Abstract
Polymorphism at the 17q21 gene locus and wheezing responses to rhinovirus (RV) early in childhood conspire to increase the risk of developing asthma. However, the mechanisms mediating this gene–environment interaction remain unclear. In this study, we investigated the impact of one of the 17q21-encoded genes, ORMDL3 (orosomucoid-like 3), on RV replication in human epithelial cells. ORMDL3 knockdown inhibited RV-A16 replication in HeLa, BEAS-2B, A549, and NCI-H358 epithelial cell lines and primary nasal and bronchial epithelial cells. Inhibition varied by RV species, as both minor and major group RV-A subtypes RV-B52 and RV-C2 were inhibited but not RV-C15 or RV-C41. ORMDL3 siRNA did not affect expression of the major group RV-A receptor ICAM-1 or initial internalization of RV-A16. The two major outcomes of ORMDL3 activity, SPT (serine palmitoyl-CoA transferase) inhibition and endoplasmic reticulum (ER) stress induction, were further examined: silencing ORMDL3 decreased RV-induced ER stress and IFN-β mRNA expression. However, pharmacologic induction of ER stress and concomitant increased IFN-β inhibited RV-A16 replication. Conversely, blockade of ER stress with tauroursodeoxycholic acid augmented replication, pointing to an alternative mechanism for the effect of ORMDL3 knockdown on RV replication. In comparison, the SPT inhibitor myriocin increased RV-A16 but not RV-C15 replication and negated the inhibitory effect of ORMDL3 knockdown. Furthermore, lipidomics analysis revealed opposing regulation of specific sphingolipid species (downstream of SPT) by myriocin and ORMDL3 siRNA, correlating with the effect of these treatments on RV replication. Together, these data revealed a requirement for ORMDL3 in supporting RV replication in epithelial cells via SPT inhibition.
Keywords: ORMDL3, asthma, rhinovirus, SPT, unfolded protein response
Asthma is one of the most prevalent chronic diseases of childhood, affecting more than 7 million children under the age of 18 years in the United States (1). The development of asthma involves contributions from multiple genes and environmental factors, such as the microbiome and viral infection (2). Aberrant wheezing responses early in life to respiratory viruses, rhinovirus (RV) in particular, predispose to subsequent development of asthma (3–5). Regarding asthma genetics, variation at the 17q21 gene locus remains one of the strongest genetic risk factors for the development of childhood asthma (6). Two genes within this locus, GSDMB (gasdermin B) and ORMDL3 (orosomucoid-like 3), are coregulated by asthma-risk–associated polymorphisms (7–9). An increasing body of evidence in murine models has implicated ORMDL3 in asthma development (10–12). ORMDL3-overexpressing transgenic mice present compelling evidence for ORMDL3 as a pathogenic factor (10): these mice manifest spontaneous airway remodeling, increased fibrosis, mucus production, inflammation, and methacholine-induced hyperresponsiveness (10). ORMDL3 has also been implicated in allergen-triggered lung disease in knockout mice and in the transgenic mouse model (10, 13).
ORMDL3 is an endoplasmic reticulum (ER)-resident transmembrane protein that has two identified functions: 1) inhibition of the SPT (serine palmitoyl CoA transferase) enzyme, the rate-limiting step in de novo sphingolipid synthesis (14); and 2) inhibition of the SERCA2b (sarco/endoplasmic reticulum Ca2+ ATPase) pump that maintains the calcium gradient between ER and cytoplasm (8, 15). Disruption of the ER–cytosolic calcium gradient induces an ER stress response known as the “unfolded protein response” (UPR) (16, 17). Both of these ORMDL3 functions appear related to abnormal airway physiology in asthma (15, 18). Mice treated with the SPT inhibitor myriocin and SPT-heterozygous mice display airway hyperresponsiveness but not airway remodeling or inflammation (19). Regarding SERCA inhibition, one study revealed decreased SERCA2b expression in airway smooth muscle cells from subjects with moderate to severe asthma (20). An ER stress pathway induced by ORMDL3 overexpression, involving the transcription factor ATF6α (activating transcription factor 6 alpha), regulates allergen-induced smooth muscle proliferation and airway hyperresponsiveness (10, 15, 21).
Investigation of the high-risk COAST (Childhood Origins of Asthma) cohort identified an intriguing interplay between 17q21 genotype, wheezing responses to RV, and asthma development. Ninety percent of the cohort children with the asthma risk genotype at 17q21, conferring increased expression of ORMDL3, who wheezed in response to RV before the age of 3 years, developed asthma in childhood. RV stimulation of peripheral blood mononuclear cells from these subjects induced further upregulation of ORMDL3 expression (3). A follow-up study in multiple blood leukocyte cell types revealed an association between 17q21 asthma-risk genotype, increased ORMDL3 expression, and greater RV-induced UPR target gene and IFN-β induction (22). However, it was unclear if there were any functional interactions responsible for the noted associations. In addition, RV replicates well in epithelial cells but not in peripheral blood leukocytes.
Currently, the role of ORMDL3 in regulating RV replication in epithelial cells is unclear. One report in the ORMDL3 transgenic mice suggested ORMDL3 overexpression limits RV infection (11). In this study, we sought to understand how ORMDL3 and its primary cellular activities modulate RV infection in human epithelial cells (see Figure E1 in the data supplement). ORMDL3 gene knockdown limited RV replication, more so in RV-A and RV-B than in RV-C species. Further exploration of ER stress and sphingolipid synthesis inhibition suggested the latter property is responsible for the effect of ORMDL3 knockdown on RV replication, as myriocin, an SPT inhibitor and ORMDL3 mimic, enhanced RV-A replication and negated the effects of ORMDL3 knockdown. Together, these results identify a requirement for ORMDL3 and implicate modulation of the sphingolipid pool in supporting RV replication in human epithelial cells.
Methods
Cells and Stimulations
Cell lines, primary epithelial cells, and RV strains are described in the data supplement. For primary cells, written consent was obtained under an internal review board–approved protocol. Epithelial cells were expanded as submerged monolayers in collagen type IV–coated flasks (#C7521; Sigma). The tauroursodeoxycholic acid (TUDCA) sodium salt (EMD Millipore) stock solution was 100 mg/ml in Dulbecco’s modified Eagle medium, tunicamycin (Sigma) stock solution was 1 mg/ml in methanol, and myriocin (Sigma) was 1 mg/ml in methanol. Cell viability using the CellTiter-Glo kit (Promega) was >93.47% for all doses of tunicamycin. For RV infections, sucrose-purified RV was added at 2 plaque-forming units (PFU) per cell for 24 to 72 hours, except for internalization studies (cells infected with 10 PFU/cell for 2 h). After infection, the media was removed and cells resuspended in TRIzol (Invitrogen).
