Abstract
Background
Several studies have reported the association between polymorphisms in Matrix metalloproteinases (MMPs) gene family and risk of Multiple sclerosis (MS). However, the results have been inconsistent and inconclusive. To resolve this issue, here we performed a systematic review and meta-analysis of the MMP-91562 C/T (rs3918242), MMP-3 (− 1612 5A/6A), and MMP-2 (− 1306 C/T) polymorphisms and susceptibility to MS.
Methods
We conducted a comprehensive systematic search in the major electronic database, including Scopus and PubMed to look up for relevant studies published before December 2019 that surveyed the association between the MMP-91562 C/T (rs3918242), MMP-3 (− 1612 5A/6A), and MMP-2 (− 1306 C/T) polymorphisms and susceptibility to MS. The level of association between the polymorphisms and susceptibility to MS in the polled analysis was determined by calculating the odds ratio (OR) and the corresponding 95% confidence interval (CI).
Results
We found 15 studies containing 2430 MS subjects and 2304 controls. A statistically significant association was observed in the all five comparisons of the MMP-91562 C/T polymorphism and MS risk as follows: dominant model (OR = 1.62, 95% CI = 1.03–2.53, P = 0.03), recessive model (OR = 2.69, 95% CI = 1.68–4.29, P < 0.001), allelic model (OR = 1.51, 95% CI = 1–2.28, P = 0.04), TT vs. CC model (OR = 3.20, 95% CI = 1.87–5.46, P < 0.001), and CT vs. CC model (OR = 1.53, 95% CI = 1.02–2.28, P = 0.04).
Conclusions
Our meta-analysis revealed significant association of MMP-9 (− 1562 C/T) Single-nucleotide polymorphism (SNP) with MS susceptibility that increased the disease risk.
Keywords: Multiple sclerosis, Central nervous system, Matrix metalloproteinases, Genetic polymorphism, Meta-analysis
Background
Multiple Sclerosis (MS) is a chronic autoimmune disease of the brain and spinal cord of the central nervous system (CNS) that is characterized by demyelination, inflammation and axonal degeneration, resulting in serious disability in young adults [1, 2]. It is estimated that above 2.5 million people in the world suffer from MS [3]. Although the main etiology of the disease remains obscure, it is thought that both genetic and environmental factors and their interactions are critical in disease development [4–6]. Especially, the role of genetic factors in the pathogenesis of MS has been established by family and twin studies. Accordingly, the heritability of MS is estimated to be about 25–80% [7–9]. As noted in recent studies, several genes, including interleukin (IL) 6, IL-12, vitamin D receptor (VDR), Signal transducer and activator of transcription (STAT) 4, Protein tyrosine phosphatase, non-receptor type 22 (PTPN22), CD40, programmed cell death (PD1/PD-L1), and Matrix metalloproteinases (MMPs) have been correlated with MS and attracted much attention to investigating more genetic factors contributing to MS risk [10–17].
MMPs are zinc-dependent endopeptidases enzymes that play an important role in many physiological and pathological processes including inflammation, invasiveness of tumor, metastasis, and angiogenesis by the degradation of the extracellular matrix and basement membrane (BM) [18, 19]. On the basis of structure and in terms of substrate specificity, MMPs are divided into six groups: collagenases (MMP-1, − 8, − 13, − 18), gelatinases (MMP-2, − 9), stromelysins (MMP-3, − 10, − 11), matrilysins (MMP-7, − 26), membrane-type MMPs (MMP-14, − 15, − 16, − 17,-24, − 25) and other non-classified MMPs (MMP-12, − 19, − 20, − 21, − 22, − 23, − 27, − 28) that is encoded by a separate gene and has a different tissue distribution and bioactive function [20–22]. MMPs have a critical role in the pathogenesis of MS by inducing migration of immune cells through the blood-brain barrier (BBB) into the CNS, which seems essential during the formation of inflammatory lesions [23].
The regulation of MMP family genes expression has not been understood, but it has been demonstrated that genomic sequence of MMP family genes is polymorphic; it is of added attention to ascertain which polymorphisms in MMP family genes have functional potentials to influence the final bioavailability of family member(s) and therefore the progression of MS [14, 24–26]. Several studies have evaluated the association between MMP family gene polymorphisms and MS risk; but the results are often inconsistent [27–29]. Therefore, we performed a meta-analysis to attain a consistent conclusion of the association between the MMP-91562 C/T (rs3918242), MMP-3 (− 1612 5A/6A), and MMP-2 (− 1306 C/T) gene polymorphisms and susceptibility to MS.
Methods
This meta-analysis was performed by sticking to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [30]. The current meta-analysis does not contain any studies with human participants or animals performed by any of the authors.
Search strategy
The initial comprehensive and systematic search was conducted in Medline, Scopus, and PubMed databases. To be assured of our search, combination of following key words and Medical Subject Headings (Mesh) terms were used: (“matrix metalloproteinase” [Mesh] OR “MMP” OR “gelatinase”) AND (“multiple sclerosis” OR “MS”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). We retrieved all studies published prior to January 2020. The references of reviews and eligible studies were cross-checked to prevent missing of any eligible study which was not identified by primary search.
Inclusion and exclusion criteria
Studies considered eligible if met the following criteria: 1) Publications evaluating the association between MMP family gene polymorphism and susceptibility to MS as main outcome; 2) Publications with case and control groups (Case-control design and cohort design); 3) Publications which report odds ratio (OR) and 95% confidence interval (CI) or crud data to calculate these items; 4) Publications with sufficient data such as genotype distribution and allele frequency. Publications like reviews, meta-analysis, case reports, book chapters, letter to editors, conference abstracts, as well as duplicates were all excluded.
Data extraction and quality assessment
We identified eligible studies by sticking to the inclusion and exclusion criteria, and to perform meta-analysis following data were extracted: the first author’s name, journal and year of publication, country of origin, ethnicity, number of subjects in the case and the control groups for each gender, mean or range of age, genotyping method, genotype counts in the case and control groups. It is noteworthy that all procedure of data extraction was performed by two authors independently and possible discrepancy was solved by consensus. Furthermore, the quality of each study was assessed using the Newcastle-Ottawa Scale (NOS) criteria [31]. Studies with scores 0–3, 4–6 or 7–9 were of low, moderate, or high-quality, respectively.
