Abstract
STUDY QUESTION
Can maternal plasma cell-free DNA (cfDNA) detect chromosomal anomalies in early pregnancy loss (EPL) and recurrent pregnancy loss (RPL)?
SUMMARY ANSWER
Genome-wide cfDNA testing can serve as an alternative to cytogenetic analysis in products of conception (POCs) in RPLs and can guide further management.
WHAT IS KNOWN ALREADY
Random chromosomal anomalies are the single most common cause for EPL and RPL. Cytogenetic analysis in POCs may be used to direct management in RPL because the detection of random chromosomal anomalies can eliminate further unwarranted testing.
STUDY DESIGN, SIZE, DURATION
This was a prospective diagnostic test study from March 2018 to January 2019 of 109 patients experiencing pregnancy loss before 14 weeks gestation at a tertiary-care academic medical center.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Blood samples were drawn for genome-wide cfDNA testing prior to chorionic villous sampling for cytogenetic analysis of POCs with both short-term cultures (STCs) and long-term cultures (LTCs). Final analysis included 86 patients with non-mosaic cytogenetic results in POCs and available cfDNA results. Aneuploidy detection rates by cfDNA testing and POC cytogenetic analysis were compared. The first 50 samples served as the Training Set to establish pregnancy loss-specific log-likelihood ratio (LLR) thresholds using receiver-operator characteristic (ROC)-like analyses. These were then used for the entire cohort.
MAIN RESULTS AND THE ROLE OF CHANCE
Seventy-eight samples (71.5%) had results available from both STC and LTC; 12 samples (11%) had a result from STC only, and 7 samples (6.4%) had a result from LTC only. A chromosomal anomaly was detected in 55/86 (64%). The rates of chromosomal anomalies were 61, 72, 73 and 44% in patients undergoing their first, second, third and ≥4th pregnancy losses, respectively. The median cfDNA fetal fraction was 5%. With standard LLR thresholds used for noninvasive prenatal screening, the sensitivity of cfDNA in detecting aneuploidy was 55% (30/55) and with a specificity of 100% (31/31). Using pregnancy loss-specific LLR thresholds, the sensitivity of cfDNA in detecting aneuploidy was 82% (45/55), with a specificity of 90% (28/31). The positive and negative likelihood ratios were 8.46 and 0.20, respectively. Fetal sex was correctly assigned in all cases.
LIMITATIONS, REASONS FOR CAUTION
Cases with a false-positive result by cfDNA analysis would not receive the indicated RPL workup. Specificity could be improved by using a fetal fraction (FF) cutoff of 4%, but this would result in exclusion of more than a quarter of cases.
WIDER IMPLICATIONS OF THE FINDINGS
cfDNA-based testing can serve as an alternative to POC cytogenetic analysis and can guide further RPL management: if cfDNA demonstrates aneuploidy, no further action is taken and if no abnormality is detected, the recommended RPL workup is performed.
STUDY FUNDING/COMPETING INTEREST(S)
Cell-free DNA testing was funded by Illumina, Inc., San Diego, CA. Y.Y. is a member of Illumina’s Clinical Expert Panel and has received travel grants. A.B. has received travel grants from Illumina. All authors have no competing interest to declare.
Keywords: cell-free DNA, noninvasive, recurrent pregnancy loss, aneuploidy, chromosome anomalies
Introduction
The etiological investigations of recurrent pregnancy loss (RPL) consists of peripheral karyotype analysis of the parents; screening for lupus anticoagulant, anticardiolipin antibodies and anti beta2-glycoprotein; sonohysterogram, hysterosalpingogram or hysteroscopy to assess the uterine cavity; and screening for thrombophilias, thyroid or prolactin abnormalities (RCOG, 2011; ASRM, 2012; ESHRE, 2018). This workup is costly and identifies an explanation in less than half of the cases (Popescu et al., 2018).
It is well established that 50–70% of early pregnancy losses (EPLs) are caused by numeric chromosomal anomalies, mostly trisomies, monosomies and polyploidy (Lathi et al., 2011; Goldstein et al., 2017; Soler et al., 2017). However, even in RPLs, random chromosomal anomalies constitute the single most common etiology (Stephenson et al., 2002; Lathi et al., 2011; Goldstein et al., 2017; Soler et al., 2017). Only ~4% are due to unbalanced chromosomal rearrangements inherited from a parent carrying a balanced chromosomal rearrangement (Stephenson and Kutteh, 2007; Jaslow et al., 2010; Tunc et al., 2016). Several studies have therefore suggested that product of conception (POC) karyotype analysis should be used to direct further management in RPLs, because the detection of random chromosomal anomalies could be cost-effective by eliminating further unwarranted testing (Bernardi et al., 2012; Petracchi et al., 2017; Popescu et al., 2018).
However, the success rate of POC karyotyping may be as low as 53% due to a 32% culture failure rate (Pauta et al., 2018) and a 15% rate of maternal cell contamination (MCC) (Lathi et al., 2014). A major problem for successful POC karyotyping, particularly for very early pregnancy losses, is actually identifying suitable material to test. The availability of POC specimens has also declined due to the increasing use of misoprostol for medical management of miscarriage. The use of chromosomal microarray (CMA) is associated with improved results (Pauta et al., 2018).