Gene Knockdown
Cells were transfected with 200 pM scrambled control or ORMDL3 siRNA (Thermofisher) using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions.
ORMDL Western Blot
ORMDL3 siRNA-transfected HeLa cells were lysed with radioimmunoprecipitation assay buffer containing protease inhibitors cocktails (Sigma). Whole-cell lysates were resolved by 12% SDS-PAGE, transferred to low-fluorescent, hydrophobic polyvinylidene difluoride membrane (GE Healthcare Amersham Biosciences) and immunoblotted with rabbit anti-ORMDL3 (Millipore) and anti–β-actin (Santa Cruz), followed by IRDye a-800CW goat anti-mouse IgG and IRDye 680RD goat anti-rabbit IgG antibodies (Licor). Proteins were visualized and quantitated with the Odyssey system (Licor). See the data supplement for more detail.
Quantitative PCR
Cells were lysed with TRIzol (Invitrogen) and processed according to the manufacturer’s instructions. RNA was treated with DNase I (Invitrogen) before reverse transcription using a Super Script kit (Thermofisher). Gene expression was quantitated by Power UP SYBR Green (Thermofisher) fluorescence detected on a StepOne real-time PCR machine (Thermofisher). Reaction efficiency and specificity were assessed using serially diluted standard curves and product melting curve analysis. Relative mRNA expression was normalized 18S rRNA using the standard ΔΔCt method. For primers, see data supplement.
RV Plaque Assay
See data supplement.
Immunofluorescence Microscopy
Infected cells on coverslips were fixed in 4% paraformaldehyde, washed with PBS, and blocked with 10% goat serum in 0.1 M, pH 7.6 Tris, 0.1% Triton X-100, and 0.2% BSA. Antibodies were used at 1:100 dilution: rabbit anti-ORMDL3 (Millipore), mouse anti–RV-A antibody (23), and β-actin rabbit monoclonal antibody labeled with Alexa Fluor 647 (Cell Signaling Technology). Secondary antibodies were goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 594 (Thermofisher). Images were acquired on a Nikon Eclipse 50i fluorescence microscope.
Flow Cytometry
Cells were incubated with CD54/ICAM-PE (Miltenyi Biotec). Flow cytometry was performed on an LSRII (BD Sioscience) using FlowJo analysis software (version 9.9.5; Tree Star Inc.).
Lipidomics
See data supplement.
Statistical Analysis
Comparisons between sample groups were conducted using a generalized linear model (see data supplement) and two-sample t test for normally distributed outcomes or nonparametric Wilcoxon rank sum test with SAS software (version 9.4; SAS Institute). Bar graphs denote means across independent experiments, and error bars denote corresponding SEs of the means. In all figures, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001.
Results
RV Infection Increases Expression of ORMDL3, Type I IFN, and UPR Indicators in HeLa Cells
We previously reported a correlation between RV infection and increased ORMDL3 and UPR target gene (HSPA5) induction in peripheral blood mononuclear cells (22). To determine if RV also increased expression of these genes in epithelial cells permissive for RV replication, we infected HeLa cells with RV-A16 (Figure 1A) (24). RV-A16 infection significantly increased ORMDL3, DDIT3 (25), spliced XBP1 mRNA (XBP1[s]), and IFNB1 and showed a trend for increased HSPA5 (BiP) (P = 0.08), suggesting a wider involvement of UPR pathways in these epithelial cells than previously noted in Bjab and THP-1 cell lines and a greater degree of target gene induction than in peripheral blood mononuclear cells (22).
Figure 1.
Rhinovirus-A16 (RV-A16) colocalizes with ORMDL3 (orosomucoid-like 3) and increases mRNA expression of ORMDL3, unfolded protein response (UPR) target genes, and type I IFN. (A) HeLa-H1 cells were infected with RV-A16 for 48 hours and then lysed in TRIzol. RNA was reverse transcribed and gene expression analyzed by quantitative PCR (qPCR) with normalization to 18S rRNA. Bars depict averages of independent experiments, with error bars denoting SEM. (N = 3–6 independent experiments for individual genes). NI = no infection. (B) HeLa-H1 cells were infected with RV-A16 for 48 hours and then fixed. The cells were costained with anti-ORMDL3 (green), anti–RV-A (red), and anti–β-actin (purple) antibodies, and nuclei were stained with DAPI. Scale bars: 10 μm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001. DDIT3 = DNA damage-inducible transcript 3 (CHOP); HSPA5 = heat shock protein family A (Hsp70) member 5 (BiP); XBP1 = X-box binding protein 1.
