Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new viral disease that has gained global attention owing to its ability to provoke community and health-care-associated outbreaks of severe infections in human populations. The virus poses serious challenges to clinical management because there are still no approved anti- SARS-CoV-2 drugs available. In this mini-review, we summarize the much updated published reports that demonstrate the mechanism of SARS-CoV-2 entry into host cells, and discuss the availability and development of attractive host-based therapeutic options for SARS-CoV-2 infections.
Keywords: SARS-CoV-2, ACE2, Entry, Proteases, Drugs
Highlights
-
•
Interplay between virus spike protein, ACE2 and host proteases in mediating entry of SARS-CoV-2 in host cells.
-
•
Association between ACE2 expression, comorbidities and severity of SARS-CoV-2 infection.
-
•
Inhibitors of endosomal acidification, TMPRSS and cathepsin proteases are candidate therapeutics of SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, also known as Coronavirus disease 2019 (COVID-19) [1], is a current pandemic viral disease that was emerged in Wuhan, China in 2019, and then has spread to nearly all of the world’s countries and territories. More than 4.5 million cases of COVID-19 have been confirmed as of 20 May, according to data compiled by researchers at Johns Hopkins University in Baltimore, USA (https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6). The particularly high infectious capacity of the virus, along with mortality rates ranging from 1% to above 5% [2,3], has caused a great deal of concerns around the world.
The infectious disease is caused by the newly discovered corona virus, SARS-CoV-2 [4]. Remarkably, the genome of SARS-CoV-2 shares ∼80% sequence homology with that of SARS-CoV, a corona virus that caused a large scale pandemic infection before 18 years ago [5], and is ∼96% identical to the bat coronavirus BatCoV RaTG13 [4]. Interestingly, it seems that both of the SARS-CoV-2 and SARS-CoV are using similar mechanism of host cell entry and that through engagement with host angiotensin converting enzyme II (ACE2) which is located on the surface of host cells, and is abundantly present in humans in the epithelia of the lung and small intestine [4,6].
2. SARS-CoV-2 entry in host cells, the essential roles of virus spike protein, ACE2 and the host proteases
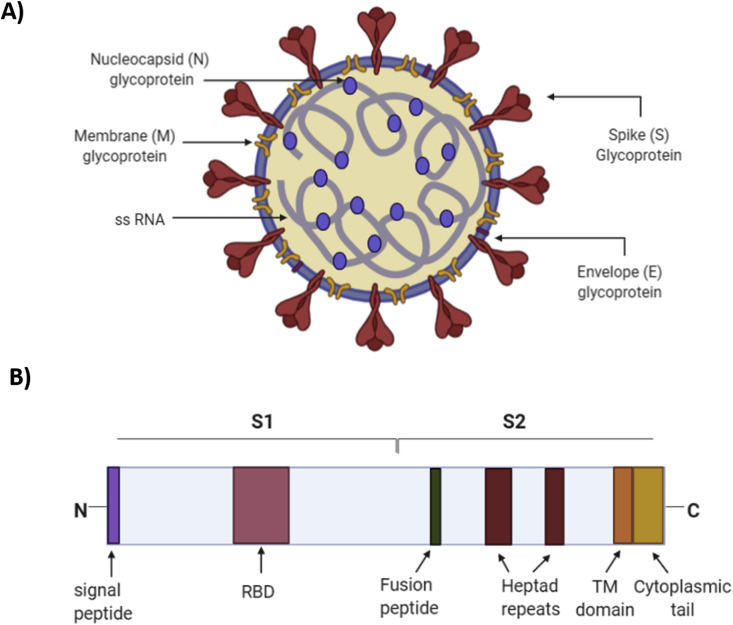
Host cell entry is the first step in the viral life cycle and constitutes a target for treatment and prevention. There are four main glycoproteins present in SARS corona viruses, which are spike (S), membrane (M) and envelope (E) and nucleocapsid (N) (Fig. 1 A). Where the M, E and N glycoproteins play a role in virus particle assembly, replication and release, the S glycoprotein is the most prominent of these since it binds to host cell receptors and fuses the viral membrane with host cell membrane, which is the processes essential for infectious entry [7,8]. The (S) glycoprotein consists of two major units, the N-terminal S1 unit, which harbours the receptor binding domain (RBD) that is necessary for attachment to host cell receptors and the C-terminal S2 unit, which contains other domains required for fusion and intracellular trafficking inside the cell (Fig. 