The current study was undertaken in order to assess whether maternal-plasma genome-wide cell-free DNA (cfDNA)-based testing can reliably detect chromosomal anomalies in random early and RPL.
Materials and Methods
Ethical approval
The study was approved by the Internal Review Board (IRB) at Hospital Clinic, Barcelona (IRB number HCB/2017/0726). All patients participating in the study gave their written informed consent.
Study design and participants
This was a prospective study performed at Hospital Clinic, Barcelona. During the study period (March 2018 to January 2019), all consecutive patients experiencing EPL were offered to participate. RPL was defined as ≥2 clinical pregnancy losses (ESHRE, 2018). Study inclusion criteria were as follows: (i) consenting patients, >18 years of age, undergoing EPL or RPL at a sonographic gestational age < 14 weeks; (ii) blood samples with sufficient volume for testing; (iii) non-mosaic cytogenetic results available from short-term culture (STC), long-term culture (LTC) or both; and (iv) available cfDNA results. Cases of confirmed mosaicism were removed from the analysis, as is common practice in validation studies (Bianchi et al., 2012; Lefkowitz et al. 2016), to allow determination of the technical capabilities of the assay. These cases were analyzed separately.
Ultrasound measurements
The standard protocol at the Hospital Clinic Barcelona for all patients experiencing EPL and RPL includes sonographic measurements of the gestational sac and the crown-rump length (CRL) when an embryonic pole is present. The clinical gestational age is estimated based on self-reported last menstrual period (LMP). The sonographic gestational age is calculated by CRL if a fetal pole was noted. Cases with an empty sac are assigned a sonographic gestational age of 5 weeks. The estimated time-from-demise is calculated as the clinical gestational age minus sonographic gestational age. Assessment for the presence of structural abnormalities is also performed after 10 weeks.
Cytogenetic analysis
As a routine practice at the Hospital Clinic Barcelona, POCs are obtained for cytogenetic analysis by chorionic villous sampling (CVS) using a Rodeck fine forceps, prior to medical or surgical evacuation of the uterus, as previously reported (Stergiotou et al., 2016; Soler et al., 2017). Cytogenetic analysis of POCs is performed at the Cytogenetic Laboratory, Hospital Clinic Barcelona, and includes karyotyping on both STC and LTC.
Blood tests
Patients with pregnancy loss usually undergo tests for complete blood count and clotting factors. For the purpose of this study, quantitative beta-human chorionic gonadotropin (beta-hCG) was also tested. An additional blood sample of 20 mL was drawn into Streck® tubes for genome-wide cfDNA testing, prior to CVS.
Cell-free DNA testing
Samples were submitted for the Verifi® Plus prenatal aneuploidy screening test at the College of American Pathologists-accredited and Clinical Laboratory Improvement Act-certified Illumina Laboratory (Verinata Health, Inc., a wholly owned subsidiary of Illumina, Inc., Redwood City, CA). Verifi® Plus involves isolation of plasma from maternal whole blood samples and preparation of sequencing libraries using TruSeq DNA Nano LP kit (Illumina, Inc., San Diego, CA). Sequencing was carried out using TruSeq SBS Kit v3-HS (50 cycles) and HiSeq 2000 instrument (96 samples-plex), with single-end reads of 36 base pairs obtained and an average of 22 million reads per sample. The sequence reads are mapped to the human reference genome (hg19) using the Bowtie software program (Langmead et al., 2009). Data are filtered to remove nonunique alignments and high-variation genomic regions. A log-likelihood ratio (LLR) score is then calculated by evaluating the likelihood of the observed sequence data under two competing hypotheses of ‘no aneuploidy present’ and ‘aneuploidy present’ for each chromosome; LLR scores consider the observed coverage and the estimated fetal fraction (FF) for the sample in question. Aneuploidy classification status for all chromosomes is then determined by comparing the LLR against a classification threshold; a score above the LLR threshold indicates the presence of aneuploidy. This test screens for the presence of aneuploidy on all 22 autosomes as well as for sex chromosome aneuploidy and reports sample FF. FF is determined using the SeqFF method developed by Kim et al. (2015). This method estimates FF by counting the number of reads aligned within specific autosomal regions and applying a weighting scheme derived from a multivariate model. FF is estimated by inferring discrete regions in the genome that are overrepresented in fetal cfDNA. Verifi® Plus uses a sample-specific quality control (QC) metric known as the individualized Fetal Aneuploidy Confidence Test (iFACT) that considers the estimated FF to determine if the system has generated sufficient sequencing coverage for each sample; samples that fail to meet this threshold do not report out a result.
