Abstract
The role CD4+ T cells play in tumor immunity is less well appreciated than the cytotoxic role of CD8+ T cells. Despite clear evidence for CD4+ T cell dependency across multiple immunotherapies, the mechanisms by which CD4+ T cells infiltrate tumors remain poorly understood. Prior studies by our group have shown in a mouse model of pancreatic cancer that systemic activation of the cell surface TNF superfamily member CD40 drives T cell infiltration into tumors and, in combination with immune checkpoint blockade, leads to durable tumor regressions and cures that depend on both CD8+ and CD4+ T cells. Here, we used single-cell transcriptomics to examine the tumor microenvironment following treatment with agonist CD40 antibody with or without immune checkpoint blockade. We show that intratumor myeloid cells produce the chemokine CCL5 in response to CD40 agonist and that CCL5 mediates an influx of CD4+ T cells into the tumor microenvironment. Disruption of CCL5 genetically or pharmacologically mitigates the influx of CD4+ but not CD8+ T cells into tumors and blunts the therapeutic efficacy of immunotherapy. These findings highlight a previously unappreciated role for CCL5 in selectively mediating CD4+ T cell tumor infiltration in response to effective immunotherapy.
Keywords: Immunology, Oncology
Keywords: Cancer immunotherapy, Chemokines, T cells
These findings demonstrate a role for CCL5 in selectively mediating CD4+ T cell tumor infiltration in response to CD40 and immune checkpoint therapy.
Introduction
CD4+ T cells play a critical role in tumor immunity and response to immunotherapy, but their mechanisms of action remain incompletely understood (1–6). Canonical functions, such as T cell help provided to professional antigen-presenting cells (APCs) during priming and production of antitumor cytokines like IFN-γ, have been well described (7–9). A recent study demonstrated, however, that spontaneous and immunotherapy-mediated antitumor responses may require CD4+ T cells in addition to CD8+ T cells, even when tumors lack MHC class II (10). These findings recall early preclinical experiments with cytotoxic T lymphocyte–associated protein 4 (CTLA-4) monoclonal antibody (mAb), in which antitumor responses were dependent not only on CD8+ T cells but on CD4+ T cells as well (4). Since then, CD4+ T cell dependency has been observed in many other cancer immunotherapeutic approaches (1–6, 11–14). In the clinic, major tumor regressions have been observed following adoptive transfer of CD4+ (without CD8+) T cells in refractory solid tumors that are unlikely to express MHC class II on cancer cells (15, 16). Therefore, further mechanistic study of CD4+ T cells in the context of immunotherapy is warranted.
The TNF superfamily member CD40 is expressed on the surface of APCs and confers cellular maturation upon ligation with CD40 ligand, which is classically expressed on activated CD4+ T cells (17). Our group has shown that systemically administered agonistic CD40 monoclonal antibody (mAb) induces intratumor T cell infiltration in a genetic mouse model of pancreatic adenocarcinoma and potentiates response to immune checkpoint blockade (18, 19). This concept has now been taken forward to a national randomized clinical trial, which is ongoing (ClinicalTrials.gov NCT03214250). Preclinically, tumor regressions with CD40 mAb require both CD8+ and CD4+ T cells. Mice depleted of CD4+ T cells fail to reject implanted pancreatic cancer cell lines despite treatment with CD40 mAb combined with chemotherapy, radiotherapy, or immune checkpoint blockade (18, 20–22). Furthermore, CD4+ but not CD8+ T cells are required for memory protection against rechallenge in mice cured of these tumors, despite the fact that these tumors do not express MHC class II (18). This antitumor response appears to be driven by a strong upregulation of cytokine production by intratumor CD4+ T cells in response to the combination of CD40 agonist and immune checkpoint blockade (18). Therefore, CD40 agonist–induced tumor immunity is a desirable system in which to study CD4+ tumor immunity. To advance this novel approach to immunotherapy (23) — with emerging promising results (24–28) — understanding cellular and molecular effects is increasingly important.
It seems clear that chemotaxis of CD4+ T cells into the tumor microenvironment is required for response to immunotherapy, including CD40 agonists, given that therapeutic response is lost upon systemic administration of the sphingosine-1-phosphate receptor antagonist, which blocks lymph node egress (2). Several studies have been performed in recent years on mechanisms of CD8+ T cell tumor infiltration, in particular implicating the CXCL9/CXCL10/CXCR3 axis, but the extent to which these mechanisms do or do not apply to CD4+ T cell tumor infiltration remain unexplored (12–14, 29, 30).
Here, we used single-cell sequencing to examine heterogeneous populations in our CD40 model in a highly dimensional and unbiased manner (31, 32). We discover a broad and consistent upregulation of the chemokine CCL5 by intratumor myeloid populations in response to CD40 activation. Blocking the CCL5/CCR5 pathway pharmacologically or genetically decreases tumor CD4+ T cell infiltration in response to CD40 agonist immunotherapy, hinders immune control of tumor outgrowth, and shortens survival. Our findings suggest a novel critical role for CCL5 in CD4+ T cell tumor chemotaxis and response to immunotherapy.
Results
Single-cell RNA-sequencing identifies intratumor immune populations.
