Highlights
-
•
Darunavir showed no in vitro antiviral activity against SARS-CoV-2 (EC50 > 100 μM).
-
•
Remdesivir demonstrated potent antiviral activity, confirming validity of the assay.
-
•
Overall, the data do not support use of darunavir for treatment of COVID-19.
Keywords: Darunavir, SARS-CoV-2, COVID-19, In vitro
Abstract
Objectives
Given the high need and the absence of specific antivirals for treatment of COVID-19 (the disease caused by severe acute respiratory syndrome-associated coronavirus-2 [SARS-CoV-2]), human immunodeficiency virus (HIV) protease inhibitors are being considered as therapeutic alternatives.
Methods
Prezcobix/Rezolsta is a fixed-dose combination of 800 mg of the HIV protease inhibitor darunavir (DRV) and 150 mg cobicistat, a CYP3A4 inhibitor, which is indicated in combination with other antiretroviral agents for the treatment of HIV infection. There are currently no definitive data on the safety and efficacy of DRV/cobicistat for the treatment of COVID-19. The in vitro antiviral activity of darunavir against a clinical isolate from a patient infected with SARS-CoV-2 was assessed.
Results
DRV showed no antiviral activity against SARS-CoV-2 at clinically relevant concentrations (EC50 > 100 μM). Remdesivir, used as a positive control, demonstrated potent antiviral activity (EC50 = 0.38 μM).
Conclusions
Overall, the data do not support the use of DRV for the treatment of COVID-19.
Introduction
In December 2019, the severe acute respiratory syndrome-associated coronavirus-2 (SARS-CoV-2) disease (COVID-19) emerged in Wuhan, Hubei Province, China (Zhu et al., 2020). The virus was subsequently identified as a coronavirus (CoV), in addition to SARS-CoV-1 and Middle East respiratory syndrome CoV (MERS-CoV) that passed from animals to humans where it can cause severe respiratory illness (Lu et al., 2020). As of March 2020, COVID-19 has spread worldwide, with the WHO declaring a global pandemic (Hoehl et al., 2020). Given the extent of the COVID-19 pandemic, there is an urgent need to identify potential treatments for the disease as well as to develop a vaccine.
As no specific antivirals for treatment of COVID-19 are available, one avenue of clinical interest is the use of human immunodeficiency virus (HIV) protease inhibitors (PIs) as a therapeutic intervention. The potential for HIV PIs as a treatment for COVID-19 is mainly based on limited virologic and clinical data on the HIV protease inhibitor lopinavir with low-dose ritonavir (as a pharmaco-enhancer; LPV/r) in patients infected with the severe acute respiratory syndrome, related to a coronavirus (SARS-CoV) (Chu et al., 2004). After demonstrating the in vitro antiviral activity of LPV against SARS-CoV-1, the clinical response of patients with SARS to a combination of LPV/r and ribavirin was examined. Patients treated with LPV/r had lower rates of adverse clinical outcomes at day 21 following the onset of symptoms than historical controls (Chu et al., 2004). However, recent data in hospitalized adults with severe confirmed COVID-19 treated with LPV/r in addition to standard care of ventilation, oxygen, vasopressor support, antibiotics, and renal-replacement therapy showed that there was no significant improvement in time to clinical improvement or mortality at day 28 compared with the standard care (Cao et al., 2020).
The HIV PI darunavir with cobicistat as a pharmaco-enhancer (DRV/c, 800/150 mg given orally once daily with food) in combination with other antiretroviral agents is approved for both treatment-naïve and -experienced patients with HIV-1 infection (Prezcobix Prescribing Information, 2020, Rezolsta Summary of Product Characteristics, 2020). The efficacy and safety profile of boosted-DRV combination therapy is well-established in the HIV setting, based on phase III clinical studies as well as real-world evidence (Tashima et al., 2014, Orkin et al., 2013, Cahn et al., 2011, Navarro and Curran, 2016).
To date, no clear clinical evidence supports the use of DRV (boosted with either ritonavir or cobicistat) in viral diseases other than HIV.
In this paper, the antiviral activity of DRV against SARS-CoV-2 was investigated in an in vitro model at clinically relevant concentrations. When DRV/c is taken at the indicated once-daily dose, the median total trough plasma concentration of DRV was 3.4 μM (1875 ng/mL). (Prezcobix Prescribing Information, 2020). The cell culture assay was shown to be suitable for antiviral assays. Productive viral infection occurs in this model with the number of SARS-CoV RNA molecules increasing continuously after infection, indicating that the virus undergoes several replication cycles (Bojkova et al., 2020). Remdesivir (GS-5734), a nucleotide analog initially developed for Ebola virus disease, has been shown to inhibit SARS-CoV-2 replication in vitro with an EC50 equal to 0,770 μM (Wang et al., 2020) and was therefore used as a positive control.
