To the Editor:
During physiologic regeneration after injury, type 2 alveolar epithelial cells (AEC2s) proliferate and then differentiate into AEC1s to restore normal alveolar architecture and function. Although mechanisms that promote AEC2 proliferation have been identified, the mechanisms by which AEC2s differentiate into AEC1s remain poorly understood. Idiopathic pulmonary fibrosis (IPF) is a progressive scarring disease that arises from ineffectual regeneration of the injured epithelium and unchecked TGF-β (transforming growth factor β) signaling (1). However, the specific manner in which alveolar regeneration goes awry in the pathogenesis of IPF is unknown. Because the IPF epithelium is characterized by hyperplasia of AECs with a transitional morphology and gene expression profile (2, 3), some investigators have speculated that the regenerative defect in IPF may be ineffectual AEC2-to-AEC1 differentiation. However, the specific defect in AEC2-to-AEC1 differentiation in IPF and its relationship to the differentiation lineage trajectory in physiologic regeneration have not been characterized. An improved understanding of the mechanisms underlying physiologic AEC1 differentiation and how it goes awry in fibrosis may ultimately lead to the development of novel therapies to promote physiologic regeneration in IPF.
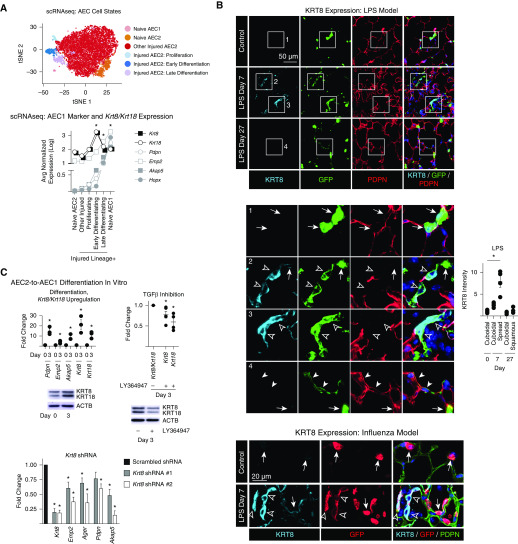
Our recent single-cell RNA sequencing study of lineage-labeled AEC2s during physiologic regeneration after lung injury induced by LPS uncovered two discrete transitional states of AEC2-to-AEC1 differentiation, early and late, which are characterized by moderate and high expression of AEC1 markers, respectively (Figure 1A), and downregulation of AEC2 markers (webtool available at https://rnabioco.github.io/lung-scrna/) (4). TGF-β signaling is highly activated during early differentiation, and subsequent TGF-β deactivation promotes late differentiation (4). Here, using previously described methods (4), we found that Krt8 (keratin 8) and Krt18 were highly upregulated during early differentiation and downregulated during late differentiation (Figure 1A). Immunostaining of lung sections from the LPS and influenza models of lung injury confirmed that although KRT8 expression was diffusely upregulated in AEC2s and AEC1s after injury (data not shown) (5), its expression was particularly high in lineage-labeled AEC2s with a partially spread, transitional morphology consistent with the early differentiation state (Figure 1B). KRT8 was then downregulated in lineage-labeled cells with a squamous, AEC1 morphology suggesting that they had fully differentiated. TGF-β activation in transitional cells was confirmed by in situ hybridization for integrin β6 (data not shown).
Figure 1.