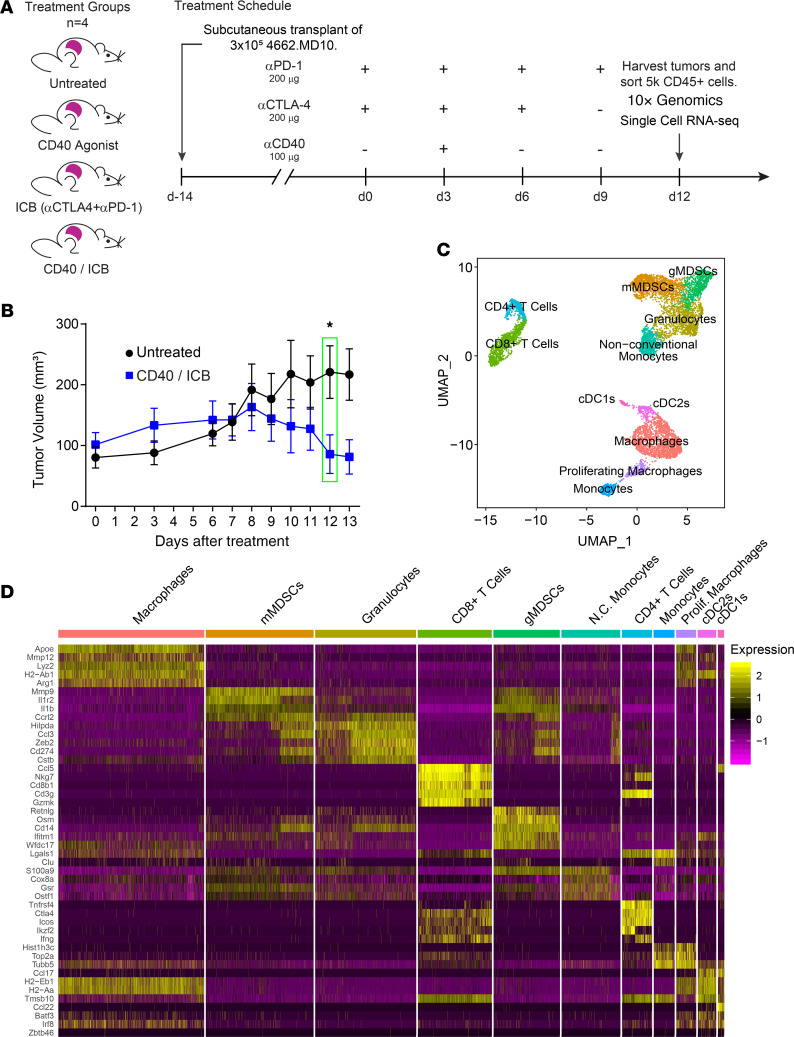
To investigate the differences in the tumor microenvironment after CD40 agonist treatment, we subcutaneously transplanted C57BL/6J mice with a clonal murine pancreatic ductal adenocarcinoma (PDAC) cell line (4226.MD10). After 14 days of tumor growth, tumor-bearing mice were randomized into groups of equal baseline tumor size and were treated with an agonist CD40 mAb, immune checkpoint blockade (ICB) with CTLA-4 and programmed cell death 1 (PD-1) mAb, both CD40 and ICB (hereafter CD40/ICB), or control mAbs (Figure 1A). Tumor growth curves comparing CD40/ICB-treated mice with untreated mice statistically diverged 12 days after the start of treatment (Figure 1B). Day 12 was therefore chosen as the optimal time point at which to query changes in the immune compartment of the tumor following therapy.
Figure 1. Single-cell RNA-sequencing identifies intratumor immune populations.
(A) Treatment of mice subcutaneously implanted with clonal KPC cell line 4662.MD10 with combination CD40 agonist and anti–CTLA-4 with anti–PD-1 (ICB). CD45+ cells were sorted for single-cell transcriptomic analysis using the 10x Genomics platform 12 days after beginning therapy. (B) Tumor growth kinetics of subcutaneously implanted mice treated as shown in A. (C) UMAP nondimensional linear reduction and clustering of immune cell populations from the tumor microenvironment merged across all treatment conditions. (D) Scaled expression of cluster-specific genes visualized by heatmap. The mean expression of each gene across all clusters was scaled to 0 with a variance of 1. n = 4 mice per treatment group (A, C, and D). n = 10 mice per group (B). Error bars indicate mean ± SEM. *P < 0.05 (Student’s 2-tailed t test). Data shown in B are representative of 2 independent experiments with 5 to 10 mice per group. gMDSC, granulocytic myeloid-derived suppressor cell; mMDSC, monocytic myeloid-derived suppressor cell.
Tumors were harvested and disaggregated on day 12 after treatment induction. Live CD45+ cells were sorted from each tumor for single-cell RNA-sequencing using the 10x Genomics pipeline. The 10x Genomics platform yielded data for approximately 5000 cells per treatment condition with an average of approximately 50,000 reads per cell (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137263DS1). In total across all 4 treatment conditions, 28,348 cells were sequenced. FASTQ files were aligned and preprocessed using 10x Genomics’ Cell Ranger software and the Seurat3 R package (Supplemental Figure 1B). To define immune populations within the tumor microenvironment, a normalized subset of approximately 2000 cells was computationally pooled from each treatment group. Graph-based clustering was then used to identify transcriptional clusters consisting of individual cell types (Figure 1C). The top conserved genes across all treatment groups were identified within each cluster (Figure 1D). Identification of canonical marker genes and comparison with the Immunological Genome Project (ImmGen) database yielded 11 distinct clusters of immune cell types. Uniform manifold approximation and projection (UMAP) nonlinear dimensional reduction revealed 3 larger metaclusters containing cells associated with distinct immune characteristics: a T cell metacluster containing CD4+ and CD8+ T cells, a “protumor myeloid” metacluster containing immune-suppressive lineages including myeloid-derived suppressor cells and granulocytes, and an “antitumor myeloid” metacluster containing monocytes, macrophages, and dendritic cells.
We next sought to determine whether differentiation of intratumor myeloid cells was affected upon treatment. To address this, single-cell myeloid clusters were subjected to a pseudotemporal analysis using the Monocle2 package in R (Supplemental Figure 2A). Monocle2 is an algorithm that aligns single cells based on gene expression along a trajectory that mirrors biological processes, such as differentiation. Cell populations from all 4 treatment conditions aligned as expected along the pseudotime trajectory. Immature myeloid-derived suppressor cells aligned earlier in pseudotime, while more terminally differentiated macrophage populations aligned later (Supplemental Figure 2B). Examination of myeloid clusters within each treatment group did not reveal any differences in their distribution along the pseudotime trajectory (Supplemental Figure 2C). Treatment with ICB, CD40 agonist, or both therefore does not appear to alter the differentiation state of myeloid cells within the tumor microenvironment.
Intratumor myeloid populations upregulate CCL5 in response to CD40 activation.
