Abstract
There are intense interests in discovering proregenerative medicine leads that can promote cardiac differentiation and regeneration, as well as repair damaged heart tissues. We have combined zebrafish embryo-based screens with cardiomyogenesis assays to discover selective small molecules that modulate heart development and regeneration with minimal adverse effects. Two related compounds with novel structures, named as Cardiomogen 1 and 2 (CDMG1 and CDMG2), were identified for their capacity to promote myocardial hyperplasia through expansion of the cardiac progenitor cell population. We find that Cardiomogen acts as a Wnt inhibitor by targeting β-catenin and reducing Tcf/Lef-mediated transcription in cultured cells. CDMG treatment of amputated zebrafish hearts reduces nuclear β-catenin in injured heart tissue, increases cardiomyocyte (CM) proliferation, and expedites wound healing, thus accelerating cardiac muscle regeneration. Importantly, Cardiomogen can alleviate the functional deterioration of mammalian hearts after myocardial infarction. Injured hearts exposed to CDMG1 display increased newly formed CMs and reduced fibrotic scar tissue, which are in part attributable to the β-catenin reduction. Our findings indicate Cardiomogen as a Wnt inhibitor in enhancing injury-induced CM proliferation and heart regeneration, highlighting the values of embryo-based small molecule screens in discovery of effective and safe medicine leads.
Keywords: small molecule screen, Wnt inhibitor, regeneration
Introduction
The heart is the first organ to function during development. Deregulation in cardiac development and growth is the hallmark of cardiovascular diseases (Olson, 2004). Myocardial infarction (MI) and ischemic heart diseases are caused by a reduction in blood flow to the myocardium and characterized by loss of cardiomyocytes (CMs), leading to lethal arrhythmias and heart failure (Braunwald, 2013). Despite advances in modern biology and medicine, understanding etiologies of ischemic heart diseases and the management of heart failure remains a major challenge. Development of novel therapeutics that can stimulate CM regeneration represents a future goal of cardiovascular medicine. Several cellular approaches have been taken to offer possible solutions in terms of replenishing CMs in diseased hearts. These include transplantation of embryonic stem cells (ESCs), bone marrow stem cells, ESC-derived CMs (Bernstein and Srivastava, 2012), or reprogramming of fibroblasts into CMs (Qian et al., 2012; Song et al., 2012). However, these studies have demonstrated modest clinical benefits and have not been able to overcome the formidable technical hurdles, such as efficacy, safety, and delivery (Chong, 2012). They highlight the need to develop small molecules that have the capacity to stimulate endogenous cardiac muscle regeneration in damaged hearts.
Phenotype-based chemical screens in zebrafish offer considerable advantages for evaluating compound efficacy on formation of target tissues and organs, leading to identification of novel compounds for generation of specific cell types and tissues (MacRae and Peterson, 2015). We previously identified Cardionogen (CDNG) small molecules as Wnt inhibitors from embryo-based phenotype screens (Ni et al., 2011). CDNG promotes heart development by antagonizing the Wnt signaling pathway, in which its treatment enhances cardiomyogenesis at late development stages (Ni et al., 2011). Wnt/β-catenin signaling regulates heart development in a temporally biphasic fashion. Activation of Wnt/β-catenin signaling induces cardiac specification at early embryogenesis, whereas late inhibition of the Wnt/β-catenin pathway is critical for cardiac differentiation and maturation (Ueno et al., 2007). Inhibition of the Wnt pathway using Wnt inhibitor pyvinium, WNT974 or ICG001, mitigates pathological remodeling and symptoms of MI (Saraswati et al., 2010; Sasaki et al., 2013; Moon et al., 2017). However, their treatments are accompanied by severe toxicities. It is not clear whether Wnt inhibitors stimulate injury-induced CM proliferation and heart regeneration. Unlike previous reported Wnt inhibitors, the application of CDNG to developing embryos does not result in overall morphological defects (Ni et al., 2011). The CDNG core structure can guide us to discover selective Wnt inhibitors in promoting cardiomyogenesis, even cardiac regeneration and repair.
In this study, we described an embryo-based screen and tested the efficacy of identified small molecules on heart regeneration and repair. We identified Cardiomogen compounds to enhance cardiomyogenesis with minimal adverse effects. Cardiomogen treatment stimulates injury-induced CM proliferation and heart regeneration in zebrafish. Mechanistically, Cardiomogen acts as a Wnt inhibitor by inhibiting β-catenin-mediated Tcf/Lef-mediated luciferase activity. Using CDMG1, infarcted hearts with reduced β-catenin levels display increased CM production and reduced fibrosis, thus improving the recovery of heart function. Our studies reveal a CDMG-mediated Wnt inhibition for myocardial regeneration and injury repair, findings that warrant further preclinical investigation.
Results
In vivo embryo-based screens identify selective cardiomyogenesis compounds
Our previous studies reveal the capacity of CDNG small molecules in enhancing zebrafish heart development and embryonic heart size (Ni et al., 2011). CDNG small molecule family contains the core motif 1,2,4-triazolo[3,4-b]-1,3,4-thiadiazole (Figure 1A) (Ni et al., 2011). To identify more potent and selective cardiomyogenesis compounds, we designed and synthesized a series of compounds, by variation of substituents at the 3 and 6 position of the core motif, to form a CDNG-analog compound library, including R1- and R1/R2-compound series. The R1 series were synthesized by holding the 3-furan group (R2) constant and varying the identity of the 6-substituent (R1) (Figure 1A; Supplementary Figure S1). The R1/R2 compound series were prepared through variations of the 3 or 6 substituents (R2 or R1) of the core motif (Figure 1A; Supplementary Figure S2).
Figure 1.

In vivo embryo-based phenotype screen identified cardiomyogenesis compounds. (A) Small molecules designed and prepared around CDNG core structure motif. (B) Schematics of embryo-based cardiac phenotype screens. (C–E) Fluorescent microscopy analyses of Tg(cmlc2:mCherry) embryos showing normal size of DMSO-treated hearts (C) and enlarged embryonic hearts treated by CDMG1 (D) or CDMG2 (E) a, atrium; v, ventricle. (F–H) cmlc2 in situ hybridization analyses showing enlarged embryonic hearts treated by CDMG1 (G) and CDMG2 (H), compared to vehicle-treated hearts (F). (I) Bar graph showing total CM number in embryos treated by CDMG1, CDMG2, or CDNG1, from the 50% epiboly stage to 48 hpf. CM numbers are quantified using Tg(cmlc2:nDsRed) embryos. Data are mean ± SEM from three hearts for each group; one-way ANOVA with Bonferroni correction: ⋆P < 0.05. (J–M) Microscopy imaging analyses displaying overall embryo morphology in DMSO-, CDMG1-, CDMG2-, or WNT974-treated animals, from the 50% epiboly stage to 72 hpf. Scale bar, 100 μm (C–H, J–M).