Statistical analysis
We used Pearson’s chi-square test in control groups to estimate Hardy–Weinberg equilibrium(HWE) for each study. In this study OR with 95% CI was used to assess the strength of the association between MMP family gene polymorphism and MS risk. The genotype model which defined for MMP-2, MMP-3, and MMP-9 were as follow: MMP-2 (Dominant model [TT + CT vs. CC], Recessive model [TT vs. CT + CC], Allelic model [T vs. C], Homozygote contrast [TT vs. CC], and Heterozygote contrast [CT vs. CC]); MMP-3 (Dominant model [6A6A + 5A6A vs. 5A5A], Recessive model [6A6A vs. 5A6A + 5A5A], Allelic model [6A vs. 5A], Homozygote contrast [6A6A vs. 5A5A], and Heterozygote contrast [5A6A vs. 5A5A]); MMP-9 (Dominant model [TT + CT vs. CC], Recessive model [TT vs. CT + CC], Allelic model [T vs. C], Homozygote contrast [TT vs. CC], and Heterozygote contrast [CT vs. CC]). Possible heterogeneity in this study was estimated by Q-test and I2 test [32, 33]. Accordingly, a P value< 0.10 of Q-test and I2 < 50% demonstrate no evidence of heterogeneity and fixed effect model (FEM) was used [34]. But, if P value> 0.10 for Q-test and I2 > 50%, then the study was considered heterogeneous and random effect model (REM) was applied [35]. Furthermore, publication bias was measured by Egger’s regression test, Begg’s adjusted rank correlation test, and visual examination of the funnel plot (P value< 0.05 was considered statistically significant) [36]. Finally, we performed sensitivity analysis to observe the impact of any individual study on the pooled OR. Statistically analyses were carried out using STATA (version 14.0; Stata Corporation, College Station, TX) and SPSS (version 23.0; SPSS, Inc. Chicago, IL).
Results
Study selection
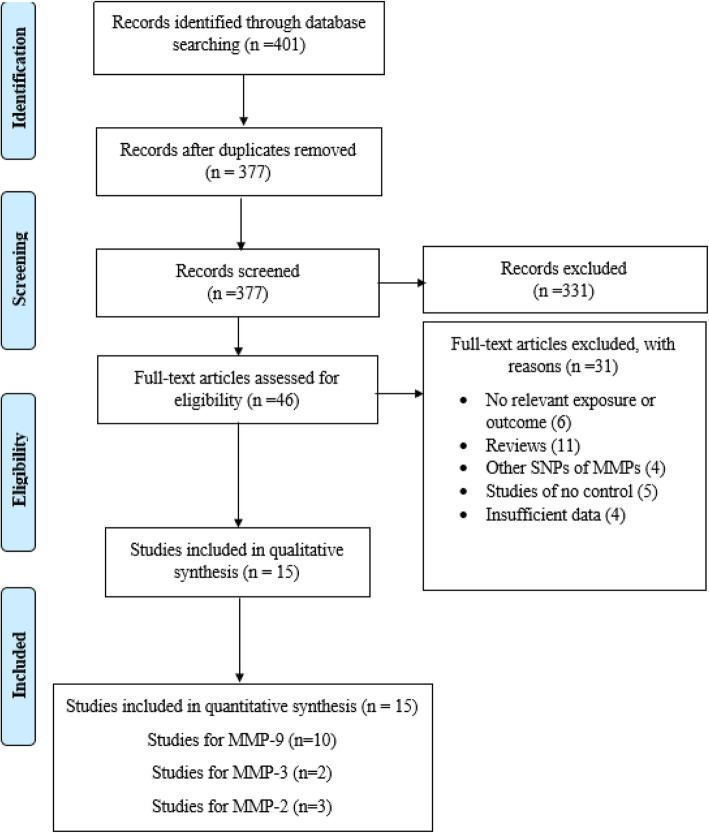
Based on aforementioned key words, primary search generated 401 studies that 24 studies were duplicates. The other 377 studies were screened according to the inclusion and exclusion criteria. Taken to gather, 331 studies were excluded by title and abstract screening and 31 studies were excluded after full text examination. Ultimately, 15 studies matched with the inclusion criteria and selected for quantitative analysis. Tables 1 and 2 summarize the characteristics and genotype frequency of the included studies. The mean age of case and control groups were between 30 and 40. All studies had good methodological score ranging from 5 to 8 and were published between 2000 to 2019. Among included studies, the Restriction fragment length polymorphism (RFLP)-PCR was used as a common genotyping model. The screening workflow and study selection process are shown in Fig. 1.
Table 1.
