complementary genomic DNA sequence is prevented, thus rendering CRISPR-Cas system inactive. Upon light-mediated deprotection of the photocaged gRNA, its base pairing capability should be restored. It has previously been reported that incorporation of photocaging groups every 5–6 bases of a DNA oligonucleotide completely abolished its hybridization with the complementary strand.36 On the other hand, gRNAs of length shorter than 16 bases do not trigger Cas9-mediated DNA cleavage, and the ones shorter than 10 bases are not able to activate gene expression via dCas9-transactivator system.37 Therefore, we envisaged that incorporating two photocaging groups within the target recognizing region of the gRNA should be sufficient to prevent CRISPR-Cas-mediated gene editing or transcription activation.
Figure 1.
Light-induced CRISPR-Cas-mediated target DNA cleavage. (a) Schematic representation of photocaged gRNAs mechanism of action. (b) Sequences and modification strategies of PC-crRNAs. AUGC are RNA bases, P is 6-nitropiperonyloxymethyl (NPOM)-caged T-DNA, asterisks denote phosphorothioate linkages, underlined are targeting sequences. (c) Kinetics of PC-crRNA light-mediated uncaging reaction monitored by reverse-phase high-performance liquid chromatography (N = 3, mean ± SD). (d) Biochemical activity assay of target DNA cleavage by noncaged (NC) or photocaged (PC) gRNAs with 365 nm light illumination (0–2 J/cm2). (e) Quantification of target DNA cleavage from gel images using Fiji ImageJ software.44 Results are expressed as individual data points overlaid with the mean ± SD (N = 3); ***p < 0.001, ns–not significant (p > 0.05) versus NC-gRNA according to one-way ANOVA analysis combined with Tukey’s (Holm-Sidak) posthoc test. Data are fitted with B-spline function.
Several nucleic acid photocaging groups have been described in the literature, varying in the activation wavelength and the type of the nucleotides they protect.38,39 In the present project, we used the commercially available nucleic acid photocaging group 6-nitropiperonyloxymethyl (N-POM) dT DNA (Figure 1a), which could be removed upon irradiation with a low intensity light of 365 nm.36 In these conditions, irradiation-induced damage to DNA is negligible, as it usually occurs at wavelengths below 315 nm.40 Despite the fact that crRNA naturally does not contain deoxynucleotides in its structure, recent studies demonstrated that virtually any site in the target region could be substituted with a deoxynucleotide, with the requirement that the RNA content is sufficient for preserving an A-form-like helical structure in the crRNA.41−43 We designed several crRNAs to target various reporter and functional genes, and their sequences and modifications are presented in Figure 1b.
First, we confirmed that PC-groups could be removed by light exposure using high-performance reverse-phase liquid chromatography (Figure 1c, SI Figure 1a,b). Complete uncaging of PC-gRNAs irrespective of their sequences could be achieved even with a 1.0 ± 0.15 J/cm2 light dose (5 min of 3.3 ± 0.5 mW/cm2 365 nm light irradiation). Importantly, virtually no single caged byproducts were detected above UV light doses of 1.0 J/cm2 (SI Figure 1b), indicating the possibility of full restoration of the gRNA function upon deprotection.
In a cell-free DNA cleavage assay, noncaged crRNA/tracrRNA duplexes were able to guide Cas9 protein to their targets and induce double-stranded breaks for a range of genes (Figure 1d,e). All photocaged gRNAs tested lost their activity, thereby confirming our hypothesis that positioning photocaging groups within the targeting regions of crRNAs prevents CRISPR-Cas-mediated DNA cleavage. In the dark, virtually no gene editing could be detected. However, exposing the reaction mixtures to the 365 nm light resulted in reactivation of target cleavage. In this assay, complete restoration of gRNA’s function was achieved already after ∼1.0 J/cm2 light dose across multiple targets (Figure 1d,e), in accordance with the liquid chromatography assay (Figure 1c, SI Figure 1).