We next conducted an in vivo embryo-based screen using transgenic zebrafish embryos Tg(cmlc2:mcherry), in which expression of red fluorescent protein (mCherry) is under the control of the cardiac myosin light chain 2 (cmlc2) promoter (Burns et al., 2005). Briefly, cmlc2-mCherry embryos were harvested from crosses and added to test wells at 5 h postfertilization (hpf), the onset of gastrulation when cardiac progenitor cells begin to form (Figure 1B) (Ni et al., 2011). Aliquots of test compounds were delivered into individual wells of plates. We examined embryonic heart size and morphology of treated embryos at 24, 48, and 72 hpf. Overall morphologies of embryos and other organs, including the anterior–posterior axis, brain, eye, and somite, were examined to determine whether overall embryogenesis was affected, providing a preliminary assessment of compound toxicity and selectivity (Supplementary Figures S1 and S2). We found that the R1-compound series failed to promote cardiomyogenesis and most of them proved toxic on embryogenesis (Supplementary Figure S1). Among the R1/R2-compound series screened (Supplementary Figure S2), we found that administration of compound 11 or 20 promoted stronger cardiomyogenesis than the original CDNG1 (Supplementary Figure S2). Their enlargement of the embryonic heart size was also validated by cmlc2-mCherry transgenic embryos (Figure 1D and E) and cmlc2 in situ hybridization (Figure 1G and H), when compared to vehicle-treated embryos (Figure 1C and F). We thus named compound 11 and 20 as Cardiomogen 1 (CDMG1) and Cardiomogen 2 (CDMG2), respectively. Like CDNG1, CDMG1 or CDMG2 treatment resulted in an increase of CM numbers (Figure 1I), without causing overall morphological defects in embryos (Figure 1J–L). However, treatment of chemicals such as compound 7 or 18 (Supplementary Figure S2), or a known porcupine/WNT inhibitor WNT974 (Liu et al., 2013), resulted in embryonic morphology defects (Figure 1M), reflective of the sensitive nature of toxicity assessment using zebrafish embryos.
We next assessed how Cardiomogen stimulates cardiogenesis by examining nkx2.5, a cardiac progenitor cell marker and cmlc2, at the lateral plate mesoderm (LPM). CDMG1 or CDMG2 treatment caused expansion of the nkx2.5-labeled cardiac progenitor cell population (Figure 2A–C) and subsequently promoted cmlc2-marked cardiac differentiation at the LPM (Figure 2E–G). Despite of the fact that WNT974 stimulated the increase of nkx2.5 and cmlc2 expression at the anterior lateral plate mesoderm (ALPM) (Figure 2D and H), its treatment caused disruption of the formation of EGFP-expressing cardiac chambers during development (Figure 2I–L). Previous studies report that Wnt signaling inhibition promotes cardiomyogenesis by repressing hematopoietic and endothelial cell fates at early embryonic stages (Naito et al., 2006). We therefore examined expression of other lateral plate cell markers, including lmo2, a hematopoietic progenitor cell marker, and flk1, an endothelial cell marker, in CDMG-treated embryos. CDMG1 treatment caused a reduction of lmo2 at the ALPM (Figure 2N), and the posterior lateral plate mesoderm (PLPM), when compared to vehicle-treated control embryos (Figure 2M and O). Furthermore, flk1 expression in the ALMP or the PLPM was slightly decreased in CDMG1-treated embryos, compared to control embryos (Figure 2Q–T). Together, our findings suggest that CDMGs promote expansion of the cardiac progenitor cell population through repressing hematopoietic and endothelial cell fates, findings that are consistent with the action mechanisms of Wnt inhibitors (Naito et al., 2006).
Figure 2.

Cardiomogen expands the cardiac progenitor cell population. (A–D) In situ hybridization analyses displaying the increased expression of nkx2.5 in the ALPM in CDMG1-, CDMG2-, or WNT974-treated embryos, compared to DMSO-treated embryos, from the 50% epiboly stage to the 5-somite stage. (E–H) Dorsal views displaying the increased cmlc2 expression at the ALPM in CDMG1-, CDMG2-, or WNT974-treated embryos, compared to DMSO-treated embryos, from the 50% epiboly stage to the 17-somite stage. (I–L) Fluorescent microscopy analyses exhibit the formation of two chambers in Tg(cmlc2:EGFP) embryos, an atrium (a, arrow) and a ventricle (v, arrow), in DMSO-treated embryos (I, K) and formation of one chamber in WNT974-treated embryos (arrow in J, L) at 72 hpf. Ventral view (I, J). Lateral view (K, L). (M–P) ISH analyses exhibiting the reduced expression of lmo2 in the ALPM (N) and the PLPM (P) in CDMG1-treated embryos, in comparison to DMSO-treated control embryos (M, O). (Q–T) CDMG1 treatment resulted in a slightly decrease of flk1 expression in the ALPM (R) and the PLPM (T), compared to DMSO-treated control embryos (Q, S), from the 50% epiboly stage to the 5-somite stage (M–T). CDMG1/CDMG2: 10 μM; WNT974: 0.5 μM; DMSO: 0.1%. Scale bar, 100 μm (A–H, I–L); 50 μm (M–T).
Cardiomogen inhibits Wnt signaling by reducing β-catenin and Tcf/Lef-mediated transcription
Because Cardiomogen compounds possess the CDNG-like core motif, we assessed whether CDMG1/CDMG2 inhibits Wnt/β-catenin signaling activity in developing embryos. Wnt signal is mediated by binding of Wnt proteins to LRP/Frizzled receptor complexes, resulting in translocation of β-catenin to the nucleus, where it activates T-cell factor/lymphoid enhancer factor (Tcf/Lef) transcription factors to promote target gene expression (Zimmerman et al., 2012). We employed Tg(TOP:GFP) embryos, in which green fluorescent protein (GFP) expression is under the control of four consensus Lef/Tcf binding sites (Dorsky et al., 2002), to measure Wnt/β-catenin activities. We observed Wnt/β-catenin activity in the midbrain of Tg(TOP:GFP) embryos by fluorescent microscopy analyses (Figure 3A and A1) (Dorsky et al., 2002). Administrating Tg(TOP:GFP) embryos with CDMG1 caused a loss of GFP fluorescence in the midbrain (Figure 3B, B1, and D). Similarly, CDMG2 treatment resulted in GFP reduction in the midbrain (Figure 3C, C1, and D).
Figure 3.

