Abstract
Temporal lobe epilepsy causes severe cognitive deficits, but the circuit mechanisms remain unknown. Interneuron death and reorganization during epileptogenesis may disrupt the synchrony of hippocampal inhibition. To test this, we simultaneously recorded from the CA1 and dentate gyrus in pilocarpine-treated epileptic mice with silicon probes during head-fixed virtual navigation. We found desynchronized interneuron firing between the CA1 and dentate gyrus in epileptic mice. Since hippocampal interneurons control information processing, we tested whether CA1 spatial coding was altered in this desynchronized circuit, using a novel wire-free miniscope. We found that CA1 place cells in epileptic mice were unstable and completely remapped across a week. This spatial instability emerged around 6 weeks after status epilepticus, well after the onset of chronic seizures and interneuron death. Finally, CA1 network modeling showed that desynchronized inputs can impair the precision and stability of CA1 place cells. Together, these results demonstrate that temporally precise intrahippocampal communication is critical for spatial processing.
Temporal lobe epilepsy (TLE) is associated with disabling cognitive deficits1,2, interneuron cell death3,4, and large-scale anatomical reorganization of limbic circuits5–8 in both human patients and rodent models. Following cell death, surviving interneurons from both the CA1 and dentate gyrus sprout new local and long-range connections, leading to altered timing and kinetics of inhibition in hippocampal slices8–12. In particular, in chronically epileptic mice, new functional inhibitory connections form within the dentate gyrus6, between the CA1 and dentate gyrus7, and even across hemispheres13. These alterations are likely to have substantial effects on spatial processing because the hippocampus relies on the precise timing of diverse interneuron subtypes that control excitatory inputs14–17. Indeed, alterations in hippocampal network activity, such as reduced power of theta oscillations18–20, have been proposed to underlie cognitive deficits in epileptic mice20,21. However, no studies have examined the synchronization of interneurons during behavior and how specific network dynamics drive memory deficits in epilepsy.
In addition, initial studies have shown degraded spatial representations of individual place cells in epileptic rodents22–25 but it remains unclear what circuit mechanisms drive these deficits and when they emerge during epileptogenesis. Critically, interneuron death occurs within days after an epileptogenic insult9, but other neuroanatomical reorganization can take several weeks to occur7. Therefore, examining the timeline of when spatial coding deficits emerge can provide new insights into the circuit processes that drive memory impairments. With the development of new calcium imaging technology26–29 we can now stably record from hundreds of neurons across weeks and determine how entire populations of neurons are altered in epileptic mice, how stable these representations are across long time spans, and how these spatial deficits develop.
To address these questions, we first used in vivo electrophysiology with silicon probes in chronically epileptic mice and found that interneurons in the dentate gyrus fired at the wrong phase of theta oscillations, which desynchronized the interneurons between the CA1 and dentate gyrus. To assess how this disrupted circuit processes spatial information we used in vivo calcium imaging with miniature microscopes to track large numbers of neurons during spatial navigation and found that spatial representations in epileptic mice were unstable across minutes and completely remapped across a week. Strikingly, this place cell instability emerged around 6 weeks after epileptogenesis and was dissociated from the onset of seizures and deficits in the precision of place cell firing. Finally, we built a CA1 network model and demonstrated that desynchronization of inputs rather than epilepsy-driven interneuron death in the CA1 can produce deficits in spatial representations. Together, these experiments indicate that disrupted hippocampal synchronization contributes to poor spatial processing in epilepsy.
Results
Epileptic mice have altered timing of inhibition in the dentate gyrus
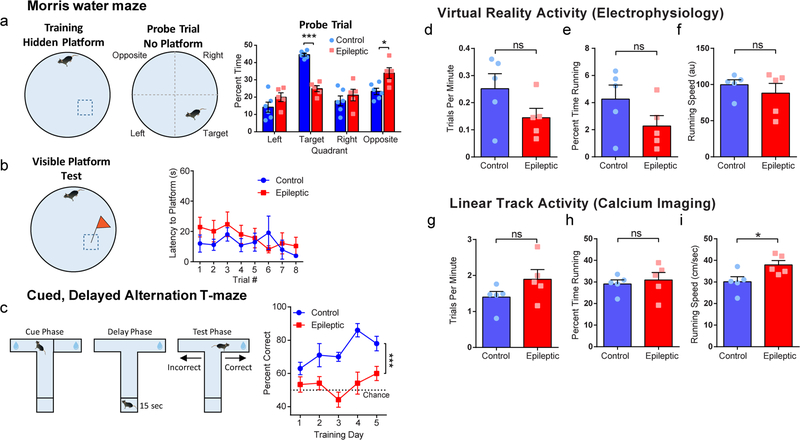
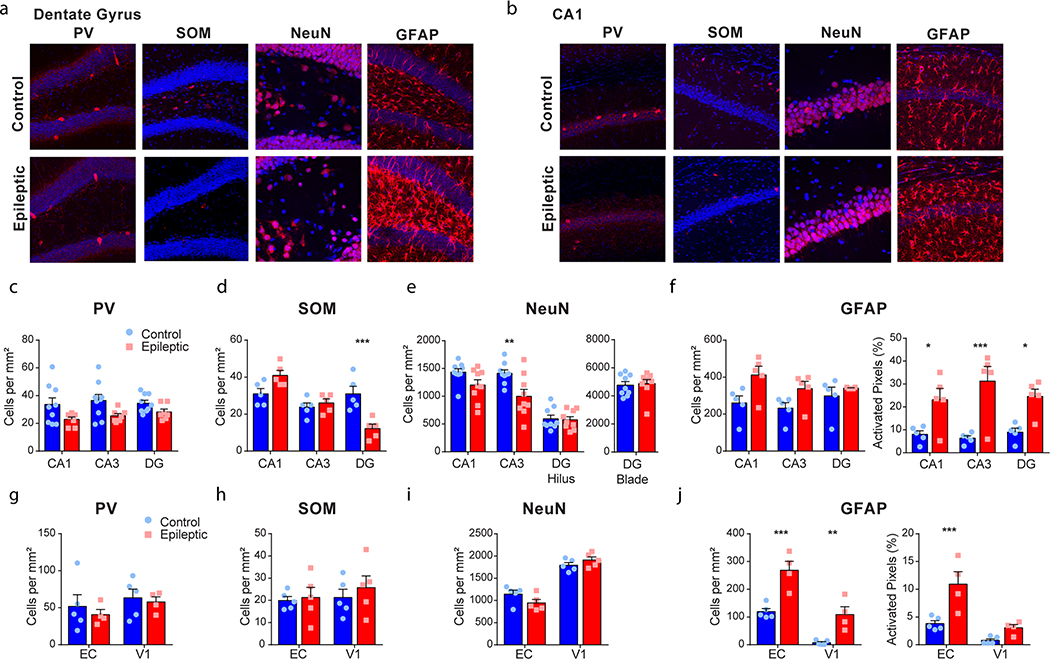
To study spatial processing in a mouse model of TLE, we used an established model in which an initial prolonged seizure induces chronic epilepsy and severe spatial memory deficits for the life of the animal2. Naive mice (around 7 weeks old) were injected with pilocarpine to induce a 2 h status epilepticus event30. All animals that recovered displayed spontaneous seizures and spatial memory deficits when tested at least 6 weeks after pilocarpine administration (Extended Data Fig. 1). Critically, although epileptic mice were impaired in the hidden platform Morris water maze, they were not impaired in the visible platform version of the water maze, indicating that their visual, motor, and motivation systems remained intact (Extended Data Fig. 1).
As interneurons are known to control information processing in the hippocampus14–17, we proposed that reorganization of the hippocampal network would lead to altered synchronization of inhibition in the hippocampus of epileptic mice. To test this idea, we used in vivo electrophysiology to capture the timing of interneuron firing. We performed silicon probe recordings in head-fixed epileptic and control mice running on a linear track in virtual reality (Fig. 1a,b and Supplementary Video 1). Following confirmation of chronic epilepsy, we implanted a headbar and trained the mice to run through a virtual linear track to record hippocampal activity during active spatial processing (see Methods). Owing to the time required to confirm spontaneous seizures, implant the headbar, and train the mice to run through virtual reality, electrophysiology recordings were performed at least 14 weeks after pilocarpine administration. After animals were trained to complete at least 100 trials within a single session, we performed acute extracellular electrophysiological recordings with 128-channel silicon probes that spanned the layers of the CA1 and dentate gyrus (Fig. 1b,c). Although there was a trend for epileptic mice to perform fewer trials and spend less time running, we found no differences (nor a trend) in running speed (Extended Data Fig. 1) and therefore constrained our analysis to periods of active locomotion.
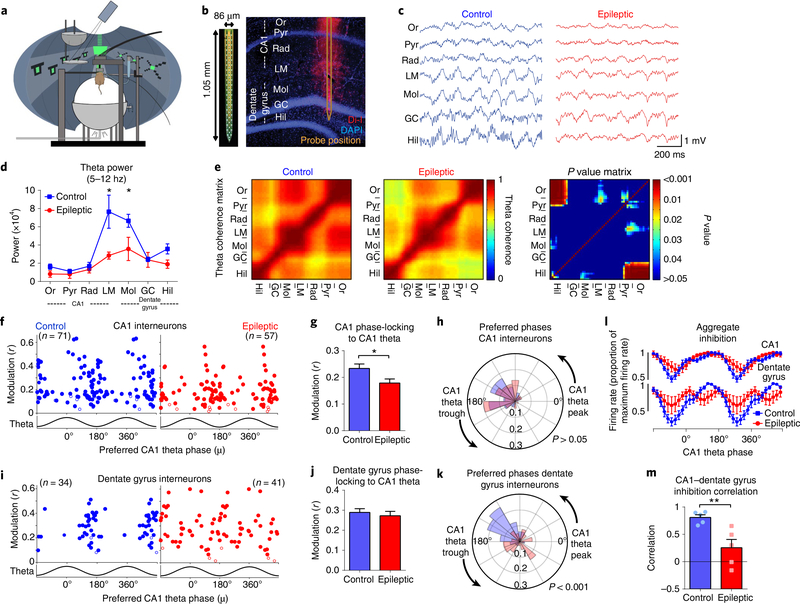
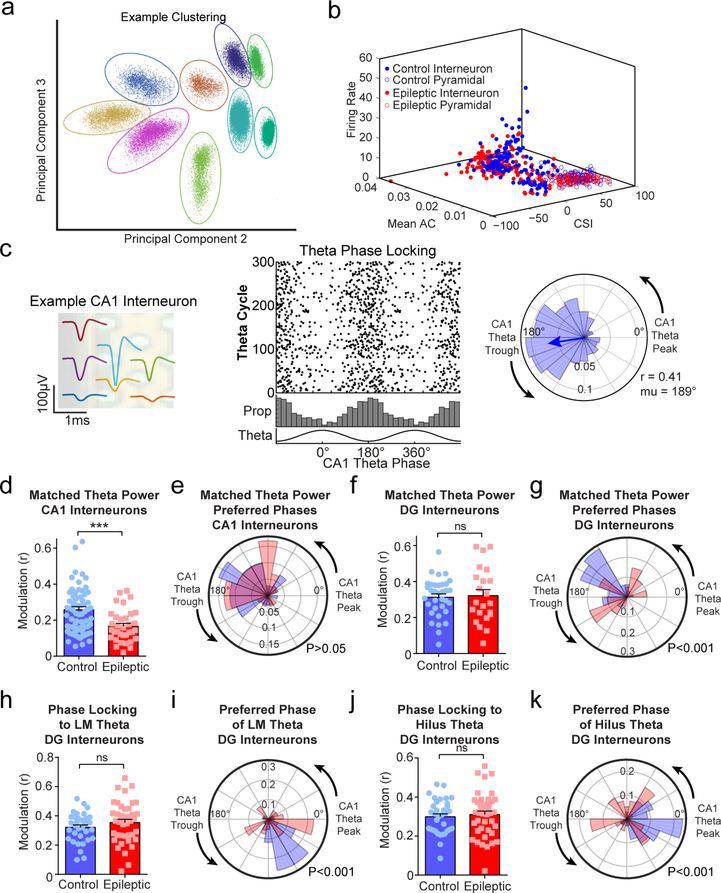
Fig. 1 |. Desynchronization of hippocampal inhibition in epileptic mice.
a, Head-fixed mice (at least 14 weeks after pilocarpine or control treatment) were trained to run through a virtual linear track for water rewards. b, During virtual navigation, silicon probes were inserted into the dorsal hippocampus, spanning the CA1 and dentate gyrus. c, Example local field potentials from each layer of the hippocampus during locomotion. d, Theta power from each hippocampal layer. Epileptic mice had reduced power in the lacunosum moleculare (LM) and molecular layer (Mol) of the dentate gyrus (repeated-measures ANOVA, FGroup × Region(6,48) = 5.30; P < 0.001; LM, P < 0.001; Mol, P = 0.03). e, Theta coherence between each channel pair along the recording probe in control (left) and epileptic (middle) mice. P value matrix (right) for each pair of recording sites (unpaired Student’s t-test for each channel pair). f, Phase-locking to CA1 theta for each interneuron in the CA1 of control and epileptic mice (Extended Data Fig. 3). Each dot represents one interneuron, and the data are double-plotted for visualization. Closed circles are significantly phase-locked (Raleigh test, P < 0.05) and open circles are not. See Methods for numbers of cells per animal. g, Mean phase modulation (r) of CA1 interneurons to CA1 theta. Interneurons in epileptic mice had reduced phase modulation (n = 71 control cells, n = 57 epileptic cells; Mann–Whitney U-test, P = 0.02). h, Rose plot of preferred firing phases of significantly phase locked CA1 interneurons. There were no differences between interneurons in control and epileptic mice (Kuiper circular test, P > 0.05). i, Phase-locking to CA1 theta for each interneuron in the dentate gyrus of control and epileptic mice. See Methods for numbers of cells per animal. j, Mean phase modulation (r) of dentate gyrus interneurons to CA1 theta. No difference was found between control and epileptic interneurons (n = 34 control cells, n = 41 epileptic cells; unpaired Student’s t-test, P = 0.57). k, Rose plot of preferred firing phases of significantly phase-locked dentate gyrus interneurons. The distribution of preferred phases in control and epileptic mice were different (Kuiper circular test, P < 0.001). l, Aggregate inhibition of all interneurons in the CA1 and dentate gyrus relative to CA1 theta phase. No differences were found in the CA1 (repeated-measures ANOVA, FGroup × Phase(17, 136) = 1.526, P = 0.09), but a significant interaction was found in the dentate gyrus (repeated-measures ANOVA, FGroup × Phase(17, 136) = 3.089, P < 0.001), indicating abnormal aggregate inhibition in the dentate gyrus. m, Correlation between CA1 and dentate gyrus aggregate inhibition. Epileptic mice had lower correlation (Pearson’s r) between CA1 and dentate gyrus aggregate firing rates than control mice (unpaired Student’s t-test, P = 0.008). GC, granule cell layer; Hil, hilus; Or, stratum oriens; Pyr, stratum pyramidale; Rad, stratum radiatum. n = 5 animals per group for all panels. Error bars represent 1 s.e.m. *P < 0.05. **P < 0.01.