Physiologic differentiation of type 2 alveolar epithelial cells (AEC2s) into AEC1s proceeds via early KRT8/KRT18hi, TGF-β (transforming growth factor β)–activated and late KRT8/KRT18lo, TGF-β–deactivated transitional cell states. (A and B) SftpcCreERT2+/−;mTmG+/− mice previously administered tamoxifen were exposed to intratracheal LPS (40 μg Escherichia coli 0111:B4) or intranasal H1N1 influenza virus (100 plaque-forming units, strain A/PR/8/34) or left untreated. At Day 7, green fluorescent protein (GFP)+ Tomato− cells isolated from naive and LPS-treated mice and GFP− Tomato+ EPCAM+ PDPN+ cells isolated from naive mice were subjected to single-cell RNA sequencing (scRNAseq). Unsupervised clustering was performed and the cell types and states shown on t-distributed stochastic neighbor embedding (tSNE) plots were identified using known markers (4). (The tSNE plot was reprinted by permission from Reference 4.) Expression of the indicated AEC1 markers and Krt8 and Krt18 is shown on the graph; n = 2 mice per group; Wilcoxon rank sum test with Bonferroni correction. *P < 0.01 for Krt8, Krt18, Pdpn, Emp2, Akap5, and Hopx in the early differentiation state compared with all other cells sequenced. *P < 0.01 for Pdpn, Emp2, Akap5, and Hopx in the late differentiation state compared with all other cells sequenced. *P < 0.01 for Pdpn, Emp2, Akap5, and Hopx in naive AEC1s compared with all other cells sequenced. (B) Lung sections were stained with the indicated antibodies. Insets 1–4 are enlarged below. Arrows indicate cuboidal AEC2s with KRT8 expression that was low or undetectable at the exposure settings used (KRT8 was detected in cuboidal AEC2s with higher exposure [data not shown]). Open arrowheads indicate lineage-labeled cells with a transitional, spread morphology but no detectable PDPN, consistent with early differentiation. These cells express high levels of KRT8. Solid arrowheads indicate lineage-labeled cells with a squamous morphology that express PDPN, consistent with late differentiation, but have low KRT8 expression. DAPI is represented by the dark blue stain. The fluorescence intensity of KRT8 was measured in the injured areas using LCmicro software (Olympus), and the average intensity of cuboidal, partially spread, or squamous cells per mouse is shown; n ≥ 4 mice/group; *P < 0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons. (C) Rat AEC2s were isolated as previously described (4) and cultured in Dulbecco’s modified Eagle medium + 10% fetal bovine serum. Quantitative PCR and Western blotting were performed. Differentiation, Krt8/Krt18 upregulation: AEC1 markers, Krt8, and Krt18 are upregulated by Day 3 of culture. Data are shown as the fold change compared with expression levels at Day 0 for each gene. TGF-β inhibition: the TGF-βRI kinase inhibitor LY364947 attenuated Krt8/Krt18 upregulation. Data are shown as the fold change compared with expression levels without the inhibitor for each gene. Krt8 shRNA: cells were transduced with lentivirus (prepared by the University of Michigan Vector Core) containing one of two distinct shRNA constructs (TRCN0000325589 and TRCN0000091874) cloned into the pLKO.1 vector (Sigma). Data are shown as the fold change compared with expression levels with the scrambled shRNA for each gene. Krt8 knockdown attenuated upregulation of AEC1 markers. Fold change was calculated by the 2−ddCt method using PPIB as the housekeeping gene. Each data point represents one experiment performed on cells isolated from one animal; the data point is the average of two technical replicates. *P < 0.05 by two-tailed t test. Mean ± SEM is shown. EPCAM = epithelial cell adhesion molecule; PDPN = podoplanin.
Keratins provide mechanical strength to epithelia but also have signaling functions such as regulating differentiation (6, 7). KRT8/KRT18 is the primary keratin pair expressed by simple epithelia, but its signaling function in the alveolar epithelium is unknown. To determine whether KRT8/KRT18 initiates AEC2-to-AEC1 differentiation, we used an in vitro assay of differentiation (Figure 1C). We previously demonstrated that TGF-β signaling is activated during early differentiation (4) and here we found that Krt8 and Krt18 were upregulated during AEC1 differentiation in culture (Figure 1C) (8), recapitulating the TGF-β–activated Krt8/Krt18hi early differentiation state observed in vivo. Inhibition of TGF-β signaling attenuated Krt8/Krt18 upregulation (Figure 1C). Finally, shRNA-mediated Krt8 knockdown attenuated AEC1 differentiation (Figure 1C). Thus, TGF-β–dependent KRT8/KRT18 expression was necessary to initiate differentiation in vitro.