Cardiomogen inhibits the Wnt signaling pathway by reducing β-catenin and Tcf/Lef-mediated transcription. (A–C) Fluorescent optics revealing strong GFP fluorescence in the midbrain in DMSO-treated Tg(TOP:GFP) embryos (A, outlined area enlarged in A1). Treatment of 10 μM CDMG1 (B) or CDMG2 (C) during 5–24 hpf eliminates the majority of GFP fluorescence in the midbrain of embryos (outlined area enlarged in B1, C1). (D) Bar chart showing the relative TOP-GFP fluorescence intensity in DMSO, CDMG1, or CDMG2 treated embryos (n = 12, 18, and 19, respectively). Data are presented as mean ± SEM; one-way ANOVA with Bonferroni correction: ⋆⋆P < 0.01. (E) CDMGs inhibit Wnt3A (20 ng/ml)-induced TOPflash activity in CGR8-ES cells. Dose-response curves represent TOPflash activities normalized to cell number (mean ± SEM; performed in quadruplicate). (F) CDMGs inhibit LRP6ICD-mediated Wnt signaling. TOPflash activity was measured and graphed (mean ± SEM; one-way ANOVA with Bonferroni correction: no statistical significance; performed in triplicate). (G) CDMGs fail to inhibit Lef/Tcf transcription that is independent of β-catenin activity. CGR8-ES cells were treated with 0.1% DMSO, 1-mM CDMG1, or 1-mM CDMG2 (F, G). Graph represents relative TOPflash activities (mean ± SEM; one-way ANOVA with Bonferroni correction: ⋆⋆P < 0.01; performed in triplicate). (H–L) Immunofluorescence analyses reveal β-catenin protein in C184 stem cells under the administration of DMSO, Chir99021, CDMG1 + Chir99021, CDMG2 + Chir99021, or CDNG1 + Chir99021. Green, β-catenin; blue, DAPI. (M) Immunoblot analyses reveal β-catenin protein levels in C184 stem cells, in the presence of Chir99021, CDNG1 + Chir99021, CDMG1 + Chir99021, or CDMG2 + Chir99021, compared to DMSO treatment. GAPDH serves as a loading control. CDMG1, CDMG2, or CDNG1: 10 μM; Chir99021: 5 μM; DMSO: 0.5% (H–M). Scale bar, 150 μm (A–C); 10 μm (H–L).
We next assessed the effects of CDMG on Wnt/β-catenin-mediated TOPflash luciferase activity in murine ESCs (CGR8-ESs) (Ni et al., 2011). TOPflash luciferase activities were measured in CGR8-ES cells treated with CDMG1 or CDMG2. We found that CDMG1 treatment caused a reduction of Wnt/β-catenin-mediated luciferase activity in a dose-dependent manner (Figure 3E). Effector concentrations for half-maximal response (EC50) to Wnt inhibition were determined for CDMG1 (11 nM) and CDNG1 (38 nM) and IWR1 (10 nM). These findings indicate that CDMG1 is a more potent Wnt antagonist than its parental compound CDNG1, and that its Wnt inhibition activity is almost indistinguishable from IWR1 (Chen et al., 2009). We next examined whether CDMG1/CDMG2 disrupted Wnt/β-catenin signaling within responding cells. GCR8-ES-TOPflash cells were transfected with LRP6ICD, a Wnt co-receptor containing intra-cellular domain constitutively activating Wnt signaling (Tahinci et al., 2007). Administration of CDMG1 or CDMG2 resulted in a marked reduction of LRP6ICD-induced TOPflash activity (Figure 3F), indicating that CDMG acts within responding cells in inhibiting Wnt signaling. Transfection of ΔNLef-VP16 can induce TOPflash luciferase activity independently of β-catenin, through a process in which ΔNLef-VP16, lacking a β-catenin-binding site of Lef1, fuses with the VP16 transactivation domain (Aoki et al., 1999). We observed that CDMG1 or CDMG2 treatment failed to inhibit ΔNLef-VP16-induced TOPflash activity in GCR8-ES cells (Figure 3G). Together, these findings suggest that Cardiomogen inhibits β-catenin-dependent Wnt signaling within responding cells.
Based on the findings, we tested whether Cardiomogen directly affects β-catenin levels within cells. C184 stem cells were administrated with a GSK3β inhibitor Chir99021 that stabilizes β-catenin (Figure 3H and I). Immunofluorescent analyses revealed a significant reduction of Chir99021-induced β-catenin in CDMG1- or CDMG2-treated C184 stem cells (Figure 3J and K), when compared to Chir99021 treatment (Figure 3I). CDNG1 treatment also resulted in a reduction of β-catenin levels (Figure 3L). Furthermore, immunoblot analyses indicated that CDMG1, CDMG2, or CDNG1 treatment causes a marked reduction of β-catenin (Figure 3M), validating the immunofluorescent analyses.
Cardiomogen administration enhances injury-induced CM production and heart regeneration in zebrafish
We hypothesized that Cardiomogen small molecules that fuel embryonic heart development as Wnt inhibitors may have the capacity to stimulate adult heart regeneration. To test the idea, we utilized zebrafish heart regeneration model, in which heart muscles are fully regenerated within 1–2 months after partial ventricular resection (Poss et al., 2002). We first assessed if CDMGs expedite zebrafish heart regeneration using fibrin and collagen scar (acid fuchsin orange G, AFOG) analyses. We performed ventricular resection surgery, subjected injured animals to CDMG1 or CDMG2 treatment, and conducted AFOG analyses at 21-day post-amputation (dpa) (Figure 4A). While injured hearts displayed a large patch of fibrin and collagen deposits at 21 dpa (Figure 4B), the amputated area in CDMG1- or CDMG2-treated hearts contained only a small collagen deposit and was covered mostly by cardiac myofiber (Figure 4C and D). Quantification analyses indicated that the greater percentage of CDMG1- or CDMG2-treated hearts expedite wound healing than vehicle-treated control hearts (Figure 4E). These findings indicate that Cardiomogen administration reduces scarring and stimulates cardiac muscle regeneration.
Figure 4.
