Abstract
Candida auris is an emerging global healthcare-associated pathogen. During July–December 2018, four patients with C. auris were identified in Victoria, Australia, all with previous overseas hospitalization. Phylogenetic analysis revealed putative transmission between 2 patients and suspected overseas acquisition in the others. Vigilant screening of at-risk patients is required.
Keywords: antimicrobial resistance, public health surveillance, Candida auris, infection control, genomics, fungi, Australia
The fungal pathogen Candida auris is an emerging global health threat associated with a range of invasive infections, most commonly candidemia; it is often resistant to multiple antifungal drugs (1). First identified in Japan in 2009, C. auris has been reported across all 6 populated continents with outbreaks in healthcare settings, particularly intensive care and high-dependence units (1,2). Four genetic lineages of C. auris with phylogeographic variation have been identified (3).
Before July 2018, only 1 case of C. auris had been reported in Australia, none in the state of Victoria (population 6.5 million) (4); no centralized surveillance or mandatory reporting has been implemented on a state or national level, and local screening policies are limited. However, Victoria has experienced large interfacility and intrafacility healthcare-associated outbreaks of other multidrug-resistant organisms (5) and has increasingly implemented genomics in both the investigation of outbreaks and routine surveillance (C.R. Lane et al., unpub. data, https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3498431).
In July 2018, C. auris was cultured from a patient hospitalized in a Victoria healthcare facility. In response, the Victoria Department of Health and Human Services (DHHS) convened an incident management team and issued a Chief Health Officer alert to all health services and laboratories recommending admission screening for patients with recent overseas hospitalization. Also recommended was consideration of C. auris in patients with cultured non–C. albicans species and risk factors for fungal infection, including diabetes mellitus and recent antimicrobial drug use. The alert specified that all C. auris and nonspeciated non–C. albicans isolates from high-risk patients be referred to Victorian Public Health laboratories for speciation and characterization and reported to the DHHS (6). We report on the use of genomics to investigate putative transmission of C. auris in Victoria during July 1–December 31, 2018.
Isolates of C. auris were referred to the Victorian Infectious Diseases Reference Laboratory, where they underwent species identification and antimicrobial susceptibility testing. All isolates were then referred to the Microbiological Diagnostic Unit Public Health Laboratory for DNA extraction, whole-genome sequencing, and bioinformatic analysis.
During July–December 2018, we identified C. auris in 4 patients. Three patients (patients 1, 2, and 4 chronologically) were identified through clinically indicated samples, whereas patient 3 was screened on transfer from an overseas healthcare facility. Patients 1 and 2 were admitted to the same hospital at specimen collection, and patients 3 and 4 were admitted to different facilities. All patients reported previous overseas hospitalization (Figure).
Figure.
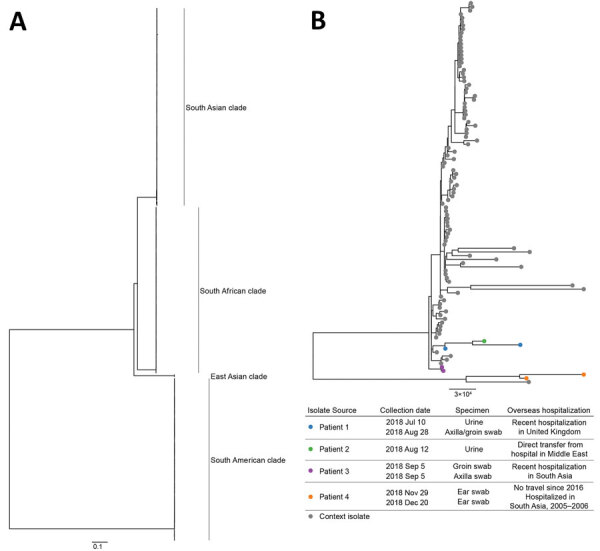
Maximum-likelihood phylogenetic trees of Candida auris isolates from Victoria, Australia, in the context of international publicly available genomes. A) Complete tree; B) South Asian clade. Isolates from 4 patients in Victoria are indicated by colored dots on the inset tree; isolate details and patient travel history are provided in the key. Scale bars indicate substitutions per site.
We obtained 7 C. auris isolates from these 4 patients and performed core genome phylogenetic analysis on all isolates (Appendix). We downloaded publicly available C. auris sequences and included those meeting quality control metrics (Appendix Table 1). Phylogenetic analysis revealed all 7 isolates fell within the previously described South Asian clade (Figure, panel A).