ORMDL3 Knockdown Significantly Inhibits RV Replication
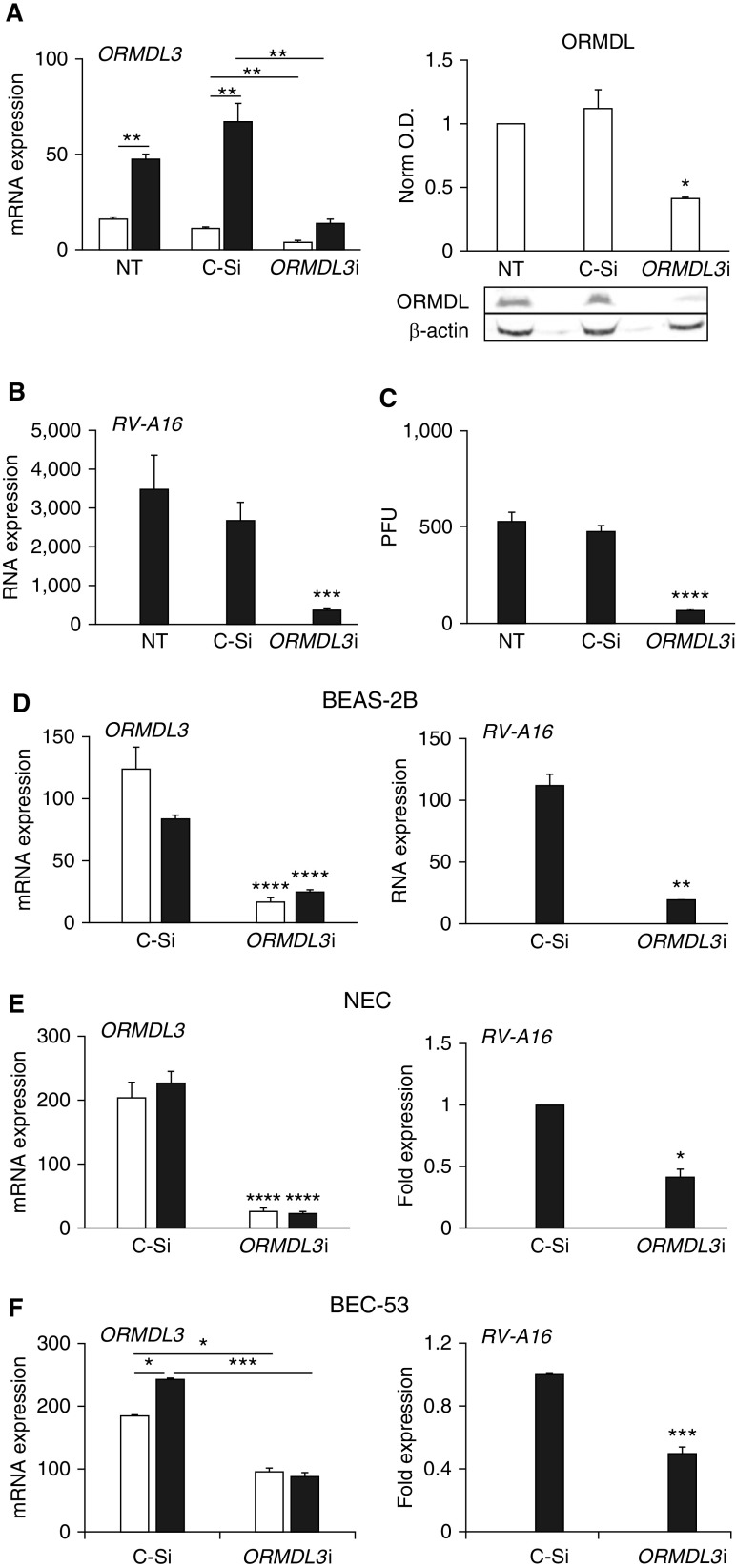
RV replicates in unique lipid organelles near the Golgi and ER (26). ORMDL3 is a transmembrane ER-resident protein (27). Immunofluorescence microscopy of RV-infected HeLa cells suggested a close spatial proximity between ORMDL3 and intracellular RV 48 hours after infection (Figure 1B). To investigate the functional relationship between ORMDL3 and RV-A16 infection, HeLa cells were knocked down for ORMDL3 and then infected with RV-A16. ORMDL3 siRNA decreased ORMDL3 mRNA by ∼75% and total ORMDL protein by greater than twofold (Figure 2A). ORMDL3 siRNA did not significantly affect ORMDL1 or ORMDL2 expression compared with control siRNA (Figure E2), although the control siRNA transfection exerted a modest nonspecific effect on all three ORMDL mRNAs compared with nontransfected untreated cells. RV-A16 infection significantly increased ORMLD3 expression in untreated and control siRNA–transfected HeLa cells (Figure 2A, left). ORMDL3 siRNA significantly decreased RV RNA levels (∼sixfold) compared with no siRNA or control siRNA treatment (Figure 2B). The RV-A16 replication decrease observed by quantitative PCR (qPCR) was confirmed by PFU assay (Figure 2C). ORMDL3 siRNA suppression of RV-A replication was tested in a time course as well (Figure E3). ORMDL3 siRNAs significantly and comparably inhibited RV-A16 replication (fold change vs. control siRNA) at 24 hours, 48 hours, and 72 hours after infection; thus, 48 hours after infection was selected for further studies. To investigate the effect of ORMDL3 knockdown in airway epithelial cells, BEAS-2B cells, originally isolated from the normal human bronchial epithelium (28), were transfected with control siRNA or ORMDL3 siRNA and then infected with RV-A16 (Figure 2D). ORMDL3 siRNA dramatically inhibited RV-A16 replication in BEAS-2B cells. To determine if these observations applied to primary human epithelial cells, nasal epithelial cell (NEC) (25) brushings were collected from patients and cultured. Bronchial epithelial cells (BEC-53) were obtained during lung transplant (29). The NECs and BECs were knocked down for ORMDL3, infected with RV-A16, and analyzed by qPCR for ORMDL3 mRNA and RV (Figures 2E and 2F). In contrast to the HeLa and BEC-53 cells, RV-A infection did not upregulate ORMDL3 expression in primary NECs. However, ORMDL3 knockdown again significantly decreased RV-A16 RNA in both NECs and BECs. We further confirmed the suppression of RV-A16 by ORMDL3 knockdown in more human lung epithelial cell lines (A549 and NCI-H358) and another primary bronchial epithelial donor cell culture (BEC-05; Figure E4). Together, these results suggest ORMDL3 expression is required for optimal RV-A16 replication in multiple epithelial cell types.
Figure 2.
ORMDL3 supports RV-A16 replication in human epithelial cells. Open bars: uninfected. Solid bars: RV-A16 infected. (A) ORMDL3 expression and protein: HeLa cells were transfected with control (C-Si) or ORMDL3 siRNA (ORMDL3i) 24 hours before infection for 48 hours. Cells were processed for qPCR and data normalized to 18S rRNA (left; N = 5). Right: cell lysates from siRNA-transfected cells were analyzed for ORMDL protein by Western blot. ORMDL optical density (O.D.) was normalized to β-actin O.D. (right; N = 3; P < 0.05 versus untreated [NT] and control siRNA). (B) Samples from A were analyzed by qPCR for RV-A16 RNA with normalization to 18S rRNA. Results are from five independent experiments. (C) A plaque-forming unit (PFU) assay from siRNA-transfected cells after 48 hours of infection. N = 3 independent experiments. P < 0.001 compared with NT or C-Si cells. (D) BEAS-2B cells were knocked down for ORMDL3, infected with RV-A16 for 48 hours, and processed as above. qPCR results were normalized to 18S rRNA. N = 4 independent experiments. (E) Primary nasal epithelial cells (NECs) were knocked down for ORMDL3 and then infected with RV-A16 for 48 hours. RNA was processed for qPCR. RV-A16 RNA was normalized to C-Si samples (set = 1). The experiment was repeated with four individual donors. (F) Primary bronchial epithelial cells from one donor (BEC-53) were knocked down for ORMDL3, infected with RV-A16 for 48 hours, and processed and normalized as in E. N = 3 independent experiments using cells from this donor. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001.