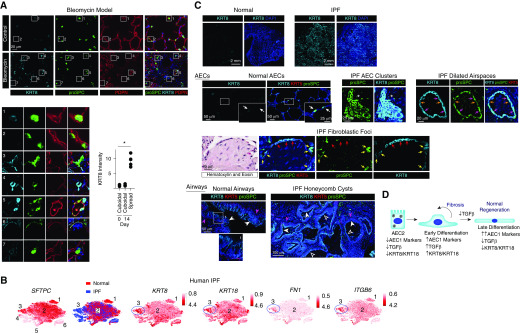
However, subsequent TGF-β deactivation promotes late AEC2-to-AEC1 differentiation (4). Because fibrosis is characterized by hyperplasia of transitional AECs with a paucity of mature AEC1s and unchecked TGF-β signaling, we hypothesized that the transitional KRT8/KRT18hi early differentiation cell state may persist in fibrosis owing to persistent TGF-β activation. We observed abundant epithelial cells with a transitional morphology and high KRT8 expression in the fibrotic lungs of mice treated with bleomycin (Figure 2A). A previous single-cell RNA sequencing study revealed a unique cell state of AEC2s present in IPF but not normal human lungs (cluster 3 in Figure 2B) (9). Here, we found that AEC2s in that cell state expressed high levels of KRT8 and KRT18 and TGF-β pathway genes (Figure 2B). Immunostaining of IPF lungs revealed hyperplastic AECs, some with a spread morphology, expressing high levels of KRT8 (Figure 2C). These cells were found in clusters in fibrotic regions, lining dilated airspaces and overlying fibroblastic foci (Figure 2C). The abundance of KRT8/KRT18hi TGF-β–activated AECs observed in both murine and human fibrotic lungs suggests that these cells may be paused or arrested in the early differentiation cell state (Figure 2D). Because TGF-β and KRT8/KRT18 are downregulated during late differentiation, TGF-β deactivation promotes late differentiation, and KRT8/KRT18 expression is TGF-β dependent, we speculate that although KRT8 upregulation initiates early differentiation, in IPF a failure of TGF-β deactivation and KRT8 downregulation may result in persistence of the early differentiation state.
Figure 2.
Alveolar epithelial cells (AECs) in the bleomycin mouse model of pulmonary fibrosis (PF) and human idiopathic PF (IPF) have a transitional morphology and express high levels of KRT8. (A) Mice were treated with 0.025 U intratracheal bleomycin. At Day 14, lung sections were stained with the indicated antibodies. Shown are insets 1–7 enlarged. Compared with cuboidal proSPC+ PDPN− cells in the control (insets 1 and 2) and bleomycin-treated (inset 7) lungs, KRT8 is upregulated in cells with a transitional morphology and varying levels of proSPC and PDPN expression in the bleomycin-treated lungs (insets 3–6). The KRT8hi cells are found in the fibrotic areas of the bleomycin-treated lungs; KRT8 expression by type 2 AECs (AEC2s) in the uninjured areas of bleomycin-treated lungs is similar to that observed in control mice (data not shown). The fluorescence intensity of KRT8 in the cuboidal and partially spread cells was measured in control lungs and the injured areas of bleomycin-treated lungs, and the average KRT8 intensity per mouse is shown; n ≥ 4 mice per group; *P < 0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons. Mean ± SEM is shown. (B) An existing dataset of single-cell RNA sequencing performed on normal human subjects and patients with PF (9) revealed the presence of six epithelial cell clusters, three of which were AEC2s. Cluster 3, which was present only in patients with IPF, displayed high expression of KRT8 and KRT18 and the TGF-β (transforming growth factor β) pathway genes FN1 (fibronectin) and ITGB6 (integrin β6). P < 10–22 for expression of these genes in cluster 3 compared with other epithelial cells. The t-distributed stochastic neighbor embedding plots were obtained from UCSC Cell Browser (https://www.nupulmonary.org/resources/?ds=fig6) on August 1, 2019. (C) Lung sections from normal human subjects (obtained from Gift of Life Michigan) or IPF explants were stained for KRT8, proSPC, and KRT5. Normal AEC2s