1B) [[8], [9], [10]]. In 2003, in Nature publication, Li and his colleagues have shown that SARS-CoV uses its (S) glycoprotein to associate with ACE2, an ectoenzyme that is expressed on surface of target host cells and known to be abundantly present in humans in the epithelia of the lung and small intestine [6,11]. However, more recent research showed that SARS-CoV- (S) binding to ACE2 is not sufficient for host cell infection, and it seems that the (S) glycoprotein needs to be further processed and cleaved at specific sites by host cell’s proteases to fuse the viral with target cell membrane [12]. The proteolytic cleavage within (S) glycoprotein exposes the internal fusion peptide, which is located directly adjacent to the cleavage site, for SARS-CoV (S) [13]. Thus, upon (S) glycoprotein cleavage, the fusion peptide can fuse into a host cell membrane, and mediates virus entry into the cell. Generally, the cysteine protease cathepsin in lysosomes has been shown to be critical for SARS-CoV entry through endocytosis. For instance, it has been demonstrated that SARS-CoV utilizes the enzymatic activity of the cathepsin L to infect ACE2-expressing cells, and the enzymatic suppression of cathepsin L led to inhibition of SARS-CoV entry and infection in cells [14,15].
Fig. 1.
General structure of SARS coronavirus. (A) The structure of SARS-CoV-2 virion, with the main set of structural glycoproteins. (B) A linear map of domain organization of SARS-coronavirus spike (S) glycoprotein, the N-terminus S1 units contains the receptor binding domain (RBD) which is required for binding of the virus to ACE2. The C-terminus S2 harbours the functional elements required for fusion. It also contains a transmembrane domain (TM) for membrane anchoring and a cytoplasmic tail for appropriate intracellular trafficking.
But, how could the binding of SARS-CoV- (S) to ACE2 mediate the release of virus component into the target cell? it is possible that the binding of ACE2 to (S) glycoprotein of SARS-CoV provokes changes in the three dimensional structure of (S) glycoprotein that might expose a cleavage site that becomes susceptible to cleavage by cathepsin L, which exposes domains required for viral fusion, and that was actually previously hypothesized [8]. In sum, these results further suggest that both ACE2 and cathepsin L play a key role in mediating SARS-CoV-2 infectivity in host cells, particularly via the endocytosis/lysosomal entry, and provides new strategies to counteract the SARS-CoV-2 entry through blocking of this pathway. Furthermore, members of type II transmembrane serine proteases (TTSP), particularly the transmembrane protease serine 2 (TMPRSS2), has been also shown to play a crucial role in activation of SARS corona infection [16], as well as the spreading of the virus within the airways in animal models of SARS-CoV [17]. Indeed, it has been demonstrated that TMPRSS2 cleaves the coronavirus (S) glycoprotein to generate unlocked, fusion-catalyzing forms at the cell surface which facilitate rapid early entry of the virus into host cells [16,18]. Just recently, it has been demonstrated that the TMPRSS2-expressing kidney epithelial cell line (VeroE6) was highly susceptible to SARS-CoV-2 infection [19], which suggests that TMPRSS2 protease is a key protease for entry of the virus into cells.