We identified putative transmission during a concurrent hospital stay between patients 1 and 2 (Figure, panel B); transmission was epidemiologically validated and reported elsewhere (7). Because both patients reported overseas hospitalization, it is unclear which constituted the index case. Isolates from patients 3 and 4 were not closely related to each other, or any other included isolates, consistent with independent overseas acquisition.
We reported results of phylogenetic analyses prospectively and concurrently to the incident management team and affected facilities. Because all patients reported overseas hospitalization, the combined analysis of genomic and epidemiologic data enabled assessment of alternative hypotheses, identifying putative local transmission between patients 1 and 2 and excluding patients 3 and 4 from the outbreak. These results justified targeted infection control measures.
In late 2015, Victoria introduced combined phylogenetic and epidemiologic surveillance for carbapenemase-producing Enterobacterales, a similar low-prevalence multiresistant organism (8). Using a search-and-destroy approach, this method enabled early identification of local transmission and complemented standard laboratory, screening, and outbreak control measures across the state. Leveraging from this work, a similar system was introduced for the control of C. auris in September 2019, with statewide mandatory notification of C. auris introduced in December 2019 (9,10). These measures address limitations in the current study, such as the inability to identify patients with C. auris because of noncompliance with screening recommendations, nonreporting, or decreased sensitivity of laboratory methods for the detection of C. auris.
A representative sample of international isolates enables inference of local transmission through local cluster identification and can indicate the source of local strains. The emergence of C. auris highlights the need for greater surveillance of nonbacterial multiresistant organisms and international data sharing. Sequences from our study were submitted to GenBank.
Our findings demonstrate the importance of proactive screening programs and of strict isolation and containment actions. Despite Australia’s geographic isolation, vigilance is necessary to ensure that patients hospitalized overseas are identified and screened for the presence of multiresistant organisms such as C. auris upon hospital admission.
Additional information about Candida auris in Australia.
Biography
Ms. Lane is an epidemiologist and PhD candidate at the University of Melbourne. Her primary research interest is the use of genomics in the surveillance of antimicrobial resistance and pathogens of public health concern.
Footnotes
Suggested citation for this article: Lane CR, Seemann T, Worth LJ, Easton M, Pitchers W, Wong J, et al. Incursions of Candida auris into Australia, 2018. Emerg Infect Dis. 2020 Jun [date cited]. https://doi.org/10.3201/eid2606.190936
References
- 1.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7:43. 10.1038/s41426-018-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Tracking Candida auris January 22, 2019: case count updated as of December 31, 2018 [cited 2019 Mar 6]. https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html
- 3.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–40. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Australian Bureau of Statistics. Quarterly population estimates (ERP), by state/territory, sex and age [cited 2019 Mar 6]. http://stat.data.abs.gov.au/Index.aspx?DataSetCode=ERP_QUARTERLY
- 5.Kwong JC, Lane CR, Romanes F, Gonçalves da Silva A, Easton M, Cronin K, et al. Translating genomics into practice for real-time surveillance and response to carbapenemase-producing Enterobacteriaceae: evidence from a complex multi-institutional KPC outbreak. PeerJ. 2018;6:e4210. 10.7717/peerj.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Victorian Department of Health and Human Services. Candida auris case detected in Victoria [cited 2019 Mar 6]. https://www2.health.vic.gov.au/about/news-and-events/healthalerts/candida-auris-case-detected-in-victoria
- 7.Worth LJ, Harrison SJ, Dickinson M, van Diemen A, Breen J, Harper SE, et al. Candida auris in an Australian healthcare facility: importance of screening high-risk patients. Med J Aust. In press 2020. [DOI] [PubMed] [Google Scholar]
- 8.Victorian Department of Health and Human Services. Victorian guideline on carbapenemase-producing Enterobacteriaceae for health services—version 2.1. Melbourne: Victorian Government; 2018. [cited 2020 Mar 23]. https://www2.health.vic.gov.au/Api/downloadmedia/%7BF1FDFD1D-C4D7-4131-A2FF-9EB24B5293E4%7D [Google Scholar]
- 9.Victorian Department of Health and Human Services. Victorian guideline on Candida auris for health services. Melbourne: Victorian Government; 2019. [cited 2020 Mar 23]. https://www2.health.vic.gov.au/Api/downloadmedia/%7BE72DE677-88A3-4AF4-8557-C20DEA78638F%7D [Google Scholar]
- 10.Parliament of Victoria. Public Health and Wellbeing Regulations 2019(Vic)(Austrl.). Parliament of Victoria; 2019 [cited 2020 Mar 23]. http://classic.austlii.edu.au/au/legis/vic/num_reg/phawr2019n135o2019412
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information about Candida auris in Australia.