Determining pregnancy loss-specific LLR thresholds
In ongoing pregnancies, even those at high risk, the likelihood of a fetal chromosome anomaly is much lower than among patients experiencing pregnancy loss. Thus, for noninvasive prenatal screening (NIPS), LLR thresholds are set relatively high to eliminate false-positive results. In contrast, in early pregnancy loss more than half of cases are expected to be aneuploid. Therefore, pregnancy loss-specific exploratory LLR thresholds were established to increase sensitivity while still maintaining a low false positive rate. These were determined using a receiver-operator characteristic (ROC)-like analyses (Supplementary Fig. S1). The first 50 samples served as the Training Set to establish a single LLR threshold for all trisomy events, a second threshold for all monosomy events and third for 45,X and 47,XXX. After applying these thresholds to the entire cohort, we slightly modified the LLR threshold for trisomy 16 to improve sensitivity without compromising specificity.
Statistical analysis
Maternal and pregnancy characteristics and data obtained from the first-trimester ultrasound examination were entered in the Statistical Package for the Social Science (SPSS) database (SPSS, Chicago, IL, USA), which was then used for statistical analyses. When missing at random, data were imputed using multiple imputations by marginal long style creating 40 subsets including all predictors, outcomes and passive variables in the analysis. The Kolmogorov–Smirnov test was used to demonstrate whether variables were normally distributed. Normally distributed variables were compared using T test and expressed as mean and standard deviation, while not normally distributed were compared using the Mann–Whitney test and expressed as median and interquartile range (IQR) or range. To determine significant differences between groups, chi-square statistics or the Fisher exact test was used to examine differences between proportions. Test for trend across ordered group was calculated by Wilcoxon-type test for trends using a nonparametric approach. Test performance was expressed as sensitivity, specificity and AUC and was compared using the DeLong test. For this proof-of-concept study, a sample size of 100 patients was chosen assuming at least 50% would have a chromosomal anomaly. A P value<0.05 was considered statistically significant. Error values shown in the text are SD.
Results
During the study period, 118 consecutive patients experiencing EPL or RPL were offered to participate in the study; 9 patients declined participation. Among the 109 consenting patients, the success rates of the cytogenetic analysis were as follows: 78 samples (71.5%) had results available from both STC and LTC; 12 samples (11%) had a result from STC only; and 7 samples (6.4%) had a result from LTC only. Of the 97 cases with cytogenetic results, there were 9 cases with mosaicism, which were not included in the final analysis. Of the remaining 88 cases, 2 did not receive cfDNA results: in one case, the tube was broken during shipping and another sample failed the presequencing DNA quantification QC threshold. The final analysis included 86 cases with complete non-mosaic cytogenetic results and available cfDNA results. The median maternal age was 37 years (range: 21–46 years). For 41 patients (48%), this was the first pregnancy loss; 25 (29%) were undergoing their second loss, 11 (13%) were having their third loss and 9 (10%) experienced four or more losses. The mean clinical gestational age was 9.6 ± 1.9 weeks (range: 5.1–13.6 weeks) and the mean sonographic gestational age was 6.4 ± 1.6 weeks (range: 5.0–11.3 weeks). The mean estimated time from embryo demise was 3.3 ± 1.9 weeks.
A chromosomal anomaly was detected in 64% (55/86). Patient characteristics among those with and without a chromosomal anomaly are presented in Table I. As expected, the rate of chromosomal anomalies increased with maternal age, from 25% (1/4) in patients 25 years of age or younger to 90% (9/10) in patients over 40 years of age (Table II). The same trend was observed when restricted to RPL patients. The rate of chromosomal anomalies was not significantly different between RPL patients (30/45, 67%) and those experiencing their first pregnancy loss (25/41, 61%; chi-square statistic 0.3014, P = 0.583) (Supplementary Table SI).
Table I.
Patient and pregnancy characteristics.
| Total (n = 86) | Chromosomally abnormal (n = 55) | Chromosomally normal (n = 31) | P value | |
|---|---|---|---|---|
| Maternal age * (years) | 37 (IQR: 5) | 38 (IQR: 5) | 35 (IQR: 8) | 0.015 |
| BMI † | 24.1 ± 4.1 | 24.7 ± 4.3 | 23.1 ± 3.4 | 0.090 |
| Recurrent loss (%) | 45 (52.3%) | 30 (54.5%) | 15 (48.4) | 0.583 |
| Clinical gestational age † (weeks) | 9.6 ± 1.9 | 9.7 ± 1.7 | 9.3 ± 2.1 | 0.355 |
| Sonographic gestational age † (weeks) | 6.4 ± 1.6 | 6.5 ± 1.5 | 6.3 ± 1.6 | 0.460 |
| Time from embryo demise † (weeks) | 3.3 ± 1.9 | 3.3 ± 1.9 | 3.2 ± 1.9 | 0.899 |
| Bleeding | 36 (41.9%) | 26 (42.3%) | 10 (32.3) | 0.175 |
| Sac volume * (cm) | 11.7 (IQR: 26) | 10.9 (IQR: 26) | 12.4 (24.5) | 0.956 |
| hCG * (mIU/mL) | 20 184 (IQR: 28752) | 18 750 (IQR: 29114) | 23 552 (IQR: 24729) | 0.590 |
| Medical management (%) | 46 (53.5%) | 31 (56.4%) | 15 (48.4%) | 0.401 |
| Fetal fraction * (%) | 5% (IQR: 4%) | 5% (IQR: 3%) | 5% (IQR: 4%) | 0.446 |
*Values are median and interquartile range (IQR)
†Values are means ± SD
Table II.