Methods
Cell culture and virus preparation
Human colon carcinoma cell line (Caco-2) cells (obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) were cultured in Minimal Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS) and containing penicillin (100 IU/mL) and streptomycin (100 μg/mL) in a 5% CO2 atmosphere at 37 °C. All culture reagents were purchased from Sigma (Hamburg, Germany).
SARS-CoV-2 was isolated from human samples and cultured in Caco-2 cells, as previously described (Hoehl et al., 2020). After one passage in Caco-2 cells, viral stocks were stored at −80 °C prior to use.
Assessment of antiviral activity by inhibition of virus-induced cytopathogenic effect
Confluent layers of Caco-2 cells were cultured at 37 °C in a 5% CO2 atmosphere for 72 h on 96 multi-well plates (50,000 cells/well). Cells were challenged with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01. The virus was added together with the compounds under investigation and incubated in MEM supplemented with 1% FBS.
DRV and remdesivir were synthesized at Johnson & Johnson. To assess in vitro antiviral activity, DRV and remdesivir, diluted in MEM without FBS, were added in 4-fold dilutions to a concentration range of 0.02 μM to 100 μM. Cells were then incubated for 48 h before the cytopathogenic effect (CPE) was visually scored by two independent laboratory technicians. Evaluation of the CPE was also done using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma-Aldrich) method according to the manufacturer's instructions. Optical densities were measured at 560/620 nm in a Multiskan Reader MCC/340 Labsystems. Three independent experiments with triplicate measurements were performed. Evaluation of inhibition of CPE using the MTT method was done for two of the three experiments. Data were analyzed by a four-parameter curve-fitting from a dose-response curve using GraphPad Prism (version 7.00) to calculate the EC50 (concentration of the compound that inhibited 50% of the infection) based on visual CPE scoring or based on the MTT method.
Assessment of cell viability
To assess the effects of the compounds on Caco–2 cell viability, cell viability was measured in confluent cell layers treated with a range of compound concentrations in the absence of virus using the Rotitest Vital (Roth) according to manufacturer’s instructions, as previously described (Bojkova et al., 2020).
All assays were performed three times independently in triplicate. From this, the CC50 (cytotoxic concentration of the compound that reduced cell viability to 50%) was calculated from a dose-response curve in GraphPad Prism (version 7.00) using four-parameter curve-fitting.
Selectivity index
The selectivity index for each of the compounds was determined as the ratio of the CC50 to the EC50.
Modeling
In general, in silico docking can be a useful approach for identifying subsets of molecules for in vitro studies and can be used to explain in vitro observations on a structural level. The coordinates of the main SARS-CoV-2 protease's crystal structure were retrieved from the PDB database (https://www.rcsb.org/structure/6lu7).
Preparing DRV and the protein for in silico molecular docking was performed with the software package MOE (Molecular Operating Environment 2019.01; Chemical Computing Group ULC, Montreal, QC, Canada, H3A 2R7, 2019). The force field used was AMBER ETH:10 with default ‘quick prep’ settings to prepare the protein complex. The original ligand was then removed. General docking settings were then altered to have 50 initial placements.
Results
Assessment of in vitro antiviral activity, cytotoxicity, and selectivity
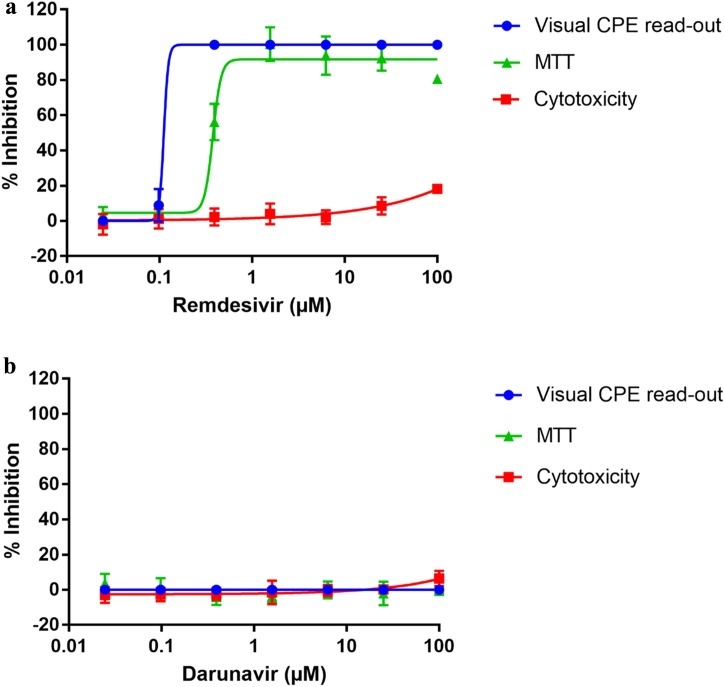
Remdesivir showed strong antiviral activity against SARS-CoV-2 with an EC50 of 0.11 μM based on visual scoring of the inhibition of CPE (Figure 1 a). In the same experiments, DRV did not show any inhibition of SARS-CoV-2 induced CPE (Figure 1b; EC50 >100 μM). Similar results were obtained using the MTT method. Remdesivir showed potent antiviral activity with an EC50 value of 0.38 μM (Figure 1a), while DRV showed no effect (EC50 >100 μM, Figure 1b). No cytotoxicity of remdesivir or DRV was observed on Caco-2 cells with CC50 values >100 μM. The selectivity index (CC50/EC50) for DRV could not be calculated (>100 μM/>100 μM for SARS-CoV-2) due to the lack of antiviral activity. In contrast, remdesivir had a selectivity index of >900 by visual CPE scoring and >260 by the MTT method, confirming a strong in vitro antiviral effect against SARS-CoV-2.