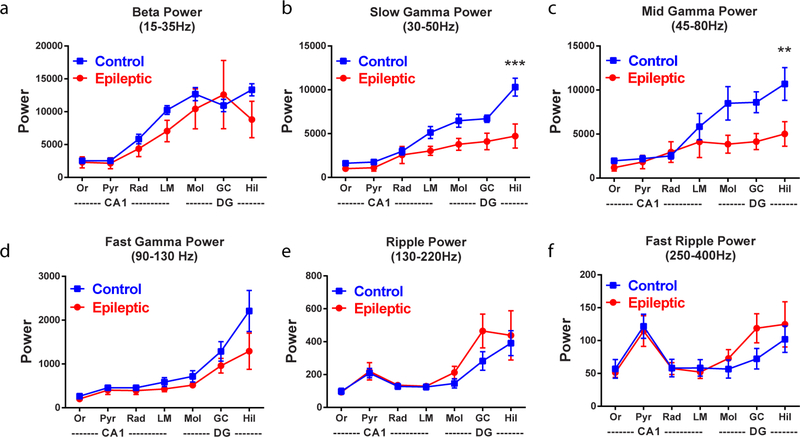
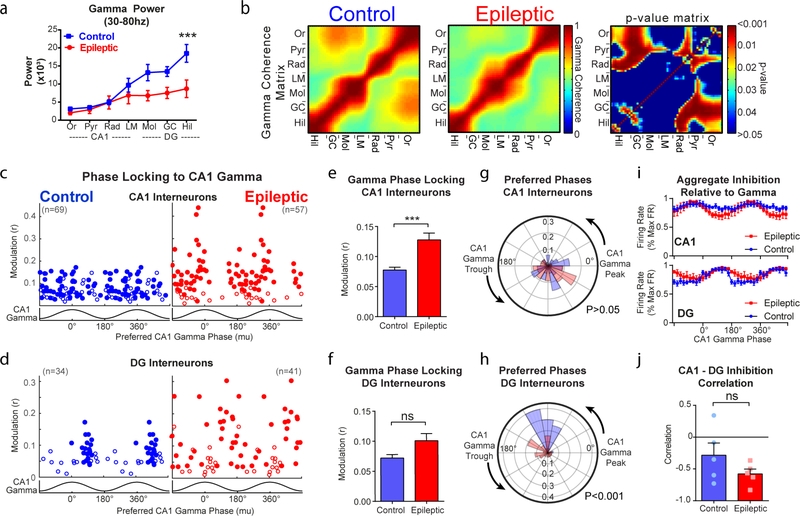
We first examined the power of local field potentials during locomotion throughout the CA1 and dentate gyrus across several frequencies (Extended Data Fig. 2). In epileptic mice, similar to previous reports18–20, there was significantly reduced theta power in the lacunosum moleculare layer of the CA1 and molecular layer of the dentate gyrus (Fig. 1d). To examine the synchrony of theta oscillations along the CA1–dentate gyrus axis, we measured the phase coherence of theta oscillations between each electrode pair and found reduced theta coherence primarily between the pyramidal layer and stratum oriens of CA1 and the dentate hilus (Fig. 1e). This reduced theta power and coherence in the epileptic hippocampus suggests a decrease in the synchronization between the CA1 and dentate gyrus, which may arise from desynchronized firing of interneurons across these regions. To test this idea, we examined the coordinated firing patterns of interneurons in both the CA1 and dentate gyrus. As previously reported14,15, individual CA1 interneurons modulate their firing rates in phase with theta oscillations (Extended Data Fig. 3), which can be measured with a modulation index (r, measuring the strength of phase locking of an individual interneuron) and preferred phase (mu, measuring the mean theta phase during which an interneuron fires). Similar to previous studies, CA1 interneurons in control animals were highly phase locked to the falling phase and trough of CA1 theta (Fig. 1f, left). However, in epileptic mice, we found that CA1 interneurons had reduced phase locking (Fig. 1f,g) but maintained a similar preferred phase to control mice (Fig. 1f,h). This reduction in phase locking was not solely due to reduced theta power as the effect was maintained when the power was matched across groups (Extended Data Fig. 3). In the dentate gyrus of control mice, interneurons were almost exclusively phase locked near the falling phase and trough of CA1 theta (Fig. 1i), indicating high synchronization of interneurons across hippocampal subregions. In the epileptic mice, individual interneurons had an equivalent magnitude of phase locking (Fig. 1j), but the preferred phases of all the neurons were highly distributed across all phases of CA1 theta (Fig. 1i,k). This observation was maintained when referenced to theta in lacunosum moleculare or the dentate hilus (Extended Data Fig. 3). Therefore, in control animals the aggregate inhibition in the CA1 and dentate gyrus is highly synchronous and peaks near the trough of CA1 theta (Fig. 1l). In the epileptic mice, the dentate gyrus inhibition is distributed across all phases of theta (Fig. 1l), which desynchronizes the two populations and leads to a reduced correlation between total inhibition in the CA1 and dentate gyrus (Fig. 1m). Together, we have now shown that, in control mice, there is exquisite synchrony in firing between CA1 and dentate gyrus interneurons, and this synchronization is likely to be critical for spatial coding. However, in epileptic mice, we discovered a decoupling in firing patterns between the CA1 and dentate gyrus. We propose that this desynchrony in the epileptic hippocampus may lead to deficits in processing spatial memory.
We also found that epileptic mice had decreased power of gamma oscillations in the dentate gyrus and decreased gamma coherence between the dentate gyrus and CA1 (Extended Data Fig. 4). To test whether individual interneurons have altered firing patterns with respect to gamma, we examined phase locking of CA1 and dentate gyrus interneurons to CA1 gamma oscillations. Similar to previous studies, CA1 interneurons in control mice only weakly phase locked to gamma oscillations; however, in epileptic mice, we found an increase in phase modulation (Extended Data Fig. 4). Nevertheless, the preferred phase was similar to controls. This increased phase locking to gamma in CA1 interneurons may represent a shift toward local inputs (gamma) and away from distal inputs31 (theta; Fig. 1g). In the dentate gyrus, we observed no difference in the magnitude of phase locking, but the preferred phase was disrupted, indicating disrupted coordination between the CA1 and dentate gyrus. At the population level, interneurons in the CA1 and dentate gyrus were negatively correlated with respect to gamma oscillations and this was similar in epileptic and control animals. Together, these data further suggest that network desynchronization in the hippocampus may provide a novel circuit mechanism underlying memory deficits in the epileptic brain.
Development of the wire-free miniscope
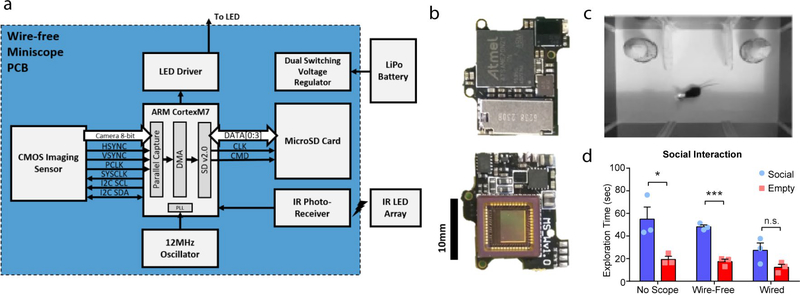
Recent developments in miniature microscope technology have allowed unprecedented access to neural signals in freely behaving animals26–29; however, all commercially available miniature microscopes use tethered cables to transmit power and data to a recording acquisition device. Although many experiments can be performed with wired miniature microscopes, wires can be prone to getting tangled or damaged, which can diminish data quality or disrupt behavior. To allow for more naturalistic behaviors and improve behavioral performance during recordings, we developed an open-source wire-free miniature microscope to perform calcium imaging without a cable tethered to the head of the animal (Fig. 2a and Extended Data Fig. 5). This wire-free miniscope saves data directly to a microSD card positioned on the miniscope and is powered by a lithium-polymer battery mounted to its side. Together, this configuration enables completely untethered and naturalistic behavior during neural recording. To test whether this novel wire-free miniscope enhances behaviorally relevant stimuli, we tested social interaction in a threechamber task with a wired miniscope26, a wire-free miniscope, or no miniscope. We found that mice preferred a social cup over an empty cup with no miniscope or with the wire-free scope, but this did not reach significance with the wired miniscope (Extended Data Fig. 5). Together, this indicates that untethered mice can perform natural behaviors better than when they are tethered by a data wire.
Fig. 2 |. Disrupted spatial coding in epileptic mice.
a, Wire-free miniscope for untethered behavior during imaging. The miniscope is powered by a lithium-polymer battery and data are saved directly to a microSD card. b, example raw images (top) and identified cells (bottom) in control and epileptic mice. Each field of view is approximately 550 μm × 550 μm. Cells were identified with CNMF-e. c, Top: animals (at least 17 weeks after pilocarpine or control treatment) were trained to run on a 2-m linear track for water reward. Bottom: 20 example cells from control and epileptic mice show spatially selective firing patterns. Black is a raster plot of the per-trial spatial neural activity rate across all trials within a session. Red represents the normalized activity rate along the track averaged across all trials. d, Normalized spatial firing rates across all cells recorded during all sessions (between four and eight sessions per animal). e, Heatmap of stability and information content in control (top) and epileptic (bottom) mice. Contours represent the 10th, 30th, 50th, 70th, and 90th percentiles. There was a shift in the distributions of epileptic mice towards lower information content and stability. f, Average information content across all cells for each animal. Epileptic mice had less information content than control mice (Mann–Whitney U-test, P = 0.03). g, Average stability across all cells for each animal. Epileptic mice had less stable cells than control mice (unpaired Student’s t-test, P = 0.02). h, Average number of active cells per session. No differences in the numbers of active cells were found (unpaired Student’s t-test, P = 0.73). i, Average activity of all cells during run and rest. No differences in neuronal activity were found (two-way ANOVA, FGroup(1,8) = 0.01, P = 0.94; run versus rest, control P = 0.01, epileptic P = 0.004). j, Normalized spatial firing rates across place cells (see Methods) recorded during all sessions (between four and eight sessions per animal). k, Average percentage of all cells that were place cells during each session. epileptic mice had fewer place cells than control mice (unpaired Student’s t-test, P = 0.03). l, Heatmap of stability and information content in control (top) and epileptic (bottom) mice in both place cells (green) and non-place cells (yellow). Contours represent the 10th, 30th, 50th, 70th, and 90th percentiles. m, Average information content across place cells for each animal. Place cells in epileptic mice had less information content than those from control mice (unpaired Student’s t-test, P = 0.02). n, Average stability across place cells for each animal. Place cells in epileptic mice were less stable than in control mice (unpaired Student’s t-test, P = 0.001). o, Average decoding error for position within each session. Epileptic mice had larger decoding position error than control mice (unpaired Student’s t-test, P = 0.03). p, Average decoding error for direction within each session. Epileptic mice had more decoding position error than control mice (Mann–Whitney U-test, P = 0.03). n = 5 per group for all panels. error bars represent 1 s.e.m. *P < 0.05.
Place cell and population coding are disrupted in epileptic mice
Our in vivo electrophysiology data revealed that epileptic mice have desynchronized interneuron and network activity in the hippocampus (Fig. 1). We proposed that this desynchronization would disrupt the encoding and maintenance of spatial information in the epileptic hippocampus. Using wire-free miniscopes, we recorded large populations of neurons and tracked them across weeks to determine how spatial representations are disrupted in epilepsy. Owing to the time required to confirm spontaneous seizures, implant the microendoscope, and train the mice to run on a linear track, these calcium imaging recordings were performed at least 17 weeks after pilocarpine administration. Critically, this time point was similar to that of the electrophysiological recordings performed (Fig. 1). To record spatial activity patterns in large populations of CA1 neurons, we injected AAV1-Syn-GCaMP6f virus into the CA1 of the dorsal hippocampus at least 7 weeks after status epilepticus. A gradient refractive index (GRIN) lens was then implanted above the CA1 and mice were trained to run on a 2 m linear track for water reward while wearing a wire-free miniscope (Fig. 2b,c, Extended Data Fig. 1, and Supplementary Video 2). Within a single 15 min session, both epileptic and control mice had spatially tuned firing patterns along the track (Fig. 2d). However, the quality of the spatial information was substantially reduced in the epileptic mice. We measured the information content (how precisely each neuron codes for spatial location) and stability (how reliably each neuron fires at that spatial location across trials, Extended Data Fig. 6) of all cells and found a substantial reduction of both measures in epileptic mice (Fig. 2e–g). To assess how well the entire population of neurons encoded position, we calculated the population vector correlation (PVC) along the track, which measures how similar firing patterns are across locations on the track. Successful spatial coding relies on having distinct firing patterns across different parts of the track. In this case, having a low correlation across locations is beneficial to distinguishing where an animal is on the track. If the neural activity across locations is similar, then the correlation will be high, indicating neural firing that is less selective to location. We found that the correlation between locations along the track was significantly higher in the epileptic group (Extended Data Fig. 6), indicating that neuronal population firing was less distinct across spatial positions. We found no differences in the overall number of cells active during each session or the activity level of neurons during running or rest (Fig. 2h,i), indicating that the deficits we observed in spatial coding were not due to changes in the activity of neurons. We next isolated place cells, which had information content and stability above chance (see Methods), and tested whether the quality of place cells, specifically, was diminished in epileptic mice (Fig. 2j). Epileptic mice had fewer place cells than control mice as a percentage of total active cells (Fig. 2k). In addition, the place cells of epileptic mice had significantly lower information content and stability within a session (Fig. 2l–n), as well as increased population vector correlation between different locations on the track (Extended Data Fig. 6). Although a linear decoder (Extended Data Fig. 7) could accurately estimate the position of the control mouse on interleaved trials, we found that the decoding accuracy was significantly impaired in the epileptic mice for both position (Fig. 2o) and running direction (Fig. 2p). Together, these findings illustrate a severely disrupted spatial code at both the single-cell and population level in the epileptic hippocampus. This suggests that the desynchronization of hippocampal interneurons (Fig. 1) may lead to an inability to create and maintain consistent mnemonic information.
Place cell stability in epileptic mice is impaired at short intervals and completely remaps after a week
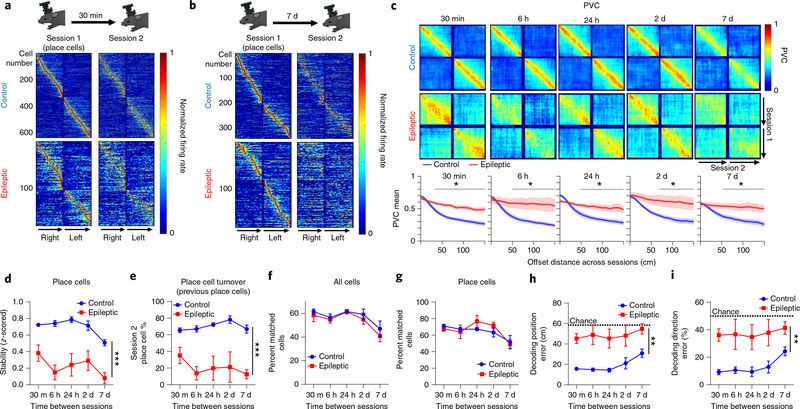
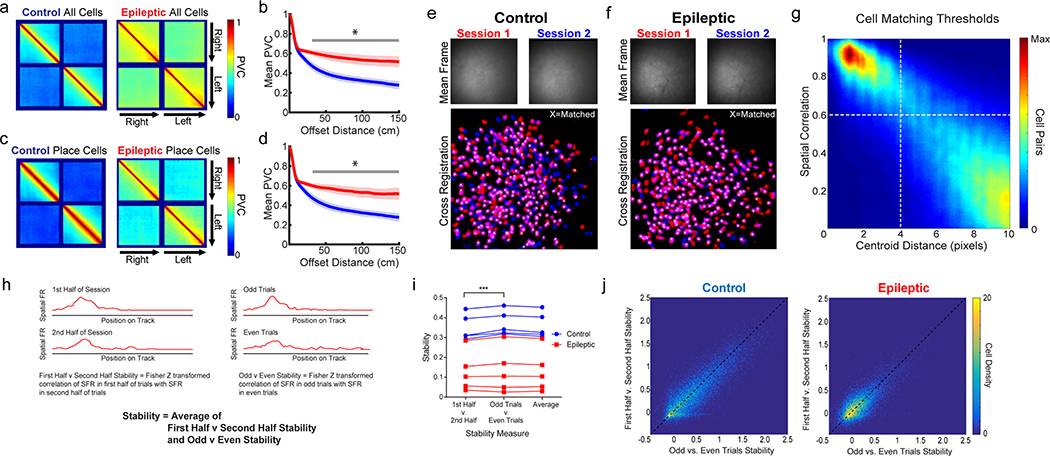
Next, we characterized the stability of place cells across minutes to days. We performed calcium imaging while animals ran on the same linear track with varying inter-session intervals of 30 min, 6 h, 24 h, 2 d, and 7 d (Fig. 3a–c and Extended Data Fig. 6). In epileptic mice, the spatial firing of a large proportion of the place cells remapped substantially when the mice returned to the same track 30 min later, whereas those in control mice remained stable (Fig. 3a). Across 7 d, a large proportion of place cells continued to fire at the same location in control mice, whereas very few neurons fired at similar locations in epileptic animals (Fig. 3b). This instability across sessions was evident in the stability of individual place cells (Fig. 3d), as well as at the population level, as epileptic mice had higher population vector correlation between large (> 66 cm) offset distances across trials than control mice (Fig. 3c). In addition, place cells in epileptic mice were less likely to meet the criteria of being a place cell again in subsequent sessions than were place cells in control mice (Fig. 3e). This was not due to turnover in the active population as cells were equally likely to be active in subsequent sessions in the epileptic mice as in control mice (Fig. 3f), even when only considering place cells (Fig. 3g), but they were not active in the same place field. The instability of place cells led to significantly worse decoding of position (location on the track) and running direction (left or right) across all time intervals in epileptic animals than in control animals (Fig. 3h,i and Supplementary Video 3). Together, these data demonstrate that epileptic mice have unstable spatial representations, even across 30 min, which continue to degrade to chance levels of remapping within a week. Furthermore, these results provide additional evidence that the inability to acquire stable spatial information is a primary contributor to place cell instability in epileptic mice.