Rate of chromosomal anomalies in all patients and in patients with RPL according to age groups (at 5-year intervals).
| Age range | All patients * | Patients with RPLs † |
|---|---|---|
| ≤25 | 1/4 (25%) | 0/0 |
| 26–30 | 3/6 (50%) | 2/4 (50%) |
| 31–35 | 11/21 (52%) | 8/14 (57%) |
| 36–40 | 31/45 (69%) | 14/20 (70%) |
| >40 | 9/10 (90%) | 6/7 (86%) |
| Total | 55/86 (64%) | 30/45 (67%) |
RPL, recurrent pregnancy loss
*Chi2 = 7.777; P value = 0.100.
Test for trend across groups rank-sum test: z = 2.66; P = 0.008.
†Chi2 = 2.3143. P value = 0.510.
Test for trend across groups rank-sum test: z = 1.49; P = 0.136.
With standard LLR thresholds commonly used for NIPS, the sensitivity of cfDNA in detecting aneuploidy in pregnancy loss was 55% (30/55) with a specificity of 100% (31/31) and an accuracy of 71% (61/86) (Table III). The first 50 cases were used as a Training Set to establish pregnancy loss-specific LLR thresholds. A uniform threshold was set at 1.5 for all autosomal trisomies, a threshold of 3.5 for all autosomal monosomies and a threshold of 0.88 for 45,X and 47,XXX. After applying these LLR thresholds to the entire cohort, the LLR threshold for trisomy 16 was reset at 1.0, to improve sensitivity without compromising specificity. Using these LLR thresholds, the sensitivity of cfDNA in detecting aneuploidy was 82% (45/55), with a specificity of 90% (28/31). The area under the curve (AUC) was 86% (79–94%), which correctly classified 85% (73/86) of the observations (Table II). The positive and negative likelihood ratios were 8.46 and 0.20 respectively. Of the 31 normal cases, 28 were correctly assigned, including 14/16 of the 46,XX cases and 14/15 of the 46,XY cases. Among the three false positive cases, two had a trisomy 4 result and one case had a result of triple trisomy for chromosomes 15, 17 and 20 (Supplementary Table SII). Fetal sex was correctly assigned in all concordant cases as well as among the 13 discordant cases.
Table III.
Detection rates for EPLs using maternal plasma cfDNA: standard log likelihood ration (LLR) thresholds vs. pregnancy loss-specific LLR thresholds.
| Karyotype | Detected by standard LLR thresholds | Detected by pregnancy loss-specific LLR thresholds |
|---|---|---|
| Trisomy 2 | 0/1 | 1/1 |
| Trisomy 4 | 0/1 | 1/1 |
| Trisomy 7 | 1/1 | 1/1 |
| Trisomy 9 | 2/2 | 2/2 |
| Trisomy 10 | 3/4 | 3/4 |
| Trisomy 11 | 1/1 | 1/1 |
| Trisomy 12 | 2/2 | 2/2 |
| Trisomy 13 | 1/2 | 2/2 |
| Trisomy 14 | 1/1 | 1/1 |
| Trisomy 15 | 3/5 | 5/5 |
| Trisomy 16*† | 4/10 | 8/10 |
| Trisomy 17 | 0/1 | 1/1 |
| Trisomy 18 | 2/2 | 2/2 |
| Trisomy 20 | 2/4 | 3/4 |
| Trisomy 21 | 2/2 | 2/2 |
| Trisomy 22 | 1/4 | 3/4 |
| Monosomy X | 4/7 | 6/7 |
| Monosomy 21 | 0/1 | 0/1 |
| Double trisomy | 1/2 | 1/2 |
| Triploidy* | 0/2 | 0/2 |
| Total | 30/55 (55%) | 45/55 (81.8%) |
EPL, early pregnancy loss
cfDNA, cell-free DNA
*In a case of 70,XXY,+16 the trisomy 16 was detected but the triploidy was not
†One case of trisomy 16 was called 47,XXX
While most of the affected cases had a single anomaly, two cases had a double trisomy: one case of 48,XX,+15,+21 was correctly detected and another case of 48,XX,+7,+22 was a false negative. An additional case had both triploidy and trisomy 16 (70,XXY,+16) wherein only trisomy 16 was detected by cfDNA. In the entire cohort, there was only one case of an unbalanced chromosomal rearrangement: an unbalanced Robertsonian translocation resulting in trisomy 15. Parental karyotyping performed in this case was negative. Of the nine mosaic cases, only one was correctly ascertained (Supplementary Table SIII).