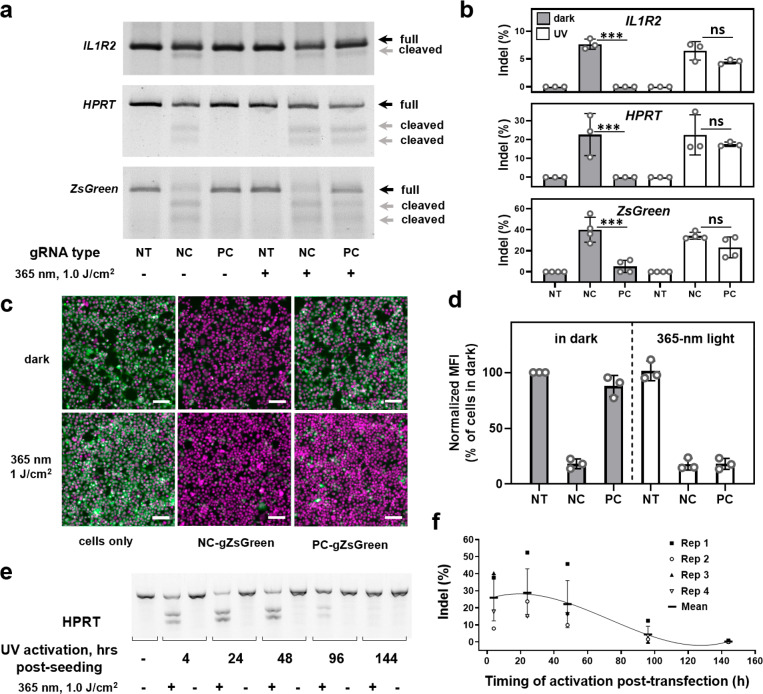
We then tested our photocaged gRNA approach on HEK293FT cells stably expressing Cas9 (Figure 2). These cells were derived from a single-cell colony, providing uniform Cas9 expression in the whole cell population. The cells were transfected with PC-gRNAs for 4 h and subsequently irradiated using a 1.0 J/cm2 UV light dose. After 2 days, the gene-editing efficiencies were evaluated in a mismatch-based assay. Exposure of cells transfected with regular noncaged gRNAs to the UV light did not affect the gene-editing efficiency. Importantly, in the absence of light irradiation, photocaging groups remained attached to the gRNA even in the complex intracellular environment, affording excellent protection from gene editing. However, when cells transfected with photocaged gRNAs were exposed to the UV light, over 70% of the gene-editing efficacy was restored (Figure 2a,b), in accordance with the in vitro results in Figure 1. Similar results were obtained when we analyzed light-controlled ZsGreen knockout at the protein level in HEK-Cas9-ZsGreens cells using fluorescence microscopy and flow cytometry (Figure 2c,d, SI Figure 2). Five days after transfection, the level of ZsGreen expression was diminished by 80% when we used uncaged gRNA (the half-life of ZsGreen protein is above 26 h45). With PC-gRNA, we observed complete recovery of gene editing after the light exposure. Despite the relatively high energy of the 365 nm UV light used to activate PC-gRNAs, overall low dose and brief exposure did not affect cell viability (SI Figure 3). This is in contrast to extended exposure to the blue light necessary to achieve dimerization and significant Cas9 activation in split-Cas9 systems, which often leads to high photocytotoxicity and low cell viability.30,46 These experiments demonstrate the utility of the photocaging strategy for light-controlled gene editing with a wide range of gRNA sequences. In principle, other types of caging groups can be installed on gRNAs, as it has been previously demonstrated that up to four different photocaging groups could be independently removed from oligonucleotides using light of longer wavelengths with better tissue penetration,38 enabling sequential gene editing/regulation at desired locations and times.
Figure 2.