ORMDL3 Knockdown Exerts Varied Effects on Different Strains of RV
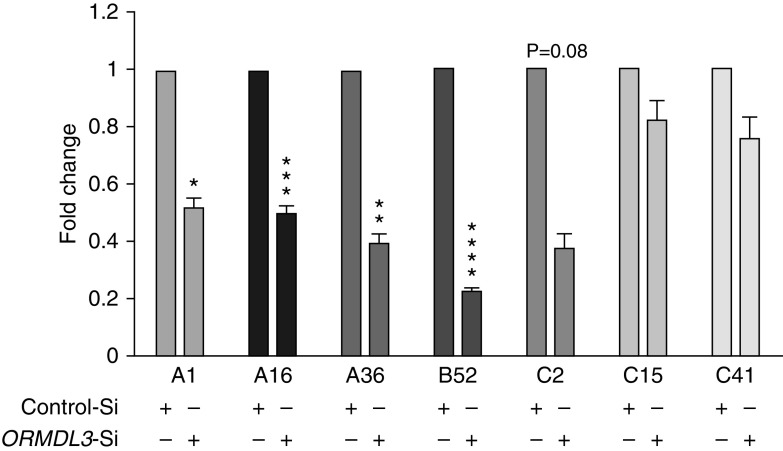
The three RV species (RV-A, RV-B, and RV-C) include more than 160 genotypes (30). RV uses three cellular membrane glycoproteins to gain entry into the host cell: ICAM-1 (intercellular adhesion molecule-1; the majority of RV-A and all RV-B), LDLR (low-density lipoprotein receptor) family members (RV-A “minor group” types), and CDHR3 (cadherin-related family member 3) (RV-C) (31). RV-A and RV-C viruses have primarily been implicated as triggers of asthma exacerbations (31). To directly compare RV with different receptor requirements in the same population of cells, we used HeLa-E8 cells expressing ICAM-1 and engineered to express CDHR3 (32). HeLa-E8 cells were transfected with ORMDL3 or control siRNA, then assessed for replication of RV-A1 (minor group, binds LDLR), RV-A16, RV-A36 (both major group), RV-B52, RV-C2, RV-C15, and RV-C41. Interestingly, ORMDL3 siRNA significantly inhibited all three RV-A strains and a representative RV-B strain. In contrast to RV-A and RV-B, only the RV-C2 showed a trend for inhibition (>60% inhibition) by ORMDL3 siRNA, whereas replication of the other two strains, RV-C15 and RV-C41, were not significantly impaired by ORMDL3 knockdown (Figure 3). The failure of ORMDL3 siRNA to impact specific RV-C strains was not related to inefficient ORMDL3 knockdown during RV-C infection or overwhelming replication by RV-C, as the species-specific effect of ORMDL3 siRNA was observed even for viruses with similar levels of replication (e.g., RV-A36 and RV-C15; Figure E5). These results suggest that ORMDL3 plays a different role in supporting RV-A versus specific RV-C infections, with more consistent inhibition of RV-A species by ORMDL3 knockdown.
Figure 3.
Different effects of ORMDL3 knockdown on replication of various RV strains. HeLa-E8 cells were transfected with ORMDL3 or control siRNA and infected with three strains of RV-A (1, 16, and 35), RV-B52, and three strains of RV-C (2, 15, and 40). Forty-eight hours later, RNA was harvested and quantitated for RV by qPCR. Levels of RV in the ORMDL3 knockdown cells were normalized to control siRNA-transfected cells (control SiRNA set = 1; N = 4 experiments). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001.
A recent report showed that ORMDL3 knockdown inhibited ICAM-1 upregulation in IL-1β–stimulated A549 respiratory epithelial cells (33). Downregulation of the surface receptor for RV-A could explain differences in effects on RV-A major group and RV-B versus RV-C replication. However, flow cytometry of siRNA-treated uninfected and RV-infected HeLa cells did not reveal any differences in surface ICAM-1 expression (Figure 4A). Therefore, ORMDL3 siRNA did not appear to be acting via alterations in RV surface receptor levels. Furthermore, ORMDL3 knockdown did not impair RV-A16 internalization 2 hours after infection (Figure 4B). Indeed, ORMDL3 knockdown slightly increased RV-A16 uptake. Together, these data suggest ORMDL3 knockdown impacts RV replication later in the viral life cycle, after cellular entry.
Figure 4.
ORMDL3 knockdown does not affect ICAM-1 expression or decrease initial RV-A16 internalization. (A) HeLa-E8 cells were uninfected or infected with RV-A16 in triplicate for 48 hours and then stained with anti–ICAM-1 or no antibody for analysis by flow cytometry. Top panel shows representative histograms (lines: uninfected; gray: RV infected). Bottom panel: Open bars are uninfected cells and solid bars are RV-16A–infected cells. gMFI = geometric mean fluorescence intensity. Histograms are representative of N = 3. (B) HeLa-E8 cells were infected with RV-A16 for 2 hours, washed, lysed, and RNA processed for qPCR detection of RV RNA. Bars are means ± SEM from N = 3 experiments. *P ≤ 0.05.
ER Stress Inhibits RV Replication
To determine the mechanism underlying the inhibition of RV-A replication, we investigated two of the major outcomes of ORMDL3 activity, ER stress (via SERCA pump inhibition and ER calcium perturbation) and SPT inhibition (Figure E1). The effect of UPR induction on RV replication is not well described, although there is one report of suppression in the setting of cystic fibrosis epithelial cells (34). To directly assess the effects of ER stress on RV replication, HeLa cells were pretreated with the UPR inducer tunicamycin (Tm; Figure 5A) or the UPR inhibitor tauroursodeoxycholic acid (TUDCA; Figure 5B) before infection with RV-A16 and RV-C15 strains. Increased ER stress significantly inhibited RV-A, but less so RV-C, replication (P = 0.11 for 1 μg/ml). The converse, UPR inhibition with TUDCA, augmented RV-A replication greater than twofold but increased RV-C by <30%. We confirmed the suppressive effects of Tm and enhancement by TUDCA of RV-A16 replication in primary bronchial epithelial cells (Figure E6). We have previously linked ER stress with augmented IFN-β production. Indeed, Tm was sufficient to induce abundant IFNB1 mRNA expression in HeLa cells. Tm pretreatment augmented RV-induced IFNB1, whereas TUDCA had the opposite effect (Figure 5C). These data raised the possibility that the process of knocking down ORMDL3 was paradoxically increasing ER stress. To test this idea, we first confirmed that the transfection reagent alone did not upregulate BiP/HSPA5 or CHOP/DDIT3 mRNA (not shown). To determine how ORMDL3 siRNA affected type I IFN and ER stress, IFN-β and UPR target genes were evaluated in knockdown cells infected with RV (Figure 5D). In the ORMDL3 siRNA knockdown cells, RV-A16 infection resulted in lower levels of IFN-β and UPR target gene induction, correlating with the decreased detectable RV. Together, these data suggested that ORMDL3 knockdown was not inhibiting RV-A replication by increasing type I IFN or ER stress.