Figure 1.
Inhibition of SARS-CoV-2 in Caco-2 cells by cytopathogenic effect assay using a visual read-out (visual CPE read-out) and MTT assay (MTT) following the addition of a) remdesivir, or b) darunavir (DRV). Cytotoxicity data (measured on uninfected cells) are also shown. Mean percent inhibition for each read-out across three independent experiments with triplicate measurements are plotted (2 independent experiments for the MTT assay). The error bars represent the standard deviation. Note: for some points, the error bars are shorter than the height of the symbol.
(a) Visual CPE-read out EC50 = 0.11 μM; MTT EC50 = 0.38 μM; CC50 >100 μM; Selectivity index = >900 (Visual CPE read-out); >260 (MTT).
(b) Visual CPE read-out EC50 > 100 μM; MTT EC50 >100 μM; CC50 >100 μM.
Modeling
In silico docking of DRV in the crystal structure of the SARS-CoV-2 main protease (3CL protease) identified five docking poses. The docking scores ranged from S = −8.6 to −8.2, showing that DRV can fit into the pocket, but these values are indicative of suboptimal binding to this protein. Visual inspection of each of these poses showed very few interactions of DRV with the active site of the protease, and the catalytic cysteine residue was not directly targeted, unlike the many strong interactions observed for DRV bound to the HIV protease (King et al., 2004).
Discussion
Current efforts to manage the COVID-19 pandemic have primarily focused on improved hygiene, quarantine of infected individuals, social distancing to limit transmission, and development of a vaccine (Kruse, 2020). Despite the expedited efforts to develop a vaccine and collaborative efforts to screen compounds in discovery and development across the broader pharmaceutical industry for activity against COVID-19, patients are in immediate need of therapeutic interventions (Bojkova et al., 2020, Baden and Rubin, 2020).
Current data on the therapeutic effect of HIV protease inhibitors in patients with COVID-19 are far from comprehensive. This study demonstrated that DRV showed no in vitro antiviral activity against SARS-CoV-2 at clinically relevant concentrations. Furthermore, structural analyses using protease structures are consistent with these data. DRV binds to the active site of the HIV virus’ dimeric aspartic protease (King et al., 2004). The crystal structure of this protease is well-elucidated. It has been shown to have an extensive hydrogen-bonding network with DRV, allowing for the potent in vitro activity (EC50 values = 1.2 to 8.5 nM) of this protease inhibitor against HIV (Prezcobix Prescribing Information, 2020, Rezolsta Summary of Product Characteristics, 2020). In contrast, the SARS-CoV-2 main protease is a cysteine protease (Protein Data Bank-code 6LU7), and while several docking poses have been found for DRV in silico models, unlike in HIV, these poses showed little interaction with the SARS-CoV-2 main protease active site. Several publications describe in silico docking experiments on the main coronavirus protease that specifically focus on or include DRV (Chang et al., 2020; Haitao and Zihe, 2020; Sang et al., 2020; Wu et al., 2020; Farag et al., 2020). Although these studies suggest DRV as a candidate for further investigation, such promising docking results could not be reproduced in our in silico docking studies. Such discrepancies can often result from in silico docking, which is primarily a useful approach for identifying subsets of molecules for in vitro activity testing. No in vitro antiviral activity of DRV against SARS-CoV-2 was found in the experiments reported here.
In this study, remdesivir demonstrated activity against SARS-CoV-2 with an EC50 of 0.38 μM, which is in line with the earlier reported remdesivir EC50 of 0.77 μM, indicating that the in vitro antiviral assay used is appropriate to assess antiviral activity against SARS-CoV-2 (Bojkova et al., 2020).