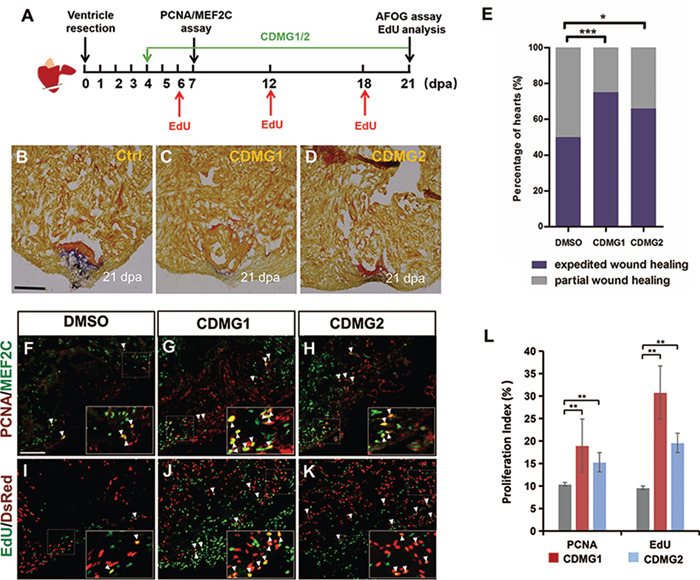
Cardiomogen stimulates injury-induced CM production and accelerates zebrafish heart regeneration. (A) Schematics of CDMG treatment procedure during cardiac regeneration. (B–D) AFOG analysis of heart sections visualizes cardiac muscle (orange), scar tissue (collagen, blue), and fibrin (red). Zebrafish were treated with 0.5% DMSO (B, Ctrl), 10 μM CDMG1 (C), or 10 μM CDMG2 (D) from 4 dpa until 20 dpa. (E) Histograms showing the wound-healing severity percentages of injured hearts treated with DMSO, CDMG1, or CDMG2. Partial wound healing (B): injury region contains large patches of fibrin and collagen deposits. Expedited wound healing (C): injury region contains largely cardiac myofiber with minimal fibrin or collagen deposits. Five treated hearts for each group were sectioned, and two sections per injured heart were chosen to score with wound healing severity. Chi-square test for measuring percentages in categorical data: ⋆⋆⋆P < 0.001, ⋆P < 0.05. (F–H) Confocal image analyses of PCNA+Mef2c+ cells (arrowheads) in 0.5% DMSO-, 10 μM CDMG1-, or 10 μM CDMG2-treated hearts at 7 dpa. Insets show high-magnification images of PCNA+Mef2c+ cells in rectangles. (I–K) Confocal image assessment of EdU+DsRed+ cells (arrowheads) in 0.5% DMSO-, 10 μM CDMG1-, or 10 μM CDMG2-treated hearts at 21 dpa. Insets show high-magnification images of EdU+DsRed+ cells in rectangles. (L) Bar charts displaying quantification of PCNA- or EdU-labeled CM proliferation index. 4264 DMSO-treated, 4189 CDMG1-treated, and 4267 CDMG2-treated CMs were analyzed from five independent hearts under each condition. Data are presented as mean ± SEM; one-way ANOVA with Bonferroni correction: ⋆⋆P < 0.01. Scale bar, 10 μm (B–D, F–K).
We next assessed the regenerative status of CMs in CDMG-treated injured hearts. Ventricle-amputated animals were subjected to CDMG1 or CDMG2 treatment from 4 to 6 dpa. CDMG- or DMSO-treated hearts were collected, sectioned, and subjected to double immunostaining at 7 dpa, using proliferating cell nuclear antigen (PCNA) antibody and myocyte enhancer factor 2C (Mef2C) antibody that recognizes nuclei of CMs (Figure 4A). We observed that PCNA-positive CMs were increased in CDMG1- or CDMG2-treated hearts (Figure 4F–H). Quantitative analyses revealed that CM proliferation index (PCNA+Mef2C+ /Mef2C+) in CDMG1-treated hearts was increased by 83% compared to vehicle-treated hearts (Figure 4L). Similarly, CDMG2-treated hearts had a 47% higher CM proliferation index than injured control hearts (Figure 4L). As an independent experimental approach to examine CM proliferation, we measured the nuclear incorporation of 5-ethynyl-2-deoxyuridine (EdU) in transgenic zebrafish Tg(cmlc2:nDsRed), where red fluorescent protein (DsRed) is expressed in nuclei of CMs (Figure 4A). We injected EdU into CDMG- or DMSO-treated wounded animals at 6, 12, and 18 dpa (Figure 4A). Immunofluorescence analysis identified that EdU incorporation index (EdU+DsRed+/DsRed+) in CDMG1- or CDMG2-treated hearts was significantly increased, when compared to vehicle-treated hearts (Figure 4I–L). Collectively, these studies indicate that Cardiomogen stimulates injury-induced CM proliferation and accelerates zebrafish heart regeneration.
To assess whether CDMG promotes heart regeneration by inhibiting Wnt/β-catenin signaling, we assessed the status of nuclear β-catenin in CDMG-treated hearts following amputation. Adult zebrafish hearts were subjected to ventricle resection surgery and treated with CDMG1 or CDMG2 from 4 to 10 dpa (Figure 5A). CDMG1/CDMG2-treated hearts were collected, sectioned, and subjected to double immunostaining using anti-β-catenin antibody and anti-myosin heavy chain (MHC) antibody that labels CMs (Figure 5A). In injured hearts, β-catenin was detectable in most nuclei of CMs, which were surrounded by MHC expression (Figure 5B, E, and H), while the majority of β-catenin remains in the cytoplasm of hearts (Figure 5B and H). We observed that β-catenin in the nucleus of CMs was significantly reduced in CDMG1-treated wounded hearts (Figure 5C, F, and I), in comparison to DMSO-treated control hearts (Figure 5B, E, and H). Similarly, CDMG2 treatment resulted in a reduction of nuclear β-catenin in CMs in injured hearts (Figure 5D, G, and J). We next conducted nuclear fraction and immunoblot analyses to examine the status of nuclear β-catenin in injured hearts treated with CDMG1/CDMG2 (Figure 5A). Notably, CDMG1 treatment caused a marked reduction of nuclear β-catenin extracted from nuclear fractions in injured hearts compared to vehicle-treated control hearts (Figure 5K and L), while total β-catenin from whole cell lysates was slightly decreased (Figure 5K and M). Similarly, nuclear β-catenin was reduced in CDMG2-treated wounded hearts (Figure 5K and L). Because CDMGs act as Wnt inhibitors by targeting β-catenin and reducing Tcf/Lef-mediated transcription in cultured cells (Figure 3), these findings indicate that the effects of CDMGs on heart regeneration are attributed, at least in part, to the inhibition of Wnt/β-catenin signaling.
Figure 5.