Characteristics of studies included in meta-analysis
| Study author | Year | Country | Ethnicity | Total cases/controls | Sex cases/controls | Genotyping method | Mean age Cases/Controls |
Quality score |
|---|---|---|---|---|---|---|---|---|
| MMP9 | ||||||||
| Nelissen et al. | 2000 | Sweden | European | 199/146 |
M = NR F=NR |
PCR-RFLP | NR/NR | 6 |
| Zivkovic et al. | 2007 | Serbia | European | 187/282 |
M = 67/140 F = 120/142 |
Touch Down PCR | 35.5 ± 10.1/40.8 ± 14.8 | 7 |
| Benesova et al. | 2008 | Czech Republic | European | 244/132 |
M = 63/45 F = 181/87 |
PCR-SSP | 38.4 ± 10.2/35.6 ± 11.7 | 7 |
| Mirowska-Guzel et al. | 2009 | Poland | European | 234/190 |
M = 66/76 F = 168/114 |
PCR-RFLP | 40.09 ± 10.19/40.09 ± 10.19 | 7 |
| Fernandes et al. | 2009 | Brazil | South-American | 158/191 |
M = 41/54 F = 117/137 |
PCR-RFLP | 38.7 ± 13/35.6 ± 9.5 | 6 |
| La Russa et al. | 2010 | Italy | European | 243/173 |
M = 96/107 F = 147/66 |
PCR-RFLP | 41.1 ± 12.2/28.5 ± 9.4 | 7 |
| Valado et al. | 2017 | Portugal | European | 169/183 |
M = 48/63 F = 121/120 |
PCR-RFLP | 41.44 ± 0.84/39.09 ± 0.96 | 6 |
| Ibrahim et al. | 2019 | Egypt | African | 50/100 |
M = 18/NR F = 32/NR |
PCR-RFLP | 32.9 ± 8.1/NR | 5 |
| Sabbagh et al. | 2019 | Iran | Asian | 100/105 |
M = 37/41 F = 63/64 |
PCR-ARMS | 42.89 ± 10.48/46.52 ± 8.90 | 5 |
| Sadr et al. | 2019 | Iran | Asian | 170/200 |
M = 121/142 F = 49/58 |
PCR-RFLP | 33.34 ± 7.91/31.88 ± 9.79 | 7 |
| MMP3 | ||||||||
| Djuric et al. | 2008 | Serbia | European | 184/236 |
M = NR F=NR |
Touch Down PCR | NR/NR | 7 |
| Rahimi et al. | 2016 | Iran | Asian | 121/106 |
M = 24/17 F = 97/89 |
PCR-RFLP | 35.3 ± 9.1/34.5 ± 11.4 | 6 |
| MMP2 | ||||||||
| Benesova et al. | 2008 | Czech Republic | European | 240/132 |
M = 60/45 F = 180/87 |
PCR-SSP | 38.4 ± 10.2/35.6 ± 11.7 | 7 |
| Aksoy et al. | 2016 | Turkey | European | 102/102 |
M = 76/75 F = 26/27 |
Taq man | 36.69 ± 8.33/35.93 ± 8.20 | 6 |
| Liutkeviciene et al. | 2018 | Lithuania | European | 26/26 |
M = NR F=NR |
Taq man | 36/34 | 5 |
Table 2.
Distribution of genotype and allele frequencies among MS patients and controls
| Study author | MS cases | Healthy control | P-HWE | MAF | ||||||||
| CC | CT | TT | C | T | CC | CT | TT | C | T | |||
| MMP9 | ||||||||||||
| Nelissen et al. | 143 | 51 | 5 | 337 | 61 | 102 | 40 | 4 | 244 | 48 | 0/973 | 0/164 |
| Zivkovic et al. | 146 | 41 | 0 | 333 | 41 | 200 | 74 | 8 | 474 | 90 | 0/716 | 0/16 |
| Benesova et al. | 191 | 50 | 3 | 432 | 56 | 87 | 42 | 3 | 216 | 48 | 0/424 | 0/182 |
| Mirowska-Guzel et al. | 128 | 103 | 3 | 359 | 109 | 136 | 50 | 4 | 322 | 58 | 0/811 | 0/153 |
| Fernandes et al. | 117 | 35 | 6 | 269 | 47 | 156 | 32 | 3 | 344 | 38 | 0/369 | 0/099 |
| La Russa et al. | 164 | 73 | 6 | 401 | 85 | 147 | 25 | 1 | 319 | 27 | 0/954 | 0/078 |
| Valado et al. | 130 | 35 | 4 | 295 | 43 | 145 | 34 | 4 | 324 | 42 | 0/247 | 0/115 |
| Ibrahim et al. | 28 | 21 | 1 | 77 | 23 | 78 | 21 | 1 | 177 | 23 | 0/751 | 0/115 |
| Sabbagh et al. | 11 | 35 | 54 | 57 | 143 | 42 | 42 | 21 | 126 | 84 | 0/087 | 0/4 |
| Sadr et al. | 96 | 60 | 14 | 252 | 88 | 163 | 33 | 4 | 359 | 41 | 0/144 | 0/103 |
| MMP2 | ||||||||||||
| Benesova et al. | 143 | 84 | 13 | 370 | 110 | 75 | 40 | 17 | 190 | 74 | 0.004 | 0.28 |
| Aksoy et al. | 40 | 56 | 6 | 136 | 68 | 77 | 25 | 0 | 179 | 25 | 0.15 | 0.123 |
| Liutkeviciene et al. | 19 | 7 | 0 | 45 | 7 | 190 | 108 | 20 | 488 | 148 | 0.38 | 0.233 |
| Study author | MS cases | Healthy control | P-HWE | MAF | ||||||||
| 5A/5A | 5A/6A | 6A/6A | 5A | 6A | 5A/5A | 5A/6A | 6A/6A | 5A | 6A | |||
| MMP3 | ||||||||||||
| Djuric et al. | 24 | 102 | 58 | 150 | 218 | 37 | 130 | 69 | 204 | 268 | 0.06 | 0.568 |
| Rahimi et al. | 3 | 66 | 52 | 72 | 170 | 1 | 67 | 38 | 69 | 143 | 0.006 | 0.675 |
P-HWE, p-value for Hardy–Weinberg equilibrium; MAF Minor allele frequency of control group
Fig. 1.