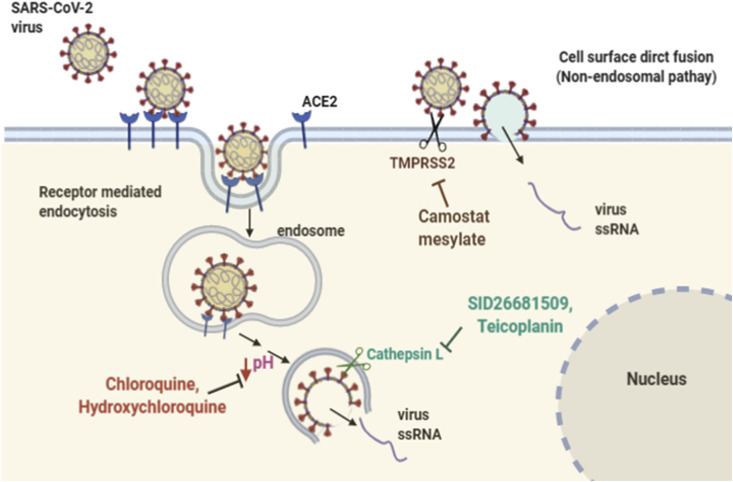
Additionally, Walls et al. just reported a notable variation in (S) glycoprotein amino acid sequence that arising from an insertion at the S1/S2 boundary that results in an “RRAR” [10], which is a well known recognition motif of furin protease, whether the presence of this multibasic cleavage site is crucial for SARS-CoV-2 entry into target cells remains to be determined. Overall, several lines of evidence have shown that the mechanism of proteolytic cleavage and activation of (S) glycoprotein, which is mediated by host cell proteases, is crucial for SARS-CoV and SARS-CoV-2 infectivity, making these enzymes potential targets for therapeutic intervention (Fig. 2 ).
Fig. 2.
Routes of SARS-CoV-2 virus entry into target cells. Binding of a viral (S) glycoprotein to ACE2 receptor can be followed by receptor mediated endocytosis, which ends up in endosomal compartment, where an increase in H+ influx into the endosome activates cathepsin L which activates viral (S) glycoprotein and facilitates viral membrane fusion and releases of ssRNA out of the endosome (left). Chloroquine and hydroxychloroquine are known to block virus infection mainly by increasing endosomal pH required for virus/cell fusion. The thiocarbazate SID26681509 and teicoplanin are inhibitors of cathepsin L. Alternatively, proteolytic cleavage of the viral (S) protein by TMPRSS2 protease on the surface of host cell can induce direct fusion of the viral and plasma membrane leading to release of the viral ssRNA into the cytoplasm (right). The camostat mesylate drug inhibits TMPRSS2 activity.
3. Molecular interaction between host ACE2 and SARS-CoV-2 virus
For SARS-CoV the first step of the entry cascade is the interaction between the (S) glycoprotein and its receptor ACE2 on the surface of host cell. As mentioned above, the (S) glycoprotein comprises two functional subunits responsible for binding to the host cell receptor (S1 subunit) and fusion of the viral with cellular membranes (S2 subunit). Just recently, the structure of the (S) protein of SARS-CoV-2 has been reported. Notably, the study showed that the SARS-CoV-2 (S) protein binds ACE2 with higher affinity than does SARS-CoV (S) protein [9]. Also, the researchers were able to calculate the binding kinetics of the SARS-CoV-2 (S) ectodomain to ACE2 with ∼15 nM affinity using surface plasmon resonance (SPR), which is about ∼10- to 20-fold higher than binding of SARS-CoV (S) to ACE2 [9]. At the same time, a very recent publication in Science, Yan et al. have demonstrated the association between SARS-CoV-2 -RBD and the full length human ACE2 at molecular levels [20]. It was not surprising to see that the overall interface between SARS-CoV-2 and ACE2 is similar to that of SARS-CoV with ACE2 [21], which is mediated mainly through polar interactions. Importantly, the study revealed key amino acid residues at the RBD of SARS-CoV-2 such as (Lys317) and (Phe486) that could be critical for the tight interaction between SARS-CoV-2 and human ACE2 [20]. We think that small molecule inhibitors that specifically designed to interfere with these key residues and could block or at least decrease affinity between the SARS-CoV-2-RBD and ACE2 would be very promising, and we might see such inhibitors in near future.
On the other side, efforts are underway to develop drugs that can bind specifically to ACE2 and block its binding to the virus. To date, no small-molecule drug has been approved for this application. Nevertheless, a biological drug called human recombinant soluble ACE2 (hrsACE2) has been developed recently and showed to be able to block the binding ACE2 and the virus [22]. By competitively binding to the virus, it prevents viral binding to the natural, membrane-bound ACE2, and thus blocks SARS-CoV-2 entry into host cells.