CDMG-treatment causes a reduction of nuclear β-catenin in injured hearts. (A) Schematics of CDMG treatment procedure during cardiac regeneration. CDMG1/CDMG2: 10 μM. (B–J) Confocal image analyses showing β-catenin in the nucleus (circles) and the cytoplasm (B–D), as well as MHC-labeled CMs (E–G) in injured hearts treated with DMSO, CDMG1, or CDMG2. H–J are merged panels of B and E, C and F, and D and G, respectively. Red: β-catenin; green: MHC; blue: DAPI. Scale bar, 100 μm (B–J). (K) Immunoblot analyses exhibit β-catenin protein extracted from nuclear extracts (NE) and total β-catenin extracted from whole-cell lysates (WL) in DMSO-, CDMG1-, or CDMG2-treated injured hearts. Tubulin and lamin B were used as loading controls for WL and NE, respectively. (L and M) Quantitative analyses of nuclear β-catenin and total β-catenin levels in DMSO-, CDMG1-, or CDMG2-treated wounded hearts. Data are presented as mean ± SEM; Student’s t-test: ⋆⋆P < 0.01, ⋆P < 0.5; performed in triplets.
Cardiomogen increases newly formed CMs and reduces fibrotic scar tissue in infarcted mouse hearts
To test whether Cardiomogen treatment enhances CM formation and reduces fibrotic scar in mouse hearts following MI, we induced MI by permanent ligation of the left anterior descending (LAD) coronary artery in mice and administrated infarcted animals with intraperitoneal injection of CDMG1 and EdU for 27 days (Figure 6A). Treated MI hearts were collected for EdU incorporation, phospho-histone H3 (Ph3) immunostaining, and Masson trichrome staining (Figure 6A), as well as cardiac function assessment using echocardiography analysis (Figure 7A–D). In agreement with previous studies (Malliaras et al., 2013), we observed the presence of EdU-positive CMs in the border zone of post-MI hearts (Figure 6B and C). Compared with vehicle-treated hearts, CDMG1 administration resulted in a marked increase of EdU-positive CMs expressing tropomyosin (TPM) in the border zone (Figure 6C–F). EdU incorporation index (EdU+TPM+ /TPM+) was increased by 2.51-fold in CDMG1-treated animals over that of vehicle-treated animals (Figure 6K). Furthermore, CDMG1-treated hearts exhibited higher percentages of TPM-labeled CMs that express Ph3 than vehicle-treated hearts in the border zone (Figure 6B, G–J, and L). In contrast, numbers of EdU incorporated or Ph3-positive CMs were not altered in the remote zone in CDMG1-treated hearts compared to vehicle-treated hearts (Figure 6B, K, and L). We did not observe the enlargement of cardiac cell size in infarcted hearts after 27-day treatment, in comparison to vehicle-treated hearts (Supplementary Figure S5). Histological analyses revealed a significant reduction of fibrotic scar tissue in CDMG1-treated infarcted mice, when compared to DMSO-treated animals (Figure 6M–O), suggestive of acceleration of the wound healing process.
Figure 6.

CDMG1 administration increases newly formed CMs and reduces fibrotic scar tissue in infarcted mouse hearts. (A) Schematic of CDMG1 treatment and EdU pulse experiments during MI. EdU was intraperitoneally injected (500 μg/animal) every 2 days post-MI. (B) Diagram of sections in post-MI hearts. IZ, infarct zone; RZ, remote zone; BZ, boarder zone. (C–F) Immunofluorescence analyses showing EdU+TPM+ CMs (arrows) in the border zone in CDMG1-treated hearts and DMSO-treated hearts. Red: EdU; green: TPM1. (G–J) Immunofluorescence images revealing an increase of Ph3+TPM+ cells in the border zone in post-MI hearts. Red: Ph3; green: TPM1. (K, L) Quantitative analyses of CM proliferation activity in CDMG1-treated MI mice. Data are presented as mean ± SEM; Student’s t-test: P represents statistical values in MI + CDMG1 group (n = 3, four sections per animal) compared to MI + DMSO group (n = 3, four sections per animal). ns, not significant. For EdU analysis, 4370 cells (infarct border zone) and 4610 cells (remote zone) in MI + DMSO group and 4361 cells (infarct border zone) and 4406 cells (remote zone) in MI + CDMG1 group were examined. For Ph3 analysis, 9244 cells (infarct border zone) and 8887 cells (remote zone) in MI + DMSO group and 8942 cells (infarct border zone) and 9142 cells (remote zone) in MI + CDMG1 group were examined. (M) The diagram of five sequential transversal sections of hearts (L1–L5, 3 μm thick) at 28-day post-MI. X: MI site. (N) Representative images of Masson trichrome staining of L1–L5 sections. Collagen-rich area (scar) stained in blue and healthy myocardium stained in red. (O) Fibrotic length percentage was calculated by dividing the midline length of total LV wall with the length of the infarcted LV wall. Data are presented as mean ± SEM; Student’s t-test: P represents the statistic value of MI + CDMG1 group (n = 4) versus MI + DMSO group (n = 4). Scale bar, 40 μm (C–J).
Figure 7.

CDMG1 ameliorates the recovery of cardiac function by reducing β-catenin following MI. (A–D) C57BL/6J mice received no (sham) or coronary artery ligation to induce MI. Representative echocardiographic M-mode images of left ventricles at 28 days post-MI are shown for respective groups of Sham + DMSO (A), Sham + CDMG1 (B), MI + DMSO (C), MI + CDMG1 (D). LVIDd, left ventricular internal dimension at end-diastole; LVIDs, left ventricular internal dimension at end-systole. (E, F) Quantitative analyses of cardiac functions for LVEF (E) and LVFS (F). Data are presented as mean ± SEM; Student’s t-test: P represents values in CDMG1 + MI group (n = 8) compared to DMSO + MI group (n = 5). Sham-operated mice were subjected to DMSO treatment (n = 5) and CDMG1 treatment (n = 5). (G) CDMG1 improves Kaplan–Meier survival curves in mice experiencing MI (n = 10). (H) Immunoblot analyses reveal β-catenin protein levels in infarcted heart tissues treated with DMSO and CDMG1. (I) Quantification analyses display a reduction of β-catenin in CDMG1-treated infarcted heart tissues under the conditions of H. Data are presented as mean ± SEM; Student’s t-test: P = 0.0156.