Flow diagram of study selection process
Meta-analysis of MMP-9 (− 1562 C/T) and risk of MS
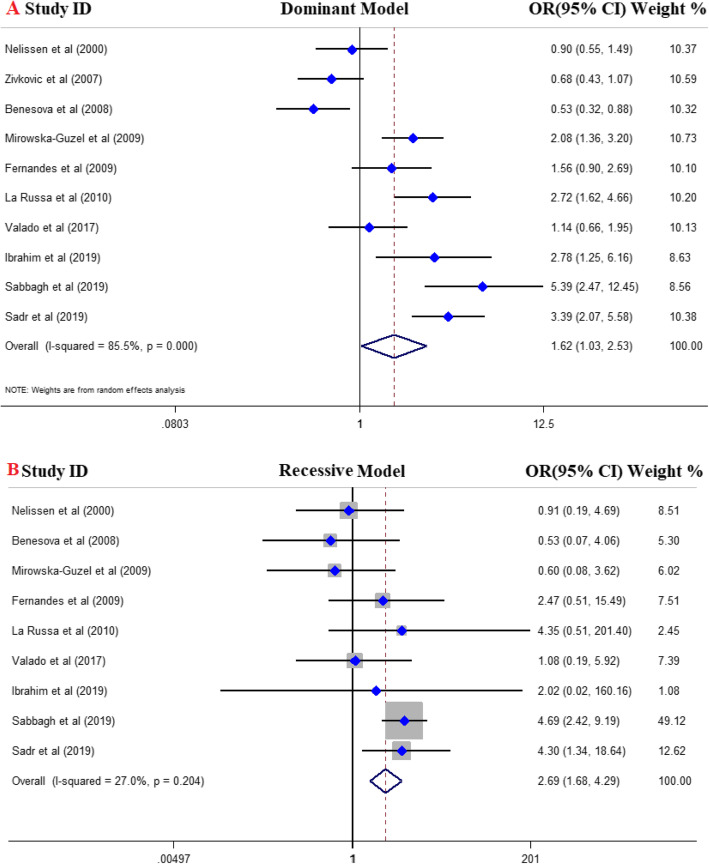
A total of 10 eligible studies with 1754 cases and 1702 controls were included in quantitative analysis. Of them, six studies were conducted in European countries [27, 29, 37–40], two studies in Asian countries [41, 42], one study was in Egypt [43] and one in Brazil [44]. The pooled OR divulged a strong positive association between MMP-9 gene rs34016235 polymorphism and risk of MS and announced this SNP as a risk factor for MS (Fig. 2). In details, dominant model (OR = 1.62, 95% CI = 1.03–2.53, P = 0.03), recessive model (OR = 2.69, 95% CI = 1.68–4.29, P < 0.001), allelic model (OR = 1.51, 95% CI = 1–2.28, P = 0.04), TT vs. CC model (OR = 3.20, 95% CI = 1.87–5.46, P < 0.001), and CT vs. CC model (OR = 1.53, 95% CI = 1.02–2.28, P = 0.04). FEM was used for recessive and homozygote compressions and REM was applied for dominant, heterozygote, and allelic models. The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Fig. 2.
Forest plot of the association between MMP-9 gene polymorphism and MS risk: A; dominant model, B; recessive model
Table 3.
Main results of pooled ORs in meta-analysis of MMP family gene polymorphism
| Subgroup | Sample size | Test of association | Test of heterogeneity | Test of publication bias (Begg’s test) | Test of publication bias (Egger’s test) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic model | Case/Control | OR | 95%CI(P-value) | I2 (%) | P | Z | P | T | P | |
| MMP9 | ||||||||||
| Overall | Dominant | 1754 / 1702 | 1.62 | 1.03– 2.53 (0.03) | 85.5 | ≤0.001 | 0.80 | 0.42 | 1.27 | 0.24 |
| Recessive | 1754 / 1702 | 2.69 | 1.68 – 4.29 (≤0.001) | 27 | 0.20 | -1.46 | 0.14 | -2.17 | 0.06 | |
| Allelic | 1754 / 1702 | 1.51 | 1- 2.28 (0.04) | 87.8 | ≤0.001 | 0.27 | 0.78 | 0.34 | 0.74 | |
| TT vs. CC | 1754 / 1702 | 3.20 | 1.87 – 5.46 (≤0.001) | 50 | 0.04 | -1.04 | 0.29 | -1.90 | 0.9 | |
| CT vs.CC | 1754 / 1702 | 1.53 | 1.02 – 2.28 (0.04) | 80.8 | ≤0.001 | 0.98 | 0.32 | 1 | 0.34 | |
| MMP3 | ||||||||||
| Dominant | 305/342 | 1.18 | 0.66-2.13 (0.57) | 0 | 0.57 | -1 | 0.31 | * | * | |
| Recessive | 305/342 | 0.57 | 0.18-1.79 (0.33) | 88.5 | ≤0.001 | 1 | 0.31 | * | * | |
| Allelic | 305/342 | 1.11 | 0.88-1.41 (0.39) | 0 | 0.91 | 1 | 0.31 | * | * | |
| 6A6A vs. 5A5A | 305/342 | 1.15 | 0.62- 2.11 (0.43) | 0 | 0.48 | -1 | 0.31 | * | * | |
| 5A6A vs.5A5A | 305/342 | 1.24 | 0.64 – 2.39 (0.52) | 0 | 0.54 | 1 | 0.31 | * | * | |
| MMP2 | ||||||||||
| Overall | Dominant | 368 /552 | 1.36 | 0.39 – 4.78 (0.07) | 90.4 | ≤0.001 | 0.52 | 0.60 | 0.04 | 0.97 |
| Allelic | 368 /552 | 1.15 | 0.36 – 3.61 (0.81) | 91.9 | ≤0.001 | 0.52 | 0.60 | 0.14 | 0.91 | |
| CT vs.CC | 368 /552 | 1.50 | 0.52 – 4.35 (0.45) | 85.9 | ≤0.001 | 0.52 | 0.60 | -0.10 | 0.93 | |
* Egger’s test was not calculable
Meta-analysis of MMP-3 (− 1612 5A/6A) and risk of MS
Two studies with 305 cases and 342 controls were included. One study was conducted in Serbia [45] and the other in Iran [46]. The results of overall population reject any association between MMP-3 gene − 1612 5A/6A SNP and risk of MS across all genotype models including; dominant model (OR = 1.18, 95% CI = 0.66–2.13, P = 0.57), recessive model (OR = 0.57, 95% CI = 0.18–1.79, P = 0.33), allelic model (OR = 1.11, 95% CI = 0.88–1.41, P = 0.39), 6A6A vs. 5A5A model (OR = 1.15, 95% CI = 0.62–2.11, P = 0.66), and 5A6A vs. 5A5A model (OR = 1.24, 95% CI = 0.64–2.39, P = 0.52). The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Meta-analysis of MMP-2 (− 1306 C/T) and risk of MS
Overall, three studies with 368 cases and 552 controls were eligible for the association between MMP-2 gene rs243865 SNP and susceptibility to MS. All three studies were conducted in Europe [38, 47, 48]. Because of TT genotype frequency of zero in both cases and controls, the recessive model and TT vs. CC model were not applicable to calculate. The results of other three models also reject association between MMP-2 gene rs243865 SNP and MS risk. The results were; dominant model (OR = 1.36, 95% CI = 0.39–4.78, P = 0.07), allelic model (OR = 1.15, 95% CI = 0.36–3.61, P = 0.81), and CT vs. CC model (OR = 1.50, 95% CI = 0.52–4.35, P = 0.45). The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Publication bias and heterogeneity
In this study, we used Egger’s regression test, Begg’s adjusted rank correlation test and visual examination of the funnel plot to measure publication bias. The results of Begg’s and Egger’s tests for MMP-91562 C/T (rs3918242), MMP-3 (− 1612 5A/6A), and MMP-2 (− 1306 C/T) gene polymorphisms showed no evidence of publication bias (Fig. 3). During our analysis, we have detected some degree of heterogeneity for three SNPs (Table 3).