and AEC1s expressed low levels of KRT8. White arrows indicate cuboidal AEC2s with KRT8 expression that was low or undetectable at the exposure settings used (KRT8 was detected in cuboidal AEC2s with higher exposure [data not shown]). In the IPF lung, there were clusters of cuboidal KRT8hi proSPC+ cells (asterisks). There were also dilated airspaces lined by KRT8hi KRT5− cells with a more spread morphology; most of these were proSPC+ (pink arrows), but some expressed lower levels of proSPC (orange arrows). The fibroblastic foci were covered KRT8hi KRT5− cells; most of these were proSPC− (red arrows), but some were proSPC+ (yellow arrows). Thus, these clusters, dilated airspaces, and fibroblastic foci contain hyperplastic AEC2s (some with a transitional morphology) that have upregulated KRT8. Luminal cells of the pseudostratified airways of normal lungs (solid white arrowheads) and simple (open white arrowheads) and pseudostratified (solid white arrowheads) honeycomb cysts of IPF lungs were KRT8hi, proSPC−, and KRT5−. The basal cells of normal airways and pseudostratified honeycomb cysts of IPF lungs were KRT5+ (solid pink arrowheads), consistent with previous reports (11) and were KRT8−. DAPI is represented by the dark blue stain. The images shown are representative of n ≥ 4 humans per group. Although lineage tracing of human cells cannot be performed, our data taken together with the literature suggest that the hyperplastic KRT8hi AECs that are proSPC+ and nearby proSPC− cells in the clusters, cysts, and fibroblastic foci are derived from AEC2s, whereas the KRT8hi luminal cells of the honeycomb cysts are airway derived. (D) During normal epithelial regeneration, AEC2s differentiate into AEC1s via two discrete transitional states: 1) early differentiation characterized by upregulation of AEC1 markers and KRT8 and activation of TGF-β signaling, and 2) late differentiation characterized by further upregulation of AEC1 markers, KRT8 downregulation, and deactivation of TGF-β signaling. TGF-β deactivation promotes late differentiation. In IPF, there is persistence of the TGF-β–activated, KRT8hi early differentiation state. PDPN = podoplanin.
Future investigation is warranted to confirm that KRT8 plays a functional role in AEC2-to-AEC1 differentiation in vivo and to dissect underlying mechanisms. Because small-molecule inhibitors can have nonspecific effects, the regulation of Krt8/Krt18 expression by TGF-β should be confirmed using genetic methods in vivo. Exactly how similar the KRT8hi cells in the animal models are to the KRT8hi cells in IPF should be assessed. We must also determine whether AECs permanently arrest or just pause in the early differentiation state, and whether a failure of KRT8 downregulation is causally related to this. Finally, although we propose that persistence of the early differentiation state may represent the ineffectual epithelial regeneration that is widely believed to promote fibrogenesis, whether KRT8/KRT18hi AEC2s activate fibroblasts should also be examined. During the preparation of this work, an unpublished preprint reported the emergence of a transitional Krt8/Krt18hi, TGF-β–activated state after lung injury induced by bleomycin (10), which is strikingly similar to the TGF-β–activated Krt8/Krt18hi transitional cell state we previously identified in the LPS model (4). The upregulation of TNF, MYCN, and NRF2 target genes reported in the bleomycin model was also observed in the LPS model (4). Although bleomycin induces fibrosis, the fibrosis eventually resolves and late differentiation, with Krt8/Krt18 downregulation and TGF-β deactivation, ensues (10).