Cardiomogen improves the recovery of heart function by reducing β-catenin after MI
Echocardiography analyses of infarcted mouse hearts (Figure 7A–D) revealed a marked increase of left ventricle ejection fraction (LVEF), a mean fraction of blood ejected from the left ventricle, with CDMG1 treatment (Figure 7B). Furthermore, the left ventricle fractional shortening (LVFS) in infarcted hearts was significantly elevated with CDMG1 administration relative to vehicle-treated infarcted animals (Figure 7B). In comparison to sham-operated animals, cardiac function measured by both LVEF and LVFS remained depressed, although to a lesser extent, in CDMG1-treated MI animals (Figure 7E and F). We also noted that CDMG1 treatment markedly reduced the MI-induced ventricular chamber dilation (Supplementary Figure S3A) and heart weight gain (Supplementary Figure S3B), indicative of a reduction of adverse cardiac remodeling, which is consistent with cardiac functional improvements in CDMG1-treated animals. As a result, CDMG1 improves survival rates in mice experiencing MI (Figure 7G). We next examined β-catenin levels in infarcted hearts. Western analyses indicated a reduction of β-catenin levels in CDMG1-treated hearts compared to vehicle-treated hearts (Figure 7H and I), confirming the inhibition of Wnt signaling in CDMG1-treated infarcted hearts. Taken together, these findings suggest that CDMG1 promotes the functional recovery of injured hearts by stimulating CM production and reducing fibrotic scarring, which are in part attributable to the β-catenin reduction in infarcted heart tissues.
Discussion
In this study, we conducted embryo-based screens and discovered promising medicine leads for treatment of MI using regenerative mechanisms. We find that Cardiomogen small molecules enhance cardiomyogenesis and heart regeneration. Myocardial infarcted hearts exposed to CDMG1 reduce fibrotic scar tissue and increase newly formed CMs in the infarct border region, resulting in improvement of cardiac function recovery. Mechanistically, Cardiomogen acts as a cardiac-specific Wnt inhibitor by repressing β-catenin in injured heart tissues. Our findings indicate the selectivity and potency of CDMG-type agents for cardiac regeneration and injury repair.
We observed that embryos treated by Wnt inhibitors such as WNT974 or IWR1 exhibited shortened embryo tails and defective cardiac chambers, despite of the fact that they have beneficial responses in heart tissue after injury. Other classes of Wnt inhibitors are also associated with severe toxicities (Saraswati et al., 2010; Liu et al., 2013). CDMG compounds identified in this study can selectively promote cardiomyogenesis with minimal adverse effects. We find that the half-maximal inhibitory concentration (IC50) of CDMG1 in causing embryo teratogenicity is much higher than that of WNT974 (Supplementary Figure S4A and B). WNT974 administration also inhibits formation of erythrocytes and other cell types and tissues (Supplementary Figure S4C–E). Thus, CDMG agents warrant further clinical investigation, demonstrating the values of embryo-based small molecule screens (Chen et al., 2012). The toxicity differences between CDMG and other Wnt inhibitors are likely due to their differences in Wnt inhibition mechanisms. CDMG might affect the selective population of Wnt-responding cells (i.e. heart and others). Second, CDMG may interfere tissue-specific modifiers that modulate Wnt pathway components in cardiac tissues. Lastly, other unknown pathways or undetermined factors might be activated after CDMG treatment to cooperate with Wnt inhibition in modulating phenotypes in other tissues. Identifying in vivo binding factor of Cardiomogen may provide insights into tissue-specific action mechanisms.
In the adult heart, Wnt ligands and Wnt inhibitors are induced in different cardiac tissues during injury (Hermans et al., 2012; Ozhan and Weidinger, 2015). Both positive and negative roles for the Wnt pathway are observed in the repair of cardiac tissue, depending on the injury model and exact stages that have been examined (Hermans et al., 2012; Ozhan and Weidinger, 2015). Nonetheless, there is a trend that improvement of the cardiac function after injury could be achieved by inhibition of Wnt/β-catenin signaling using Wnt inhibitors. Overexpression of endogenous Wnt inhibitors sFrp1, Dkk3, and IGFBP4 or administration of chemical Wnt inhibitors alleviates the functional deterioration of myocardial infarcted hearts (Barandon et al., 2003; Hermans et al., 2012; Bao et al., 2015; Wo et al., 2016). Recent studies report that administration of Porcupine inhibitors not only reduces cardiac fibrosis and adverse remodeling, but also can enhance CM proliferation and heart regeneration after MI (Moon et al., 2017; Yang et al., 2017). Our studies find that Cardiomogen compounds reduce Wnt/β-catenin signaling and increase newly formed CMs in vertebrate hearts after injury. This is consistent with mechanisms that promote generation of CMs by β-catenin depletion after MI or enhance proliferation of resident CMs upon treatment of Porcupine inhibitors following injury (Zelarayan et al., 2008; Malliaras et al., 2013; Yang et al., 2017). The source of injury-induced CMs in adult hearts is predominantly derived from pre-existing CMs (Kikuchi et al., 2010; Porrello et al., 2011). Like porcupine inhibitors, Cardiomogen administration also reduces fibrotic scar tissue and accelerates wound healing in injured zebrafish and murine hearts. These effects might be achieved by targeting Wnt-mediated cardiac fibrosis mechanisms, in which epicardial or endothelial cells undergo epithelial-to-mesenchymal transition and adopt fibroblastic features (Aisagbonhi et al., 2011; Misztani et al., 2016). Hence, Cardiomogen compounds, with novel structures and minimal toxicities, act as Wnt inhibitors for heart development and regeneration, which may ultimately aid in design of therapeutic approaches for promoting cardiac repair following injury.
Materials and methods
Animal maintenance
All experimental procedures with animals were approved by the Institutional Animal Care and Use Committee at Fudan University, Shanghai. Zebrafish (Danio rerio, AB line) used in this study was raised and maintained according to standard procedures (Westerfield, 2000). Transgenic lines Tg(cmlc2:EGFP), Tg(cmlc2:mCherry), and Tg(cmlc2:nDsRed) used in this study were previously described (Burns et al., 2005; Jin et al., 2014). Adult male C57BL/6J mice (3 months old) were used in animal experiments. Murine experiments were performed according to the recommendation in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Chemical synthesis and treatment
Chemical compound 1a–1j (Supplementary Figure S1) and compound 5–9, 11, 15, 16, 20, and 23 (Supplementary Figure S2) were designed by TPZ and synthesized by Vanderbilt Chemical Synthesis Core. These chemicals were dissolved in DMSO (10 mM) and their identities were confirmed by 1H NMR. Compound 1k and 1l (Supplementary Figure S1) and compound 17 (001-858-140), compound 18 (001-858-147), compound 19 (001-858-143), compound 21 (001-846-733), compound 22 (000-811-240), and compound 24 (002-749-613) in Supplementary Figure S2 were purchased from MolPort.com and dissolved in DMSO (10 mM) as stocks. CHIR99021 was purchased from Sigma and dissolved in DMSO (20 mM). All non-aqueous reactions were performed in flame-dried flasks under an atmosphere of argon. The microwave assisted reactions were perfumed with Biotage Initiator. The compounds were purified by flash chromatography (Combi-flash Rf). Analytical thin-layer chromatography was carried out on E. Merck silica gel 60 F254 plates and visualized using UV. All solvents and chemicals were purchased from Sigma.