Fig. 3 |. Disrupted stability of place cells across sessions.
a,b, Animals ran on the same linear track with inter-session intervals of 30 min, 6 h, 24 h, 2 d, or 7 d. Normalized spatial firing rates of place cells on session 1 that were also active on the second session 30 min (a) or 7 d (b) later. Cells are sorted by the peak firing rate from the first session only. c, Top: PVC across sessions for place cells recorded at each time interval. Bottom mean PVC across animals. For all time points, epileptic mice had higher PVC (less distinct firing patterns) than control mice at offset distances above 34–66 cm (30 min, P < 0.05 for 34–150 cm; 6 h, P < 0.05 for 50–150 cm; 24 h, P < 0.05 for 50–150 cm; 2 d, P < 0.05 for 66–150 cm; 7 d, P < 0.05 for 52–150 cm). Shading indicates 1 s.e.m. d, Stability across sessions at each time interval. epileptic mice had lower stability than control mice across all time points (two-way ANOVA, FGroup(1,7.75) = 26.75, P = 0.001; post hoc for all time points, P < 0.05). e, Percentage of cells that were place cells in session 1. epileptic mice had fewer place cells than control mice across all time intervals (two-way ANOVA, FGroup(1,8.27) = 34.8, P < 0.001; post hoc for all time points, P < 0.05). f, Percentage of all cells active during session 1 that were active again during session 2 at each time interval. There were no differences between groups (two-way ANOVA, FGroup(1,4.2) = 0.008, P = 0.93). g, Percentage of place cells active during session 1 that were active again during session 2 at each time interval. There were no differences between groups (two-way ANOVA, FGroup(1,5.48) = 0.635, P = 0.46). h, Decoding position error across sessions. epileptic mice had higher error than control mice across all intervals (two-way ANOVA, FGroup(1,8.11) = 17.85, P = 0.003; post hoc for all time points, P < 0.05). i, Decoding direction error across sessions. Epileptic mice had higher error than control mice across all intervals (two-way ANOVA, FGroup(1,8.13) = 11.57, P = 0.009; post hoc for all time points, P < 0.05). n = 5 per group for all panels. error bars represent 1 s.e.m. *P < 0.05. **P < 0.01. ***P < 0.001.
Instability in spatial coding emerges around 6 weeks after status epilepticus and is decoupled from seizure onset and deficits in spatial precision
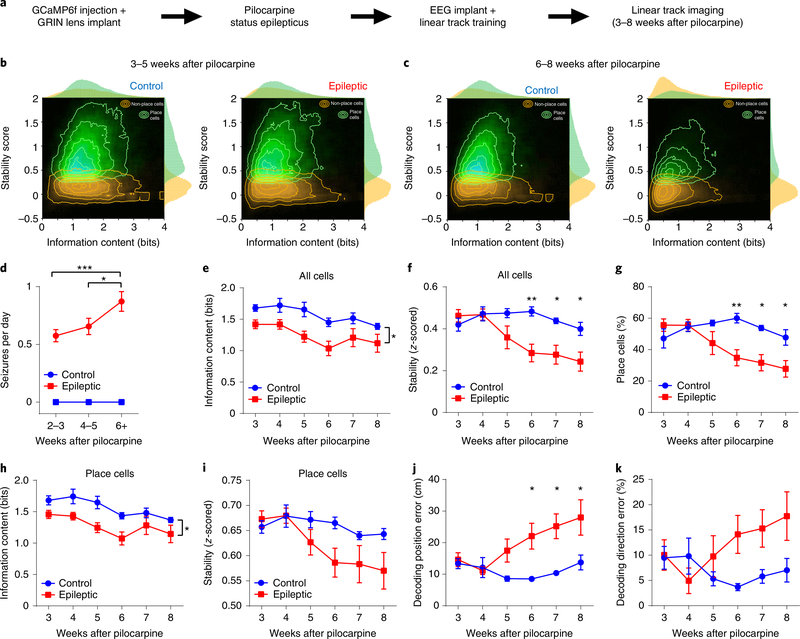
Pilocarpine-induced status epilepticus produces a progressive increase in seizures over many weeks, but how spatial coding deficits develop with the progression of seizures is unknown. To investigate the development of spatial coding deficits following pilocarpine treatment, we recorded epileptic and control animals using miniscopes on a linear track 3–8 weeks following status epilepticus, together with chronic electroencephalogram (EEG) recordings beginning around 2.5 weeks after pilocarpine treatment (Fig. 4a–c, n = 5 control, n = 7 epileptic). All pilocarpine-treated animals that were recorded had spontaneous seizures from 2.5 to 3 weeks after pilocarpine treatment, and seizures progressively increased across the course of the experiment (Fig. 4d). We found that the average information content across all recorded cells was reduced in the epileptic mice throughout the 3–8 weeks after pilocarpine treatment (Fig. 4e). Conversely, the stability of spatial representations remained normal 3–5 weeks after pilocarpine treatment but deteriorated into a large deficit beginning 6 weeks after pilocarpine treatment (Fig. 4f). Similarly, the percentage of place cells was not reduced until 6 weeks after pilocarpine treatment (Fig. 4g). When we isolated only place cells we observed the same pattern, with a consistent decrease in spatial information and an interaction between stability over time and the two groups (Fig. 4h,i). Finally, we tested whether our decoder could estimate the location of the animal from neural activity. We found that the epileptic mice had no deficit in decoding performance 3–5 weeks after pilocarpine treatment, but a deficit emerged 6–8 weeks after pilocarpine treatment (Fig. 4j,k). Together, these data establish a clear dissociation between the information content and the stability of spatial representations during the development of epilepsy. Critically, these animals were already having seizures 3–4 weeks after pilocarpine treatment, but the stability of place coding was normal. Therefore, this instability in spatial coding was not driven solely by the development of chronic seizures. In addition, interneuron loss generally occurs in the first 72 h after pilocarpine treatment9 and this loss of interneurons is therefore unlikely to mediate the progressive development of unstable spatial coding.
Fig. 4 |. Spatial instability emerges 6 weeks after pilocarpine treatment.
a, To examine spatial coding in the weeks after status epilepticus, we injected GCaMP6f virus and implanted a GRIN lens before status epilepticus. Immediately after status epilepticus, we trained animals to run on the linear track and implanted a wireless EEG device to monitor seizures. We performed calcium imaging on a linear track 3–8 weeks after pilocarpine treatment. b, Heatmap of stability and information content of all recorded neurons 3–5 weeks after pilocarpine treatment. c, Heatmap of stability and information content of all recorded neurons 6–8 weeks after pilocarpine treatment. Note the large difference in stability in the epileptic group across the development of epilepsy. d, Seizures recorded during the weeks after pilocarpine-induced status epilepticus (n = 3 control, n = 4 epileptic animals). Seizures in the epileptic group increased across the weeks after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,5) = 172, P < 0.001; post hoc test, 2–3 weeks versus 6+ weeks, P = 0.007, 4–5 weeks versus 6+ weeks, P = 0.04). e, Information content of all recorded cells was consistently reduced in epileptic mice 3–8 weeks after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 9.572, P = 0.01; FGroup × Week(5,50) = 0.435, P = 0.82). f, The stability of all recorded cells was reduced in epileptic mice 6–8 weeks after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 6.165, P = 0.03; FGroup × Week(5,50) = 5.622, P < 0.001, *P < 0.05, **P < 0.01). g, The percentage of recorded cells classified as place cells was reduced in epileptic mice 6–8 weeks after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 5.401, P = 0.04; FGroup × Week(5,50) = 6.654, P < 0.001, *P < 0.05, **P < 0.01). h, Information content in place cells was consistently reduced in epileptic mice (two-way repeated-measures ANOVA, FGroup(1,10) = 6.959, P = 0.02; FGroup × Week(5,50) = 0.719, P = 0.61). i, The stability of place cells was reduced across weeks and showed a significant interaction between group and time after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 1.872, P = 0.20; FWeek(5,50) = 6.86, P < 0.001; FGroup × Week(5,50) = 3.01, P = 0.02). j, Decoding position error was significantly increased in epileptic mice 6–8 weeks after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 6.657, P = 0.03; FGroup × Week(5,50) = 3.659; P = 0.007; *P < 0.05). k, Decoding direction error showed a significant interaction between group and time after pilocarpine treatment (two-way repeated-measures ANOVA, FGroup(1,10) = 1.922, P = 0.20; FGroup × Week(5,50) = 3.55, P < 0.01). Error bars represent 1 s.e.m. n = 5 control, n = 7 epileptic animals for all panels except d.
Hippocampal desynchronization, rather than loss of interneurons, can drive poor place coding in a CA1 network model
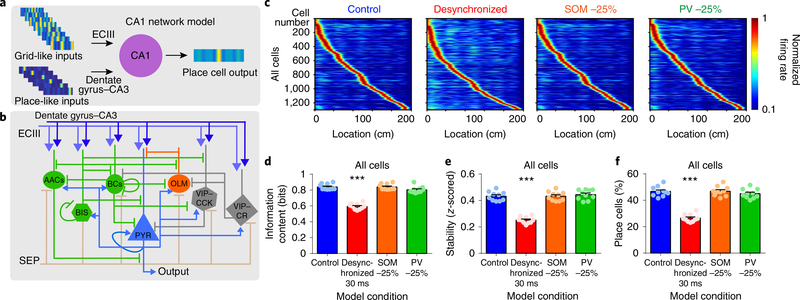
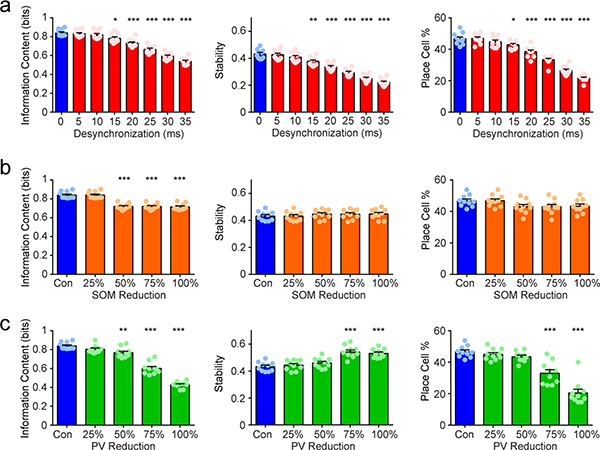
Our electrophysiological findings (performed 14 or more weeks after pilocarpine treatment, well after the onset of deficits in stability and spatial memory) demonstrate that synchronization mechanisms in the epileptic hippocampus are severely impaired, with abnormal firing patterns in this circuit being critical for spatial coding. To test how hippocampal desynchronization can impair spatial processing, we simulated place cell formation in a CA1 network model that receives grid-like inputs from entorhinal cortex layer 3 (ECIII) and place-like inputs from the dentate gyrus–CA3 (Fig. 5a,b, see Methods). Briefly, the CA1 network includes morphologically reduced compartmental models for pyramidal cells and different types of interneurons (axoaxonic, basket, bistratified, oriens-lacunosum moleculare (OLM), vasoactive intestinal peptide-cholecystokinin cells (VIP–CCK), and vasoactive intestinal peptide–calretinin (VIP–CR); Fig. 5b and Supplementary Table 1) and was validated against experimental data, similar to previous work32. CA1 pyramidal model neurons produce place-like activity that resembles experimental data (Fig. 5c). We first modeled how the epileptic CA1 processes spatial information. As our model was limited to the CA1 subregion, we simulated interneuron desynchronization (Fig. 1) as a disruption in the timing of inputs from ECIII and dentate gyrus–CA3 converging in CA1. We did so by adding Gaussian noise (μ = 0 ms, σ = 30 ms) to the temporal offset between these two input streams and measured the spatial output of pyramidal neurons. We found that desynchronization reduced spatial information, stability, and the number of place cells (Fig. 5c–f and Extended Data Fig. 8), recapitulating our experimental findings. In addition, as interneuron loss has also been reported in the CA1 area of epileptic mice9,10 (Extended Data Fig. 9), we tested the effect of reducing parvalbumin-positive (PV+) or somatostatinpositive (SOM+) interneuron populations. We found that removal of 25% of PV+ or SOM+ interneurons had no effect on spatial coding (Fig. 5c–f).
Fig. 5 |. Desynchronization disrupts place cell coding in a CA1 network model.
a, Schematic of the CA1 network model. Grid-like inputs from ECIII and place-like inputs from the dentate gyrus–CA3 are processed by CA1 and output place cell representations in pyramidal model neurons (PYR). b, Detailed model connectivity including excitatory inputs from ECIII (grid-like) and dentate gyrus–CA3 (place-like); and inhibitory inputs from medial septum (SEP), PV+ interneurons (axoaxonic cells (AACs), basket cells (BCs) and bistratified cells (BIS)), SOM+ interneurons (OLM) and VIP interneurons (VIP–CCK, VIP–CR). We tested the effects of decreasing PV+ interneurons, SOM+ interneurons and desynchronization, which have all been reported in chronic epilepsy. c, Normalized spatial firing rates of pyramidal model neurons for each condition tested. d, For all pyramidal cells, information content was reduced only in the condition with desynchronization (one-way ANOVA, F(3,36) = 96.09, P < 0.001). e, For all pyramidal cells, stability was reduced only in the condition with desynchronization (one-way ANOVA, F(3,36) = 78.11, P < 0.001). f, The percentage of excitatory cells that were classified as place cells was reduced in the desynchronization group, whereas groups with decreased PV+ or SOM+ did not differ from control (one-way ANOVA, F(3,36) = 80.07, P < 0.001). n = 10 simulated animals with 130 pyramidal cells per group for all panels. Error bars represent 1 s.e.m. ***P < 0.001, all comparisons against the control condition.