Light-induced CRISPR-Cas-mediated gene editing in HEK293FT cells. (a) Representative gel images of mutation detection assay of target genes (IL1R2, HPRT, and ZsGreen) in nontreated cells (NT) or in cells transfected with noncaged (NC) or photocaged (PC) gRNAs followed by 365 nm light illumination for 5 min (1.0 J/cm2). (b) Quantification of target gene mutation rates from gel images using Fiji ImageJ software. Results are expressed as individual data points overlaid with the mean ± SD (N = 3–4); ***p < 0.001, ns–no significant difference (p > 0.05) between indicated groups according to one-way ANOVA analysis combined with Tukey’s (Holm-Sidak) posthoc test. (c) Light-induced ZsGreen protein knockout in HEK-Cas9-ZsGreen cells. Merged fluorescence images of ZsGreen signal (green) and Hoechst-stained nuclei (magenta) for cells transfected with noncaged or photocaged gZsGreen in the absence or presence of 1.0 J/cm2 of UV light illumination; images were taken 5 days postirradiation. Scale bar 100 μm. (d) ZsGreen mean fluorescence intensity quantification using flow cytometry analysis 5 days postirradiation. Results are expressed as individual data points overlaid with the mean ± SD (N = 3). (e) Representative gel image of temporally controlled HPRT gene editing using PC-gRNA. HEK-Cas9 cells were transfected for 4 h with PC-gHPRT and were either kept in the dark or exposed to 1.0 J/cm2 of UV light at the indicated time points after starting the transfection. (f) Quantification of HPRT mutation rates from gel images using Fiji ImageJ software. Results are expressed as individual data points overlaid with the mean ± SD (N = 4).
After confirming the possibility of light-controlled gene editing using photocaged gRNAs, we sought to define the time frame of possible activation of the gene editing, which would be determined by intracellular stability of the photocaged gRNA and its dilution upon cell division. In order to answer this question, HEK-Cas9 cells were transfected with the noncaged or photocaged gRNA targeting of the HPRT gene and subsequently light-activated after specific time points (4–144 h after starting the transfection; Figure 2e,f and SI Figure 4). As expected, the gene-editing activity of the noncaged gRNA remained unchanged with time because of the permanent character of the mutations and equal division rate of mutated and unmutated cells (SI Figure 4). Interestingly, the gene-editing efficacy of photocaged gRNA was not drastically reduced even when it was activated 48 h after the transfection, which indicates its high intracellular stability. Mutations were still detectable even when the gRNA was activated 4 days after the transfection. Moreover, the prolonged incubation of photocaged gRNA inside the cells did not induce significant gene editing in the absence of light irradiation. Similarly to the on-target activity, photocaged gRNA cleaved predicted off-target sequences only after UV irradiation (SI Figure 5). In cells, we could not detect gene editing on the predicted off-target sites using the mismatch-based assay, indicating low off-target activity of the chosen gRNA. Overall, these experiments illustrate one of the advantages of our photocaged gRNA approach for light-controlled gene editing over existing systems that suffer from the stronger leakage of the gene-editing activity in the absence of light irradiation.21,22,25,27−30
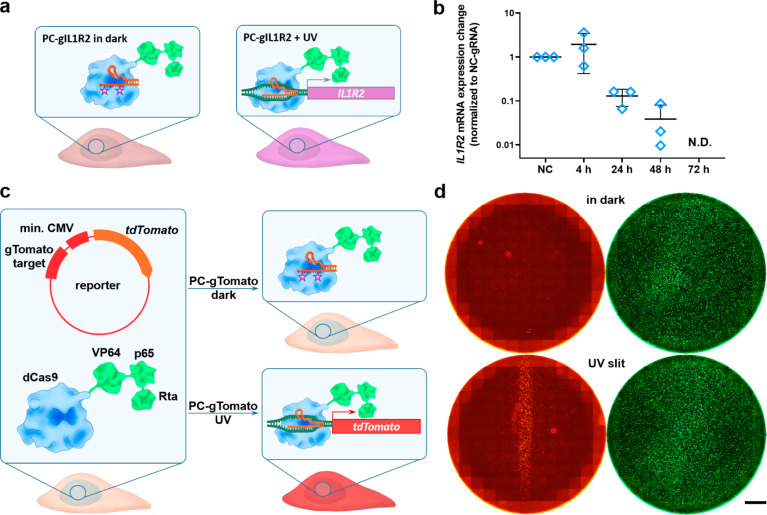
The photocaged gRNAs could be used not only for gene editing but also to achieve spatiotemporal control over gene transcription activation (Figure 3). To illustrate this, we used the same gIL1R2 sequence that was used in the gene-editing experiments (targeting a region upstream of the IL1R2 gene) in transcription activation experiments (Figures 2a and 3a,b). HEK293FT cells were engineered to constitutively express nuclease-deficient Cas9 fused to previously described V64-p65-Rta tripartite transcription activator (dCas9-VPR) acting as a gRNA-controlled transcription factor.47 Single transfection of HEK-dCas9-VPR cells with gIL1R2 resulted in a prolonged IL1R2 mRNA expression (up to 3 days), with the maximum expression at 24 h post-transfection (SI Figure 6). In contrast, the IL1R2 mRNA was not detected after 40 cycles of qPCR in nontransfected cells or in cells transfected with the