RNA in situ hybridization and quantification of cardiac cell number
Whole-mount in situ hybridizations were carried out as described using antisense ribonucleotide probes for nkx2.5, flk1, cmlc2 (Jia et al., 2007), and lmo2, hbe1 (Gansner et al., 2017). Tg(cmlc2:nDsRed) line was employed to quantify CM number during development using confocal microscopy analyses (Ni et al., 2011).
TOPflash luciferase reporter assay
TOPflash assays were performed as described (Ni et al., 2011). The murine CGR8-ES cells were transfected with constructs of TOPflash reporter using Lipofectamine 3000 (Life Tech) and seeded into 96-well plates at sub-confluency. Wnt3A (20 ng/ml) together with CDMG1 (0.1 nM–10 μM), CDMG2 (0.1 nM–10 μM), or IWR1 (0.1 nM–10 μM) were added into the medium for 24 h. Luciferase activities were measured by Steady-Glo Luciferase Assay (Promega) and normalized to viable cell number using the CellTiter-Glo Assay (Promega). All graphs were made in Prism 5 (GraphPad Software) with nonlinear regression fit to a sigmoidal dose-response curve.
Cell culture, immunoblot, and immunofluorescence analyses
C184 stem cell line was cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma). CGR8-ES cells were cultured in DMEM containing 10% fetal bovine serum (FBS), 20 mM L-glutamine, 50 mM β-mercaptoethanol, and 100-units/ml leukemia inhibitory factor (ESGRO-Chemicon). Chir99021 (5 μM) together with CDNG1 (10 μM), CDMG1 (10 μM), or CDMG2 (10 μM) were added into media. After 3-h treatment, cells were harvested and incubated in ice-cold radio immunoprecipitation assay (RIPA) cell lysis buffer containing protease inhibitors. The proteins were separated by SDS-polyacrylamide gel electrophoresis, probed with antibodies against β-catenin (Santa Cruz Biotechnology, 1:200), β-actin (Sigma, 1:500), Tubulin (Abcam, 1:500), and lamin B2 (Abcam, 1:500) and detected by horseradish peroxidase–conjugated antibodies. Image J measured the band intensity for quantification. C184 stem cells were treated with Chir99021 (5 μM) together with CDNG1 (10 μM), CDMG1 (10 μM), or CDMG2 (10 μM). After 3-h treatment, cells were subjected to immunostaining of β-catenin antibodies and 4 ,6-diamidino-2-phenylindole (DAPI) staining. Secondary antibodies used include AlexaFluor Alexa 488 (Invitrogen).
,6-diamidino-2-phenylindole (DAPI) staining. Secondary antibodies used include AlexaFluor Alexa 488 (Invitrogen).
Amputation surgery, AFOG, histology, and nuclear fraction analysis of zebrafish hearts
Adult zebrafish (4–12 months of age) was used for ventricular resection surgery as previously described (Poss et al., 2002). Isolated injured hearts were fixed in 4% paraformaldehyde, equilibrated in 30% sucrose and embedded in optimal cutting temperature compound, and sectioned at 10 μm. The section slides were fixed and subjected to immunostaining of various antibodies. Primary antibodies used in this study include anti-Mef2c (rabbit; Santa Cruz), anti-PCNA (mouse; Sigma), anti-β-catenin (Abcam; 1:100), and anti-MHC (MF20, DSHB; 1:100). Secondary antibodies include goat anti-mouse AlexaFluor 488 (Invitrogen), goat anti-mouse AlexaFluor 555 (Invitrogen), goat anti-rabbit AlexaFluor 488 (Invitrogen), and goat anti-rabbit AlexaFluor 555 (Invitrogen). AFOG analyses were performed as previously described (Poss et al., 2002). Nuclear proteins were extracted from injured zebrafish hearts using a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime). Protein concentration was determined using the bicinchoninic acid assay kit (Thermo Fisher Scientific).
CM proliferation index analyses in amputated zebrafish hearts
CM regeneration experiments were performed as previously described (Fang et al., 2013). For each heart, Z-stacks were acquired from 10 μm-thick sections using Zeiss Axio Observer. Three sections of each heart containing large regenerate areas were chosen for Apotome imaging. PCNA+Mef2c+ nuclei and Mef2c+ nuclei, as well as nDsRed+EdU+ and nDsRed+ nuclei, were counted by ImageJ2x software. Click-iT® EdU Alexa-Fluor-488 and 555 Imaging Kit (Invitrogen) was used for EdU detection. Confocal images were obtained using a Zeiss LSM 710 microscope.
MI and echocardiography methods in mice
MI was produced in male C57BL/6J mice at 16 weeks of age (25–30 g) by ligation of the LAD coronary artery as previously described (Tarnavski et al., 2004). Briefly, mouse was anesthetic by 2% isoflurane inhalation. A fixed suture using 7-0 silk at 2–3 mm from LAD origin was performed to induce permanent ligation. Sham-operated models were similarly subjected without LAD tied. Echocardiograms to assess systolic function were performed using M-mode and 2D measurements as described previously (Tarnavski et al., 2004). End diastole was defined as the maximal left ventricle diastolic dimension and end systole was defined as the peak of posterior wall motion. LVIDd and LVIDs were measured. LVEF was calculated by the cubic method: LVEF (%) = {(LVIDd)3 − (LVIDs)3} (LVIDd)3 × 100. LVFS was calculated by FS (%) = (LVIDd − LVIDs) / LVIDd × 100. Data were measured and analyzed by an experienced operator, who was blinded to treatment.
Measurement of interstitial fibrosis, CM proliferation, and size in post-MI hearts
EdU detection was performed using a commercially available kit (Invitrogen), according to the manufacturer's instructions. For immunostaining of Ph3, the 3 μm-thick sections were blocked with 10% normal goat serum for 60 min and then treated with antibody against Ph3 (CST, 3475S) and TPM (Sigma, T9283) overnight at 4°C. The sections were washed with phosphate buffered saline (PBS) thrice and incubated with secondary antibody (1:200; Invitrogen) for 1 h in the dark at 37°C. Nuclei were labeled with DAPI (Sigma). Images of stained tissues were captured by Olympas confocal laser scanning microscope (FluoView 1000, Japan). Proliferation indice were calculated by dividing the numbers of CM nuclei by the numbers of Ph3+ or EdU+ CMs using the Image J software.