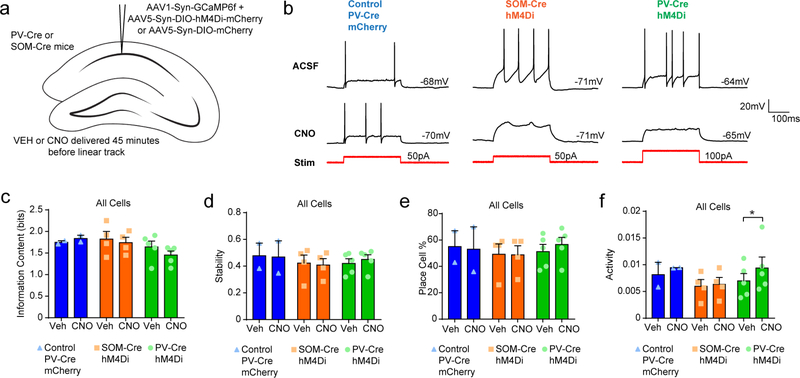
We next sought to experimentally validate the predictions of our model by inhibiting PV+ or SOM+ interneurons while mice ran on a linear track (Extended Data Fig. 10). To accomplish this, we used PV-Cre and SOM-Cre mice and injected both AAV5-SynDIO-hM4Di-mCherry (or AAV5-Syn-DIO-mCherry as control) and AAV1-Syn-GCaMP6f into the CA1. We then used a miniscope to image CA1 neurons and trained mice to run on a linear track. We administered vehicle or clozapine-N-oxide (CNO) (5 mg kg−1, intraperitoneal) 45 min before training, which has been shown to decrease interneuron firing by around 20% in vivo33. As predicted by our model, we found no differences in the information content, stability, or number of place cells when animals received CNO (Extended Data Fig. 10) compared to vehicle. Finally, we validated that the hM4Di was effective at reducing inhibition by measuring reduced responses to stimulation of PV+ and SOM+ interneurons in the presence of CNO in vitro. In addition, as would be expected by disinhibition, we found increased firing rates of neurons in vivo during behavior when PV+ interneurons were inhibited with CNO (Extended Data Fig. 10).
We next extended our model to conditions of increasingly large reductions in CA1 interneuron signaling (Extended Data Fig. 8). We examined 25%, 50%, 75%, and 100% reductions in PV+ and SOM+ interneurons. For SOM+ interneurons we found that high levels of interneuron disruption (≥ 50% reduction) decreased information content, with no effect on stability or place cell percentage. For PV+ interneurons, we found a progressive decrease in information content and place cell percentage at a 50% or greater reduction, and increased stability at a 75% or greater reduction. Our model therefore predicted that only extreme loss of PV+ and SOM+ interneurons, beyond what has been observed in the epileptic CA1, leads to alterations in place cell coding. Together, these results indicate that desynchronization of inputs (rather than CA1 interneuron loss) is the primary driver of place cell instability in our model and probably contributes to poor spatial processing in TLE.
Discussion
Memory deficits are a core feature of temporal lobe epilepsy and understanding how cognitive impairments arise is a major priority for the epilepsy field34. Here we used in vivo electrophysiology and in vivo calcium imaging, together with network modeling, to characterize how chronic epilepsy disrupts spatial processing in the hippocampus. Our data suggest that desynchronization in the hippocampus of epileptic mice contributes to severe spatial processing deficits. Furthermore, we found deficits in the stability of spatial representations within trials and across short time intervals, indicating disrupted encoding of spatial representations. Strikingly, these deficits in stability emerged 6 weeks after pilocarpine treatment, well after the onset of spontaneous seizures and deficits in spatial information content. This timeline of instability does not match the loss of interneurons in the dentate gyrus or CA1, which occurs in the first few days after epileptogenesis9. Our results indicate that CA1 place cell instability is driven by a distinct circuit mechanism that is independent from seizure onset and interneuron loss, and separate from mechanisms driving reduced precision of spatial coding in epileptic mice. Finally, our modeling results further indicate that desynchronization, rather than CA1 interneuron loss, is likely to drive place cell instability in epilepsy. We found that modest levels of CA1 interneuron loss, similar to those seen in our epileptic mice, had no effect on the information content or stability of spatial representations in our model. We validated this hypothesis by expressing hM4Di in PV+ and SOM+ interneurons and saw no change in spatial representations in the presence of CNO. However, our modeling suggests that larger levels of interneuron loss can reduce spatial information while having either no effect on (SOM+ loss) or even increasing (PV+ loss) stability. This increased stability in the model may be due to increasing the reliance on inputs that are more consistent than inhibition. These hypotheses are currently difficult to test experimentally given the relatively low levels of inhibition provided by hM4Di expression and may require new tools (such as ultrapotent chemogenetics35) to fully test the predictions of the model. These results may also reconcile previous seemingly contradictory findings showing that inhibiting CA1 interneurons can either reduce information content36 or have little to no effect on spatial information15. Our modeling results suggest that the amount of suppression may be the critical factor in determining whether CA1 interneurons alter spatial coding.
It is also important to note that our modeling and interneuron manipulations were restricted to the CA1. In epileptic mice, we found the greatest damage (~60% loss) to SOM+ interneurons in the dentate gyrus. Although our model was restricted to the CA1, it did indicate that this level of interneuron loss can cause deficits in spatial information content. In addition, as these interneurons are lost in the first few days after pilocarpine treatment, they may better match the timeline of deficits in information content. Therefore, although our data indicate that CA1 interneuron loss is unlikely to be a driving factor in spatial coding deficits, interneuron loss in the dentate gyrus may contribute to deficits in spatial information content.
Spatial memory relies on the precise timing of interneurons that shape excitatory information processing through the hippocampus. We have now discovered that in epileptic mice the timing of inhibitory firing is disrupted and that, without properly timed excitation and inhibition, this circuit is unable to appropriately process spatial information. These findings suggest that it is the timing rather than amount of inhibition that is relevant for spatial coding deficits. This is consistent with recent work showing that, during the transition to seizures, hippocampal interneuron firing remains intact but the timing of inhibition is altered37,38. Interneuron dysfunction and disrupted spatial coding is also a common feature of other rodent models of diseases with cognitive deficits (such as Alzheimer’s disease39,40, schizophrenia41, aging42, and Down syndrome43) and it will therefore be important to explore how interneuron timing may be altered in these models. In future studies, it will be critical to identify the specific mechanisms that desynchronize this circuit to precisely target these mechanisms with new therapeutics for treating cognitive deficits associated with epilepsy.
Methods
Animals
All experimental protocols were approved by the Chancellor’s Animal Research Committee of the University of California, Los Angeles, or the Institutional Care and Use Committee of the Icahn School of Medicine at Mount Sinai, in accordance with the US National Institutes of Health (NIH) guidelines. For the pilocarpine experiments, 7-week-old male C57B6 mice from Charles River Laboratories were group housed (2–5 per cage) on a 12 h light–dark cycle for all experiments. Male mice were used because they had a lower mortality rate during preliminary studies, which has also been reported before44. For all pilocarpine experiments, naive animals were randomly assigned to either the pilocarpine or control groups before epileptogenesis. For hM4Di experiments, PV-Cre (The Jackson Laboratory, 008069) mice or SOM-Cre (The Jackson Laboratory, 013044) mice, aged 3–6 months were used. PV-Cre mice were randomly assigned to either hM4Di or control virus groups. Blinding was not performed during data collection because there are clear behavioral differences in epileptic and control animals. In addition, chronically epileptic animals have high morbidity and water restriction was closely monitored in the epileptic mice to prevent death.
Viral construct
AAV1.Syn.GCaMP6f.WPRE.SV40 virus (titer 4.65 × 1013 GC per ml) was purchased from Penn Vector Core. AAV5-Syn-DIO-hM4Di-mCherry (44362, titer 1.2 × 1013 genome copies (GC) per ml) and AAV5-Syn-DIO-mCherry (50459, titer 1.5 × 1013 GC per ml) were purchased from Addgene.
Drugs
CNO (Enzo Life Sciences) was made in a stock solution of 20 mg ml−1 DMSO and then diluted in saline to the desired concentration (0.5 mg ml−1 CNO). Vehicle consisted of saline with a similar proportion of DMSO. CNO (5 mg kg−1) or vehicle was injected (intraperitoneally) 45 min before imaging on the linear track.
Epileptogenesis and monitoring
Seven-week-old mice were randomly assigned to receive pilocarpine or control injections. All mice first received 1 mg kg−1 intraperitoneal injections of scopolamine methyl bromide (Sigma-Aldrich, CAS 155–41-9) to reduce the peripheral effects of pilocarpine. After 30 min, pilocarpine-treated mice received 275 mg kg−1 intraperitoneal injections of pilocarpine hydrochloride (Sigma-Aldrich, CAS 54–71-7) and all animals were monitored continuously for seizures. After 45 min, pilocarpine-treated mice that did not show behavioral seizures received additional pilocarpine injections of 50–100 mg kg−1. The beginning of status epilepticus was recorded as the time of the final behavioral seizure before entering continuous seizure activity. After 2 h of status epilepticus, all mice received 10 mg kg−1 intraperitoneal injections of diazepam and were closely monitored for 48 h to ensure recovery. Pilocarpine-treated mice that did not enter status epilepticus or showed continued negative health effects 48 h after pilocarpine were removed from the experiment. Our typical success rate for inducing epilepsy with pilocarpine is around 50%, including animals that die during status epilepticus and in the days and weeks following. A small proportion (around 10–15%) of epileptic animals were unable to perform the linear track or virtual reality tasks, presumably owing to ongoing seizures, and were removed from the experiments. To confirm chronic epilepsy in animals without EEG implants (Figs. 1–3), mice were video monitored in their home cage and seizures were recorded by a blind observer. In all experiments, spontaneous seizures were confirmed in all epileptic mice.
Video-EEG
For experiments examining the development of place cell stability (Fig. 4), a subset of animals (n = 3 control, n = 4 epileptic) were implanted with a wireless EEG device (ETA-F10, Data Sciences International) into the subcutaneous pocket posterior to the neck and continuously recorded in the home cage. EEG leads were placed between the dura and skull, with one lead above the frontal cortex and one lead over the contralateral parietal cortex. Meloxicam (1 mg kg−1) was administered both sduring surgery and for 2 d after surgery together with ampicillin (20 mg kg−1) for 7 d after surgery. Data were acquired with a telemetry system (Data Sciences International) running Ponemah v6 software and analyzed using Neuroscore v3.2.9 software (Data Sciences International). Seizures were identified from the EEG by blind human observers and confirmed by video review.
Morris water maze
Morris water maze45 was performed with a circular tub (4 ft in diameter) filled with a combination of water and white paint. For hidden platform training, a platform (10 cm in diameter) was submerged in one quadrant and start locations were randomized to one of four equidistant locations. Animals, at least 6 weeks after pilocarpine or control treatment, were trained three times per day for 6 d. In each trial, mice were given 60 s to find the platform. If mice found the platform earlier than 60 s, the trial ended then. If mice failed to find the platform, the trial terminated at 60 s. After each trial, mice were put on the platform for 15 s. Animals were given a probe trial, 1 d after the last training session, with the platform removed and animals were allowed to swim freely for 60 s. For the visible platform test, a black cover that extended above the water line was placed on top of the platform and two flags were affixed to the sides of the platform extending 14 cm above the water. The location of the platform was moved to a random location for each trial. Animals were tested for 2 d with four trials per day. For all sessions, animal location was tracked by an overhead camera running Actimetrics WaterMaze software v. 3.21 or a webcam analyzed offline with EthoVision XT 13.0 software.
Cued, delayed alternation T-maze
T-maze experiments were performed similar to previously described46. Before the experiment, mice at least 6 weeks after pilocarpine or control treatment were water restricted and maintained at a body weight of around 85% of their initial weight. Mice were handled for 3 min per day for 5 d and were then trained for 5 d. On each training day, mice received up to 20 trials within a limit of 45 min. Each trial consisted of three phases: cue phase, delay phase, and test phase (Extended Data Fig. 1). During the cue phase, animals began at the end of the long arm and one side of the T-maze was closed and one side remained open. Animals ran to the open arm to drink the 10 μl water reward, and then ran back to the starting point and received a second 10 μl water reward. During the delay phase, animals were confined to the starting location for 15 s while the maze was cleaned with 7% ethanol. For the test phase, the animal was released and allowed to choose one arm to visit. If the animal chose the side opposite the cue, it received a 10 μl water reward. If it chose the same side as the cue, no reward was presented. The animal then ran back to the starting point, received another water reward, and was confined to the start zone until the next trial began 30 s later.
Immunohistochemistry
For immunohistochemistry, epileptic mice were at least 19 weeks after pilocarpine treatment, and control mice were of a similar age. Mice were transcardially perfused with 4% PFA, followed by 24 h after fixation in the same solution. Free-floating 50 μm coronal sections were prepared using a vibratome and stored in 1 × PBS. Sections underwent three 10 min washes in 1 × PBS, followed by 2 h of blocking in PBS with 0.3% Triton X-100 and 3% normal goat serum. Next, sections were incubated with anti-rabbit primary antibody in PBS with 3% normal goat serum and 0.3% Tween-20 overnight at 4 °C (PV antibody, Thermo Fisher Scientific (Invitrogen), PA1933, at 1:400; GFAP antibody, Thermo Fisher Scientific, NB300141, at 1:2,000; SOM antibody, Thermo Fisher Scientific (Millipore), MAB354, at 1:300; NeuN antibody, Millipore Sigma, ABN78, at 1:2,000). For the SOM staining, tissue was incubated with the primary antibody for 96 h. After four 15 min washes in PBS, slices were incubated in secondary antibody in PBS (1:500 goat anti-rabbit polyclonal Alexafluor 555, Thermo Fisher Scientific, A21429) for 2 h at room temperature (22 °C) while covered in foil. Sections were then washed again four times with PBS for 15 min per wash. DAPI (Thermo Fisher Scientific, D1306, at 1:5,000) was included in with the second wash. Sections were left in PBS overnight and mounted on slides using ProLong Gold mounting solution (Thermo Fisher Scientific, P36934) the following day. Images were taken using a Zeiss LSM780 confocal microscope at ×20, ×40 (NeuN only), or ×63 (NeuN only). Settings were held constant across animals within a data set and cells were either counted manually by a blind experimenter (for PV, SOM, and NeuN), or with an unbiased automated cell counter (for GFAP, see https://github.com/ZachPenn/CellCounting). For each animal in each staining condition 2–4 images were quantified and an average was taken for each animal. Additional validation information can be found in the Nature Research Reporting Summary.
In vivo electrophysiology surgeries
For in vivo electrophysiology recordings, all mice underwent two stereotaxic surgeries. First, a stainless steel headbar was fixed onto the skull of the mouse. The mouse skull and headbar were each stereotaxically aligned and the headbar was lowered near the skull and fixed onto the skull of the mouse using cyanoacrylate glue and dental cement (Lang Dental). Dental cement was built up around the perimeter, creating a ring around the exposed skull, and filled with Kwik-Sil (World Precision Instruments). A final layer of dental cement was applied above the Kwik-Sil and dental cement perimeter. Carprofen (5 mg kg−1) was administered both during surgery and for 2 d after surgery together with amoxicillin (0.25 mg ml−1) for 7 d after surgery.
A craniotomy and ground implantation was performed 1 d before recording. First, the most superficial layer of dental cement was drilled off and the Kwik-Sil was removed. Mice were stereotaxically aligned and an approximately 2 mm × 1 mm craniotomy was performed over the target recording site (−2.1 mm anteroposterior, ±1.5 mm mediolateral). A second craniotomy was performed above the cerebellum, and a silver reference wire (Warner Instruments) was slipped between the skull and dura and fixed with dental cement. The exposed skull was covered with Kwik-Sil and the mouse was allowed to recover overnight.