The median FF was 5% (range <1 to 28%), the highest level detected in a case of triploidy. Among the 73 correctly classified cases, the median FF was 5% (IQR: 3%), compared to 4% (IQR: 2%) among the 13 incorrectly classified ones (Supplementary Table SII, Mann–Whitney U test, P = 0.169). Performance was evaluated with different possible FF cutoffs (<1, <2, <3 and <4%), below which cases would be excluded (Supplementary Table SIV). No improvement in sensitivity was noted for any of these thresholds compared to using no threshold at all. An improvement was noted in specificity only when a threshold of <4% FF was employed. This however resulted in exclusion of over one-quarter of samples. Similarly, we found no clinically useful cutoff value for beta-hCG levels, patient BMI or sac size. The sensitivity, specificity and AUC were similar in patients with and without bleeding at the time of testing (sensitivity 81 vs. 83%; specificity 91 vs. 90%; AUC: 85 vs. 87%, respectively; DeLong test: P = 0.877).
Discussion
In this study, 109 patients experiencing early pregnancy loss were evaluated for chromosomal anomalies by cytogenetics of POCs and maternal plasma cfDNA analysis. Of these, 86 patients met the inclusion criteria. The rate and type of chromosomal anomalies in this cohort concurs with other similar studies, as did the sex distribution, indicating that it is appropriately representative (Jaslow et al., 2010, Lathi et al., 2011; Bernardi et al., 2012; Tunc et al., 2016; Goldstein et al., 2017; Soler et al., 2017; Popescu et al., 2018). Cell-free DNA testing achieved a sensitivity of 82% and a specificity of 90%, an overall accuracy of 85%. This rate compares favorably with that of routine cytogenetic analysis of POCs (Pauta et al., 2018). It is of note that pregnancy loss-specific LLR thresholds had to be established to achieve this rate of detection.
To the best of our knowledge, this is the first systematic study of its kind. A few small studies evaluated the levels of fetal cfDNA in EPLs. One study found that levels of fetal cfDNA and total cfDNA were significantly higher in patients with pregnancy loss (both euploid and aneuploid) in comparison with the normal controls (Lim et al., 2013). Another prospective study examined whether cfDNA can be used for diagnosis in nonviable pregnancies at all gestational ages and found that 76% had FFs within the detectable range (>3.7%) (Clark-Ganheart et al., 2015). They recommended that cfDNA be used only after 8 weeks of gestation when FF is expected to be above their suggested cutoff in most cases. In that study, however, prior cytogenetic analysis or analysis of POCs was only available in 38% of cases. In contrast, we found no clinically useful cutoff for exclusion of low FF cases and cfDNA testing yielded results even before 8 weeks and at low FFs. This is in accordance with previous publications demonstrating that genome-wide sequencing for cfDNA analysis has >80% sensitivity for trisomies even in low FF samples (Artieri et al., 2017).
The high rate of successful genetic POC analysis (97/109, 89%) achieved here was through a combination of meticulous CVS prior to uterine evacuation, followed by cytogenetic analysis of both STC and LTC. This degree of lab effort is not characteristic of most routine centers, and the realistic success rate of karyotype analysis of POCs may be as low as 53% (Pauta et al., 2018). Other limitations of cytogenetic analysis of POCs include failed culture due to contamination or the extraction of nonviable tissue. This may be overcome, to some extent, by the use of SNP-based chromosomal microarray analysis (CMA) that obviates the need for culture while concurrently detecting MCC (Liu et al., 2015; Pauta et al., 2018). Consistent with this, the aforementioned meta-analysis showed that CMA improved POC analysis success rates by 27% (from 68 to 95%). However, the incremental yield of CMA in detecting pathogenic submicroscopic variants undetectable by karyotyping was only 2% (106/5507) (Pauta et al., 2018).
Regardless of the technology used for POC analysis, obtaining a POC sample is challenging: some patients miscarry before a proper sample is obtained. Moreover, many patients with an early missed abortion now undergo medical management using misoprostol, rather than surgical extraction (Wu et al., 2017). Indeed, in our series, 55% of the patients were managed medically. For such cases, a tissue-collection kit has been developed but its success rate in obtaining a proper sample is lower than that of surgical extraction (84 vs 100%, respectively) (Kucherov et al., 2018).
The etiological investigations of RPLs have traditionally not centered on the abortus. Except for karyotyping both parents to screen for chromosomal rearrangements, most have focused on defects in the female partner, namely congenital or acquired uterine abnormalities, autoimmune factors, endocrine imbalances and thrombophilia (Jaslow et al., 2010, Stephenson and Kutteh, 2007). More recently, several groups challenged this approach and promoted the concept that RPL workup should be guided by genetic analysis of POCs (Bernardi et al., 2012; Kutteh 2015; Petracchi et al., 2017; Popescu et al., 2018). Their suggested model stipulates that genetic analysis of POCs be performed in the second and subsequent pregnancy loss: if aneuploidy is demonstrated, no further evaluation is needed; if an unbalanced chromosomal rearrangement (such as a translocation or inversion) is detected, parental karyotyping is performed; and if no chromosomal aberration is detected and MCC has been ruled out, then the American Society for Reproductive Medicine (ASRM) RPL workup should be performed. With this approach, a definitive or probable cause for RPL could be identified in 95% of cases, compared with 45% using the classical approach (Popescu et al., 2018). This approach has been shown to produce cost savings of around $1100 per case by eliminating unnecessary investigations (Wolf and Horger, 1995; Bernardi et al., 2012; Petracchi et al., 2017). Finally, Popescu et al. (2018) calculated that if POC genetic analysis was used to guide workup, the total cost to make a diagnosis would be $1879.16 per patient whereas if the ASRM RPL evaluation were performed for all RPL patients, the total cost to make a diagnosis would be $3866.84 per patient.