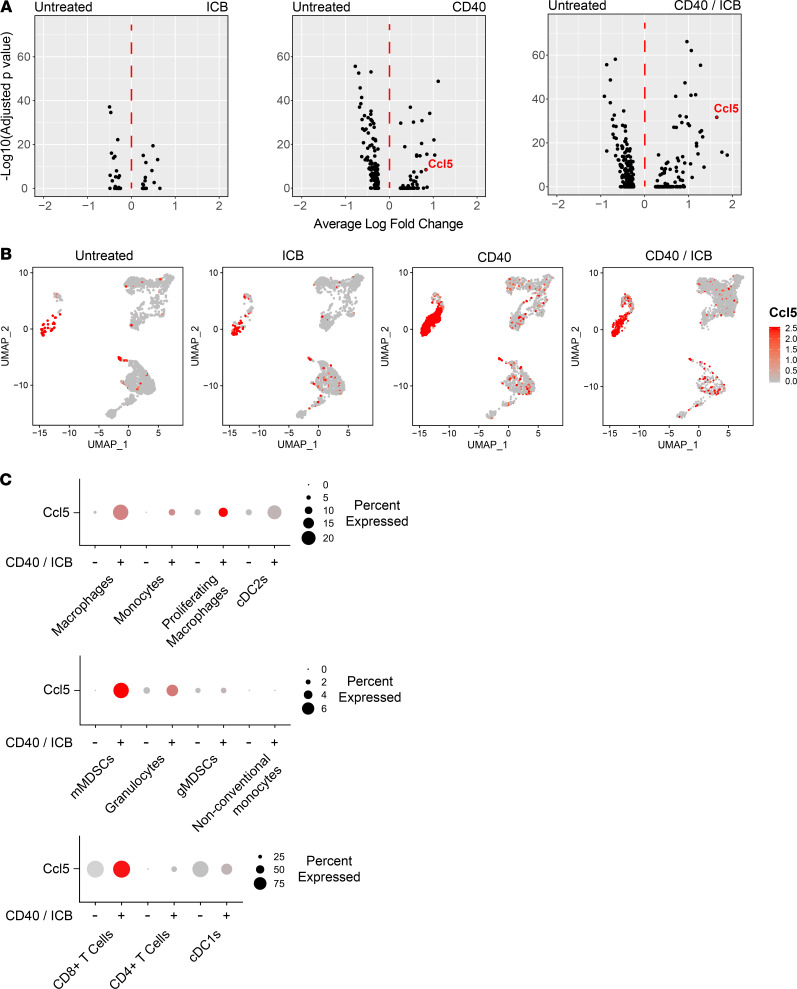
We next sought to query transcriptional changes within each cluster as a function of treatment. Differential gene expression analysis was used to compare gene expression in cell clusters isolated from CD40/ICB-treated versus untreated tumors, beginning with the numerically predominant macrophages. After filtering for genes that achieved an adjusted P value less than 0.05, we ranked genes based on absolute value of fold change in expression. The top 40 differentially expressed genes by adjusted P value in macrophages from CD40/ICB-treated tumors compared with untreated tumors can be found in Supplemental Table 1. This list of genes was then intersected with genes known to be associated with T cell trafficking. The most upregulated of these genes was Ccl5 (Figure 2A). Differential gene expression analysis of macrophages from CD40 agonist–treated versus untreated tumors also yielded Ccl5. Notably, macrophages from tumors treated with ICB alone did not upregulate Ccl5. The chemokine CCL5, also known as RANTES, is a T cell chemoattractant that has been best described for its critical roles in immune control of viral infections (33). The role of CCL5 in cancer remains incompletely examined, as it has been associated with both antitumor and protumor functions, including T regulatory cell attraction, progression and metastasis, tumor-associated macrophage function, and the indirect modulation of both CD8+ chemoattraction and repulsion (30, 34–38).
Figure 2. Antitumor myeloid populations upregulate Ccl5 transcripts after CD40 activation.
Differential gene expression analysis was performed on immune cell clusters from the tumor microenvironment as resolved by UMAP nonlinear dimensional reduction shown in Figure 1. (A) Volcano plot of differentially expressed genes in macrophages as a function of treatment. (B) Expression of Ccl5 overlaid onto UMAP clusters. Color intensity scale represents average number of Ccl5 transcripts per Ccl5+ cell. (C) Proportion of cells positive for reads of Ccl5 gene transcript in immune clusters from untreated versus combination-treated (CD40/ICB) tumors. Size of circle indicates proportion of cells within a cluster positive for Ccl5 transcript. Color intensity scale represents average number of Ccl5 transcripts per Ccl5+ cell.
To examine whether other cell clusters upregulated Ccl5 in response to CD40 agonist treatment, a heatmap of Ccl5 expression was overlaid onto the UMAP visualization of our graph-based clustering (Figure 2B). The macrophage, proliferating macrophage, monocyte, and type 2 conventional DC clusters all increased Ccl5 expression following CD40/ICB treatment — on the basis of both the proportion within each cluster expressing Ccl5 as well as the average expression of Ccl5 per cell (Figure 2C). In contrast, Ccl5 expression remained insignificant within the granulocyte, monocytic myeloid-derived suppressor cell, granulocytic myeloid-derived suppressor cell, and nonconventional monocyte populations, as none of these clusters expressed Ccl5 in more than 6% of their cells even following CD40 agonism. The proportion of cells within the CD8+ T cell and type 1 conventional DC clusters that expressed Ccl5 remained unchanged from baseline, though the average expression of Ccl5 per cell increased among CD8+ T cells (Figure 2C).