Figure 5.
UPR induced by low-dose tunicamycin (Tm) inhibits RV replication. (A) HeLa-E8 cells were pretreated with vehicle or 0.5 μg/ml or 1 μg/ml Tm 1 hour before 48-hour infection with RV-A16 (black bars) or RV-C15 (gray bars). qPCR results were normalized to vehicle-treated controls. N = 5 for RV-A infections and N = 3 for RV-C. (B) HeLa-E8 cells were pretreated with 0.5 mg/ml or 1 mg/ml tauroursodeoxycholic acid (TUDCA) 1 hour before infection and analyzed as in A; N = 4. (C) Samples from A and B were assessed for IFN-β (IFNB1) by qPCR; N = 3. (D) HeLa-E8 cells transfected with control (C-Si) or ORMDL3 siRNA (ORMDL3i) were infected with RV-A16 for 48 hours before RNA harvest. IFN-β (IFNB1) and UPR target genes (HSPA5/BiP, DDIT3/CHOP) were assessed by qPCR with normalization to 18S rRNA. N = 3 experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001.
SPT Inhibition Enhances RV-A16 Replication
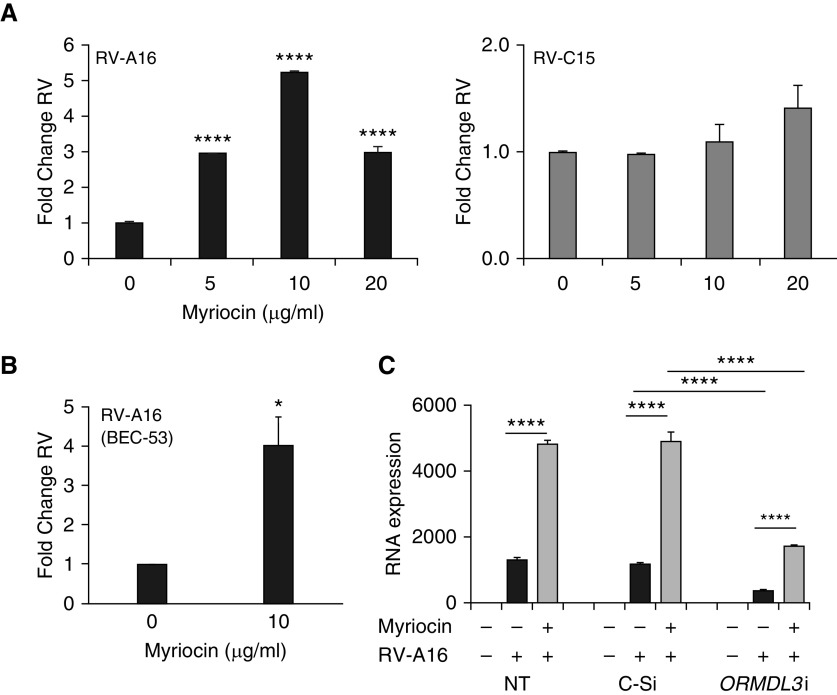
In light of these results, we next focused on the other major activity of ORMDL3, SPT inhibition. Overexpressing ORMDL3 induces ER stress (22, 35); therefore, it would be difficult to tease apart conflicting effects of ER stress and SPT inhibition using ORMDL3 transfection. Myriocin is a highly “specific” SPT inhibitor (19) and hence inhibits de novo sphingolipid synthesis (36). In this way, it acts as an ORMDL3 mimic without inducing ER stress (data not shown). HeLa-E8 cells were treated with different concentrations (5, 10, and 20 μg/ml) of myriocin for 3 hours and then infected with RV-A16 or RV-C15 (Figure 6). Myriocin at 5 μg/ml, and even more so at 10 μg/ml, significantly increased RV-A16 replication but not RV-C15 replication (Figure 6A). Myriocin also increased RV-A16 replication in primary bronchial epithelial cells (Figures 6B and E6). Furthermore, myriocin was able to restore RV-A replication suppressed by ORMDL3 siRNA (Figure 6C). Together, these data suggested the effect of ORMDL3 knockdown on RV-A replication is mediated through increased SPT activity.
Figure 6.
Myriocin, an SPT inhibitor, enhances RV-A replication and counteracts ORMDL3 knockdown. HeLa-E8 cells were treated with different concentrations of myriocin for 3 hours and then infected with RV-A16 (black bars) or RV-C15 (gray bars) for 48 hours. RV RNA levels were quantified by qPCR, with normalization to vehicle control (N = 5). (B) Primary bronchial epithelial cells (BEC-53) were treated with 10 μg/ml myriocin before infection with RV-A16 for 48 hours. RV levels were quantified by qPCR (N = 3). (C) HeLa-E8 cells were not transfected or were transfected with control or ORMDL3 siRNA the day before myriocin treatment (10 μg/ml) and RV infection. Black bars are RV only; gray bars are myriocin + RV. Samples were processed for RNA, which was quantitated by qPCR with normalization to 18S rRNA (N = 7). *P ≤ 0.05 and ****P ≤ 0.001.