Fig. 3.
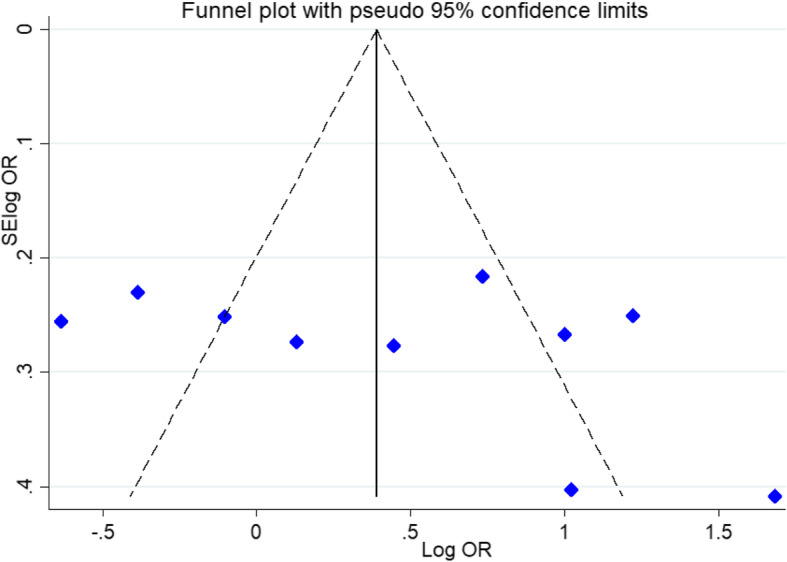
Begg’s funnel plot for publication bias test. Each point represents a separate study for the indicated association
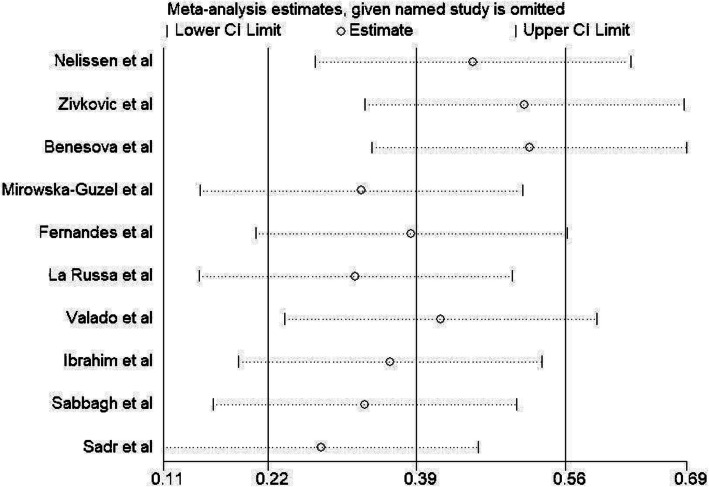
Sensitivity analysis
Here, we performed sensitivity analysis only for MMP-9 gene rs3918242 SNP in order to determine whether sequential omission of the eligible studies affect the final pooled OR. As shown in (Fig. 4), the result was not changed, confirming the stability of our meta-analytical result.
Fig. 4.
Sensitivity analysis in present meta-analysis investigates the individual influence of studies on pooled results
Discussion
To date, several surveys have been conducted addressing the possible association between polymorphisms of the MMP gene family, including MMP-91562 C/T (rs3918242), MMP-3 (− 1612 5A/6A), and MMP-2 (− 1306 C/T) and risk of MS, resulting in inconsistent and inconclusive results. Through resolving the limitation of insufficient statistical power and small sample size in individual studies, meta-analysis studies confer a beneficial approach to settle the problem of conflict. In order to meet this need, here in this meta-analysis we included the most comprehensive and up-to-date original works to obtain exact approximation with respect to association between MMP gene polymorphisms and MS risk.
Previously, large genome-wide association study (GWAS) did not reveal that MMP9 polymorphisms to be significantly associated with the risk for MS [49]. Additionally, a meta-analysis performed on the case-control studies from Europe and one Brazilian study and did not indicate a significant association of MMP9 (− 1562 C/T) SNP with and MS susceptibility [50]. The number of included studies in the meta-analysis was limited and inclusion of further recent studies in Asian populations, like Iran and Egypt, led to significant association of MPP9 polymorphism with MS susceptibility. On the other hand, the role of variations in the MMP9 gene may differ in various populations. Reports have indicated that the T allele was underrepresented only in female MS patients [37, 38]. It seems that differences in the genetic originality of subjects in different populations as well as the gender of the MS patients are involved in determining the function of MMP gene variations in the etiopathogenesis of MS.
MMPs are enzymes that have proteolytic function and have been attributed with numerous implications in tissue remodeling and development. MMPs, especially MMP-9 (also known as gelatinase B), have been observed to be the primary enzymes involved in the degradation of the BBB in MS setting [23, 51]. The major function of MMP-9 is to degrade extracellular matrix (ECM) and myelin basic protein (MBP), hence mediating the recruitment of the inflammatory cells into the involved CNS in MS disease [52–55]. MS patients have shown increased cerebrospinal fluid (CSF) and serum levels of MMP-9 [56, 57]. MMP-9 level has been implied to be a proper marker for the evaluation of clinical type and severity of the disability in MS patients [58].