negative control gRNA. This indicates a sequence-specific mechanism of IL1R2 upregulation through CRISPR-Cas activation and confirms the extremely low levels of endogenous IL1R2 expression in HEK293FT cells. After verifying the sequence-specificity of transcription activation, we next explored the possibility of light-controlled transcriptional activation using photocaged gRNAs. HEK-dCas9-VPR cells were transfected with the noncaged or photocaged gIL1R2, light-activated after specific time points (4–72 h after starting the transfection), and the IL1R2 mRNA expression levels were analyzed 1 day after the irradiation (Figure 3a,b). The IL1R2 mRNA was detected in cells activated up to 48 h post-transfection and remained below the limit of detection for all the samples kept in the dark. Cells treated with the PC-gIL1R2 and exposed to the UV light 72 h post-transfection had CT values between 35 and 40, which is typically considered to be below the reliable detection threshold. Interestingly, in contrast to the gene-editing experiments (Figure 2c), the efficacy of transcriptional activation by photocaged gRNA was significantly reduced already at 24 h post-transfection, which indicates a higher dependency of transcription activation on the intracellular gRNA concentration (Figure 3b).
Figure 3.
Spatiotemporal control of gene expression using PC-gRNA. (a) Schematic representation of the experimental setup for the IL1R2 transcription activation. (b) Light-mediated expression of silent IL1R2 gene in HEK293FT cells. HEK-dCas9-VPR cells were transfected with the NC- or PC-gIL1R2 and were either kept in the dark or exposed to the UV light at 4–72 h post-transfection. The IL1R2 mRNA expression levels were determined 24 h postirradiation and were normalized to the NC-gIL1R2 treated samples (NC). The GAPDH mRNA expression level served as a reference. N.D.–not detected. (N = 3, mean ± SD). (c) Schematic representation of the experimental setup for visualization of transcription activation. HEK-dCas9-VPR cells were cotransfected with PC-gTomato and a reporter plasmid encoding tdTomato under minimal CMV promoter and containing the gRNA binding site. The uncaging of the PC-gTomato with the UV light leads to its binding to the complementary sequence in the reporter plasmid and enhances the tdTomato expression. (d) HEK-dCas9-VPR cells cotransfected with PC-gTomato, the tdTomato reporter plasmid, and a GFP plasmid as a transfection control, irradiated through a photomask. Transfection in a 24-well plate, tdTomato (red), GFP (green). Scale bar 2 mm.
One of the key advantages of the light-controlled gene expression over other stimuli-responsive systems (e.g., based on small-molecule regulators) is the possibility of spatial resolution. To illustrate this point, we have used a previously described reporter system based on the expression of tdTomato fluorescent protein under the control of the minimal CMV promoter.48 Upstream of this cassette a gRNA-binding site based on an AAVS1 sequence was installed. By sequence-specific binding of the gRNA to the AAVS1 sequence the dCas9-VPR fused transcription factor will be brought closer to the transcription start site (TSS) of the tdTomato open reading frame (ORF), thereby enhancing its transcription and protein synthesis (Figure 3c). Cells were transfected with a mixture of PC-gTomato, tdTomato reporter plasmid, and a plasmid encoding GFP as a transfection control (Figure 3c,d). After a 4-h transfection, the cells were UV-irradiated through a photomask to expose only a 1.5 mm strip of cells (1.0 J/cm2). One day after the illumination, cells were fixed and imaged using a fluorescence microscope. We observed a confined tdTomato expression pattern, corresponding to the irradiation area. The GFP signal was evenly distributed throughout the well, indicating that the cells were transfected uniformly, and that the tdTomato fluorescence was indeed triggered by light-irradiation. The cells kept in the dark showed virtually no tdTomato expression. We went even further and used a 375 nm programmable laser setup to draw triangular shapes on top of the transfected cells, and similarly, we observed enhanced tdTomato expression in the irradiated area (SI Figure 7). The cells uniformly expressed GFP, confirming the successful transfection in the whole well.