In conclusion, the lack of in vitro antiviral activity of DRV against SARS-CoV-2 does not support the use of DRV for the treatment of COVID-19. Hence, DRV (boosted with either ritonavir or cobicistat) should remain solely for the treatment of patients with HIV infection.
Funding source
Funding for this study was provided by Janssen Pharmaceuticals.
Ethical approval
As this was an in vitro study, no ethical approval was required.
Data sharing
The data sharing policy of Janssen Pharmaceutical Companies of Johnson & Johnson is available at https://www.janssen.com/clinical-trials/transparency. As noted on this site, requests for access to the study data can be submitted through the Yale Open Data Access (YODA) Project site at http://yoda.yale.edu.
Conflict of interest
Sandra De Meyer, Christophe Buyck, Ellen Van Damme, Marnix Van Loock, and Brian Woodfall are employees and could be stock owners of Johnson and Johnson. Sandra Ciesek received research funding from Janssen for this research.
Acknowledgments
Medical writing support for the development of this manuscript was provided by Patrick Hoggard of Zoetic Science, an Ashfield company, part of UDG Healthcare plc; this support was funded by Janssen Pharmaceuticals. We thank Lena Stegmann for technical support by antiviral assays.
References
- Baden L.R., Rubin E.J. Covid-19 — the search for effective therapy. N Engl J Med. 2020;382(19):1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S. SARS-CoV-2 infected host cell proteomics reveal potential therapy targets, [Under review] Nature. 2020 doi: 10.1038/s41586-020-2332-7. https://www.researchsquare.com/article/rs-17218/v1 Preprint available at: [DOI] [PubMed] [Google Scholar]
- Cahn P., Fourie J., Grinsztejn B., Hodder S., Molina J.M., Ruxrungtham K. Week 48 analysis of once-daily vs. twice-daily darunavir/ritonavir in treatment-experienced HIV-1-infected patients. AIDS. 2011;25(7):929–939. doi: 10.1097/QAD.0b013e328345ee95. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-C, Tung Y-A, Lee K-H, Chen T-F, Hsiao Y-C, Chang H-C, et al. Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints (www.preprints.org) 2020. 10.20944/preprints202002.0242.v1. [DOI]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag A., Wang P., Ahmed M., Sadek H. 2020. Identification of FDA approved drugs targeting COVID-19 virus by structure-based drug repositioning.https://chemrxiv.org/articles/Identification_of_FDA_Approved_Drugs_Targeting_COVID-19_Virus_by_Structure-Based_Drug_Repositioning/12003930/1 [Google Scholar]
- Haitao Y, Zihe R. 2020. Available at Shanghai Institute of Materia Medica. http://www.simm.ac.cn/xwzx/kydt/202001/t20200125_5494417.html.
- Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020;382(13):1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N.M., Prabu-Jeyabalan M., Nalivaika E.A., Wigerinck P., de Béthune M.P., Schiffer C.A. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J Virol. 2004;78(21):12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Research. 2020;9(January):72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Juan L., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J., Curran A. Profile of once-daily darunavir/cobicistat fixed-dose combination for the treatment of HIV/AIDS. HIV AIDS (Auckl) 2016;8:175–182. doi: 10.2147/HIV.S56158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin C., DeJesus E., Khanlou H., Stoehr A., Supparatpinyo K., Lathouwers E. Final 192-week efficacy and safety of once-daily darunavir/ritonavir compared with lopinavir/ritonavir in HIV-1-infected treatment-naïve patients in the ARTEMIS trial. HIV Med. 2013;14(1):49–59. doi: 10.1111/j.1468-1293.2012. [DOI] [PubMed] [Google Scholar]
- Prezcobix prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/205395s001lbl.pdf. [Accessed 27 March 2020].
- Rezolsta Summary of product characteristics. https://www.ema.europa.eu/en/documents/product-information/rezolsta-epar-product-information_en.pdf. [Accessed 27 March 2020].
- Sang P., Tian S., Meng Z., Yang L. Insight derived from molecular docking and molecular dynamics simulations into the binding interactions between HIV-1 protease inhibitors and SARS-CoV-2 3CLpro. ChemRxiv. 2020 https://chemrxiv.org/articles/Insight_Derived_from_Molecular_Docking_and_Molecular_Dynamics_Simulations_into_the_Binding_Interactions_Between_HIV-1_Protease_Inhibitors_and_SARS-CoV-2_3CLpro/11932995/1 Preprint. [Google Scholar]
- Tashima K., Crofoot G., Tomaka F.L., Kakuda T.N., Brochot A., Van de Casteele T. Cobicistat-boosted darunavir in HIV-1-infected adults: week 48 results of a Phase IIIb, open-label single-arm trial. AIDS Res Ther. 2014;11:39. doi: 10.1186/1742-6405-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]