Studies have established that two functional SNPs in the promoter region MMP-9 gene, namely rs3918242 and rs3222264, impress the expression of this gene [39, 44, 59, 60]. In rs3222264 SNP, the − 90 position is involved in double strand DNA opening and exerted by transcription factors and DNA regulatory proteins. In vitro experiments indicated that the C–1562 T SNP play a role in blocking the nuclear repressor protein binding to the promoter region in which this SNP is located, leading to upregulation of MMP-9 expression [61].
Our meta-analysis indicated significant association of MMP-9 (− 1562 C/T) SNP and risk of MS. Interestingly, all genetic model comparisons, including dominant model (OR = 1.62), recessive model (OR = 2.69), allelic model (OR = 1.51), TT vs. CC model (OR = 3.20), and CT vs. CC model (OR = 1.53), increased the risk of MS susceptibility. The previous meta-analysis by Li et al. in 2017 included 6 studies comprising 1265 MS patients and 1104 controls. The meta-analysis did not show any association of MMP-9 (− 1562 C/T) SNP with MS risk [50]. However, our meta-analysis, by including 10 studies for MMP-9 (− 1562 C/T) SNP containing 1757 MS subjects and 1702 controls, indicated an increased risk of MS by all genetic models of this polymorphism. Furthermore, we evaluated the possible association of MMP-3 (− 1612 5A/6A) and MMP-2 (− 1306 C/T) SNPs and risk of MS to attain more comprehensive conclusion of MMP gene polymorphisms and MS risk. However, we did not find association of these SNPs with MS risk, possibly due to small sample size and little number of studies, which need further evaluations in the future.
Although MMP-3 (− 1612 5A/6A) and MMP-2 (− 1306 C/T) SNPs were not associated with MS risk according to our meta-analysis, the role of genetic interactions and haplotypes should not be neglected. In a study, the MMP-9 T allele was not associate with MS risk, however, a synergism was identified between MMP-9 C and MMP-7 G alleles in increasing MS risk by 1.5 times. Furthermore, there was 3.13 times increased MS risk in association with the haplotype MMP-9 T, MMP-7 G, and MMP-2 C (TGC) in comparison to the CAG haplotype [28]. Therefore, further studies may disclose the association on MMP-3 (− 1612 5A/6A) and MMP-2 (− 1306 C/T) SNPs with MS risk in the haplotype analysis.
Although we tried to carry out the most comprehensive meta-analysis of the MMP gene SNPs and the risk of MS, a number of limitations of this meta-analysis study should be remarked. First, the number of studies and sample size for MMP-3 (− 1612 5A/6A) and MMP-2 (− 1306 C/T) SNPs in this meta-analysis was relatively small to conclude the role of these SNPs and MS risk. Second, we only searched for articles published in the English language and a number of potential data might be neglected. Third, the current meta-analysis was according to crude analysis of the genetic polymorphisms, and the adjusting the analysis by gender, age, and other environmental factors were not taken into consideration. Fourth, we detected some degrees of heterogeneity for the analyzed SNPs, that might stem from difference in genetic stratification and ethnicity, variety in the environmental factors in different populations, and the detection methods.
Conclusion
Taken all the evidence into conclusion, this was the first and most comprehensive evaluation of the MMP gene family SNPs in association with MS. Unlike previous meta-analysis, our study detected significant association of MMP-9 (− 1562 C/T) SNP with increased risk of MS. Nonetheless, other polymorphisms were not associated, perhaps due to little sample size. Hence, we acknowledge the further studies with respect to evaluation of other MMP gene SNPs in association with MS, particularly in a haplotype analysis. Furthermore, the role of other factors, like age, gender, and environmental factors in the analysis ahead will hopefully shed further light on the bona fide implication of MMP gene polymorphisms and susceptibility to MS.
Acknowledgements
Bahman Razi would like to thanks Mrs. Soraya Moghimi for all her support.
Authors ‘contributions
DI, AKH and MM originated the study, acquired data. BR, FB and KL performed statistical analysis, interpreted data, drafted the manuscript. SA and SHT revised the manuscript. All authors read and approved the final manuscript.
Abbreviations
- MMP
Matrix metallo proteinases
- MS
Multiple sclerosis
- CNS
Central nervous system
- CI
Confidence interval
- VDR
Vitamin D receptor
- STAT
Signal transducer and activator of transcription
- PTPN22
Protein tyrosine phosphatase, non-receptor type 22
- OR
Odds ratio
- SNP
Single-nucleotide polymorphism
- PRISMA
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- NOS
Newcastle–Ottawa scale
- HWE
Hardy–Weinberg equilibrium
Funding
Not applicable.