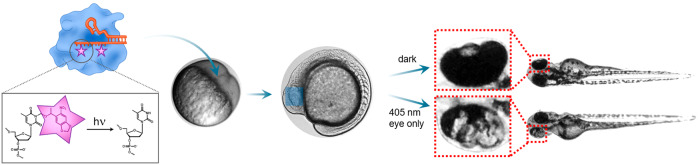
Given the tight control over the gRNA function and rapid uncaging process, we envisioned that our photocaged gRNA approach could be particularly useful for developmental biology research. Indeed, UV-responsive photocaged molecules are widely used in this field;49−51 however, the resulting gene regulation is reversible, and therefore, light-activatable systems based on CRISPR-Cas would be an important addition to the gene-editing toolbox. To this end, we tested a photocaged gRNA targeting the slc45a2 gene (responsible for the pH homeostasis of melanosomes required for melanin production52) in developing zebrafish embryos as a model system.53 Loss-of-function mutations in the slc45a2 gene led to albino-like phenotype in the retinal pigment epithelium and in the body of the developing embryo, which can be observed already starting from 24 h post fertilization (hpf).52 We used Cas9 protein for the experiments because it has previously demonstrated better gene-editing capabilities compared with Cas9 in mRNA format.54 This was further confirmed by our preliminary test of protein Cas9 versus mRNA Cas9 in zebrafish embryos (SI Figure 8).
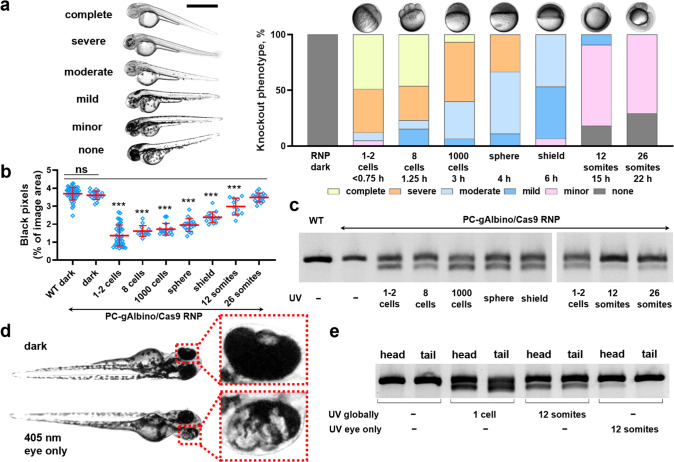
First, we verified that the UV light at a dose of 2.0 J/cm2 did not decrease the embryo viability or affect its development (SI Figure 9), in accordance with previously reported studies.55 For temporally resolved gene editing, zebrafish embryos were microinjected with Cas9/PC-gRNA complex at 1–2 cell stage and subjected to the UV light irradiation at various developmental stages.56 The embryos were imaged 2 days after the irradiation and assessed for slc45a2 gene mutation using a mismatch-based assay (Figure 4). As expected, the PC-gAlbino/Cas9-microinjected embryos exhibited no detectable slc45a2 gene mutation in the absence of light irradiation neither phenotypically nor at the genomic DNA level. In contrast, UV light exposure of the PC-gAlbino/Cas9-microinjected embryos led to the slc45a2 gene mutations and the albino-like phenotype, while the efficacy of the gene knockout decreased with delayed irradiation (Figure 4). Automated image analysis could detect the statistically significant difference in total pigment content in embryos irradiated up until 12 somite stage compared to wild-type embryos (Figure 4b). We could also detect the gene editing phenotypically by the decrease in pigment content in embryos irradiated at up to 26-somite stage using scoring-based system (Figure 4a), which was also confirmed by the mutation detection PCR-based assay (Figure 4c). This indicates that the PC-gRNA/Cas9 complex exhibited residual activity inside the zebrafish embryos even at the 26-somite stage after single injection (22 hpf).
Figure 4.