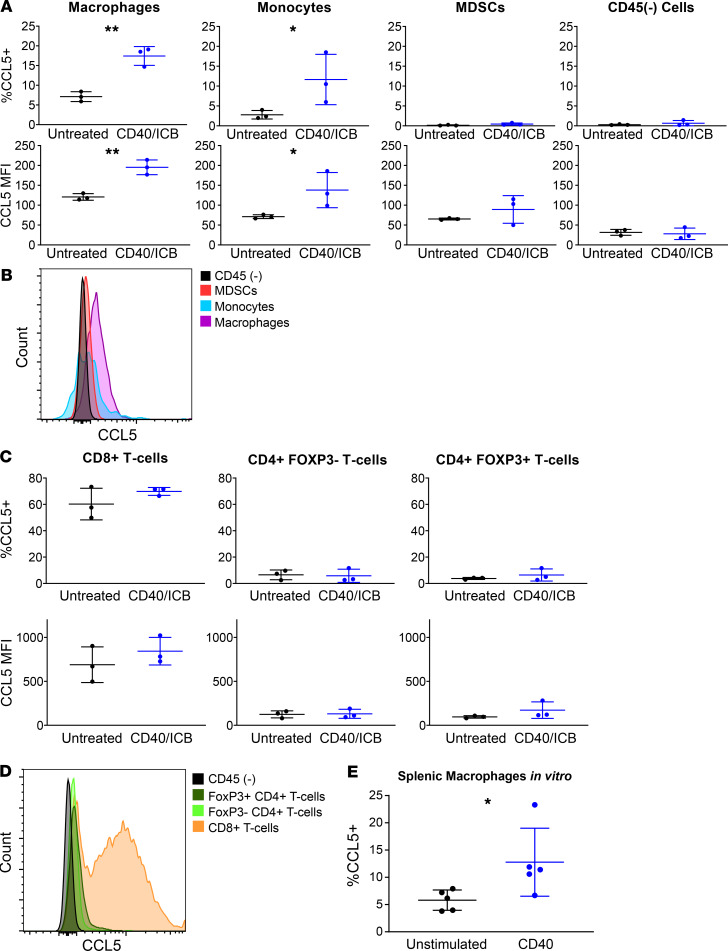
To examine CCL5 induction at the protein level, 4662.MD10 tumor cells were subcutaneously implanted into C57BL/6J mice. Mice were then treated with CD40/ICB and sacrificed on day 12 after treatment induction. Tumors were harvested for flow cytometric analyses, and cell subsets were gated according to the schema outlined in Supplemental Figure 4A. Consistent with our single-cell transcriptomic analysis, macrophages increased expression of CCL5 in response to treatment (Figure 3, A and B). Monocytes also increased expression of CCL5 in response to treatment (Figure 3, A and B). MDSCs did not express CCL5 in either the untreated or treated settings, nor did the CD45– compartment, composed of tumor cells, stroma, and fibroblasts (Figure 3, A and B). In the T cell compartment at baseline, relatively high CCL5 expression was observed in CD8+ T cells, and relatively low CCL5 expression was observed in CD4+ T cells, both FoxP3+ and FoxP3– subpopulations (Figure 3, C and D). Consistent with single-cell transcriptomic analysis, the proportion of all T cell subsets expressing CCL5 did not change as a result of treatment (Figure 3, C and D). The magnitude of CCL5 expression also remained unchanged in T cells from CD40/ICB-treated tumors.
Figure 3. CCL5 is upregulated by antitumor myeloid populations following CD40/ICB therapy.
Female C57BL/6J mice were subcutaneously transplanted with 3 × 105 4662.MD10 cells and treated with CD40/ICB as shown in Figure 1A. Flow cytometric analysis of tumors was then performed on day 12 following initiation of therapy. Gating scheme for flow cytometric analysis is shown in Supplemental Figure 4A. (A and B) Expression of CCL5 in intratumor macrophages, monocytes, myeloid-derived suppressor cells (MDSCs), and the CD45(–) compartment from untreated versus CD40/ICB-treated mice. (C and D) Expression of CCL5 in intratumor CD8+ T cells, CD4+ T cells, and FoxP3+ T regulatory cells from untreated versus CD40/ICB-treated mice. (E) Proportion of CCL5-expressing macrophages. Splenic macrophages were isolated and cultured for 24 hours either unstimulated or stimulated with cross-linked anti-CD40 mAb. n = 3 mice per group. *P ≤ 0.05, and **P ≤ 0.01 (1-tailed Student’s t test). Data shown are representative of 3 independent experiments with 3 to 5 mice per group (A–D). n = 5 mice per group. *P ≤ 0.05 (paired, 1-tailed Student’s t test). Data shown are representative of 3 independent experiments with 3 to 5 biological replicates (E).
To determine whether CD40 agonism can directly induce CCL5 expression, F4/80+ splenic macrophages were isolated from C57BL/6J mice and cultured for 24 hours with cross-linked CD40 agonist mAb. Macrophages cultured with CD40 agonist significantly upregulated CCL5 compared with unstimulated controls as quantified by flow cytometry (Figure 3E). Having supported our findings at the protein level, we next set out to interrogate the functional relevance of CCL5 in the context of CD40/ICB immunotherapy.
CCL5 mediates treatment efficacy.
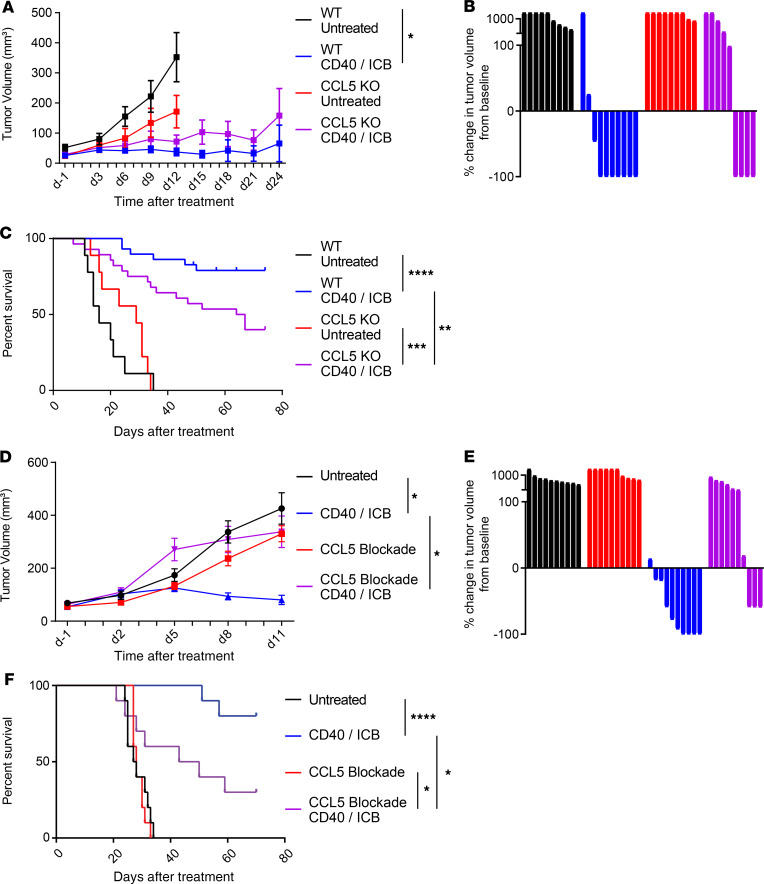
To determine whether CCL5 is required for response to CD40 agonism, we implanted syngeneic CCL5–genetic knockout mice (B6.129P2-Ccl5tm1Hso/J) with 4662.MD10 and compared tumor growth kinetics and survival with those of C57BL/6J WT controls. A subset then received CD40/ICB while another subset was left untreated, as described in Supplemental Figure 5A. Additionally, we observed that the 4662.MD10 tumor cell line expresses MHC class I but not MHC class II following IFN-γ treatment in vitro (Supplemental Figure 4B). Tumors in WT mice responded as expected to treatment with CD40/ICB, both in terms of tumor growth retardation (Figure 4A) and rate of tumor regressions (Figure 4B). In CCL5-KO mice, however, while the treatment effect of CD40/ICB-treated mice remained directionally true, the effect was no longer observed at the statistically significant level relative to untreated CCL5-KO controls (Figure 4, A and B). Over the time course of the entire experiment (75 days), CD40/ICB-treated CCL5-KO mice bearing tumors exhibited statistically worse long-term survival than tumor-bearing WT controls (Figure 4C), consistent with a potential role of CCL5 in mediating response to CD40/ICB immunotherapy.