4. Association between ACE2 expression, comorbidities and severity of SARS-CoV-2 infection
ACE2 is an integral membrane protein with catalytic activity that plays a central role in regulation of the renin-angiotensin system (RAS), a signalling pathway that acts as a homeostatic regulator of vascular function [23]. The main biochemical function of ACE2 is the degradation of Angiotensin II, a major vasoactive peptide in the RAS system and acting as a potent vasoconstrictor through its receptor angiotensin II type-I receptor (AT1R), resulting in the formation of angiotensin 1–7, which opposes the action of Angiotensin II [24]. It has been shown that the renal expression of ACE2 is decreased in patients with type-2 diabetes with overt nephropathy [25]. However, blockade of RAS with either ACE inhibitors or Ang II receptor blockers (ARBs) may increase ACE2 expression as it has been demonstrated in murine models. Indeed, Huang and co-workers have demonstrated that ACE inhibitors were able to upregulate the levels of ACE2 mRNA and proteins in injured rat liver [26]. Furthermore, Ferrario et al. have demonstrated that the administration of agents that either inhibiting the synthesis of circulating Angiotensin II (ACE inhibitors) or blocking the activity of Angiotensin II at the AT1 receptor induced an increase in cardiac ACE2 mRNA expression in rats [27]. In addition, more recent data showed that olmesartan, an ARB and antihypertensive drug, increased renal expression and urinary excretion of ACE2 in hypertensive patients [28], which suggests that upregulation of ACE2 may also occur in humans.
People older than 60 years of age and those with comorbid conditions, such as diabetes, hypertension and cardiovascular disease (CVD), seem to be involved in a severe course of illness and may suffer from serious/life threatening COVID [[29], [30], [31], [32]]. It is still unclear how these comorbidities contribute to the higher risk of SARS-CoV-2 infection. Importantly, patients with comorbidities such as diabetes, hypertension are often treated with ACE inhibitors. In fact, ACE inhibitors have been the first line of treatment against hypertension for decades, and are widely used in the clinical management of patients affected by hypertension and diabetes [24]. Despite the potential role of ACE inhibitors as inducers of ACE2 expression, the treatment was not assessed before with corona viral infections, and whether the treatment is associated with severity of SARS-CoV-2 needs to be assessed. So, there is a possibility that treatment of diabetes and hypertension patients with ACE inhibitors and/or ARBs drugs may speed-up viral entry and host’s pulmonary tissue colonization which subsequently increases the risk of developing severe and fatal SARS-CoV-2 infection [3,33,34]. Nevertheless, we still wait for additional evidences coming from studies conducted on humans to validate such association.
5. Blockers of endosomal acidification and inhibitors of TMPRSS and cathepsin proteases are potential therapeutics of SARS-CoV-2
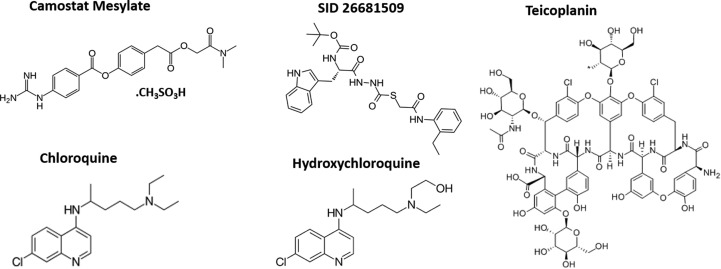
Chloroquine (QC), a 4-aminoquinolines (Fig. 3 ) that was identified in 1934, has been the most successful single drug for the treatment and prophylaxis of malaria [35,36]. The drug was very effective before resistant strains of malaria parasite began to emerge in the 1960s [36]. QC is a weak base that causes a rapid and substantial increase in the lysosomal pH. Since acidification is an essential factor for endosome maturation and function, QC suppresses the endosome maturation and blocks the pathway at intermediate stages of endocytosis [37], which results in failure of further transport of virus particles to its ultimate releasing site (Fig. 2). However, QC could also interfere with the terminal glycosylation of ACE2 [38], the cellular receptors of SARS-CoV-2, which is another potential mechanism by which QC inhibits the virus infection. Importantly, QC was previously described as a potent and an effective antiviral agent for SARS-CoV, as it was effective in inhibiting the spread of SARS CoV in culture cells that either treated with QC prior to or after SARS CoV infection [38].