Based on our results and these cost-effectiveness analyses, we suggest an alternative, modified algorithm for RPL evaluation, guided by cfDNA results rather than karyotyping or CMA analysis of POCs (Fig. 1): if cfDNA in the second and subsequent RPL demonstrates aneuploidy, no further action is taken; and if no abnormality is detected, then the recommended RPL workup is performed. While speculative as regards this cohort, cfDNA testing can also detect unbalanced rearrangements such as unbalanced reciprocal translocations, provided they are of a substantial size (Lefkowitz et al., 2016; Van Opstal et al., 2017; Wapner et al., 2015; Hu et al., 2019). When detected, this would be the only direct prompt for parental karyotyping. If an unbalanced rearrangement is not detected by cfDNA testing, parental karyotyping would be performed, at any rate, as a part of the recommended RPL workup, and thus a carrier parent would not be missed. For trisomies involving the acrocentric chromosomes (13, 14, 15, 21 and 22), one could still consider the rare occurrence of an unbalanced Robertsonian translocation, which may be detected on parental karyotyping, but these would account for ~0.5% of cases (Soler et al., 2017). A result compatible with other random trisomies (such as the common trisomy 16) would clearly obviate parental karyotyping.
Figure 1.
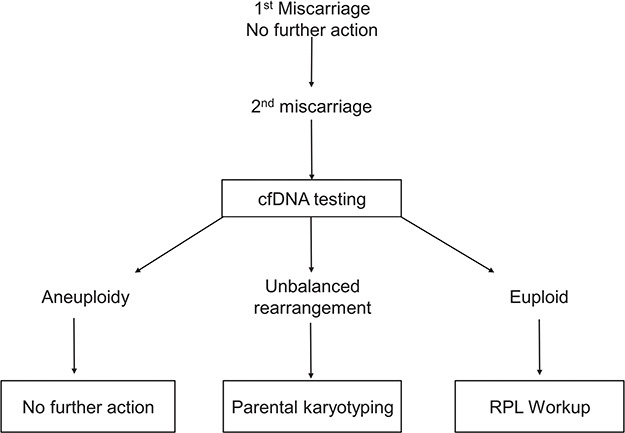
Proposed algorithm for recurrent pregnancy loss workup based on cfDNA results. cfDNA, cell-free DNA; RPL, recurrent pregnancy loss.
In our series, a positive result for a chromosomal aberration was given in 48 cases (56%), 45 of which were true positives, for a positive predictive value (PPV) of 94%. This implies that in more than half of the cases, the RPL workup would be averted, because a definite cause for miscarriage had been revealed. Conversely, in those with a negative result, the yield of standard RPL workup would expectedly be higher.
Nonetheless, the approach suggested in this proof-of-concept study has several limitations: an important implication is that cases with a false-positive result by cfDNA analysis would not receive the indicated RPL workup, and a biological explanation for the loss may not be identified. A larger cohort may promote additional refinement of chromosome-specific LLR thresholds to further improve cfDNA test performance. Specificity could be improved by using an FF cutoff of 4%, but in this case more than a quarter of the cases would be excluded.
This approach is also unable to discriminate true fetal monosomy X from low-grade, maternal monosomy X mosaicism, which increases with maternal age (Machiela et al., 2016). This is a potential source of false positive results in as much as 8.6% of sex chromosome anomalies reported by NIPS (Wang et al., 2014). This issue may be overcome by employing paired-end sequencing, which enables determination of cfDNA fragment length, as fetal cfDNA fragments are generally shorter than the corresponding maternal ones (Yu et al., 2014). This methodology allows deduction of FF as well as improvement in the sensitivity and specificity of autosomal aneuploidy as well as of monosomy X. Another limitation of genome-wide cfDNA sequencing is that it does not detect triploidy, the incidence of which was 3.4% in our series but may account for 8% of cases (Soler et al., 2017). Such missed cases of triploidy however would undergo the standard RPL workup with no contribution to cost-savings. Despite these limitations, cfDNA testing could prove to be of value particularly in cases where medical management is used and in the absence of POC sample for cytogenetic analysis.
Finally, cfDNA testing may also be used in sporadic EPLs. While not significantly impacting further clinical management, it is likely to have a positive influence on patient well-being. The psychological impact of EPL is often neglected. Farren et al. have recently found evidence of significant depression and anxiety in the first month following EPL in women (Farren et al., 2018). For some patients, knowing that the cause for loss is chromosomal rather than maternal may provide comfort. In summary, genome-wide cfDNA-based screening provides a noninvasive approach for determining whether fetal aneuploidy could explain the loss in patients experiencing early or RPL.