Figure 4. CCL5 is required for treatment efficacy.
(A) Three hundred thousand 4662.MD10 cells were subcutaneously implanted into C57BL/6J or B6.129P2-Ccl5tm1Hso/J (CCL5-KO) mice. Mice were treated with CD40/ICB as shown in Supplemental Figure 5A. Tumor growth kinetics shown over the course of treatment. (B) Change in tumor volume of mice from A on day 24 (or most recent available) compared with day 0. (C) Survival of mice of mice from A from each treatment group. (D) Three hundred thousand 4662.MD10 cells were subcutaneously implanted into C57BL/6J mice that were then treated with CD40/ICB and/or CCL5-blocking antibody as shown in Supplemental Figure 5B. Tumor growth kinetics shown over the course of treatment. (E) Change in tumor volume of mice from D on day 16 (or most recent available) compared with day 0. (F) Survival of mice from D from each treatment group. (A–B): n = 10 mice per group. (C): combined results of 2 identical experiments with n = 10 mice per group. (D–F): n = 10 mice per group. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 (1-way ANOVA with Tukey’s honestly significant difference posttest in A and D; log-rank test in C and F). Data shown are representative of 2 independent experiments with 10–20 mice per group.
T cells from CCL5-KO mice are known to have baseline defects with potential but unknown compensation during development (39). To rule out this potential confounder, we used a pharmacological inhibitor of CCL5 given just before therapy in WT mice to extend our observations. C57BL/6J mice were subcutaneously injected with 4662.MD10 and were treated with CD40/ICB, anti-CCL5, both, or neither, according to the schema in Supplemental Figure 5B. CCL5 blockade alone did not affect tumor growth, or affect the rate of tumor progression, compared to control antibody (Figure 4, D and E). Although CD40/ICB showed major tumor growth delay and high rate of tumor regressions, these effects were abrogated with the addition of anti-CCL5 to CD40/ICB treatment. Additionally, tumor-bearing mice treated with anti-CCL5 and CD40/ICB had significantly worse long-term survival compared with those treated with CD40/ICB alone (Figure 4F).
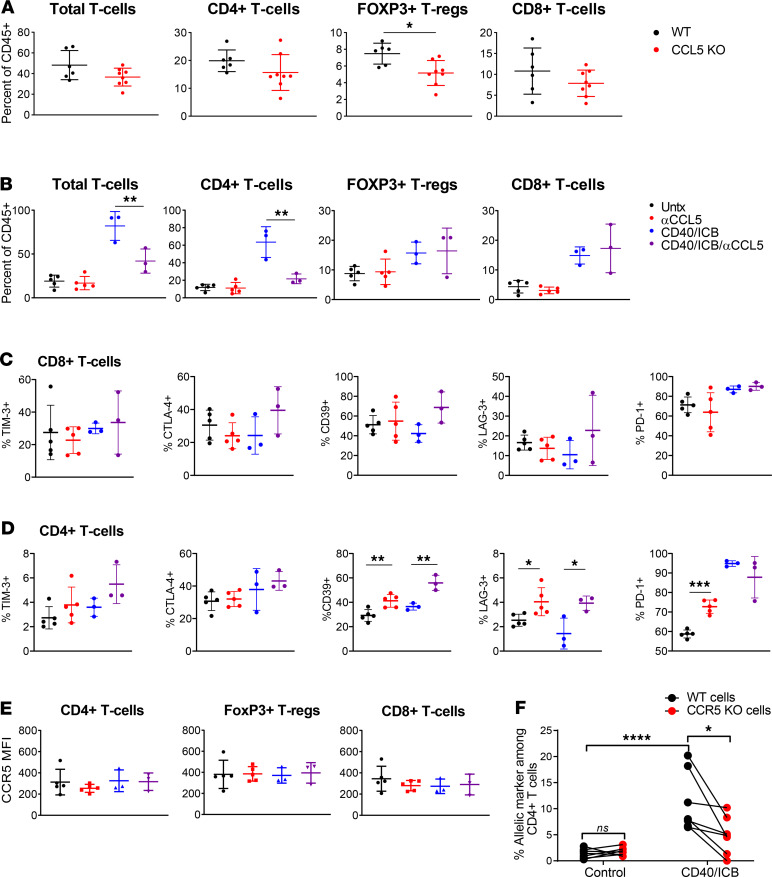
We next sought to determine which immune cell types mediated this treatment dependency on CCL5. We used flow cytometry to compare the T cell content of untreated tumors in WT and CCL5-KO mice 16 days after subcutaneous implantation with 4662.MD10. CCL5-KO mice had statistically significantly lower proportions of FoxP3+CD4+ T cells among all CD45+ cells in the tumor compared with WT, although no differences were otherwise found in total T cell, FoxP3–CD4+ T cell, or CD8+ T cell quantity (Figure 5A). We next examined the effect of CCL5 blockade on the tumor microenvironment of tumor-bearing WT mice, with or without CD40/ICB. In contrast to CCL5-KO mice, WT tumor-bearing mice treated with anti-CCL5 did not have altered T cell content compared with untreated mice at day 12 posttreatment (Figure 5B). Treatment with CD40/ICB, as expected, increased the percentage of total T cells, CD4+ T cells, and CD8+ T cells (20). The addition of anti-CCL5 to CD40/ICB at the same time point, however, decreased total T cell infiltration and abrogated the FoxP3–CD4+ T cell influx in response to therapy. Notably, CCL5 blockade did not affect the proportion of FoxP3+CD4+ T cells or CD8+ T cells. These T cell infiltration dynamics were also observed at the level of cells per unit volume of tumor, indicating that CCL5 modulates absolute infiltration and is not simply repolarizing the immune infiltrate (Supplemental Figure 6). FoxP3–CD4+ and CD8+ T cells in the tumor microenvironment were further examined for expression of T cell activation markers. None of these markers changed in CD8+ T cells in any treatment or control condition (Figure 5C). In contrast, a number of changes were observed in CD4+ T cells in both the untreated and CD40/ICB-treated settings. Anti-CCL5 treatment alone increased the percentage of CD4+ T cells expressing CD39, lymphocyte activating 3 (LAG-3), and PD-1, and CD40/ICB decreased the percentage of cells expressing LAG-3 and markedly increased PD-1+ cells compared with untreated controls (Figure 5D). The addition of anti-CCL5 to CD40/ICB also increased the percentage of CD4+ T cells expressing CD39, restored the percentage of cells expressing LAG-3, and did not affect PD-1, compared with CD40/ICB treatment without anti-CCL5.