Fig. 3.
Chemical structure of candidate drugs of SARS-CoV-2 infection.
Chloroquine is considered to be a relatively well-tolerated and safe drug. However, serious side effects such as retinopathy, neuromyopathy and bone-marrow toxicity may occur, especially if used for long-term prophylaxis [36]. Hydroxychloroquine (HCQ), a derivative of CQ (Fig. 3), was first produced in 1946 by introducing a hydroxyl group into CQ and was shown to be much less toxic than CQ in animals [39]. Notably, the HCQ is still used to treat autoimmune diseases, such as rheumatoid arthritis and systemic lupus erythematosus. Just recently, Liu et al. have demonstrated that Both CQ and HCQ can efficiently inhibit SARS-CoV-2 infection in monkey kidney VeroE6 cells in vitro [40]. Interestingly, a small sample size clinical study has been recently done in France, showed that HCQ treatment is effective in treatment of SARS-CoV-2 infected patients [41]. Taking these studies together, the CQ and HCQ are candidate drugs for treatment of SARS-CoV-2. However, clinical trials coming from different locations worldwide are needed to assess the effectiveness and safety of using these drugs against SARS-CoV-2 infection.
On the other hand, it is well established that human cell proteases cathepsin L and TMPRSS are essential for viral entry into cells. It has been shown that inhibition of cathepsin L by teicoplanin, a glycopeptide antibiotic, blocked the entry of SARS corona pseudotyped viruses in host cells [42,43]. A recent study by Zhang et al. demonstrated that teicoplanin could inhibit the entry of SARS-CoV-2 in cells in vitro [44]. Also, suppression of cathepsin L by Oxocarbazate drug has been claimed to block SARS-CoV and Ebola pseudotype virus entry in human cells [45]. Notably, a very recent study by Ou et al. has described that treatment of HEK 293 cells expressing human ACE2 with thiocarbazate SID26681509 (Fig. 3), a potent and selective inhibitor of human cathepsin L and was previously shown to inhibit propagation of malaria parasite Plasmodium falciparum and Leishmania major in vitro [46], led to a marked decrease in SARS-CoV-2 (S) pseudovirions entry into the cells [47]. Also, SID 26681509 was reported to be non-toxic to human aortic endothelial cells and zebrafish in a live organism assays at 100 μM [43]. Similarly, TMPRSS2 is also required for (S) glycoprotein priming and is essential for SARS virus entry. Indeed, in a very recent study, Hoffmann and co-workers showed that SARS-CoV-2 can hijack TMPRSS2 protease for priming of (S) glycoprotein. It was very promising to see the camostat mesylate drug (Fig. 3), which is a known suppressor of TMPRSS2 activity, inhibits SARS-CoV-2 infection in the human lung cancer cell line (Calu-3) [48]. In sum, targeting proteases which are involved in activation of SRAS-CoV-2 glycoprotein, specifically cathepsin L and TMPRSS2 proteases could be a potential therapy for SRAS-CoV-2 infection.
6. Conclusion
SARS CoV-2 infection is currently a big threat to global health. It is obvious from the current cumulative published data that the SARS-CoV-2 virus enters into cells via at least two main distinct pathways: one is induced by TMPRSS2 at the cell surface and the other is mediated by ACE2 -endosomal pathway. Potential inhibitors of these pathways are currently available and showed promising results in vitro. However, clinical studies are required to determine the efficacy of such drugs in preventing and treating SARS-CoV-2 infection as well as to assess the safety of using these drugs in humans.
Author contribution
Ismail Mahmoud: Conception, designing and writing of the manuscript.
Yazun Jarrar: Conception and writing of the manuscript.
Walhan Alshaer, Said Ismail: Conception and revising the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgment
This work was supported by The Hashemite University, Jordan.