Virtual reality training and recording
Mice were water restricted and maintained at a body weight of around 85% of their initial weight 1 week after headbar surgery. Mice were checked daily for signs of dehydration, fatigue, or infection and were immediately given access to water if any signs of distress were observed. This water deprivation schedule produces robust task motivation and no adverse health effects across at least 4 months47. Mice were handled for 5 min each day for 3 d and habituated to head-fixation on a spherical treadmill, locked to rotate on only one axis, for 3–5 d. Next, mice were trained to lick from a water tube by allowing a 5 μl water reward during each lick. Once mice reliably licked from the water tube, they were immediately trained to run along the virtual reality linear track. The virtual reality apparatus was similar to that previously described48 and consisted of a projector (Epson), a flat mirror, a circular mirror, and a toroidal screen that surrounded the animal (Fig. 1a), and the display was warped to match the curved screen with NTHUSIM software. The virtual linear track was created with Platinum Arts Studio Sandbox and modified to trigger water delivery and record location and reward data through a custom data-acquisition device. Mice were trained to run forwards to reach the end of the track and receive a water reward. Mice were first trained on a short track until they completed more than 100 trials within 45 min, and were then moved to a longer track. This was repeated until animals consistently ran on the longest track (total training was approximately 5–20 d). A 5 μl water reward was dispensed at the end of the virtual track through a metal tube controlled by a solenoid valve (Lee Company). Mice were deemed ready for recording after five consecutive and consistent training sessions. On the day of the recording, mice were head-fixed onto the treadmill, the Kwik-Sil covering the craniotomy was removed and replaced with buffered artificial cerebrospinal fluid (ACSF), and the mouse was aligned to the micromanipulator. A 128-channel silicon probe49 was slowly lowered into the hippocampus and the surface of the exposed brain was covered with mineral oil. After at least 60 min to allow the brain to settle, the recording began and continued for 180 min.
Silicon probe data was collected as previously described49. Signals were amplified (gain = 200), filtered (0.1–8,000 Hz), and multiplexed (32 channels per output wire) by custom headstages using integrated electronic circuits (Intan Technologies, RHA-2164B). Analog signals were simultaneously digitized on National Instruments data-acquisition devices (USB-6356) using Labview 2012 software (National Instruments). Each channel was sampled at a rate of 25 kHz. Licking and reward times were also recorded simultaneously on the same data-acquisition device. Licking behavior was monitored with an infrared beam lickometer (Island Motion). Trial start times, position, and reward times were also saved through a custom recording software connected via ethernet to the virtual reality software.
In vivo electrophysiology analysis
All data analysis was carried out with custom MATLAB scripts. Data files were first demultiplexed into continuous signals. In epileptic mice, there was a trend towards reduced numbers of trials performed and a reduced proportion of time spent running (Extended Data Fig. 1); however, there was no such trend in running speed. We therefore restricted all analysis to times of locomotion. Interictal spikes were also removed. For local field potential (LFP) analysis, signals were bandpass filtered using zero-phase digital filtering and phase relationships were measured using the Chronux toolbox50. Sublayers of the hippocampus were identified from stereotaxic coordinates and electrophysiological markers, including peak theta and ripple power51, theta phase shifts51, gamma coherence52, dentate spike phase reversal53,54, and the density of single units. A subset was confirmed with post-hoc histology. Spike sorting was performed using PyClust software55. All channels from each sublayer of the hippocampus were background subtracted and bandpass filtered at 600–6,000 Hz, and putative spikes were extracted from signals at least 4 s.d. from the mean. Using peak amplitude, trough amplitude, and principal components, single units were isolated by manual clustering. Putative interneurons were identified based on firing rate, mean autocorrelogram, and complex spike index54–56. For phase-locking analysis, LFPs were filtered and the Hilbert transform was used to determine the phase of each spike. For each interneuron, phase angles from each spike were then used to calculate a Raleigh’s r-value and mean phase, and the significance of phase locking was tested with a Raleigh’s test for non-uniformity. Group differences in the distribution of preferred phases was assessed with the Kuiper test using the CircStats toolbox in MATLAB57. Owing to the relatively high variability of phase preferences and low numbers of recorded cells we collapsed across animals with the following number of cells per animal (Fig. 1f,i): control CA1, 16, 9, 15, 18, 13; epileptic CA1, 6, 19, 10, 9, 13; control dentate gyrus, 7, 4, 3, 14, 6; epileptic dentate gyrus, 3, 13, 6, 12, 7. To confirm that the observed desynchronization between the CA1 and dentate gyrus in epileptic mice was not skewed by differences in the number of cells recorded we performed the aggregate firing analysis and cross-region correlations across animals (Fig. 1l,m).
Calcium imaging surgeries
For all surgeries, mice were anesthetized with 1.5–2.0% isoflurane and placed into a stereotactic frame (David Kopf Instruments). Lidocaine (2%, Akorn) was applied to the sterilized incision site as an analgesic, and subcutaneous saline injections were administered throughout each surgical procedure to prevent dehydration.
For calcium imaging experiments, all mice underwent two stereotaxic surgeries. First, mice were unilaterally injected with 500 nl of AAV1.Syn.GCaMP6f. WPRE.SV40 virus at 50–120 nl min−1 in the dorsal CA1 (−2.1 mm anteroposterior relative to bregma, +2.0 mm mediolateral from bregma, and 1.65 mm ventral from the skull surface) using a Nanoject microinjector (Drummond Scientific). After a week of recovery, mice underwent a GRIN lens implantation surgery. A skull screw was fastened to the skull and a craniotomy 2 mm in diameter was performed above the viral injection site. The cortical tissue above the targeted implant site was carefully aspirated using 27 gauge and 30 gauge blunt needles. Buffered ACSF was constantly applied throughout the aspiration to prevent desiccation of the tissue. The aspiration ceased after partial removal of the corpus callosum and full termination of bleeding, at which point a GRIN lens (2 mm diameter, 4.79 mm length, 0.25 pitch, 0.50 numerical aperture, Grintech) was stereotaxically lowered to the targeted implant site (−1.35 mm dorsoventral from the skull surface relative to the most posterior point of the craniotomy). Cyanoacrylate glue and dental cement was used to seal and cover the exposed skull, and Kwik-Sil covered the exposed GRIN lens. Carprofen (5 mg kg−1) and dexamethasone (0.2 mg kg−1) were administered during surgery and for 7 d after surgery together with amoxicillin (0.25 mg ml−1) in the drinking water. Animals were anesthetized again 3 weeks later, and a miniature microscope locked onto an aluminum baseplate was placed on top of the GRIN lens. After searching the field of view for in-focus blood vessels and cells, the baseplate was cemented into place and the miniscope was unlocked and detached from the baseplate. A plastic cap was locked into the baseplate to prevent debris build-up.
For experiments tracking the development of place cell instability (Fig. 4), we used a modified timeline to allow for imaging exactly 3 weeks after pilocarpine treatment. We first injected GCaMP6f virus and implanted a GRIN lens above the CA1 during a single surgery. We induced status epilepticus with pilocarpine 2 weeks later, and started habituation and training on the linear track after 6 d. We then attached the baseplate and implanted a wireless EEG transmitter into the animals to record seizure activity in the home cage between trials.
For experiments inhibiting PV+ and SOM+ interneurons (Extended Data Fig. 10), we performed two virus injections. First, we injected 200 nl of AAV5-Syn-DIO-hM4Di-mCherry or AAV5-Syn-DIO-mCherry directly into the pyramidal layer of the CA1. We then injected 500 nl of AAV1-Syn-GCaMP6f into the CA1 at the same coordinates as previous experiments and implanted the GRIN lens as described above.
Wire-free miniscope
The wire-free miniscope is a modified version of the wired miniscope described previously26, with the additional features of being battery powered and logging imaging data onto onboard removable memory. The optics, Delrin housing, and excitation light source are the same between the wired and wire-free miniscope but the complementary metal oxide semiconductor (CMOS) imaging sensor electronics and data acquisition electronics were redesigned and packaged onto a single printed circuit board (PCB) to enable wire-free operation. All electrical components were chosen to minimize power consumption and weight. The main components of the wire-free PCB are a low-power CMOS imaging sensor (Teledyne E2V, JADE EV76C454), ARM Cortex M7 microcontroller (Atmel, ATSAME70N21A), and microSD card mount (Molex, 0475710001). The PCB also includes necessary voltage regulators, an SD card voltage translation transceiver (Texas Instruments, TXS0206AYFPR), custom excitation LED current-driving circuitry, battery connector, and 12 MHz oscillator. A single-cell lithium-polymer battery (Power Stream, GM041215, 45 mAH, 1.1 g) was attached to the side of the miniscope and connected to the wire-free PCB during operation. The upper limit on the length of recording sessions is determined by the size and efficiency of the battery. Storage space is not a limiting factor as microSD cards can now store up to 1 TB of data (~147 h of continuous recording). Using 45 mAh batteries we can reliably record for at least 30 min. Overall, the wire-free miniscope weighs 4–5 g, depending on the imaging sensor and battery used. Following power-up, the microcontroller (MCU) accesses configuration information, programmed by custom PC software, in a specific memory block of the microSD card that holds recording parameters such as recording length, frame rate, excitation LED intensity, imaging resolution, and imaging exposure length. Once configuration is complete, the MCU waits 5 s before beginning recording, at which time the excitation LED and onboard status LED turn on. At the end of the configurable recording duration, the excitation LED and onboard status LED turn off and the MCU terminates data logging onto the microSD card. Offline synchronization of the miniscope recording with a behavioral camera is achieved through detecting the on and off event of the status LED of the miniscope.
Pixel values from the CMOS imaging sensor are clocked into a three-frame circular buffer in the MCU using its parallel capture interface and direct memory access (DMA) controller to minimize processor overhead. Once an imaging frame has been received by the MCU it is written in raw format into incremental memory blocks of the microSD card together with a footer containing timing and error flag information. Wire-free miniscope data were recorded at 20 frames per s at a resolution of 320 pixels by 320 pixels using ×2 pixel subsampling. Running at this data rate, the entire system consumes approximately 300 mW of power during recording.
Linear track imaging
Before the experiment, mice were water restricted and maintained at a body weight of around 85% of initial weight. Mice were handled for 5 min per day for 3 d and habituated to the miniscope for at least 10 d with the weight and duration of the sham microscope progressively increasing each day until mice comfortably sustained the full miniscope for 15 min sessions. A wire-free miniscope, powered by a single-cell, 45 mAh lithium-polymer battery, was affixed to the baseplate of the mouse. Before the beginning of each imaging session, a sample 15 s imaging video was recorded to ensure the proper field of view and the linear track cleaned with 70% ethanol. Data were locally saved onto a microSDHC card (SanDisk). At the beginning of the linear track recording session, the miniscope was powered on and the mouse was placed on one end of the 2 m track. Mice ran back and forth along the track as experimenters pipetted 10 μl water reward at each end of the track. Behavior was recorded with an overhead webcam (Logitech). After 15 min, the microscope powered off and the mouse was taken off the track and returned to its cage. For initial characterization (Figs. 2,3), imaging sessions were performed every 2 d for 8–10 sessions, followed by shorter interval imaging sessions occurring 24 h, 6 h, and 30 min apart. For experiments tracking the development of place cell instability (Fig. 4), we used a wired miniscope as previously described26 to reduce the time needed to habituate animals to the heavier wire-free miniscope.
Linear track analysis
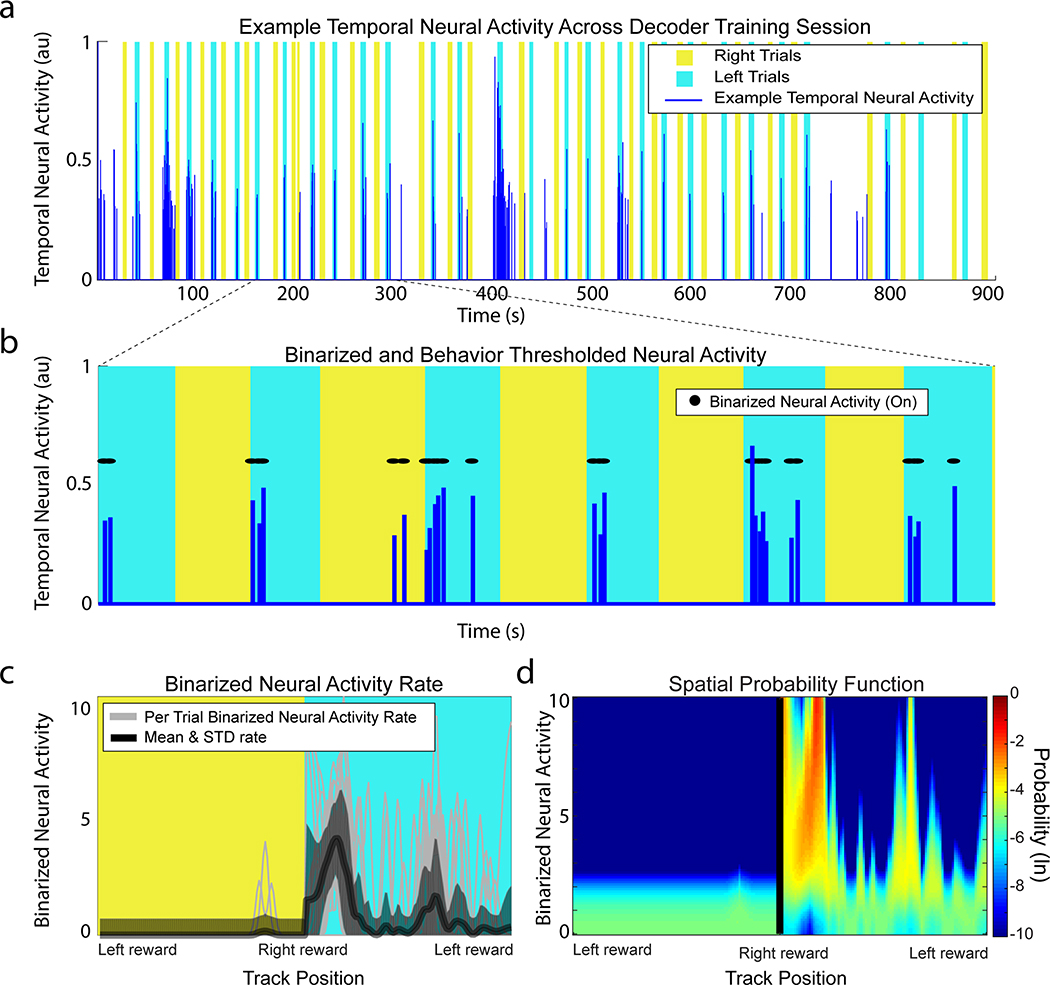
Wire-free miniscope data were extracted from microSD cards and saved as uncompressed 8 bit AVI video files for processing and analysis. Animal position was tracked with an overhead behavioral camera that was synchronized offline with the miniscope data using MATLAB scripts or ezTrack software58 (available at https://github.com/denisecailab/ezTrack). A fast Fourier transform-based image registration algorithm (courtesy of Dario Ringach, David Geffen School of Medicine, UCLA) was used to correct for frame to frame translational shifts of the brain during animal movement. Constrained non-negative matrix factorization for endoscopic recordings (CNMF-E)59 was used to identify and extract the spatial shapes and fluorescent calcium activity of individual neurons. Fast non-negative deconvolution60 was used to deconvolve the extracted fluorescent activity of each neuron. The resulting measure can be interpreted as the probability of a neuron being active at each frame scaled by an unknown constant. For the sake of brevity, we refer to this measure as the ‘temporal neural activity’ of a cell. Because the temporal neural activity of each cell is scaled by an unknown number that varies across cells we calculated the activity of each cell as the proportion of frames with temporal neural activity above 0.