Figure 5. CCL5 is required for CD4+ T cell infiltration following CD40/ICB.
(A) Enumeration of T cell populations by flow cytometry in tumors of untreated CCL5-KO and WT control mice on day 16 postimplantation. (B) Enumeration of T cell populations in tumors of mice treated with combination CD40/ICB with or without anti-CCL5 day 12 postimplantation, as outlined in Supplemental Figure 5B. (C) Expression of T cell activation markers on CD4+ T cells from B. (D) Expression of T cell activation markers on CD8+ T cells from B. (E) Expression of CCR5 on CD8+ T cells, CD4+ T cells, and FoxP3+ T regulatory cells from B. (F) Enumeration of adoptively transferred WT or CCR5-KO CD4+ T cells identified by flow cytometry in tumors of mice treated with combination CD40/ICB relative to untreated mice. (A): n = 6 C57BL/6J and n = 8 CCL5-KO mice. (B–E): n = 3–5 C57BL/6J mice each group. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 (2-tailed Student’s t test in A–E; 2-tailed paired Student’s t test in F). Data shown are representative of 2 independent experiments with at least 5 mice per group.
The best-characterized receptor for CCL5 is CCR5 (40). CCR5 expression on intratumor T cells was confirmed by flow cytometry but did not change as a function of either CD40/ICB treatment or CCL5 blockade (Figure 5E). To determine whether CD4+ T cell trafficking to the tumor after CD40/ICB was mediated by CCR5, an equal mixture of CCR5-KO and WT CD4+ T cells was adoptively transferred into tumor-bearing mice 13 days after tumor implantation. Mice were then treated with CD40/ICB according to the schema in Supplemental Figure 5B and sacrificed 7 days later to compare the ability of CCR5-KO T cells (as distinguished by an allelic marker) to traffic to the tumor relative to WT control T cells. Tumors of untreated mice contained equal proportions of CCR5-KO and WT CD4+ T cells, but tumors from CD40/ICB-treated mice contained more than twice as many WT CD4+ T cells on average than CCR5-KO cells (Figure 5F). Thus, CCR5 is at least partially responsible for CD4+ T cell tumor infiltration in response to CD40/ICB immunotherapy.
Discussion
CD4+ T cells are critical mediators of tumor immunity, but mechanisms of intratumor CD4+ T cell chemotaxis remain incompletely understood. Our group has previously demonstrated that CD40 agonism drives CD4+ T cell tumor influx and synergizes with ICB in a CD4+ and CD8+ T cell–dependent manner. Here, we report that the chemokine CCL5 is broadly induced in myeloid populations after treatment with agonist CD40 mAb. Using a suite of genetic and pharmacological experiments in vivo, we show that CCL5 mediates CD4+ T cell tumor influx via CCR5 following CD40 therapy. The effect of CCL5 is selective for CD4+ T cells, not CD8+ T cells. Therapeutic benefit is substantially diminished in the absence of CCL5. Overall, our results demonstrate a previously unappreciated role for CCL5 that underlies the therapeutic adaptive immune response to CD40 agonist.
Given the diverse nature of CD40 expression, it has long been appreciated that the activity of CD40 agonist is likely pleiotropic. CD40 agonism has been shown to have antitumor effects on a number of CD40-expressing myeloid cell types. Macrophages have been shown to remodel tumor stroma after CD40 agonist treatment (24). Monocytes have been shown to degrade fibrosis and enhance the effects of chemotherapy upon CD40 activation (41). We have also observed that the antitumor efficacy of CD40 agonist requires conventional type 1 DCs, the subset of DCs uniquely capable of antigen cross-presentation (18, 20, 42). However, because of past technological limitations, it has been difficult to query all CD40-expressing cell types simultaneously following treatment. The recent emergence of single-cell RNA-sequencing allows us to examine these pleiotropic effects in a highly dimensional and unbiased manner for the first time. Our single-cell transcriptomic analysis reveals an upregulation of the chemokine Ccl5 across a broad range of myeloid cells following CD40 agonism. This is shown to critically and selectively mediate CD4+ T cell chemotaxis and immune control of tumor outgrowth following therapy. Thus, we demonstrate that the antitumor effects of CD40 agonism are largely dependent on the upregulation of a single chemokine.
The current understanding of CCL5 in cancer posits that the chemokine is generally a negative prognostic marker and attracts FoxP3+ T regulatory cells and tumor-associated macrophages to the tumor (34, 37, 43, 44). Consistent with these prior studies, we observed fewer T regulatory cells in the tumors of CCL5-KO mice. When CD40 agonist was administered, however, the primary effect of CCL5 in our system was the promotion of CD4+ (FoxP3–) T cell infiltration into the tumor. CCL5 blockade also increased the expression of CD39, LAG-3, and PD-1 in intratumor CD4+ T cells with no effect on CD8+ T cells, suggesting a role for CCL5 in maintaining CD4+ T cell activation in the tumor microenvironment. Thus, we show a strikingly different role of CCL5 in tumor immune biology following CD40 agonism. Rather than attracting protumor T regulatory cells at baseline, CCL5 plays a critical antitumor role in FoxP3–CD4+ T cell chemotaxis following CD40 agonism. Additionally, in at least one other model, CCL5 derived from the tumor cells has been shown to indirectly enable chemoattraction of CD8+ T cells by way of CXCL9 (36). In our system, however, CCL5 did not modulate CD8+ T cell infiltration and was not produced by any of the nonhematopoietic tumor components. This differential effect is particularly interesting given the comparable expression levels of CCR5 between CD4+ and CD8+ T cells in our system. It may be that different homing receptors are functionally more important in different T cell subsets than others, for example, CCR5 dominating CD4+ T cell homing and CXCR3 dominating CD8+ T cell homing. Alternatively, there may be additional chemokine/chemokine receptor interactions at play in CD8+ T cells acting in opposition to the effect of CCL5/CCR5 in this system or differentially modulating T cell egress from the tumor. Our data, therefore, highlight the context-dependent nature of CCL5 in tumor immunology.