Spatiotemporal control of gene editing in zebrafish embryos. (a) Temporally resolved light-mediated gene editing in zebrafish embryos microinjected with PC-gAlbino and Cas9 protein and globally exposed to the 365 nm light at indicated developmental stages. The total number of embryos exhibiting each knockout phenotype category was counted for each treatment group (N = 11–41). The representative images of each knockout phenotype are shown on the left side. Dark–no UV irradiation, only ambient light. Scale bar 1 mm. (b) Automated image analysis of impaired pigment formation in slc45a2 mutated embryos using Fiji ImageJ software. Results are expressed as individual data points overlaid with the mean ± SD (N = 11–49); ***p < 0.001, ns–not significant (p > 0.05) versus wild-type embryos according to one-way ANOVA analysis combined with Tukey’s (Holm-Sidak) posthoc test. (c) Mismatch-based mutation detection PCR assay of slc45a2 gene in embryos microinjected with Cas9 protein and PC-gAlbino and globally irradiated with 365 nm UV light at various developmental stages. (N = 5 embryos per sample). (d) Representative photographs of zebrafish embryos microinjected with PC-gAlbino and Cas9 protein and either exposed to 405 nm laser light at 12-somite stage locally in the eye area (bottom image) or kept in the ambient light (dark) as a negative control (upper image). (e) Mismatch-based mutation detection assay of slc45a2 gene in embryos microinjected with Cas9 protein and PC-gAlbino, and locally irradiated in the eye area using 405 nm laser. The assay was run on the head and tail parts separately for each individual fish, and 3–5 embryos were combined for each treatment group.
We next explored the possibility of spatially restricted gene editing in zebrafish embryos using photocaged gRNAs. The embryos were microinjected at 1–2 cell stage and were left in the dark until they reached the 12-somite stage, at which the head and the tail of the embryo are clearly visible. Subsequently, one eye of the embryo was irradiated using a 405 nm laser in a confocal microscope setup. Two days after the treatment, we observed mosaic pigment expression in the eye of the locally irradiated embryos (Figure 4d). We analyzed mutations in the slc45a2 gene in the head and tail parts of the same embryos separately and could detect the mutations only in the head (Figure 4e). Globally irradiated embryos showed uniform mutations in both head and tail, and embryos kept in the dark (i.e., without UV irradiation) did not exhibit mutations detectable in our assay, confirming the possibility for localized gene editing using photocaged gRNAs. To the best of our knowledge, this is the first demonstration of light-controlled CRISPR-Cas-based system in developing embryos resulting in spatiotemporally resolved gene editing.
In conclusion, we have developed a photoswitchable CRISPR-Cas gene editing system based on modification of the gRNAs with photocaging groups. This approach relies on a straightforward solid-phase synthesis of the photocaged gRNAs, with simpler purification and characterization processes in comparison to engineering a light-responsive protein. We have achieved gene editing as well as transcription activation in cells upon brief exposure to the UV light, whereas photocaged gRNAs showed virtually no detectable activity in the absence of light irradiation. Finally, we have applied this system to spatiotemporally control gene editing in vivo, enabling the use of this strategy for developmental biology and tissue engineering applications.49−51
Acknowledgments
Authors are grateful to Ms. Melisse Chee and Ms. Sirli Treumuth for plasmid preparations, to Mr. Stephen M. Rothery for his help with the automated image analysis, and to Dr. Akemi Nogiwa-Valdez for her support with data management. We thank the Central Biomedical Services (CBS) facility at Imperial College London for their support in zebrafish maintenance and husbandry. The Facility for Imaging by Light Microscopy (FILM) at Imperial College London is partially funded by the Wellcome Trust (grant 104931/Z/14/Z) and BBSRC (grant BB/L015129/1). Financial support from the Swiss National Science Foundation (E.M.-O., grant P2EZP3_172181), Swedish Research Council (E.M.-O., C.L.G., M.M.S.; grant VR 4-478/2016), Aker Scholarship (H.H.), National Heart and Lung Institute Foundation Scholarship (I.S.), the United Kingdom Regenerative Medicine Platform “Acellular/Smart Materials − 3D Architecture” (J.P.W., M.M.S.; grant MR/R015651/1), British Heart Foundation Centre of Research Excellence (L.B.; grant RE/18/4/34215), and Whitaker International Program (C.L.G.) is highly acknowledged. Raw data are available on request from rdm-enquiries@imperial.ac.uk.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01093.