Spatial neural activity rates were calculated using 2-cm-wide spatial bins and a speed threshold of greater than 10 cm s−1. Temporal neural activity and occupancy of the animal were spatially binned and then smoothed using a Gaussian kernel with σ = 5 cm. The binned neural activity was divided by the binned occupancy to calculate the spatial neural activity rate of each cell.
The information content of the spatial neural activity rate map (in bits)61 of a single neuron was defined as:
where ti represents the occupancy time spent in the i-th bin and λi represents the neural activity rate in the i-th bin. The significance of the spatial information was calculated using a circular trial shuffling procedure. For each trial, the position of the animal was circularly shifted by a random amount and then the spatial activity rate map and information content was calculated across all shuffled trials. This procedure was repeated 500 times to determine a significance measure for the spatial activity rate map of each neuron. The significance measure is the percentile of the true information content value within the distribution of the information content values from the 500 randomly shuffled sessions.
The stability of the spatial activity rate map of each single neuron was calculated by taking the Fisher Z-score of the Pearson correlation coefficient between the spatial activity rate maps at two time points (Extended Data Fig. 6). Within-session stability was calculated by averaging the stability measure of odd versus even trials with the stability measure of first half versus the second half of the trials (Extended Data Fig. 6). We found that stability in odd versus even trials was slightly higher than from the first half versus the second half of the trials, consistent with findings that CA1 populations can represent temporal information across a trial62. The same circular trial shuffling procedure mentioned above was applied to calculate the significance of within-session stability.
The PVC for a population of N neurons over a track length L was defined as:61
where and are the spatial activity rate maps of neuron j at two different time points. PVC curves were generated by calculating the mean PVC value for all PVC bins with a Δx difference in positions between the two time points being compared. Δx was varied from 0 to 150 cm to generate the full PVC curve.
Place cell criteria
For a cell to be classified as a place cell it must satisfy the three following criteria: the information content of the neuron is above chance (P < 0.05) based on the circularly shuffled distribution, the within-session stability of the neuron is above chance (P < 0.05) based on the circularly shuffled distribution, and the spatial activity rate map of the neuron has consecutive bins spanning at least 10 cm with activity rates in at least the 95th percentile of binned activity rates of circular trial shuffled spatial activity rate maps.
Matching cells across sessions
First, field-of-view shifts between sessions were corrected by performing rigid image registration between the mean frame of two sessions. For all pairs of cells across two sessions, the centroid distance and Pearson correlation of spatial footprints was calculated. Pairs with a centroid distance that was less than or equal to 4 pixels (~7 μm) and with a spatial footprint correlation that was greater than or equal to 0.6 were counted as the same cell. More information can be found in Extended Data Fig. 6.
Bayesian decoder
A Bayesian decoder was constructed to evaluate how well neural activity estimated the location of the animal based on an independent training set of neural activity (Extended Data Fig. 7). For within-session decoding, the training data set was generated from three-quarters of the trials evenly distributed across the session and then the decoder was applied to the remaining one-quarter of interspersed trials. For across-session decoding, the training data set was generated from the entire session of the first time point and then the decoder was applied to the entire session of the second time point. All decoders used the top 100 neurons with the highest within-session stability in the training session and that showed up in both training and decoding sessions.
Temporal neural activity (deconvolved fluorescence activity) was first binarized for both the training data set and decoding data set. For each neuron, time points with zero probability of neural activity were kept at zero; all other time points were set to one. Data points where the animal was in a reward zone or when the speed of the animal was less than 10 cm s−1 were excluded from the training and decoding data sets. For the training data set, the binarized spatial neural activity rate per trial per neuron was calculated. Binarized neural activity and occupancy were spatially binned along the length of the track, smoothed using a Gaussian kernel (σ = 5 cm), and then the binned activity was divided by the binned occupancy. For each neuron in the training set, the mean and s.d. of the per-trial binarized spatial neural activity rate were calculated per spatial bin. Bins with an s.d. below the mean per bin s.d. of that neuron were raised to the mean s.d. The training data set for each neuron resulted in a Gaussian distribution for each spatial bin along the track, which was used as the probability function in the decoder.
To estimate the position of the animal at time t using the neural activity of N neurons present in both the training data set and decoding data set the decoder I was defined as:
where occ(x) is the occupancy per spatial bin x from the training data set. μi(x) is the mean and σi(x) is the s.d. of the binarized neural activity rate at spatial bin x of neuron i from the training data set. Si(t) is the mean binarized neural activity rate of neuron i from the decoding data set over time t ± Δt, where Δt = 0.125 s.
The CA1 network model
Place cell formation was simulated using a CA1 network model consisting of biophysical neurons (Hodgkin-Huxley formalism) with reduced morphology32. The model was based on the structure and the connectivity data previously described63–66 and included 130 principal neurons (pyramidal cells) and six types of interneurons: two axoaxonic cells, eight basket cells, two bistratified cells, two OLMs, one VIP–CCK cell and four VIP/CR cells, as previously detailed32. Each pyramidal cell model consisted of 27 apical and basal dendritic compartments, whereas interneurons were simulated with simplistic dendritic complexity. The synaptic properties of the CA1 network connections are listed in Supplementary Table 1. All neuronal models included Na+, delayed rectifier K+, and leak channels. Pyramidal cells contained A-type and M-type K+ channels, accompanied by L-type, R-type, and T-type Ca2+ channels, as well as Ih current and Ca2+-dependent medium and slow after-hyperpolarization K+ conductances. Axoaxonic cells, basket cells, bistratified cells, and VIP/CCK cells also contained A-type K+, L-type and N-type Ca2+ currents, and Ca2+-dependent K+ and Ca2+dependent and voltage-dependent K+ conductances. OLMs included A-type K+ and Ih currents, and VIP/CR similarly comprised D-type K+, N-type Ca2+, and Ca2+-dependent K+ channels. Channel parameters are the same as previously reported32. All single-cell models were validated against experimental data with respect to passive and active properties, such as input resistance, membrane time constant, and rheobase current32. Axoaxonic cells contact pyramidal cells on their axons, basket cells and VIP/CCK on cell bodies, bistratified cells on medial apical and basal dendrites, and OLMs on distal apical dendrites. VIP/CR contact OLMs and basket cells, providing the network with the disinhibitory circuitry. Principal neurons also received excitatory background activity on their dendrites, in the form of AMPA, NMDA, GABAA, or GABAB synaptic currents. All interneurons received excitation via AMPA synapses, whereas pyramidal cells received excitation via AMPA and NMDA receptors in their apical and basal dendrites. Inhibitory synapses consisted primarily of GABAA receptors, except for pyramidal cell dendrites, which had both GABAA and GABAB receptors.
External synaptic input to the network was provided by ECIII, CA3 Schaffer collaterals, and medial septum. ECIII and CA3 excited both pyramidal cells and interneurons, whereas afferents from the medial septum contacted interneurons with inhibitory synapses. This input was modeled so as to reproduce theta-like activity at 8 Hz. ECIII provided grid-like synaptic inputs, whereas the CA3 inputs were place-like, produced by the application of a spatial filter (see below) onto ECIII inputs. The hallmark properties that the network model reproduced in line with experimental data were: the formation of place cells along a one-dimensional linear track67, the increased number of place cells encoding reward location (enrichment) in the presence of a simulated reward zone41, and the decrease in enrichment following removal of VIP+ model cells32.
Place cell formation and simulated experiments
A single place field in the CA1 emerged from the convergence of eight grid cells differing in their size and phase and six place cells. Grid-like inputs (ECIII) were simulated as simple summation of three sinusoidal waves68, whereas place-like inputs (CA3) were formed using a spatial filter to the grid-like inputs in the following way: first, the spikes from the eight grid cells were pooled in a single vector consisting of two non-overlapping categories, the infield (Nin) and outfield (Nout) spikes. For a specific theoretical place field:
where are the spike times of ECIII from j-th neuron (j = 1,…,8), xcenter is the theoretical place-field center and I xleft and xright are the borders of place cell to the left and right of its center, respectively. Both xleft and xright are set to 6 cm, thus every spike that occurred in place field center ± 6 cm is considered as an infield spike, otherwise it was included in outfield category. In order to produce place cell’s properties, that is, higher mean firing rate inside the place field and low mean firing rate outside69, we randomly sampled more spikes from the infield category. Specifically, each place-like input was formed using a random amount of infield spike times (Gaussian μ = 0.16,σ = 0.02) and outfield spike times (Gaussian μ = 0.016,σ = 0.02).
where N(·) denotes the Gaussian distribution. Using this approach, we increased the signal-to-noise ratio for each place cell. Subsequently, six similar place-like inputs were produced to have the same characteristics as those found in CA3 (ref. 69). This process was repeated for all predefined place fields and the CA3 input production from ECIII input ensured its theta-rhythm-locked firing70.
Each pyramidal cell received the convergent octal of grid-like inputs (from ECIII) to its distal apical dendrites and the convergent hexal of place-like inputs (from CA3) to its medial apical and basal dendrites. Roughly 50% of pyramidal cells had reduced synaptic weights (10% of their control values) for ECIII and CA3 inputs, representing the silent place cells reported in CA1 (ref. 71). To cover the linear track (200 cm in length) with place cells, we simulated 41 theoretical place cells located at places from 0 to 200 cm with a step of 5 cm. Thus, 328 inputs from ECIII and 246 from CA3 provided the network with external excitatory input.
To simulate the changes observed in epileptic animals9,10,72, OLM (SOM+) interneurons were reduced by 25%, axoaxonic, basket, and bistratified (PV+) interneurons were reduced by 25%, and the ECIII and CA3 inputs were desynchronized by ± 30 ms (Fig. 5c–f). Desynchronization of 30 ms was modeled by shifting the spike timings in ECIII inputs by a normal distribution with mean μ = 0 ms and s.d. σ = 30 ms. The network model was calibrated via slight modifications in its synaptic weights to produce place cells consistent with the experimental evidence reported here (Fig. 2). In addition, to examine how changes in interneuron signaling alter spatial coding we performed simulations with progressively increased levels of interneuron reduction and desynchronization levels (Extended Data Fig. 8).
Analysis of modeling data
For each pyramidal cell with a place field, we calculated the information content, stability, and peak firing rate. Information content and stability were calculated similarly to the experimental place cell data (see Linear track analysis above). Occupancy information was circularly shuffled by a random amount for each of the trials, and the information content and stability index of the resulting rate map were computed. This procedure was repeatedly and independently applied to generate two null distributions of 500 information content values and 500 stability index values. A cell was considered to have significant spatial information content, stability index, or both, if the information content and stability index of its true rate map exceeded that of 99% of the values in the corresponding null distribution. A place cell was defined as a cell that had field size within 5 and 25 cm and a peak firing rate greater than 1 Hz.
Each ‘virtual animal’ represented a CA1 model network with a slightly different connectivity profile, drawn at random while keeping the same connection probabilities. For each virtual animal we simulated ten repetition trials in which the stimulation was different owing to input and path navigation noise. Linear track navigation was simulated so that the virtual animal remained in each position (cm) for 50 ms (Gaussian with μ = 50 ms, σ = 30 ms). To account for the experimentally reported random stops, we introduced a random amount of simulated stops (0–10) at various locations (uniformly distributed), whereby the occupancy time at the stop bin and its neighbors (±1 cm) was increased to 200 ms (Gaussian with μ = 200 ms, σ = 50 ms). Both the number and duration of random stops were different for every trial. Thus, the simulation time for every trial ranged between 10,000 ms (with no stops) and 12,000 ms (with ten random stops). The simulation time step was 0.1 ms. Simulations were performed in a high-performance computing cluster with 312 cores under the CentOS 6.7 Linux operating system. The CA1 network model and all single cells were simulated in the NEURON (7.6.2) simulation environment73, and the grid-like and place-like inputs, the background noise, and the analysis were constructed using python (2.7.15) custom-made software under the Anaconda Distribution (5.3.1) platform.
In vitro electrophysiology
In vitro electrophysiology was performed 2–3 weeks after virus injection. Mice were anesthetized by isoflurane through inhalation followed by rapid decapitation. The brain was dissected and 350 μm coronal acute brain slices were prepared on a vibratome in ice-cold cutting solution containing 135 mM NMDG, 10 mM d-glucose, 4 mM MgCl2, 0.5 mM CaCl2, 1 mM KCl, 1.2 mM KH2PO4, and 20 mM HEPES (305–310 mOsm, pH 7.35). Slices were then transferred to a recovery chamber with sucrose-based ACSF (sACSF) containing 55 mM sucrose, 85 mM NaCl, 25 D-glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, 4 mM MgCl2, and 26 mM NaHCO3 (300–305 mOsm, bubbled with 95% O2 and 5% CO2). The recovery chamber was kept in a 34 °C water bath for 30 min and then moved to room temperature.
Slices were kept in sACSF at room temperature for at least 30 min before recording. Then, acute brain slices were placed in a submerged chamber circulating with ACSF containing 126 mM NaCl, 10 mM D-glucose, 2 mM MgCl2, 2 mM CaCl2, 2.5 mM KCl, 1.25 mM NaHPO4, 1.5 mM sodium pyruvate, 1 mM l-glutamine and 26 mM NaHCO3 (300–305 mOsm, bubbled with 95% O2 and 5% CO2). All recordings were done at room temperature. Patch electrodes (4–6 MΩ) were filled with an intracellular solution containing 127.5 mM k-methanesulfonate, 10 mM HEPES, 5 mM KCl, 0.6 mM EGTA, 2 mM MgCl2, 2 mM Mg-ATP, 0.3 mM Na-GTP, and 5 mM Na-phosphocreatine.
Cells were visualized under an upright microscope (Nikon Eclipse FN1) with a differential interference contrast camera (DAGE-MTI IR-1000) and objective-coupled LEDs (Prizmatix). For current-clamp recordings, cells were held at −70 mV, and 500 ms current steps from 50 pA to 300 pA at 50 pA increments were injected to evoke action potentials. Data were digitized at 10 kHz and low-pass filtered at 5 kHz using Multiclamp 700B and pClamp 10 (Axon Instruments), and further analyzed using AxoGraph (version 1.7.0). Access resistance was monitored throughout the recording and cells with access resistance that changed more than 20% were discarded. Recordings were done before and after perfusing CNO (20 μM for SOM+ and 10 μM for PV+ interneurons).
Statistics
Statistics were performed using MATLAB (Mathworks), SPSS (IBM), and GraphPad Prism software. Statistical significance was assessed by twotailed unpaired Student’s t-tests, Mann-Whitney U-tests, one-way analysis of variance (ANOVA), or two-way ANOVA, where appropriate. Normality was assessed using the Kolmogorov–Smirnov test for normality. For non-normal distributions, non-parametric Mann-Whitney tests were performed. Significant main effects or interactions were followed up with post-hoc testing using Bonferroni corrections where applicable. Significance was declared at P < 0.05, with a precise P value stated in each case, and all tests were two-sided. For circular tests, group differences in the distribution of preferred phases were assessed with the Kuiper test. For calcium imaging in pilocarpine-treated mice, minimum sample sizes were determined by preliminary data using a wired miniature microscope showing more than 80% power with n = 5 per group. For all other experiments, sample size was not predetermined but our sample sizes are similar to those reported in previous publications15,26,74,75. Owing to poor behavior (< 10 trials run), one session from one control mouse and four sessions from one epileptic mouse were removed from Figs. 2,3. One pilocarpine-treated mouse was removed because there were no seizures identified during video monitoring or experimental tests, a pre-established criterion for inclusion in the study. One control animal was removed from Fig. 4 owing to seizure-like activity, a pre-established exclusion criterion for this study.