Availability of data and materials
All data that support the conclusions of this manuscript are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Imani D, et al. Association of nod-like receptor protein-3 single nucleotide gene polymorphisms and expression with the susceptibility to relapsing–remitting multiple sclerosis. Int J Immunogenet. 2018;45(6):329–336. doi: 10.1111/iji.12401. [DOI] [PubMed] [Google Scholar]
- 2.Campbell GR, Worrall JT, Mahad DJ. The central role of mitochondria in axonal degeneration in multiple sclerosis. Mult Scler J. 2014;20(14):1806–1813. doi: 10.1177/1352458514544537. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, et al. Fingolimod (FTY720): first approved oral therapy for multiple sclerosis. J Pharmacol Pharmacother. 2011;2(1):49. doi: 10.4103/0976-500X.77118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamińska J, et al. Multiple sclerosis-etiology and diagnostic potential. Postepy Hig Med Dosw (Online) 2017;71:551–563. doi: 10.5604/01.3001.0010.3836. [DOI] [PubMed] [Google Scholar]
- 5.Emamnejad R, et al. Circulating mesenchymal stem cells, stromal derived factor (SDF)-1 and IP-10 levels increased in clinically active multiple sclerosis patients but not in clinically stable patients treated with beta interferon. Mult Scler Relat Disord. 2019;35:233–238. doi: 10.1016/j.msard.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Aslani S, et al. Epigenetic modifications and therapy in multiple sclerosis. NeuroMolecular Med. 2017;19(1):11–23. doi: 10.1007/s12017-016-8422-x. [DOI] [PubMed] [Google Scholar]
- 7.Hawkes C, Macgregor A. Twin studies and the heritability of MS: a conclusion. Mult Scler J. 2009;15(6):661–667. doi: 10.1177/1352458509104592. [DOI] [PubMed] [Google Scholar]
- 8.Kuusisto H, et al. Concordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twins. Eur J Neurol. 2008;15(10):1106–1110. doi: 10.1111/j.1468-1331.2008.02262.x. [DOI] [PubMed] [Google Scholar]
- 9.Fagnani C, et al. Twin studies in multiple sclerosis: a meta-estimation of heritability and environmentality. Mult Scler J. 2015;21(11):1404–1413. doi: 10.1177/1352458514564492. [DOI] [PubMed] [Google Scholar]
- 10.Vandenbroeck K, et al. High-resolution analysis of IL-6 minisatellite polymorphism in Sardinian multiple sclerosis: effect on course and onset of disease. Genes Immun. 2000;1(7):460. doi: 10.1038/sj.gene.6363706. [DOI] [PubMed] [Google Scholar]
- 11.Smolders J, et al. The relevance of vitamin D receptor gene polymorphisms for vitamin D research in multiple sclerosis. Autoimmun Rev. 2009;8(7):621–626. doi: 10.1016/j.autrev.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Matesanz F, et al. Protein tyrosine phosphatase gene (PTPN22) polymorphism in multiple sclerosis. J Neurol. 2005;252(8):994–995. doi: 10.1007/s00415-005-0795-y. [DOI] [PubMed] [Google Scholar]
- 13.Sokolova EA, et al. Association of SNPs of CD40 gene with multiple sclerosis in Russians. PLoS One. 2013;8(4):e61032. doi: 10.1371/journal.pone.0061032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong VW, et al. Elevation of matrix metalloproteinases (MMPs) in multiple sclerosis and impact of immunomodulators. J Neurol Sci. 2007;259(1–2):79–84. doi: 10.1016/j.jns.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Javan MR, et al. Downregulation of immunosuppressive molecules, PD-1 and PD-L1 but not PD-L2, in the patients with multiple sclerosis. Iran J Allergy Asthma Immunol. 2016;15(4):296–302. [PubMed] [Google Scholar]
- 16.Javan M-R, et al. Molecular analysis of interleukin-25 exons 1 and 2 and its serum levels in Iranian patients with multiple sclerosis. Am J Clin Exp Immunol. 2014;3(2):91. [PMC free article] [PubMed] [Google Scholar]
- 17.Javan MR, et al. An interleukin 12 B single nucleotide polymorphism increases IL-12p40 production and is associated with increased disease susceptibility in patients with relapsing-remitting multiple sclerosis. Neurol Res. 2017;39(5):435–441. doi: 10.1080/01616412.2017.1301623. [DOI] [PubMed] [Google Scholar]
- 18.Yoon S-O, et al. Roles of matrix metalloproteinases in tumor metastasis and angiogenesis. J Biochem Mol Biol. 2003;36(1):128–137. doi: 10.5483/bmbrep.2003.36.1.128. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro SD. Matrix metalloproteinase degradation of extracellular matrix: biological consequences. Curr Opin Cell Biol. 1998;10(5):602–608. doi: 10.1016/s0955-0674(98)80035-5. [DOI] [PubMed] [Google Scholar]
- 20.Chintala SK, Tonn JC, Rao JS. Matrix metalloproteinases and their biological function in human gliomas. Int J Dev Neurosci. 1999;17(5–6):495–502. doi: 10.1016/s0736-5748(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 21.Cui N, Hu M, Khalil RA. Biochemical and biological attributes of matrix metalloproteinases. In Progress in molecular biology and translational science. Vol. 147. Academic Press; 2017. p. 1–73. [DOI] [PMC free article] [PubMed]
- 22.Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177–183. doi: 10.3109/14756366.2016.1161620. [DOI] [PubMed] [Google Scholar]
- 23.Yong VW, et al. Metalloproteinases in biology and pathology of the nervous system. Nat Rev Neurosci. 2001;2(7):502. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye S. Polymorphism in matrix metalloproteinase gene promoters: implication in regulation of gene expression and susceptibility of various diseases. Matrix Biol. 2000;19(7):623–629. doi: 10.1016/s0945-053x(00)00102-5. [DOI] [PubMed] [Google Scholar]
- 25.Kanamori Y, et al. Correlation between expression of the matrix metalloproteinase-1 gene in ovarian cancers and an insertion/deletion polymorphism in its promoter region. Cancer Res. 1999;59(17):4225–4227. [PubMed] [Google Scholar]
- 26.dos Reis ST, et al. Genetic polymorphisms of matrix metalloproteinases: susceptibility and prognostic implications for prostate cancer. J Urol. 2009;181(5):2320–2325. doi: 10.1016/j.juro.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Mirowska-Guzel D, et al. Association of MMP1, MMP3, MMP9, and MMP12 polymorphisms with risk and clinical course of multiple sclerosis in a polish population. J Neuroimmunol. 2009;214(1–2):113–117. doi: 10.1016/j.jneuroim.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Rahimi Z, et al. Functional promoter polymorphisms of MMP-2 C-735T and MMP-9 C-1562T and their synergism with MMP-7 A-181G in multiple sclerosis. Immunol Investig. 2016;45(6):543–552. doi: 10.1080/08820139.2016.1180303. [DOI] [PubMed] [Google Scholar]