HPLC analysis of uncaging of PC-crRNA; ZsGreen protein knockout in HEK-Cas9-ZsGreen cells; UV light (365 nm) cytotoxicity assay on HEK-Cas9 cells; time-dependency of HPRT gene editing in HEK-Cas9 cells transfected with NC-gHPRT; biochemical activity assay of off-target DNA cleavage by NC- or PC-gIL1R2 with 365 nm light illumination (0–2 J/cm2); time-dependent expression of the IL1R2 gene in HEK293FT cells; spatial control over gene expression using PC-gRNA; comparison of gene-editing efficacy in zebrafish embryos using RNP and mRNA format for Cas9 delivery; UV light toxicity on zebrafish embryos (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ginn S. L.; Amaya A. K.; Alexander I. E.; Edelstein M.; Abedi M. R. Gene therapy clinical trials worldwide to 2017: an update. J. Gene Med. 2018, 20, e3015 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- Cooper M. L.; et al. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia 2018, 32, 1970–1983. 10.1038/s41375-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophinni Y.; Inoue M.; Kotaki T.; Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci. Rep. 2018, 8, 7784. 10.1038/s41598-018-26190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoasii L.; et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018, 362, 86–91. 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 2017, 127, 2719–2724. 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–23. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P.; et al. RNA-guided human genome engineering via Cas9. Science (Washington, DC, U. S.) 2013, 339, 823–826. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A.; Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science (Washington, DC, U. S.) 2014, 346, 1258096. 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Hsu P. D.; Lander E. S.; Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–78. 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D. E.; et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018, 46, 792–803. 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P.; Parlak M.; Kuscu C.; Bandaria J.; Mir M.; Szlachta K.; Singh R.; Darzacq X.; Yildiz A.; Adli M. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat. Commun. 2017, 8, 1–10. 10.1038/ncomms14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.-C.; et al. Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res. 2019, 47, e13–e13. 10.1093/nar/gky1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K.; et al. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell 2016, 63, 293–305. 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Fukuda A.; et al. Spatiotemporal dynamics of OCT4 protein localization during preimplantation development in mice. Reproduction 2016, 152, 417–430. 10.1530/REP-16-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. M.; Pattanayak V.; Thompson D. B.; Zuris J. A.; Liu D. R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015, 11, 316–318. 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; et al. Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets. Nat. Biomed. Eng. 2019, 3, 126. 10.1038/s41551-018-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J.; Borchardt E. K.; Brown K.; Asokan A.; Deiters A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015, 137, 5642–5645. 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieke C.; Rohrbach F.; Gottschalk A.; Mayer G.; Heckel A. Light-controlled tools. Angew. Chem., Int. Ed. 2012, 51, 8446–8476. 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- Nihongaki Y.; Yamamoto S.; Kawano F.; Suzuki H.; Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015, 22, 169–174. 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Jain P. K.; et al. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem., Int. Ed. 2016, 55, 12440–12444. 10.1002/anie.201606123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; et al. Near-infrared upconversion–activated CRISPR-Cas9 system: A remote-controlled gene editing platform. Sci. Adv. 2019, 5, eaav7199. 10.1126/sciadv.aav7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein L. R.; Gersbach C. A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015, 11, 198–200. 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y.; Kawano F.; Nakajima T.; Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015, 33, 755–760. 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- Gao Y.; et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 2016, 13, 1043–1049. 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y.; et al. CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 2017, 14, 963–966. 10.1038/nmeth.4430. [DOI] [PubMed] [Google Scholar]