Supplementary data
Supplementary data are available at Human Reproduction online.
Supplementary Material
Acknowledgements
The authors thank Dr Sucheta Bhatt, Kirsten Curnow, Victoria Corey, Sung Kim and Diem Huynh from Illumina and Sandra Oms and Miriam Muñoz from Hospital Clinic Barcelona. The authors thank Illumina for providing funding for cfDNA testing.
Authors’ roles
Y.Y.: substantial contributions to conception and design, acquisition of data, analysis and interpretation of data; drafting the article and revising it critically for important intellectual content; and final approval of the version to be published. M.P.: substantial contributions to conception and design, acquisition of data and analysis and interpretation of data; revising it critically for important intellectual content; and final approval of the version to be published. C.B.: substantial contributions to acquisition of data revising it critically for important intellectual content; and final approval of the version to be published. A.S.: substantial contributions to acquisition of data; revising it critically for important intellectual content; and final approval of the version to be published. V.B.: substantial contributions to acquisition of data; revising it critically for important intellectual content; and final approval of the version to be published.C.I.: substantial contributions to acquisition of data; revising it critically for important intellectual content; and final approval of the version to be published. F.P.y.M.: substantial contributions to acquisition of data; revising it critically for important intellectual content; and final approval of the version to be published. R.M.-P.: substantial contributions to acquisition of data, and analysis and interpretation of data; revising it critically for important intellectual content; and final approval of the version to be published. A.B.: substantial contributions to conception and design, acquisition of data, analysis and interpretation of data; drafting the article and revising it critically for important intellectual content; and final approval of the version to be published.
Funding
Cell-free DNA testing was funded by Illumina, Inc., San Diego, CA.
Conflict of interest
Y.Y. is a member of Illumina’s Clinical Expert Panel and has received travel grants. A.B. has received travel grants from Illumina. Other authors have no conflicts of interest.
References
- Artieri CG, Haverty C, Evans EA, Goldberg JD, Haque IS, Yaron Y, Muzzey D. Noninvasive prenatal screening at low fetal fraction: comparing whole-genome sequencing and single-nucleotide polymorphism methods. Prenat Diagn 2017;37:482–490. [DOI] [PubMed] [Google Scholar]
- ASRM American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril 2012;98:1103–1111. [DOI] [PubMed] [Google Scholar]
- Bernardi LA, Plunkett BA, Stephenson MD. Is chromosome testing of the second miscarriage cost saving? A decision analysis of selective versus universal recurrent pregnancy loss evaluation. Fertil Steril 2012;98:156–161. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, MatErnal BLood IS Source to Accurately diagnose fetal aneuploidy (MELISSA) Study Group . Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 2012;119:890–901. [DOI] [PubMed] [Google Scholar]
- Clark-Ganheart CA, Fries MH, Leifheit KM, Jensen TJ, Moreno-Ruiz NL, Ye PP, Jennings JM, Driggers RW. Use of cell-free DNA in the investigation of intrauterine fetal demise and miscarriage. Obstet Gynecol 2015;125:1321–1329. [DOI] [PubMed] [Google Scholar]
- ESHRE European Society of Human Reproduction and Embryology guideline: recurrent pregnancy loss. Hum Reprod Open 2018;1–12. [Google Scholar]
- Farren J, Mitchell-Jones N, Verbakel JY, Timmerman D, Jalmbrant M, Bourne T. The psychological impact of early pregnancy loss. Hum Reprod Update 2018;24:731–749. [DOI] [PubMed] [Google Scholar]
- Goldstein M, Svirsky R, Reches A, Yaron Y. Does the number of previous miscarriages influence the incidence of chromosomal aberrations in spontaneous pregnancy loss? J Matern Fetal Neonatal Med 2017;30:1–5. [DOI] [PubMed] [Google Scholar]
- Hu H, Wang L, Wu J, Zhou P, Fu J, Sun J, Cai W, Liu H, Yang Y. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 8141 single pregnancies. Hum Genomics 2019;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaslow CR, Carney JL, Kutteh WH. Diagnostic factors identified in 1020 women with two versus three or more recurrent pregnancy losses. Fertil Steril 2010;93:1234–1243. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hannum G, Geis J, Tynan J, Hogg G, Zhao C, Jensen TJ, Mazloom AR, Oeth P, Ehrich M et al. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat Diagn 2015;35:810–815. [DOI] [PubMed] [Google Scholar]
- Kucherov A, Atrio J, Williams Z. Patient-controlled tissue collection for genetic testing after early pregnancy loss: a pilot study. Prenat Diagn 2018;38:204–209. [DOI] [PubMed] [Google Scholar]