The mechanisms underlying AEC2-to-AEC1 differentiation during physiologic regeneration and the manner in which alveolar regeneration may go awry during the pathogenesis of fibrosis have remained fundamental unanswered questions in the field. Here, we demonstrate that physiologic AEC2-to-AEC1 differentiation proceeds via an early differentiation state characterized by TGF-β–dependent KRT8/KRT18 upregulation and a late differentiation state characterized by TGF-β deactivation and KRT8/KRT18 downregulation. This regenerative lineage trajectory appears to be conserved in three injury models, which is a significant finding with therapeutic implications. However, in fibrosis, likely owing to persistent TGF-β activation, regenerating AEC2s persist in the KRT8/KRT18hi early differentiation state. These findings may ultimately lead to novel therapies to promote physiologic regeneration and suppress fibrogenesis in IPF.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Jay Hesselberth for thoughtful discussions, and Steven Huang, Eric White, Thomas Lanigan, Abigail Chinn, and Carol Wilke for technical assistance.
Footnotes
Supported by NIH R01 HL131608 (R.L.Z.) and R01 DK47918 (M.B.O.).
Author Contributions: Conception and design: R.L.Z. Data analysis and acquisition: P.J., R.G.d.R., S.M.H., S.J.G., K.A.R., B.B.M., M.B.O., and R.L.Z. Interpretation of data: R.G.d.R., K.A.R., B.B.M., M.B.O., K.M.R., and R.L.Z. Drafting or revision of the manuscript: B.B.M. and R.L.Z. Final approval of the manuscript: all authors.
Originally Published in Press as DOI: 10.1164/rccm.201909-1726LE on February 19, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Blackwell TS, Tager AM, Borok Z, Moore BB, Schwartz DA, Anstrom KJ, et al. Future directions in idiopathic pulmonary fibrosis research: an NHLBI workshop report. Am J Respir Crit Care Med. 2014;189:214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med. 1998;157:1301–1315. doi: 10.1164/ajrccm.157.4.9707039. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riemondy KA, Jansing NL, Jiang P, Redente EF, Gillen AE, Fu R, et al. Single cell RNA sequencing identifies TGFβ as a key regenerative cue following LPS-induced lung injury. JCI Insight. 2019;5:123637. doi: 10.1172/jci.insight.123637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodcock-Mitchell JL, Burkhardt AL, Mitchell JJ, Rannels SR, Rannels DE, Chiu JF, et al. Keratin species in type II pneumocytes in culture and during lung injury. Am Rev Respir Dis. 1986;134:566–571. doi: 10.1164/arrd.1986.134.3.566. [DOI] [PubMed] [Google Scholar]
- 6.Omary MB, Ku NO, Strnad P, Hanada S. Toward unraveling the complexity of simple epithelial keratins in human disease. J Clin Invest. 2009;119:1794–1805. doi: 10.1172/JCI37762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lähdeniemi IAK, Misiorek JO, Antila CJM, Landor SK, Stenvall CA, Fortelius LE, et al. Keratins regulate colonic epithelial cell differentiation through the Notch1 signalling pathway. Cell Death Differ. 2017;24:984–996. doi: 10.1038/cdd.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paine R, Ben-Ze’ev A, Farmer SR, Brody JS. The pattern of cytokeratin synthesis is a marker of type 2 cell differentiation in adult and maturing fetal lung alveolar cells. Dev Biol. 1988;129:505–515. doi: 10.1016/0012-1606(88)90396-x. [DOI] [PubMed] [Google Scholar]
- 9.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strunz M, Simon LM, Ansari M, Mattner LF, Angelidis I, Mayr CH, et al. Longitudinal single cell transcriptomics reveals Krt8+ alveolar epithelial progenitors in lung regeneration [preprint] bioRxiv. 2019 [accessed 2020 May 7]. Available from: https://www.biorxiv.org/content/10.1101/705244v1.full. [Google Scholar]
- 11.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.