References
- 1.Jiang S., Shi Z., Shu Y. A distinct name is needed for the new coronavirus. Lancet. 2020;395(10228):949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention [published online ahead of print, 2020 feb 24] J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.2648. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Patel A.B., Verma A. COVID-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? [published online ahead of print, 2020 mar 24] J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4812. 10.1001/jama.2020.4812. [DOI] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten C., Günther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83(15):7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang I.C., Bosch B.J., Li F. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281(6):3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glowacka I., Bertram S., Müller M.A. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93(6) doi: 10.1128/JVI.01815-18. e01815-18. Published 2019 Mar 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88(2):1293-1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 22.Monteil V., Kwon H., Prado P. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905-913. doi: 10.1016/j.cell.2020.04.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuiri S., Hemmi H., Arita M. Expression of ACE and ACE2 in individuals with diabetic kidney disease and healthy controls. Am. J. Kidney Dis. 2008;51(4):613–623. doi: 10.1053/j.ajkd.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 26.Huang M.L., Li X., Meng Y. Upregulation of angiotensin-converting enzyme (ACE) 2 in hepatic fibrosis by ACE inhibitors. Clin. Exp. Pharmacol. Physiol. 2010;37(1):e1–e6. doi: 10.1111/j.1440-1681.2009.05302.x. [DOI] [PubMed] [Google Scholar]
- 27.Ferrario C.M., Jessup J., Chappell M.C. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111(20):2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 28.Furuhashi M., Moniwa N., Mita T. Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 2015;28(1):15–21. doi: 10.1093/ajh/hpu086. [DOI] [PubMed] [Google Scholar]
- 29.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study [published online ahead of print, 2020 Feb 24] [published correction appears in Lancet Respir Med. 2020 Apr;8(4):e26] Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study [published correction appears in Lancet. 2020b Mar 28;395(10229):1038] [published correction appears in Lancet. 2020 Mar 28;395(10229):1038] Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Qi T., Liu L. Clinical progression of patients with COVID-19 in Shanghai, China [published online ahead of print, 2020 Mar 19] J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan W.J., Ni Z.Y., Hu Y. Clinical characteristics of coronavirus disease 2019 in China [published online ahead of print, 2020 feb 28] N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommerstein R., Kochen M.M., Messerli F.H., Gräni C. Coronavirus disease 2019 (COVID-19): do angiotensin-converting enzyme inhibitors/angiotensin receptor blockers have a biphasic effect? J. Am. Heart Assoc. 2020;9(7) doi: 10.1161/JAHA.120.016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slater A.F. Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacol. Ther. 1993;57(2–3):203–235. doi: 10.1016/0163-7258(93)90056-j. [DOI] [PubMed] [Google Scholar]
- 36.Schlitzer M. Malaria chemotherapeutics part I: history of antimalarial drug development, currently used therapeutics, and drugs in clinical development. ChemMedChem. 2007;2(7):944–986. doi: 10.1002/cmdc.200600240. [DOI] [PubMed] [Google Scholar]
- 37.Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5(1) doi: 10.1002/prp2.293. Published 2017 Jan 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent M.J., Bergeron E., Benjannet S. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. Published 2005 Aug 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McChesney E.W. Animal toxicity and pharmacokinetics of hydroxychloroquine sulfate. Am. J. Med. 1983;75(1A):11–18. doi: 10.1016/0002-9343(83)91265-2. [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Cao R., Xu M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. Published 2020 Mar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautret P., Lagier J.C., Parola P. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial [published online ahead of print, 2020 Mar 20] Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousefi B., Valizadeh S., Ghaffari H., Vahedi A., Karbalaei M., Eslami M. A global treatments for coronaviruses including COVID-19 [published online ahead of print, 2020 May 11] J. Cell. Physiol. 2020 doi: 10.1002/jcp.29785. 10.1002/jcp.29785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou N., Pan T., Zhang J. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291(17):9218-9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Ma X., Yu F., Liu J., Zou F., Pan T., Zhang H. 2020. Teicoplanin Potently Blocks the Cell Entry of 2019-nCoV. bioRxiv. [DOI] [Google Scholar]
- 45.Shah P.P., Wang T., Kaletsky R.L. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 2010;78(2):319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah P.P., Myers M.C., Beavers M.P. Kinetic characterization and molecular docking of a novel, potent, and selective slow-binding inhibitor of human cathepsin L. Mol. Pharmacol. 2008;74(1):34–41. doi: 10.1124/mol.108.046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou X., Liu Y., Lei X. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. Published 2020 Mar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor [published online ahead of print, 2020 mar 4] Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]