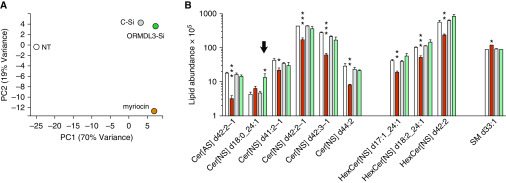
In the murine ORMDL3 transgenic model, ORMDL3 overexpression exerted specific effects on individual circulating ceramide species (37). We hypothesized that certain ORMDL3-regulated sphingolipid species might be detrimental to RV replication. To define the effect of myriocin and ORMDL3 siRNA on the sphingolipid milieu in our experimental system, we took a discovery lipidomics approach (Figures 7 and E7). HeLa cells were untreated, treated with myriocin, or transfected with control or ORMDL3 siRNA. We identified 453 distinct lipid species. Average interreplicate coefficient of variation was 22.6%. Perhaps not unexpectedly, transfection of control siRNA with the cationic lipid-based transfection reagent profoundly affected the lipid status of the cell, making this an essential comparator for the specific effects of ORMDL3 siRNA. The diverse lipid perturbations from these treatments are evident in the principal components analysis (Figure 7A). In assessing overall effects of the treatments on sphingolipids (Figure E7), about one-third were downregulated by myriocin and one-third upregulated by all three treatments compared with no treatment. However, myriocin and ORMDL3 siRNA disparately affected specific species of sphingolipids, particularly among the hexosylceramides (bar in Figure 7B) and one of the ceramide species (arrows in Figures 7B and E7). The sphingolipid species with the greatest increase during ORMDL3 knockdown was Cer(4) d18:0_24:1 (∼threefold; P = 0.037; Figure 7B). Spingosine-1 phosphate was not identified in this unbiased lipidomics analysis.
Figure 7.
Lipidomics analysis of HeLa myriocin treatments and siRNA transfections. HeLa cells were treated in triplicate with 10 μg/ml myriocin or transfected with control or ORMDL3 siRNA for 72 hours. Cells were pelleted and processed for analysis by lipid mass spectrometry. (A) Principal components analysis of the means for the different treatment groups. (B) Sphingolipids displaying at least one significant difference between a treatment group (myriocin or ORMDL3 siRNA) and the untreated controls when adjusted for false discovery rate (q-values designated with asterisks) and at least a 0.7-fold change. The eight ceramide species significantly impacted by control siRNA alone are not shown. Note the log scale. Arrow: ceramide species significantly increased by ORMDL3 siRNA. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.005. Cer[AS] = ceramide species α-hydroxy fatty acid sphingosine; Cer[NS] = ceramide species non-hydroxy fatty acid sphingosine; HexCer[NS] = hexosylceramide non-hydroxy fatty acid sphingosine species; PC = principal compontent; SM = sphingomyelin.
Discussion
In this study, we sought to gain a greater understanding of the interaction between ORMDL3 expression and RV infection in human epithelial cells. ORMDL3 knockdown reproducibly suppressed RV-A replication in HeLa cells, primary nasal and bronchial epithelium and airway epithelial cell lines. RV-B and RV-A major and minor group strains were comparably affected, despite having different surface receptors for attachment (ICAM-1 and LDLR). Indeed, surface staining of the ICAM-1 receptor and internalization studies suggest ORMDL3 expression is important later on in the viral life cycle. Compared with the RV-A viruses, ORMDL3 siRNA decreased only one of the three RV-C strains tested (RV-C2). C15 and C41 strains are closer phylogenetically, with C2 belonging to a different clade (32, 38, 39); however, the reason for the difference in effect of ORMDL3 knockdown is not clear. The results from pharmacologic manipulation of ER stress and use of the SPT inhibitor myriocin suggest the SPT inhibition activity of ORMDL3 is more likely to play a role in supporting RV replication than its effects on UPR induction.
The decrease in UPR and IFN genes observed with ORMDL3 siRNA may reflect both direct contributions from decreased ORMDL3 expression as well as indirect effects related to reduced RV-A replication. Selectively inhibiting the UPR with TUDCA enhanced RV-A replication. Interestingly, UPR inhibition did not significantly impact RV-C15, suggesting RV-A may be more sensitive to infection-associated ER stress. Pharmacologically induced ER stress negatively affected replication of both RV-A and less so RV-C viruses. This effect may be mediated by the increased type I IFN observed in these treatments. Our results are consistent with a report in cystic fibrosis epithelial cells, in which pharmacologically enhanced UPR suppressed RV replication (34). Our results with exogenous tunicamycin are also consistent with the implications of a previous report examining the interaction between ORMDL3 and RV infection in a mouse model. Song and colleagues reported decreased RV viral load in the ORMDL3 overexpressing transgenic mouse model (11). ORMDL3 overexpression has also been shown by this group to cause induction of a UPR signaling pathway (ATF6) and increased expression of CXCL10 in vitro and in vivo (35). Infection of these ORMDL3 transgenic mice resulted in much greater type I IFN production and IFN-response genes, likely causing the decrease in RV load. It should be noted, however, that RV does not replicate well in any wild-type or transgenic mice. What our study highlights is that the converse does not necessarily hold: in our study, decreased ORMDL3 did not augment RV replication.
The results of UPR manipulation in this study suggested ORMDL3 knockdown must influence RV through a mechanism other than ER stress modulation, namely altered SPT activity. In support of this idea, the greater impact of ORMDL3 knockdown on RV-A versus RV-C viruses was consistent with the observation that the SPT inhibitor myriocin augmented RV-A16, but not RV-C15, replication. The capacity of myriocin to counteract ORMDL3 siRNA also supports the idea that the RV-suppressing effect of ORMDL3 knockdown is related to decreased SPT inhibition (myriocin achieves enzymatic “epistasis”). The role of ORMDL3 in regulating SPT has been contentious in the literature. There have been reports that ORMDL3 knockdown alone is insufficient to impact SPT (40). However, other studies have shown an effect of ORMDL3 on specific sphingolipid entities: ORMDL3−/− epithelial cells had an increase in sphingosine-1-phosphate, a product of SPT activity, and ORMDL3 transgenic mice had decreased sphinganine and ceramide 24:0 in serum (37, 41). In a study by Oyeniran and colleagues (12), ORMDL3 siRNA increased C:24 ceramide species. In liver cells, ORMDL3 siRNA increased dihydrosphingosine and multiple dihydroceramide species. Thus, the effect of decreased ORMDL3 may be specific to different cell types and degree of gene expression change (knockdown vs. knockout, timing, and compensatory mechanisms). In our experiments in HeLa cells, ORMDL3 knockdown exerted a limited effect on the sphingolipid pool, significantly increasing Cer(5) d18:0_24:1 (∼threefold; Figure 7) and less significantly Cer[NS](5) d 18:0_16:0 (twofold; uncorrected P < 0.05). Decreases in multiple ceramide and hexosylceramide species on myriocin treatment are consistent with expectations for an inhibitor of de novo sphingolipid synthesis. However, one sphingomyelin species paradoxically increased with myriocin treatment (SM 33:1), perhaps reflecting compensatory salvage pathways.