The cardiac cell size was examined using WGA staining (Invitrogen, W11261). Briefly, heart sections were washed with PBS three times and incubated using Alexa Fluor 488 conjugated WGA according to the manufacturer's instructions. Masson’s trichrome staining was performed to assess interstitial fibrosis. Briefly, hearts from each group were embedded in paraffin after conventional processing (alcohol dehydration) and further processed for histology.
Statistical analysis
All experimental data are presented as the mean ± standard error. Statistical analyses were performed by one-way analysis of variance (ANOVA) followed by Bonferroni correction and Shapiro–Wilk normality test (for comparison of more than two groups) or Student's t-test (for comparison of two groups).
Supplementary Material
Acknowledgements
We acknowledge Guozhen Wu for assistance in fish care. We are grateful to members of our laboratories for comments on the manuscript and helpful discussions.
Funding
This research was supported in part by grants from the National Natural Science Foundation of China (NSFC31530044, NSFC31471357, and NSFC31172173 to T.P.Z.).
Conflict of interest: none declared.
References
- Aisagbonhi O., Rai M., Ryzhov S., et al. (2011). Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Dis. Model Mech. 4, 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M., Hecht A., Kruse U., et al. (1999). Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc. Natl Acad. Sci. USA 96, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M.W., Cai Z., Zhang X.J., et al. (2015). Dickkopf-3 protects against cardiac dysfunction and ventricular remodelling following myocardial infarction. Basic Res. Cardiol. 110, 25. [DOI] [PubMed] [Google Scholar]
- Barandon L., Couffinhal T., Ezan J., et al. (2003). Reduction of infarct size and prevention of cardiac rupture in transgenic mice overexpressing FrzA. Circulation 108, 2282–2289. [DOI] [PubMed] [Google Scholar]
- Bernstein H.S., and Srivastava D. (2012). Stem cell therapy for cardiac disease. Pediatr. Res. 71, 491–499. [DOI] [PubMed] [Google Scholar]
- Braunwald E. (2013). Research advances in heart failure: a compendium. Circ. Res. 113, 633–645. [DOI] [PubMed] [Google Scholar]
- Burns C.G., Milan D.J., Grande E.J., et al. (2005). High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263–264. [DOI] [PubMed] [Google Scholar]
- Chen B., Dodge M.E., Tang W., et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ren X., Liang F., et al. (2012). Characterization of two novel small molecules targeting melanocyte development in zebrafish embryogenesis. Pigment Cell Melanoma Res. 25, 446–453. [DOI] [PubMed] [Google Scholar]
- Chong J.J. (2012). Cell therapy for left ventricular dysfunction: an overview for cardiac clinicians. Heart Lung Circ. 21, 532–542. [DOI] [PubMed] [Google Scholar]
- Dorsky R.I., Sheldahl L.C., and Moon R.T. (2002). A transgenic Lef1/β-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol. 241, 229–237. [DOI] [PubMed] [Google Scholar]
- Fang Y., Gupta V., Karra R., et al. (2013). Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl Acad. Sci. USA 110, 13416–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansner J.M., Leung A.D., Superdock M., et al. (2017). Sorting zebrafish thrombocyte lineage cells with a Cd41 monoclonal antibody enriches hematopoietic stem cell activity. Blood 129, 1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans K.C.M., Daskalopoulos E.P., and Blankesteijn M.W. (2012). Interventions in Wnt signaling as a novel therapeutic approach to improve myocardial infarct healing. Fibrogenesis Tissue Repair 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., King I.N., Chopra S.S., et al. (2007). Vertebrate heart growth is regulated by functional antagonism between Gridlock and Gata5. Proc. Natl Acad. Sci. USA 104, 14008–14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Ni T.T., Sun J., et al. (2014). Prostaglandin signalling regulates ciliogenesis by modulating intraflagellar transport. Nat. Cell Biol. 16, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Werdich A.A., et al. (2010). Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Pan S., Hsieh M.H., et al. (2013). Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl Acad. Sci. USA 110, 20224–20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae C.A., and Peterson R.T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14, 721–731. [DOI] [PubMed] [Google Scholar]
- Malliaras K., Zhang Y., Seinfeld J., et al. (2013). Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol. Med. 5, 191–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztani M., Wu J.C., and Nusse R. (2016). Fibrosis of the neonatal mouse heart after cryoinjury is accompanied by Wnt signaling activation and epicardial-to-mesenchymal transition. J. Am. Heart Assoc. 5, e002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhou H., Zhang L.S., et al. (2017). Blockade to pathological remodeling of infarcted heart tissue using a porcupine antagonist. Proc. Natl Acad. Sci. USA 114, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A.T., Shiojima I., Akazawa H., et al. (2006). Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl Acad. Sci. USA 103, 19812–19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T.T., Rellinger E.J., Mukherjee A., et al. (2011). Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem. Biol. 18, 1658–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N. (2004). A decade of discoveries in cardiac biology. Nat. Med. 10, 467–474. [DOI] [PubMed] [Google Scholar]
- Ozhan G., and Weidinger G. (2015). Wnt/β-catenin signaling in heart regeneration. Cell Regen. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., et al. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Wilson L.G., and Keating M.T. (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C.I., et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswati S., Alfaro M.P., Thorne C.A., et al. (2010). Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling. PLoS One 5, e15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Hwang H., Nguyen C., et al. (2013). The small molecule Wnt signaling modulator ICG-001 improves contractile function in chronically infarcted rat myocardium. PLoS One 8, e75010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.J., Luo X., et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahinci E., Thorne C.A., Franklin J.L., et al. (2007). Lrp6 is required for convergent extension during Xenopus gastrulation. Development 134, 4095–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnavski O., McMullen J.R., Schinke M., et al. (2004). Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics 16, 349–360. [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., et al. (2007). Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book (4th edn). Eugene, OR: University of Oregon Press. [Google Scholar]
- Wo D., Peng J., Ren D.N., et al. (2016). Opposing roles of Wnt inhibitors IGFBP-4 and Dkk1 in cardiac ischemia by differential targeting of LRP5/6 and β-catenin. Circulation 134, 1991–2007. [DOI] [PubMed] [Google Scholar]
- Yang D., Fu W., Li L., et al. (2017). Therapeutic effect of a novel Wnt pathway inhibitor on cardiac regeneration after myocardial infarction. Clin. Sci. 131, 2919–2932. [DOI] [PubMed] [Google Scholar]
- Zelarayan L.C., Noack C., Sekkali B., et al. (2008). β-Catenin downregulation attenuates ischemic cardiac remodeling through enhanced resident precursor cell differentiation. Proc. Natl Acad. Sci. USA 105, 19762–19767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman Z.F., Moon R.T., and Chien A.J. (2012). Targeting Wnt pathways in disease. Cold Spring Harb. Perspect. Biol. 4, pii: a008086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