Although we predict that the effect of CCL5 in this system is source agnostic, it may be that cells beyond the antitumor myeloid lineages meaningfully contribute to this phenotype. In terms of CCL5 derived from CD8+ T cells or conventional type 1 DCs, there were far fewer of these cells than macrophages in the PDAC tumors at baseline (Supplemental Figure 3), and neither lineage increased the proportion of cells expressing CCL5 as a function of CD40/ICB treatment. Nevertheless, both lineages are strong producers of CCL5 at the cell-by-cell level and could contribute to this phenotype on that basis. Therefore, an important future direction to understand the specific effect of myeloid-derived CCL5 would be testing CD40/ICB in myeloid-specific CCL5-KO systems.
Our findings raise several additional preclinical questions of interest for future studies. This study was performed in a subcutaneously transplanted model of pancreatic cancer, which facilitated single-cell analysis. T cell trafficking to the pancreas in orthotopic or autochthonous models in response to CD40/ICB therapy may operate under different biology. Whether our findings extend to other priming-deficient cancers beyond PDAC is also of significant interest. In addition, while we have no evidence to support a role for CCL5 beyond attracting CD4+ T cells to the tumor, we cannot eliminate that possibility. Finally, although our results demonstrate the dominance of CCR5 in this system, chemokine/chemokine receptor interactions are notoriously complex, and supplemental or compensatory roles for other CCL5 receptors may exist.
Past manipulations of the CCL5/CCR5 signaling axis in patients with cancer have been dominated by the use of CCR5 antagonists to mitigate T regulatory cell and tumor-associated macrophage infiltration (38, 45, 46). CCR5 inhibition has also been used in attempts to sensitize tumors to chemotherapy and prevent metastasis and shows promise as a means of preventing visceral graft-versus-host disease in patients with cancer after allogenic bone marrow transplant (47–50). Our finding that T regulatory cell content is reduced in tumors implanted into CCL5-KO mice corroborates these findings and supports the use of these inhibitors at baseline before immunotherapy. However, our findings suggest further that the use of CCR5 antagonists may be harmful with continued use once an immune response is initiated. This may have immediate clinical relevance for at least 2 ongoing clinical trials (ClinicalTrials.gov NCT03631407, NCT03274804) combining the CCR5 small-molecule inhibitors maraviroc and vicriviroc with the PD-1 inhibitor pembrolizumab. Moving forward, our data should inform the optimal combinations in which CCR5 inhibitors are administered.
CD40 agonist immunotherapies are currently being tested in the clinic (ClinicalTrials.gov NCT03214250, NCT02588443) (23, 25, 26). Early results are promising, especially in combination with PD-1 inhibitors. A particularly promising trial was recently performed in which patients with pancreatic adenocarcinoma received a CD40 agonist mAb (APX005M) in addition to standard-of-care gemcitabine/nab-paclitaxel chemotherapy (26). The overall response rate was 54.2%, compared with historical controls of 18% with standard-of-care chemotherapy alone. Moving forward, CCL5 can be evaluated as a potential biomarker of response to CD40 agonism in these clinical studies. Finally, our findings also provide rationale for enhancing CD40 agonist or other cancer immunotherapies through ectopic delivery of CCL5 using CCL5-expressing oncolytic viruses or intratumor injection of recombinant CCL5.
Methods
Animal studies.
Mice were housed under specific pathogen–free conditions in a barrier facility. C57BL/6J mice were purchased from The Jackson Laboratory; B6.129P2-Ccl5tm1Hso/J (CCL5-KO) mice were purchased from The Jackson Laboratory or bred in-house. Tumor cell lines were derived from spontaneous tumors in the KPC (LSL-KrasG12D/+ LSL-Trp53R172H/+ Pdx-1-Cre) mouse model of PDAC (51) as previously described (52). 4662 is a polyclonal KPC cell line, and 4662.MD10 is a clonal KPC cell line derived from 4662. Cell culture was performed in DMEM supplemented with 10% FBS, l-glutamine, and gentamicin.
Transplanted tumors were generated by injecting 3 × 105 cells in serum-free DMEM subcutaneously into the right flank. Tumors were then allowed to grow for 14 days (average size, 30–60 mm3). Mice were assigned to groups such that average tumor volume at baseline did not vary by treatment condition. Tumors were measured every 3 days by caliper. Tumor volumes were calculated using the formula (L × W2)/2, where L is the longer diameter and W is the diameter perpendicular to L. For survival studies, mice were deemed to have reached endpoint when their tumor exceeded 500 mm3. Mice that died suddenly or developed large tumor ulcerations were censored from survival studies on the day of death or euthanasia.
In vivo antibody studies.
Mice were treated intraperitoneally with ICB (anti–PD-1: RMP1-14; Bio X Cell; 200 μg/dose on days 0, 3, 6, 9, and 12 and anti–CTLA-4: 9H10; Bio X Cell; 200 μg/dose on days 0, 3, and 6) and CD40 agonist (FGK45; Bio X Cell; endotoxin free; 100 μg/dose) on day 3. For CCL5 blockade studies, mice were treated intraperitoneally with anti-CCL5 blocking antibody (PeproTech; 32 μg/dose on days –1, 2, 5, 8, and 11) or polyclonal rabbit isotype control (PeproTech; 32 μg/dose on days –1, 2, 5, 8, and 11).
Tissue processing and flow cytometry.
Mice were sacrificed on day 12 posttreatment. The entire tumor was dissected, washed in DMEM-F12 and 10% FBS, minced into small fragments, and digested in DMEM-F12 with 1 mg/mL collagenase and protease inhibitor (MilliporeSigma C6079) for 30 minutes at 37°C. Cells were then filtered through a 70-μm cell strainer then 40-μm strainer. Tissue-derived cells were washed with PBS before viability stain with LIVE/DEAD Fixable Aqua (Invitrogen, Thermo Fisher Scientific, L34957) for 20 minutes at room temperature. Samples were then washed with FACS Buffer (PBS with 0.2% BSA + 2 mM EDTA) before being stained for surface markers for 30 minutes at 4°C. Samples were then fixed and permeabilized using the eBioscience fixation/permeabilization kit (Thermo Fisher Scientific, 88-8824-00) and stained intracellularly overnight at 4°C. Flow cytometry antibodies can be found in Supplemental Table 2. Samples were collected on an LSR Fortessa flow cytometer (BD Biosciences). Data were analyzed using FlowJo v10 (Tree Star).