- 29.Nelissen I, et al. Polymorphism analysis suggests that the gelatinase B gene is not a susceptibility factor for multiple sclerosis. J Neuroimmunol. 2000;105(1):58–63. doi: 10.1016/s0165-5728(00)00189-2. [DOI] [PubMed] [Google Scholar]
- 30.Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 31.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 32.Huedo-Medina TB, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11(2):193. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 35.Begg Colin B., Mazumdar Madhuchhanda. Operating Characteristics of a Rank Correlation Test for Publication Bias. Biometrics. 1994;50(4):1088. [PubMed] [Google Scholar]
- 36.Egger M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Živković M, et al. Matrix metalloproteinase-9− 1562 C/T gene polymorphism in Serbian patients with multiple sclerosis. J Neuroimmunol. 2007;189(1–2):147–150. doi: 10.1016/j.jneuroim.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Benešová Y, et al. Matrix metalloproteinase-9 and matrix metalloproteinase-2 gene polymorphisms in multiple sclerosis. J Neuroimmunol. 2008;205(1–2):105–109. doi: 10.1016/j.jneuroim.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 39.La Russa A, et al. Single nucleotide polymorphism in the MMP-9 gene is associated with susceptibility to develop multiple sclerosis in an Italian case-control study. J Neuroimmunol. 2010;225(1–2):175–179. doi: 10.1016/j.jneuroim.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Valado A, et al. Multiple sclerosis: association of gelatinase B/matrix metalloproteinase-9 with risk and clinical course the disease. Mult Scler Relat Disord. 2017;11:71–76. doi: 10.1016/j.msard.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Sabbagh S, et al. Association study between functional polymorphisms of MMP9 gene promoter and multiple sclerosis susceptibility in an Iranian population. Iran J Public Health. 2019;48(9):1697. [PMC free article] [PubMed] [Google Scholar]
- 42.Sadr NKS, et al. Matrix Metalloproteinase-9 gene polymorphisms in south-west Iranian multiple sclerosis (MS) patients. Russ J Genet. 2019;55(10):1266–1272. [Google Scholar]
- 43.Ibrahim I, et al. Matrix metalloproteases 9 rs3918242 gene polymorphism and serum vit D in MS Egyptian patients. Mult Scler Relat Disord. 2019;32:103–106. doi: 10.1016/j.msard.2019.04.029. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes KS, et al. Matrix metalloproteinase-9 genotypes and haplotypes are associated with multiple sclerosis and with the degree of disability of the disease. J Neuroimmunol. 2009;214(1–2):128–131. doi: 10.1016/j.jneuroim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Djurić T, et al. Association of the MMP-3 5A/6A gene polymorphism with multiple sclerosis in patients from Serbia. J Neurol Sci. 2008;267(1–2):62–65. doi: 10.1016/j.jns.2007.09.037. [DOI] [PubMed] [Google Scholar]
- 46.RAHIMI Z, ABBASI A, RAHIMI Z. Functional promoter polymorphism of matrix metalloproteinase (MMP)-3 5A/6A and its interaction with MMP-7 a-181G polymorphism in multiple sclerosis. Biharean Biologist. 2016;10(2):137–140. [Google Scholar]
- 47.Liutkevičienė R, et al. Association of MMP-2 (−1306 C/T) gene polymorphism with predisposition to optic neuritis and optic neuritis together with multiple sclerosis. Medicina. 2018;54(2):29. doi: 10.3390/medicina54020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksoy D, et al. Analysis of MMP2-1306C/T and TIMP2G-418C polymorphisms with relapsing remitting multiple sclerosis. J Investig Med. 2016;64(6):1143–1147. doi: 10.1136/jim-2016-000111. [DOI] [PubMed] [Google Scholar]
- 49.Nischwitz S, et al. MS susceptibility is not affected by single nucleotide polymorphisms in the MMP9 gene. J Neuroimmunol. 2015;279:46–49. doi: 10.1016/j.jneuroim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Li J, et al. Association study of MMP-9− 1562C/T gene polymorphism with susceptibility to multiple autoimmune diseases: a meta-analysis. Arch Med Res. 2017;48(1):105–112. doi: 10.1016/j.arcmed.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Waubant E. Biomarkers indicative of blood-brain barrier disruption in multiple sclerosis. Dis Markers. 2006;22(4):235–244. doi: 10.1155/2006/709869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woessner JF., Jr Matrix metalloproteinases and their inhibitors in connective tissue remodeling. FASEB J. 1991;5(8):2145–2154. [PubMed] [Google Scholar]
- 53.Chandler S, et al. Matrix metalloproteinases degrade myelin basic protein. Neurosci Lett. 1995;201(3):223–226. doi: 10.1016/0304-3940(95)12173-0. [DOI] [PubMed] [Google Scholar]
- 54.Gijbels K, et al. Gelatinase B is present in the cerebrospinal fluid during experimental autoimmune encephalomyelitis and cleaves myelin basic protein. J Neurosci Res. 1993;36(4):432–440. doi: 10.1002/jnr.490360409. [DOI] [PubMed] [Google Scholar]
- 55.Proost P, Vandamme J, Opdenakker G. Leukocyte gelatinase B cleavage releases encephalitogens from human myelin basic protein. Biochem Biophys Res Commun. 1993;192(3):1175–1181. doi: 10.1006/bbrc.1993.1540. [DOI] [PubMed] [Google Scholar]
- 56.Gijbels K, et al. Gelatinase in the cerebrospinal fluid of patients with multiple sclerosis and other inflammatory neurological disorders. J Neuroimmunol. 1992;41(1):29–34. doi: 10.1016/0165-5728(92)90192-n. [DOI] [PubMed] [Google Scholar]
- 57.Paemen L, et al. Evaluation of gelatinases and IL-6 in the cerebrospinal fluid of patients with optic neuritis, multiple sclerosis and other inflammatory neurological diseases. Eur J Neurol. 1994;1(1):55–63. doi: 10.1111/j.1468-1331.1994.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 58.Benešová Y, et al. Matrix metalloproteinase-9 and matrix metalloproteinase-2 as biomarkers of various courses in multiple sclerosis. Mult Scler J. 2009;15(3):316–322. doi: 10.1177/1352458508099482. [DOI] [PubMed] [Google Scholar]
- 59.Shimajiri S, et al. Shortened microsatellite d (CA) 21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455(1–2):70–74. doi: 10.1016/s0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- 60.Zhang B, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99(14):1788–1794. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 61.Zhang B, et al. Genetic variation at the matrix metalloproteinase-9 locus on chromosome 20q12. 2–13.1. Hum Genet. 1999;105(5):418–423. doi: 10.1007/s004390051124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the conclusions of this manuscript are included within the article.