Sphingolipids regulate multiple steps in viral replication, from receptor binding and endocytosis to replication and budding of new particles (42). Rhinoviruses enter cells via ceramide and GM1-rich lipid rafts (43). Acid sphingomyelinase favors rhinovirus replication by generating ceramides (44). However, too much ceramide may adversely affect membrane fluidity. One way to interpret our results is that a decrease in overall ceramide or hexosylceramide levels is beneficial to RV-A replication, and elevation of particular ceramides, those possessing 18 or 24 carbon acyl chains, could be problematic for replication. This idea is also consistent with a report on host lipid changes resulting from RV infection, as assessed via lipidomics (45). In that study, Ceranib 1, a ceramidase inhibitor (decreases sphingosine and increases ceramides) markedly suppressed RV-1A replication. This report also identified a marked decrease in sphingosine-1-phosphate synthesis during RV infection, again suggesting decreased SPT activity may favor RV replication.
One would not expect the consequences of SPT modulation to be isolated to immediate lipid metabolites in the sphingolipid pathway, because sphingolipid synthesis is part of a greater interacting network of lipid regulation. Therefore, some of the effects of myriocin versus ORMDL3 siRNA treatment may reflect changes in other lipid species. Formation of RV replication domains at the ER-Golgi interface requires lipid remodeling through phosphatidylinositol 4-phosphate (PI4P)–cholesterol “counter currents” (26). Thus, generation of specific phosphatidylinositol substrates could aid or hamper this process. The lipidomic study from Nguyen and colleagues identified increases in shorter chain phosphoinositols (C32 and C34), and myriocin treatment also increased these species (Figure E8) (45). ORMDL3 siRNA also increased 16 chain species of phosphoglycerides, but the significance of these findings is not yet clear. Finally, RV-A but not RV-C binds short 12-carbon fatty acid–like molecules known as “pocket factor” (46). Although the role of pocket factor in replication is somewhat contentious, different effects of ORMDL3 versus myriocin on the generation of pocket factor could affect RV-A over RV-C. Future studies will investigate which aspect of the RV life cycle is impacted by these opposing manipulations.
The goal of this study was to determine how differences in ORMDL3 expression regulate RV infection. The data presented here identify a previously unknown role for ORMDL3 and its target SPT in regulating RV-A replication in human epithelial cells. Putting these data into context with previous reports (Figure E9), optimal RV replication may require a balance between helpful sphingolipid pool modulation by ORMDL3 and the predilection of ORMDL3 to cause ER stress. When ORMDL3 expression is low, the negative impact of specific ceramide generation may outweigh the benefit of decreased ER stress. In the setting of high ORMDL3 expression, increased ER stress and type I IFN production may predominate (11). Regarding asthma susceptibility, our results suggest decreased ORMDL3 expression (seen in children less likely to develop asthma [3]) may be protective by limiting the extent of RV replication in epithelial cells. On the other hand, increased ORMDL3 may exacerbate illness through UPR-augmented cytokine responses to viral infection (22). It will be interesting to see how these opposing roles for ORMDL3 in RV infection play out in human subjects.
Supplementary Material
Footnotes
Supported by U.S. National Institutes of Health (NIH)-U.S. National Institute of Allergy and Infectious Diseases grant R21AI121808-01; NIH–U.S. National Heart, Lung, and Blood Institute grant P01HL07083; and NIH-P41 grant GM108538.
Author Contributions: Y.L. performed experiments, analyzed data, wrote the initial draft, put together figures, and participated in the draft revisions. Y.A.B. made critical reagents, provided experimental design input, and edited the manuscript. J.C.E. performed all the statistical analysis outside of the lipidomics. T.H. performed experiments and read and approved the manuscript. N.A.Z. performed an experiment, drafted the related figure, wrote the related methods and legend, and read the manuscript. J.W.T. and C.L. performed select experiments and read and approved the manuscript. P.S.F. provided primary cells for a figure, participated in the experimental troubleshooting, and edited and approved the manuscript. T.R.R. performed the lipidomics experiment, wrote associated methods, and read the manuscript. K.A.O. provided figures for the lipidomics experiment and edited associated methods, results, and figure legends. J.E.G. provided funding for N.A.Z., provided intellectual contribution to the related experiment, and approved the manuscript. J.C. provided funding for lipidomics, supervision of T.R.R. and K.A.O., and intellectual input into lipidomics and approved the manuscript. S.K.M. provided funding and supervision of P.S.F., provided intellectual input into the related experiment, and edited the manuscript. J.E.G. provided funding for the project, critical intellectual input in the experimental design, and troubleshooting and data interpretation and edited the manuscript. J.A.S. provided funding for project, project conception, design, trouble shooting, and data analysis and wrote the manuscript and edited the figures.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0237OC on February 20, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Sullivan PW, Ghushchyan V, Navaratnam P, Friedman HS, Kavati A, Ortiz B, et al. The national cost of asthma among school-aged children in the United States. Ann Allergy Asthma Immunol. 2017;119:246–252, e1. doi: 10.1016/j.anai.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Coleman L, Laing IA, Bosco A. Rhinovirus-induced asthma exacerbations and risk populations. Curr Opin Allergy Clin Immunol. 2016;16:179–185. doi: 10.1097/ACI.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 3.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyvärinen MK, Kotaniemi-Syrjänen A, Reijonen TM, Korhonen K, Korppi MO. Teenage asthma after severe early childhood wheezing: an 11-year prospective follow-up. Pediatr Pulmonol. 2005;40:316–323. doi: 10.1002/ppul.20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Midulla F, Pierangeli A, Cangiano G, Bonci E, Salvadei S, Scagnolari C, et al. Rhinovirus bronchiolitis and recurrent wheezing: 1-year follow-up. Eur Respir J. 2012;39:396–402. doi: 10.1183/09031936.00188210. [DOI] [PubMed] [Google Scholar]
- 6.Ono JG, Worgall TS, Worgall S. 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr Res. 2014;75:165–170. doi: 10.1038/pr.2013.186. [DOI] [PubMed] [Google Scholar]