- Kutteh WH. Novel strategies for the management of recurrent pregnancy loss. Semin Reprod Med 2015;33:161–168. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathi RB, Gray Hazard FK, Heerema-McKenney A, Taylor J, Chueh JT. First trimester miscarriage evaluation. Semin Reprod Med 2011;29:463–469. [DOI] [PubMed] [Google Scholar]
- Lathi RB, Gustin SL, Keller J, Maisenbacher MK, Sigurjonsson S, Tao R, Demko Z. Reliability of 46,XX results on miscarriage specimens: a review of 1,222 first-trimester miscarriage specimens. Fertil Steril 2014;101:178–182. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, Almasri E, Hogg G, Angkachatchai V, Zhao C, Grosu DS et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am J Obstet Gynecol 2016;215:227 e221–227 e216. [DOI] [PubMed] [Google Scholar]
- Lim JH, Kim MH, Han YJ, Lee DE, Park SY, Han JY, Kim MY, Ryu HM. Cell-free fetal DNA and cell-free total DNA levels in spontaneous abortion with fetal chromosomal aneuploidy. PLoS One 2013;8:e56787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Song L, Cram DS, Xiong L, Wang K, Wu R, Liu J, Deng K, Jia B, Zhong M et al. Traditional karyotyping vs copy number variation sequencing for detection of chromosomal abnormalities associated with spontaneous miscarriage. Ultrasound Obstet Gynecol 2015;46:472–477. [DOI] [PubMed] [Google Scholar]
- Machiela MJ, Zhou W, Karlins E, Sampson JN, Freedman ND, Yang Q, Hicks B, Dagnall C, Hautman C, Jacobs KB et al. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat Commun 2016;7:11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauta M, Grande M, Rodriguez-Revenga L, Kolomietz E, Borrell A. Added value of chromosomal microarray analysis over karyotyping in early pregnancy loss: systematic review and meta-analysis. Ultrasound Obstet Gynecol 2018;51:453–462. [DOI] [PubMed] [Google Scholar]
- Petracchi F, Paez C, Igarzabal L. Cost-effectiveness of cytogenetic evaluation of products of conception by chorionic villus sampling in recurrent miscarriage. Prenat Diagn 2017;37:282–288. [DOI] [PubMed] [Google Scholar]
- Popescu F, Jaslow CR, Kutteh WH. Recurrent pregnancy loss evaluation combined with 24-chromosome microarray of miscarriage tissue provides a probable or definite cause of pregnancy loss in over 90% of patients. Hum Reprod 2018;33:579–587. [DOI] [PubMed] [Google Scholar]
- RCOG Royal College of Obstetricians & Gynaecologists. The investigation and treatment of couples with recurrent first trimester and second-trimester miscarriage. 2011. https://www.rcog.org.uk/globalassets/documents/guidelines/gtg_17.pdf.
- Soler A, Morales C, Mademont-Soler I, Margarit E, Borrell A, Borobio V, Munoz M, Sanchez A. Overview of chromosome abnormalities in first trimester miscarriages: a series of 1,011 consecutive chorionic villi sample karyotypes. Cytogenet Genome Res 2017;152:81–89. [DOI] [PubMed] [Google Scholar]
- Stephenson M, Kutteh W. Evaluation and management of recurrent early pregnancy loss. Clin Obstet Gynecol 2007;50:132–145. [DOI] [PubMed] [Google Scholar]
- Stephenson MD, Awartani KA, Robinson WP. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: a case-control study. Hum Reprod 2002;17:446–451. [DOI] [PubMed] [Google Scholar]
- Stergiotou I, Borobio V, Bennasar M, Gonce A, Mula R, Nuruddin M, Soler A, Borrell A. Transcervical chorionic villus sampling: a practical guide. J Matern Fetal Neonatal Med 2016;29:1244–1251. [DOI] [PubMed] [Google Scholar]
- Tunc E, Tanriverdi N, Demirhan O, Suleymanova D, Cetinel N. Chromosomal analyses of 1510 couples who have experienced recurrent spontaneous abortions. Reprod Biomed Online 2016;32:414–419. [DOI] [PubMed] [Google Scholar]
- Van Opstal D, Maarle MC, Lichtenbelt K, Weiss MM, Schuring-Blom H, Bhola SL, Hoffer MJV, Huijsdens-van Amsterdam K, Macville MV, Kooper AJA et al. Origin and clinical relevance of chromosomal aberrations other than the common trisomies detected by genome-wide NIPS: results of the TRIDENT study. Genet Med 2018;20:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, Han X, Hu W, Ma D, Cram D et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem 2014 Jan;60:251–259. [DOI] [PubMed] [Google Scholar]
- Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, Wayham N, Ryan A, Banjevic M, Lacroute P et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol 2015;212:332 e331–332 e339. [DOI] [PubMed] [Google Scholar]
- Wolf GC, Horger EO 3rd.. Indications for examination of spontaneous abortion specimens: a reassessment. Am J Obstet Gynecol 1995;173:1364–1368. [DOI] [PubMed] [Google Scholar]
- Wu HL, Marwah S, Wang P, Wang QM, Chen XW. Misoprostol for medical treatment of missed abortion: a systematic review and network meta-analysis. Sci Rep 2017;7:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SC, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, Akolekar R, Leung TY, Go AT, Vugt JM et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A 2014;111:8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