- Zhou X. X.; et al. A single-chain photoswitchable CRISPR-Cas9 architecture for light-inducible gene editing and transcription. ACS Chem. Biol. 2018, 13, 443–448. 10.1021/acschembio.7b00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter F.; Fonfara I.; Bouazza B.; Schumacher C. H.; Bratovic M.; Charpentier E.; Moglich A. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res. 2013, 44, 10003–10014. 10.1093/nar/gkw930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J.; et al. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E6722–E6730. 10.1073/pnas.1802448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N. T.; He L.; Martinez-Moczygemba M.; Huang Y.; Zhou Y. Rewiring Calcium Signaling for Precise Transcriptional Reprogramming. ACS Synth. Biol. 2018, 7, 814–821. 10.1021/acssynbio.7b00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.; Deiters A. Conditional Control of CRISPR/Cas9 Function. Angew. Chem., Int. Ed. 2016, 55, 5394–5399. 10.1002/anie.201511441. [DOI] [PubMed] [Google Scholar]
- Toettcher J. E.; Gong D.; Lim W. A.; Weiner O. D. Chapter seventeen – Light Control of Plasma Membrane Recruitment Using the Phy–PIF System. Methods Enzymol. 2011, 497, 409–423. 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–9. 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Kelley M. L.; Strezoska Ž.; He K.; Vermeulen A.; Smith A. v. B. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol. 2016, 233, 74–83. 10.1016/j.jbiotec.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Young D. D.; Edwards W. F.; Lusic H.; Lively M. O.; Deiters A. Light-triggered polymerase chain reaction. Chem. Commun. 2008, 17, 462–464. 10.1039/B715152G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S.; et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 2015, 12, 1051–1054. 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Correia A.; Seyfried P.; Heckel A. Wavelength-selective uncaging of oligonucleotides. Curr. Protoc. nucleic acid Chem. 2014, 57, 6.11.1–6.11.32. 10.1002/0471142700.nc0611s57. [DOI] [PubMed] [Google Scholar]
- Fichte M. A. H.; et al. Three-dimensional control of DNA hybridization by orthogonal two-color two-photon uncaging. Angew. Chem., Int. Ed. 2016, 55, 8948–8952. 10.1002/anie.201603281. [DOI] [PubMed] [Google Scholar]
- Rastogi R. P.; Kumar A.; Tyagi M. B.; Sinha R. P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J Nucleic Acids 2010, 2010, 592980. 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartje Z. J.; Barkau C. L.; Rohilla K. J.; Ageely E. A.; Gagnon K. T. Chimeric Guides Probe and Enhance Cas9 Biochemical Activity. Biochemistry 2018, 57, 3027–3031. 10.1021/acs.biochem.8b00107. [DOI] [PubMed] [Google Scholar]
- Yin H.; et al. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity-supporting info. Nat. Chem. Biol. 2018, 14, 311–316. 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimo N.; Chatterjee P.; Jacobson J. M.. Chimeric CRISPR guides enhance Cas9 target specificity. bioRxiv, 2017. 10.1101/147686. [DOI] [Google Scholar]
- Schindelin J.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–82. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards B.; et al. Stable expression ofAnthozoa fluorescent proteins in mammalian cells. Cytometry 2002, 48, 106–112. 10.1002/cyto.10117. [DOI] [PubMed] [Google Scholar]
- Godley B. F.; et al. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J. Biol. Chem. 2005, 280, 21061–21066. 10.1074/jbc.M502194200. [DOI] [PubMed] [Google Scholar]
- Chavez A.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P.; et al. Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A.; et al. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and xenopus embryos. J. Am. Chem. Soc. 2010, 132, 15644–15650. 10.1021/ja1053863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. T.; et al. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater. 2015, 14, 352–360. 10.1038/nmat4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambridge S. B.; et al. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat. Methods 2009, 6, 527–531. 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- Dooley C. M.; et al. Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigm. Cell Melanoma Res. 2013, 26, 205–217. 10.1111/pcmr.12053. [DOI] [PubMed] [Google Scholar]
- Vejnar C. E.; Moreno-Mateos M. A.; Cifuentes D.; Bazzini A. A.; Giraldez A. J. Optimized CRISPR-Cas9 system for genome editing in zebrafish. Cold Spring Harb. Protoc. 2016, 2016, 856–870. 10.1101/pdb.prot086850. [DOI] [PubMed] [Google Scholar]
- Kotani H.; Taimatsu K.; Ohga R.; Ota S.; Kawahara A. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS One 2015, 10, e0128319 10.1371/journal.pone.0128319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q.; Svoboda K.; Tiersch T. R.; Monroe W. T. Photobiological effects of UVA and UVB light in zebrafish embryos: evidence for a competent photorepair system. J. Photochem. Photobiol., B 2007, 88, 137–146. 10.1016/j.jphotobiol.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B.; Ballard W. W.; Kimmel S. R.; Ullmann B.; Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.