Wire-free miniscope availability
All design files, software, parts lists, and tutorials necessary to build and implement the wire-free miniscope are available at www.miniscope.org.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The experimental data that support the findings of this study are available from Peyman Golshani (pgolshani@mednet.ucla.edu) or Tristan Shuman (tristan. shuman@mssm.edu) upon reasonable request.
Code availability
The software and codes related to the CA1 network model and its analysis are available from the Poirazi lab (poirazi@imbb.forth.gr) on reasonable request. The model is available on ModelDB, accession number 256311. Data analysis scripts are available on reasonable request from Peyman Golshani (pgolshani@mednet.ucla.edu) or Tristan Shuman (tristan.shuman@mssm.edu).
Extended Data
Extended Data Fig. 1 |. Spatial memory deficits in epileptic mice.
a. Morris water maze task. Mice (at least 6 weeks after pilocarpine or control treatment) were trained to find a hidden platform for 6 days at random start locations. The next day, mice were given a probe trial to assess learning. On the probe trial, control mice spent more time in the target quadrant than epileptic mice, while epileptic mice spent more time in the opposite quadrant (n = 6 Control, n = 5 Epileptic, 2-way ANOVA FGroupXQuadrant(3,36) = 14.6, P < 0.001, Training Quadrant Post-hoc: P < 0.001, Opposite Quadrant Post-hoc: P = 0.02). Mice were trained to find a visible platform with a flag extending above the water. There were no differences between the control and epileptic mice on this task (n = 5 Control, n = 8 Epileptic, 2-way ANOVA FGroup(1,11) = 0.424, P = 0.53). c. Mice were trained on a cued, delayed alternation T-maze task for 5 days. On each trial, animals were cued to one direction (Cue Phase), returned to the start position for a 15 s delay (Delay Phase), and then released for the Test Phase. If the animal went to the opposite side it received a water reward. Epileptic mice performed worse than control mice (n = 5 Control, n = 6 Epileptic, 2-way ANOVA FGroup(1,45) = 44.98, P < 0.001). d–f. Activity during virtual reality recordings. No differences were found in trials per minute (n = 5 per group, Unpaired t-test, P = 0.16), percent time running (n = 5 animals per group, Unpaired t-test, P = 0.14), or running speed (n = 5 animals per group, Mann-Whitney test, P = 0.94). Error bars represent 1 S.E.M. *P < 0.05. g-i. Activity during calcium imaging on the linear track. No differences were found in the number of trials per minute (n = 5 animals per group, Mann-Whitney test, P = 0.15) or percent time running (n = 5 animals per group, Unpaired t-test, P = 0.64). epileptic mice ran faster than control mice on the linear track (n = 5 animals per group, Unpaired t-test, P = 0.03). Error bars represent 1 S.E.M. *P < 0.05, ***P < 0.001.
Extended Data Fig. 2 |. Local field potential power across frequencies.
a–f. LFP power in beta, slow gamma, mid-gamma, fast gamma, ripple and fast ripple frequencies throughout CA1 and DG. No differences were found in beta (FGroupXRegion(6,48) = 1.23, P = 0.31, FGroup(1,8) = 0.47, P = 0.51), fast gamma (FGroupXRegion(6,48) = 1.88, P = 0.10, FGroup(1,8) = 2.03, P = 0.19), ripple (FGroupXRegion(6,48) = 0.93, P = 0.48, FGroup(1,8) = 0.67, P = 0.44), or fast ripple (FGroupXRegion(6,48) = 1.38, P = 0.24, FGroup(1,8) = 0.24, P = 0.64) power. Epileptic mice had reduced slow gamma and mid-gamma power in the hilus of DG (Slow Gamma: FGroupXRegion(6,48) = 6.76, P < 0.001, FGroup(1,8) = 7.31, P = 0.03, Hilus: P < 0.001; Mid-gamma: FGroupXRegion(6,48) = 4.49, P = 0.001, FGroup(1,8) = 3.48, P = 0.09, Hilus: P = 0.01). n = 5 animals per group for all graphs. error bars represent 1 S.E.M. **P < 0.01, ***P < 0.001.
Extended Data Fig. 3 |. Theta phase locking changes are not caused by decreased power or specific to reference location.
a. Example clustering of single units from a single channel set in one animal using principal components, peak amplitude, and trough amplitude. b. Units were characterized as putative interneurons based on complex spike index (CSI), mean autocorrelogram (Mean AC), and mean firing rate. N = 5 Control, N = 5 Epileptic animals. c. Example phase locking in a CA1 interneuron. Top, mean waveforms from each recorded channel within the pyramidal layer. Middle, spike raster of 300 theta cycles during running, with the proportion of spikes occurring during each phase of theta. Bottom, Rose plot of firing phase relative to theta. This cell has a modulation index (r-value) of 0.41 and preferred firing phase (mu) of 189°. d–g. Theta cycles were subsampled to match power between control and epileptic mice. This did not change the results as CA1 interneurons in epileptic mice were less phase locked than in control mice (d; n = 71 Control cells, n = 34 Epileptic cells, Unpaired t-test, P < 0.001), while their preferred phase was not different (e; Kuiper circular test, P > 0.05). There were no differences in the modulation of DG interneurons (f; n = 34 Control cells, n = 22 Epileptic cells, Unpaired t-test, P = 0.80) but there were differences in the preferred phase of DG interneurons (g; Kuiper circular test, P < 0.001). h, i. Phase locking of DG interneurons to theta in the lacunosum moleculare. No difference was found in modulation (h; n = 34 Control cells, n = 41 Epileptic cells, Unpaired t-test, P = 0.25) but the distributions of preferred phases differed between the groups (i; Kuiper circular test, P < 0.001). j-k. Phase locking of DG interneurons to the theta in hilus of DG. No difference was found in modulation (j; n = 34 Control cells, n = 41 Epileptic cells, Unpaired t-test, P = 0.64) but the distributions of preferred phases differed between the groups (k; Kuiper circular test, P < 0.001). N = 5 animals per group for all panels. Error bars represent 1 S.E.M. ***P < 0.001.
Extended Data Fig. 4 |. Altered gamma synchronization in epileptic mice.
a. Gamma power throughout CA1 and DG. Epileptic mice had reduced gamma power in the hilus of DG (2-way RM ANOVA, FGroupXRegion(6,48) = 5.26, P < 0.001, Hilus: P < 0.001). b. Gamma coherence between each channel pair along the recording probe in control (left) and epileptic (middle) mice. Right, p-value matrix for each pair of recording sites (unpaired t-tests, uncorrected for multiple comparisons). c. Phase locking to CA1 gamma for each interneuron in CA1 of control and epileptic mice. each dot represents one interneuron and the data is double plotted for visualization. Closed circles were significantly phase locked (Raleigh test, P < 0.05) and open circles were not. d. Phase locking to CA1 gamma for each interneuron in DG of control and epileptic mice. e. Mean phase modulation (r) of CA1 interneurons to CA1 gamma. Interneurons in epileptic mice had increased phase modulation (n = 69 Control cells, n = 57 Epileptic cells, Mann-Whitney test, P < 0.001). f. Mean phase modulation (r) of DG interneurons to CA1 gamma. Interneurons in epileptic mice had no change in phase modulation (n = 34 Control cells, n = 41 Epileptic cells, Mann-Whitney test, P = 0.42). g. Rose plot of preferred firing phases of significantly phase locked CA1 interneurons. There were no differences between interneurons in control and epileptic mice (Kuiper circular test, P > 0.05). h. Rose plot of preferred firing phases of significantly phase locked DG interneurons. The distribution of preferred phases in control and epileptic mice were different (Kuiper circular test, P < 0.001). i. Aggregate inhibition of all interneurons in CA1 and DG relative to CA1 gamma phase (2-way RM ANOVA, CA1: FGroupXPhase(17,136) = 1.22, P = 0.25; DG: FGroupXPhase(17,136) = 2.196, P = 0.007). j. Pearson’s correlation between CA1 and DG aggregate inhibition relative to gamma. No difference was found between epileptic and control animals (Unpaired t-test, P = 0.19). All data in this figure came from 5 animals per group. Error bars represent 1 S.E.M. *P < 0.05, ***P < 0.001, ns: not significant.
Extended Data Fig. 5 |. Wire-free Miniscope design and behavior.
a. Schematic of wire-free Miniscope printed circuit board (PCB). b. Picture of top and bottom of wire-free sensor board. This camera sensor board attaches to the Miniscope body, which contains all optic components. c. The wire-free Miniscope can be used to probe naturalistic behaviors including social interaction. We tested social behavior with the wire-free Miniscope compared to no Miniscope or a wired version. d. During a social interaction test mice with either no Miniscope or the wire-free Miniscope chose a social cup over an empty cup, while mice with a wired Miniscope showed no significant preference (n = 3 per group, Unpaired t-tests, No Scope: P = 0.03, Wire-Free: P < 0.001, Wired: P = 0.10). Error bars represent 1 S.E.M. *P < 0.05, ***P < 0.001.
Extended Data Fig. 6 |. Stability of spatial representations.
a. Population Vector Correlation (PVC) of all cells in Control (left) and Epileptic (right) mice across all sessions recorded. b. Mean PVC as a function of offset distance (from the diagonal) across animals. Epileptic mice had higher PVC (that is, less distinct firing patterns) across distances of 30–150 cm (2-way RM ANOVA, FGroup(1,8) = 10.39, P = 0.01, post hoc P < 0.05 for each bin from 30–150 cm). c. Population Vector Correlation (PVC) of place cells in Control (left) and Epileptic (right) mice across all sessions recorded. d. Mean PVC as a function of offset distance (from the diagonal) across animals. Epileptic mice had higher PVC (that is, less distinct firing patterns) across distances of 32–150 cm (2-way RM ANOVA, FGroup(1,8) = 10.19, P = 0.01, post hoc P < 0.05 for each bin from 32–150 cm). N = 5 animals per group for all panels. Shading represents 1 S.E.M. *P < 0.05. e, f. Example cross registration in a Control (e) and Epileptic (f) mouse. Top, Aligned mean frame from each session (∼ 550 um x 550 um). Bottom, overlaid cells from each session with white X indicating matched cells. g. Spatial correlation and centroid distance were calculated for all cell pairs. Dotted lines indicate thresholds used as matching criteria. All matched cells had spatial correlation ≥ 0.6 and centroid distance ≤ 4 pixels (∼ 7 um). h. We assessed within-session stability of spatial representations in two ways. We first examined stability of the first half of trials against the second half of trials. We then examined stability of odd trials versus even trials. In both cases, we used the Fisher z-transformed correlation of the spatial firing rates between trials. We report stability as the average of the two different stability measures. i. We found a slight but significant difference between the two stability measures as the stability of odd/even trials was higher than in the first/second half of trials (2-way RM ANOVA FMeasurement(2,16) = 13.59, P < 0.001; post hoc tests: ***P < 0.001). j. The two stability measures were highly correlated in both control (Pearson’s correlation: All Cells: r = 0.83; Place Cells: r = 0.75) and epileptic (Pearson’s correlation: All Cells: r = 0.73; Place Cells: r = 0.72) neurons. N = 5 animals per group for all panels.
Extended Data Fig. 7 |. Example processing of one training session neuron for Bayesian decoding.
a. For each frame, temporal neural activity is calculated and classified into rightward trials, leftward trials, or non-trial times. b. Temporal neural activity is binarized and non-trial times are removed. c. The per trial binarized neural activity rate is calculated for rightward and leftward trials. d. The spatial probability function is constructed for each cell. A Gaussian distribution is first generated for each spatial bin using the mean and standard deviation of the binarized neural activity rate. The overall distribution is normalized across binarized neural activity rate rows and this data is entered into the Bayesian decoder for each cell. See Supplementary Video 3 for example decoding across all cells.
Extended Data Fig. 8 |. CA1 network model reveals that high levels of interneuron disruption can reduce information content, but not stability.
a. We tested how increasing amounts of desynchronization altered information content, stability, and place cell percent in our CA1 network model. We found significant reductions in all metrics with desynchronization of 15–35 ms (1-way ANOVA, Information Content: F(7,72) = 77.84, P < 0.001; Stability: F(7,72) = 70.02, P < 0.001; Place cell percent: F(7,72) = 99.44, P < 0.001; post hoc tests: *P < 0.05, **P < 0.01, ***P < 0.001). b. We next tested how reducing somatostatin-expressing neurons altered spatial coding in our model. We found that reducing SOM by 50–100% reduced information content (1-way ANOVA, F(4,45) = 38.01, P < 0.001, post hoc tests: ***P < 0.001) but not stability (1-way ANOVA, F(4,45) = 0.564, P = 0.69) or place cell percent (1-way ANOVA, F(4,45) = 2.369, P = 0.067). c. We next tested how reducing parvalbumin-expressing neurons altered spatial coding in our model. We found that reducing PV by 50–100% reduced information content (1-way ANOVA, F(4,45) = 134, P < 0.001, post hoc tests: **P < 0.01, ***P < 0.001) but also increased stability at 75–100% PV reduction (1-way ANOVA, F(4,45) = 18.26, P < 0.001, post hoc tests: ***P < 0.001). We also found that place cell percent was lower at 75–100% PV reduction (1-way ANOVA, F(4,45) = 41.78, P < 0.001, post hoc tests: ***P < 0.001).
Extended Data Fig. 9 |. Cell death and gliosis in epileptic mice.
a, b. Example immunohistochemistry staining for parvalbumin (PV), somatostatin (SOM), NeuN, and glial fibrillary acidic protein (GFAP) in dentate gyrus (a) and CA1 (b) of control and epileptic mice. For epileptic mice, tissue was collected at least 19 weeks after pilocarpine. c. PV staining was reduced in epileptic mice (2-way RM ANOVA, FGroup(1,14) = 7.086, P = 0.02, post hoc for CA1, CA3, DG: P > 0.05). N = 9 Control, N = 7 Epileptic. d. SOM staining was reduced in the DG of epileptic mice (2-way RM ANOVA, FGroup x Region(2,16) = 12.12, P < 0.001, post hoc for DG: P < 0.001; CA1, CA3: P > 0.05). N = 5 Control, N = 5 Epileptic. e. NeuN staining was reduced in CA3 of epileptic mice (2-way RM ANOVA, FGroup(1,16) = 5.581, P = 0.03, post hoc for CA3: P = 0.002; CA1, DG Hilus: P > 0.05; DG Blade: Unpaired t-test, t = 0.302, P = 0.77). N = 9 Control, N = 9 epileptic. f. Because GFAP expression can be altered in the soma or processes we analyzed both the cell counts and percent of activated pixels within each image. We did not detect a difference in the number of GFAP + neurons (2-way RM ANOVA, FGroup(1,8) = 4.60, P = 0.06), however we did find increased percent of activated pixels in CA1, CA3, and DG (2-way RM ANOVA, FGroup(1,8) = 14.15, P = 0.005, post hoc for all regions: P < 0.05). N = 5 Control, N = 5 Epileptic. g–i. In entorhinal cortex and area V1 we found no differences between groups in PV (2-way RM ANOVA, FGroup(1,7) = 0.371, P = 0.56, N = 5 Control, N = 4 Epileptic), SOM (2-way RM ANOVA, FGroup(1,8) = 0.815, P = 0.39, N = 5 Control, N = 5 Epileptic), or NeuN (2-way RM ANOVA, FGroup(1,8) = 0.288, P = 0.61, N = 5 Control, N = 4 Epileptic). j. We did find an increased number of GFAP positive neurons in EC and V1 (2-way RM ANOVA, FGroup(1,7) = 432.1, P < 0.001; post hoc: EC, P < 0.001; V1, P < 0.01, N = 5 Control, N = 4 Epileptic), and increased percent of activated pixels for GFAP in EC (2-way RM ANOVA, FGroup(1,7) = 31.1, P < 0.001; post hoc: EC, P < 0.001; V1, P = 0.31, N = 5 Control, N = 4 Epileptic). EC, entorhinal cortex; DG, dentate gyrus; Error bars denote 1 S.E.M. All sample sizes are by animal. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Fig. 10 |. Chemogenetic inhibition of PV + or SoM + interneurons does not alter spatial coding in CA1.
a. To selectively inhibit PV + and SOM + interneurons, we used PV-Cre and SOM-Cre mice and injected a cre-dependent virus expressing hM4Di (AAV5-Syn-DIO-hM4Di-mCherry) or control virus (AAV5-Syn-DIO-mCherry). We also injected virus to express GCaMP6f in all neurons (AAV1-Syn-GCaMP6f) to allow for calcium imaging. We then trained mice to run on a linear track and delivered CNO (5 mg/kg) or Vehicle (VeH) 45 min prior to imaging on the track. b. To confirm that CNO was effective in reducing firing in vitro we used whole-cell recordings of hippocampal mCherry + interneurons in acute brain slices and applied ACSF or ACSF with CNO during stimulation. Interneurons expressing hM4Di had reduced spiking to stimulation (middle, right) while control virus did not reduce spiking. c. We found no differences in information content between VEH and CNO in any of the groups examined (Paired t-tests – PV: t(4) = 1.769, P = 0.15; SOM: t(3) = 0.717, P = 0.53). d. We found no differences in stability between VEH and CNO in any of the groups examined (Paired t-tests – PV: t(4) = 2.047, P = 0.11; SOM: t(3) = 0.672, P = 0.55). e. We found no differences in place cell percent between VEH and CNO in any of the groups examined (Paired t-tests – PV: t(4) = 2.29, P = 0.08; SOM: t(3) = 0.169, P = 0.88). f. We found increased Activity with CNO compared to VEH in the PV-hM4Di mice, but not SOM mice (Paired t-tests – PV: t(4) = 2.897, P = 0.04; SOM: t(3) = 0.96, P = 0.41). N = 2 PV-Cre mCherry animals, N = 4 SOM-Cre hM4Di animals, N = 5 PV-Cre hM4Di animals. Error bars denote 1 S.E.M. *P < 0.05.