- 7.Zhao CN, Fan Y, Huang JJ, Zhang HX, Gao T, Wang C, et al. The association of GSDMB and ORMDL3 gene polymorphisms with asthma: a meta-analysis. Allergy Asthma Immunol Res. 2015;7:175–185. doi: 10.4168/aair.2015.7.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das S, Miller M, Broide DH. Chromosome 17q21 genes ORMDL3 and GSDMB in asthma and immune diseases. Adv Immunol. 2017;135:1–52. doi: 10.1016/bs.ai.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 10.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192:3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song DJ, Miller M, Beppu A, Rosenthal P, Das S, Karta M, et al. Rhinovirus infection of ORMDL3 transgenic mice is associated with reduced rhinovirus viral load and airway inflammation. J Immunol. 2017;199:2215–2224. doi: 10.4049/jimmunol.1601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyeniran C, Sturgill JL, Hait NC, Huang WC, Avni D, Maceyka M, et al. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J Allergy Clin Immunol. 2015;136:1035–1046, e6. doi: 10.1016/j.jaci.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loser S, Gregory LG, Zhang Y, Schaefer K, Walker SA, Buckley J, et al. Pulmonary ORMDL3 is critical for induction of alternaria-induced allergic airways disease. J Allergy Clin Immunol. 2017;139:1496–1507, e3. doi: 10.1016/j.jaci.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siow D, Sunkara M, Dunn TM, Morris AJ, Wattenberg B. ORMDL/serine palmitoyltransferase stoichiometry determines effects of ORMDL3 expression on sphingolipid biosynthesis. J Lipid Res. 2015;56:898–908. doi: 10.1194/jlr.M057539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Miller M, Unno H, Rosenthal P, Sanderson MJ, Broide DH. Orosomucoid-like 3 (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca2+ oscillations in asthma. J Allergy Clin Immunol. 2018;142:207–208, e6. doi: 10.1016/j.jaci.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs J, Agellon LB, Michalak M. Ca(2+) homeostasis and endoplasmic reticulum (ER) stress: an integrated view of calcium signaling. Biochem Biophys Res Commun. 2015;460:114–121. doi: 10.1016/j.bbrc.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 18.Ono JG, Worgall TS, Worgall S. Airway reactivity and sphingolipids-implications for childhood asthma. Mol Cell Pediatr. 2015;2:13. doi: 10.1186/s40348-015-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, et al. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5:186ra67. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 20.Mahn K, Hirst SJ, Ying S, Holt MR, Lavender P, Ojo OO, et al. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci USA. 2009;106:10775–10780. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unno H, Miller M, Rosenthal P, Beppu A, Das S, Broide DH. ATF6α regulates airway hyperreactivity, smooth muscle proliferation, and contractility. J Allergy Clin Immunol. 2018;141:439–442, e4. doi: 10.1016/j.jaci.2017.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YP, Rajamanikham V, Baron M, Patel S, Mathur SK, Schwantes EA, et al. Association of ORMDL3 with rhinovirus-induced endoplasmic reticulum stress and type I Interferon responses in human leucocytes. Clin Exp Allergy. 2017;47:371–382. doi: 10.1111/cea.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosser AG, Brockman-Schneider R, Amineva S, Burchell L, Sedgwick JB, Busse WW, et al. Similar frequency of rhinovirus-infectible cells in upper and lower airway epithelium. J Infect Dis. 2002;185:734–743. doi: 10.1086/339339. [DOI] [PubMed] [Google Scholar]
- 24.Amineva SP, Aminev AG, Gern JE, Palmenberg AC. Comparison of rhinovirus A infection in human primary epithelial and HeLa cells. J Gen Virol. 2011;92:2549–2557. doi: 10.1099/vir.0.031302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roulin PS, Lötzerich M, Torta F, Tanner LB, van Kuppeveld FJ, Wenk MR, et al. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 28.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 29.Schroth MK, Grimm E, Frindt P, Galagan DM, Konno SI, Love R, et al. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol. 1999;20:1220–1228. doi: 10.1165/ajrcmb.20.6.3261. [DOI] [PubMed] [Google Scholar]
- 30.Basnet S, Palmenberg AC, Gern JE. Rhinoviruses and their receptors. Chest. 2019;155:1018–1025. doi: 10.1016/j.chest.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bochkov YA, Gern JE. Rhinoviruses and their receptors: implications for allergic disease. Curr Allergy Asthma Rep. 2016;16:30. doi: 10.1007/s11882-016-0608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Willis-Owen SAG, Spiegel S, Lloyd CM, Moffatt MF, Cookson WOCM. The ORMDL3 asthma gene regulates ICAM1 and has multiple effects on cellular inflammation. Am J Respir Crit Care Med. 2019;199:478–488. doi: 10.1164/rccm.201803-0438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schögler A, Caliaro O, Brügger M, Oliveira Esteves BI, Nita I, Gazdhar A, et al. Modulation of the unfolded protein response pathway as an antiviral approach in airway epithelial cells. Antiviral Res. 2019;162:44–50. doi: 10.1016/j.antiviral.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA. 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worgall TS. Sphingolipids, ORMDL3 and asthma: what is the evidence? Curr Opin Clin Nutr Metab Care. 2017;20:99–103. doi: 10.1097/MCO.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 37.Miller M, Tam AB, Mueller JL, Rosenthal P, Beppu A, Gordillo R, et al. Cutting edge: targeting epithelial ORMDL3 increases, rather than reduces, airway responsiveness and is associated with increased sphingosine-1-phosphate. J Immunol. 2017;198:3017–3022. doi: 10.4049/jimmunol.1601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmenberg AC, Gern JE. Classification and evolution of human rhinoviruses. Methods Mol Biol. 2015;1221:1–10. doi: 10.1007/978-1-4939-1571-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, et al. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134:332–341. doi: 10.1016/j.jaci.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhakupova A, Debeuf N, Krols M, Toussaint W, Vanhoutte L, Alecu I, et al. ORMDL3 expression levels have no influence on the activity of serine palmitoyltransferase. FASEB J. 2016;30:4289–4300. doi: 10.1096/fj.201600639R. [DOI] [PubMed] [Google Scholar]
- 41.Miller M, Rosenthal P, Beppu A, Gordillo R, Broide DH. Oroscomucoid like protein 3 (ORMDL3) transgenic mice have reduced levels of sphingolipids including sphingosine-1-phosphate and ceramide. J Allergy Clin Immunol. 2017;139:1373–1376, e4. doi: 10.1016/j.jaci.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waheed AA, Freed EO. The role of lipids in retrovirus replication. Viruses. 2010;2:1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi T, Suzuki T. Function of membrane rafts in viral lifecycles and host cellular response. Biochem Res Int. 2011;2011:245090. doi: 10.1155/2011/245090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grassmé H, Riehle A, Wilker B, Gulbins E. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. J Biol Chem. 2005;280:26256–26262. doi: 10.1074/jbc.M500835200. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen A, Guedán A, Mousnier A, Swieboda D, Zhang Q, Horkai D, et al. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J Lipid Res. 2018;59:1671–1684. doi: 10.1194/jlr.M085910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossmann MG. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.