Supplementary Material
Acknowledgements
We thank K. Maguire, J. Lou, A. Fariborzi, J. Daneshrad, S. Ghiaee, R. Manavi, C. Araradian, M. Song, B. Wei, C. Zhou, A. Meyer, H. Chen, J. Davis, N. Abduljawad, J. Hodson, I. Bachmutsky, L. Zilbermintz, H. Karbasforoushan, J. Friedman, T. Kotze, D. McCoy, K. Casale and E. Goldblatt (all of UCLA); and N. Berryman, G. Condori, M. Abdelmageed, C. Rosado, and B. Nunez (all of Mount Sinai) for excellent technical assistance and help with experiments. This work was supported by VA Merit Award 1 I01 BX001524-01A1, U01 NS094286, R01MH101198, R01 MH105427, U54 HD87101, R01NS099137, and NSF Neurotech Hub 1700408 to P.G.; a David Geffen School of Medicine Dean’s Fund for development of open-source miniaturized microscopes to B.S.K., A.J.S., and P.G.; a CURE Epilepsy Taking Flight Award, an American Epilepsy Society Junior Investigator Award, R03 NS111493, a Cellular Neurobiology Training Grant T32 NS710133, and an Epilepsy Foundation Postdoctoral Research Training Fellowship to T.S.; Neurobehavioral Genetics Training Grant T32 NS048004 and Neural Microcircuits Training Grant T32 NS058280 to D.A.; DP2 MH122399, a Klingenstein-Simons Fellowship, a McKnight Memory and Cognitive Disorder Award, a NARSAD Young Investigator Award, a Fay/Frank Seed Grant Program award, a One Mind Rising Star Research Award, National Research Service Award F32 MH97413, and Behavioral Neuroscience Training Grant T32 MH15795 to D.J.C.; DP1 MH104069 to B.S.K.; a McKnight Technological Innovations in Neuroscience Award to S.C.M.; and Dr. Miriam and Sheldon G. Adelson Medical Research Foundation funding to A.J.S. S.C., I.P. and P.P. were supported by the European Research Council Starting Grant dEMORY (GA 311435) and the Fondation Sante.
Footnotes
Competing interests
The authors declare no competing interests.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s4159-3019-0559-0.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41593-019-0559-0.
Supplementary information is available for this paper at https://doi.org/10.1038/s41593-019-0559-0.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bell B, Lin JJ, Seidenberg M & Hermann B The neurobiology of cognitive disorders in temporal lobe epilepsy. Nat. Rev. Neurol 7, 154–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleen JK, Scott RC, Lenck-Santini PP & Holmes GL in Jasper’s Basic Mechanisms of the Epilepsies (eds Noebels JL, Avoli M, Rogawski MA, Olsen RW & Delgado-Escueda AV) 915–929 (Oxford Univ. Press, 2012). [Google Scholar]
- 3.Henshall DC & Meldrum BS in Jasper’s Basic Mechanisms of the Epilepsies Noebels JL, Avoli M, Rogawski MA, Olsen RW & Delgado-Escueda AV) 362–376 (Oxford Univ. Press, 2012). [PubMed] [Google Scholar]
- 4.Sloviter RS Decreased hippocampal inhibition and a selective loss of interneurons in experimental epilepsy. Science 235, 73–76 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Houser CR et al. Altered patterns of dynorphin immunoreactivity suggest mossy fiber reorganization in human hippocampal epilepsy. J. Neurosci 10, 267–282 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W et al. Surviving hilar somatostatin interneurons enlarge, sprout axons, and form new synapses with granule cells in a mouse model of temporal lobe epilepsy. J. Neurosci 29, 14247–14256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Z et al. A reorganized GABAergic circuit in a model of epilepsy: evidence from optogenetic labeling and stimulation of somatostatin interneurons. J. Neurosci 33, 14392–14405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi M & Buckmaster PS Reduced inhibition of dentate granule cells in a model of temporal lobe epilepsy. J. Neurosci 23, 2440–2452 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinocourt C, Petanjek Z, Freund TF, Ben-Ari Y & Esclapez M Loss of interneurons innervating pyramidal cell dendrites and axon initial segments in the CA1 region of the hippocampus following pilocarpine-induced seizures. J. Comp. Neurol 459, 407–425 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Cossart R et al. Dendritic but not somatic GABAergic inhibition is decreased in experimental epilepsy. Nat. Neurosci 4, 52–62 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Hirsch JC et al. Deficit of quantal release of GABA in experimental models of temporal lobe epilepsy. Nat. Neurosci 2, 499–500 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Goldberg EM & Coulter DA Mechanisms of epileptogenesis: a convergence on neural circuit dysfunction. Nat. Rev. Neurosci 14, 337–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christenson Wick Z, Leintz CH, Xamonthiene C, Huang BH & Krook-Magnuson E Axonal sprouting in commissurally projecting parvalbumin-expressing interneurons. J. Neurosci 95, 2336–2344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varga C, Golshani P & Soltesz I Frequency-invariant temporal ordering of interneuronal discharges during hippocampal oscillations in awake mice. Proc. Natl Acad. Sci. USA 109, E2726–E2734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Royer S et al. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat. Neurosci 15, 769–775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klausberger T et al. Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature 421, 844–848 (2003). [DOI] [PubMed] [Google Scholar]
- 17.van Dijk MT & Fenton AA On how the dentate gyrus contributes to memory discrimination. Neuron 98, 832–845 (2018). e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauviere L et al. Early deficits in spatial memory and theta rhythm in experimental temporal lobe epilepsy. J. Neurosci 29, 5402–5410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inostroza M, Brotons-Mas JR, Laurent F, Cid E & de la Prida LM Specific impairment of “what-where-when” episodic-like memory in experimental models of temporal lobe epilepsy. J. Neurosci 33, 17749–17762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuman T, Amendolara B & Golshani P Theta rhythmopathy as a cause of cognitive disability in TLE. Epilepsy Curr. 17, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Pigozzi D et al. Altered oscillatory dynamics of CA1 parvalbumin basket cells during theta-gamma rhythmopathies of temporal lobe epilepsy. eNeuro 3, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X et al. Seizure-induced changes in place cell physiology: relationship to spatial memory. J. Neurosci. 23, 11505–11515 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenck-Santini PP & Holmes GL Altered phase precession and compression of temporal sequences by place cells in epileptic rats. J. Neurosci 28, 5053–5062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenck-Santini PP & Scott RC Mechanisms responsible for cognitive impairment in epilepsy. Cold Spring Harb. Perspect. Med 5, pii: a022772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou JL, Shatskikh TN, Liu X & Holmes GL Impaired single cell firing and long-term potentiation parallels memory impairment following recurrent seizures. Eur. J. Neurosci 25, 3667–3677 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Cai DJ et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh KK et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberti WA, Perkins LN, Leman DP & Gardner TJ An open source, wireless capable miniature microscope system. J. Neural Eng 14, 045001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbera G et al. Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information. Neuron 92, 202–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curia G, Longo D, Biagini G, Jones RS & Avoli M The pilocarpine model of temporal lobe epilepsy. J. Neurosci. Methods 172, 143–157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Stein A & Sarnthein J Different frequencies for different scales of cortical integration: from local gamma to long range alpha/theta synchronization. Int. J. Psychophysiol 38, 301–313 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Turi GF et al. Vasoactive intestinal polypeptide-expressing interneurons in the hippocampus support goal-oriented spatial learning. Neuron 101, 1150–1165 (2019). e1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cichon J & Gan WB Branch-specific dendritic Ca2+ spikes cause persistent synaptic plasticity. Nature 520, 180–185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley MS, Jacobs MP, Lowenstein DH & Stewards NEB The NINDS epilepsy research benchmarks. Epilepsia 50, 579–582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnus CJ et al. Ultrapotent chemogenetics for research and potential clinical applications. Science 364, eaav5282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grienberger C, Milstein AD, Bittner KC, Romani S & Magee JC Inhibitory suppression of heterogeneously tuned excitation enhances spatial coding in CA1 place cells. Nat. Neurosci 20, 417–426 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Grasse DW, Karunakaran S & Moxon KA Neuronal synchrony and the transition to spontaneous seizures. Exp. Neurol 248, 72–84 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Miri ML, Vinck M, Pant R & Cardin JA Altered hippocampal interneuron activity precedes ictal onset. eLife 7, e40750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mably AJ, Gereke BJ, Jones DT & Colgin LL Impairments in spatial representations and rhythmic coordination of place cells in the 3xTg mouse model of Alzheimer’s disease. Hippocampus 27, 378–392 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Cacucci F, Yi M, Wills TJ, Chapman P & O’Keefe J Place cell firing correlates with memory deficits and amyloid plaque burden in Tg2576 Alzheimer mouse model. Proc. Natl Acad. Sci. USA 105, 7863–7868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaremba JD et al. Impaired hippocampal place cell dynamics in a mouse model of the 22q11.2 deletion. Nat. Neurosci 20, 1612–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenzweig ES & Barnes CA Impact of aging on hippocampal function: plasticity, network dynamics, and cognition. Prog. Neurobiol 69, 143–179 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Raveau M et al. Alterations of in vivo CA1 network activity in Dp(16)1Yey Down syndrome model mice. eLife 7, e31543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 44.Buckmaster PS & Haney MM Factors affecting outcomes of pilocarpine treatment in a mouse model of temporal lobe epilepsy. Epilepsy Res. 102, 153–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris R Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11, 47–60 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA & Gordon JA Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature 464, 763–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo ZV et al. Procedures for behavioral experiments in head-fixed mice. PLoS One 9, e88678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey CD, Collman F, Dombeck DA & Tank DW Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shobe JL, Claar LD, Parhami S, Bakhurin KI & Masmanidis SC Brain activity mapping at multiple scales with silicon microprobes containing 1,024 electrodes. J. Neurophysiol 114, 2043–2052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bokil H, Andrews P, Kulkarni JE, Mehta S & Mitra PP Chronux: a platform for analyzing neural signals. J. Neurosci. Methods 192, 146–151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lubenov EV & Siapas AG Hippocampal theta oscillations are travelling waves. Nature 459, 534–539 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Schomburg EW et al. Theta phase segregation of input-specific gamma patterns in entorhinal-hippocampal networks. Neuron 84, 470–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karlsson KA & Blumberg MS Temperature-induced reciprocal activation of hippocampal field activity. J. Neurophysiol 91, 583–588 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Senzai Y & Buzsaki G Physiological properties and behavioral correlates of hippocampal granule cells and mossy cells. Neuron 93, 691–704 (2017). e695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willers B Multimodal sensory contributions to hippocampal spatiotemporal selectivity. Doctoral Dissertation, University of California Los Angeles. (2013). [Google Scholar]
- 56.Csicsvari J, Hirase H, Czurko A, Mamiya A & Buzsaki G Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving Rat. J. Neurosci. 19, 274–287 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berens P CircStat: a MATLAB toolbox for circular statistics. J. Stat. Softw 10.18637/jss.v031.i10 (2009). [DOI] [Google Scholar]
- 58.Pennington ZT, et al. ezTrack: an open-source video analysis pipeline for the investigation of animal behavior. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/592592v1 (2019). [DOI] [PMC free article] [PubMed]
- 59.Zhou P et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogelstein JT et al. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol 104, 3691–3704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravassard P et al. Multisensory control of hippocampal spatiotemporal selectivity. Science 340, 1342–1346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mau W et al. The Same Hippocampal CA1 Population Simultaneously Codes Temporal Information over Multiple Timescales. Curr. Biol 28, 1499–1508.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bezaire MJ, Raikov I, Burk K, Vyas D & Soltesz I Interneuronal mechanisms of hippocampal theta oscillations in a full-scale model of the rodent CA1 circuit. eLife 5, e18566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cutsuridis V, Cobb S & Graham BP Encoding and retrieval in a model of the hippocampal CA1 microcircuit. Hippocampus 20, 423–446 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Cutsuridis V & Poirazi P A computational study on how theta modulated inhibition can account for the long temporal windows in the entorhinal-hippocampal loop. Neurobiol. Learn. Mem 120, 69–83 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Konstantoudaki X, Papoutsi A, Chalkiadaki K, Poirazi P & Sidiropoulou K Modulatory effects of inhibition on persistent activity in a cortical microcircuit model. Front. Neural Circuits 8, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danielson NB et al. Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area CA1. Neuron 91, 652–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solstad T, Moser EI & Einevoll GT From grid cells to place cells: a mathematical model. Hippocampus 16, 1026–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Leutgeb S, Leutgeb JK, Treves A, Moser MB & Moser EI Distinct ensemble codes in hippocampal areas CA3 and CA1. Science 305, 1295–1298 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Buzsaki G Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Epsztein J, Brecht M & Lee AK Intracellular determinants of hippocampal CA1 place and silent cell activity in a novel environment. Neuron 70, 109–120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith BN & Dudek FE Short- and long-term changes in CA1 network excitability after kainate treatment in rats. J. Neurophysiol 85, 1–9 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Hines ML & Carnevale NT The NEURON simulation environment. Neural Comput. 9, 1179–1209 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Ziv Y et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci 16, 264–266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ewell LA, Fischer KB, Leibold C, Leutgeb S & Leutgeb JK The impact of pathological high-frequency oscillations on hippocampal network activity in rats with chronic epilepsy. eLife 8, e42148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data that support the findings of this study are available from Peyman Golshani (pgolshani@mednet.ucla.edu) or Tristan Shuman (tristan. shuman@mssm.edu) upon reasonable request.