In vitro stimulation assay.
Spleens from 5 female C57BL/6J mice were isolated, and macrophages were enriched by magnet-assisted cell sorting column using the F4/80 positive selection kit (Miltenyi Biotec 130-110-443). Macrophages were cultured in a 96-well plate overnight in an incubator at 37°C in DMEM with 10% FBS, l-glutamine, and gentamicin and stimulated with cross-linked CD40 agonist (FGK45; Bio X Cell; endotoxin free). Cells were stained for CCL5 by flow cytometry the following day as described above.
Single-cell RNA-sequencing library generation.
Five thousand live CD45+ cells were isolated from each tumor by FACS using the 100-μm nozzle on a BD Biosciences FACSAria II. Sorted cells were then barcoded and used to generate single-cell RNA libraries with the droplet-based 10x Genomics Chromium platform according to the manufacturer’s protocol. Library quality was verified with an Agilent BioAnalyzer and LifeTech QuBit fluorimeter. Libraries were then sequenced as 150-bp paired-end reads on an Illumina HiSeq4000 to a depth of approximately 312 million read pairs.
Library alignment, barcode assignment, and unique molecular identifier counting.
10x Genomics’ Cell Ranger Single-Cell Software Suite v. 3.1.0 was used to perform sample demultiplexing, barcode processing, and single-cell 3′ counting from the generated FASTQ files. The “count” function was used to align samples to the mm10 Mus musculus genome, filter cells, and quantify reads. The resulting analysis files were aggregated per treatment group using the “aggr” function, which performs between-sample normalization and sample merging. These combined data sets were used as input into Seurat v3.0 on R v. 3.6.1 (53, 54).
Preprocessing.
Cells that contained reads for more than 2500 or less than 200 genes were excluded as doublets or empty wells, respectively. Cells that contained reads for which more than 5% aligned to mitochondrial genes were excluded as dead cells. Data were normalized with a scale factor of 104. Highly variable genes between cells were identified using variance stabilizing transformation (“vst”), which directly models mean-variance relationships within single-cell data sets. The number of cells in each treatment group was then reduced to 2072 cells. Batch correction within treatment groups was performed using the “FindIntegrationAnchors” and “IntegrateData” functions, generating a “batch-corrected” expression matrix. Cells across all treatment groups were then integrated into a single data set using the same functions (i.e., “FindIntegrationAnchors” and “IntegrateData”).
Linear dimensional reduction and clustering.
The fully merged data set was linearly transformed using the “ScaleData” function such that the mean expression of a given gene across all cells was 0 and the variance of that gene across all cells was 1. Linear dimensional reduction was then performed using principal component analysis. Based on the distribution of P values per principal component, the first 20 principal components were used to cluster cells using the “FindNeighbors” and “FindClusters” functions, which implement shared nearest neighbor modularity optimization-based clustering. This was performed using a chosen resolution of 0.5, yielding 16 total clusters. Nonlinear dimensional reduction was then performed using UMAP to visualize clusters in 2-dimensional space.
Cluster identification.
To identify cell type within a given cluster, the “FindConservedMarkers” function was used to identify genes for which expression was conserved across treatment groups. This function performs differential gene expression testing for each treatment group and combines the P values using meta-analysis methods from the MetaDE R package. Cell type identities were then assigned to clusters based on identification of canonical cell markers and characterization of top conserved genes using the MyGeneSet tool from ImmGen. Clusters that comprised contaminating nonimmune populations (i.e., tumor cells and fibroblasts) were removed. Scaled expression of conserved marker genes was used for heatmap representation.
Differential gene expression analysis.
A Wilcoxon rank-sum test was used to identify differentially expressed genes between 2 treatment groups within a given cluster. The fold change in expression and adjusted P value for each gene were used for volcano plot representation using the ggplot2 R package. After filtering for genes with an adjusted P value < 0.05, genes were then ranked based on highest to lowest absolute value of fold change.
Pseudotime analysis.
Myeloid clusters identified using Seurat (as described above) were used as input to the Monocle v. 2.4.0 R package (55). Genes expressed in 10 or more cells were ranked based on differential analysis between clusters. Genes with a q value less than 0.01 were used for downstream pseudotemporal analysis. Dimensionality reduction was done using the DDRTree method. Cells were ordered along a pseudotime trajectory with the orderCells function and visualized in 2-dimensional space.
Data availability.
Data were deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus database (GSE150176).
Statistics.
Comparison of 2 groups was performed using 2-tailed Student’s t test unless otherwise indicated. Tumor growth curves were analyzed by 2-way ANOVA, with Tukey’s multiple comparisons of means as a post hoc test to assess differences between any 2 groups. Survival curves were compared using log-rank (Mantel-Cox) test. Statistical analyses were performed in Prism 7 (GraphPad) or Excel (Microsoft). P < 0.05 was considered statistically significant, and *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Study approval.
All mouse experiments were performed at the Perelman School of Medicine of the University of Pennsylvania in accordance with university IACUC and University Laboratory Animal Resources approvals and regulations.
Author contributions
APH, JHL, KTB, and RHV designed experiments. APH, JHL, and SIK performed experiments. JHL performed informatics. All authors interpreted data. APH, JHL, and RHV wrote the manuscript, which all authors helped edit. RHV supervised the study.
Supplementary Material
Acknowledgments
We thank the Next Generation Sequencing Core at the University of Pennsylvania for assistance and advice. This work was supported by National Cancer Institute grants R01 CA229803, P30 CA016520, and P01 CA210944 (to RHV); the Parker Institute for Cancer Immunotherapy (to RHV and KTB); and the Roy and Diana Vagelos Scholars Program in the Molecular Life Sciences at the University of Pennsylvania (to SIK).
Version 1. 04/23/2020
In-Press Preview
Version 2. 05/21/2020
Electronic publication
Footnotes
Conflict of interest: RHV reports having received consulting fees or honoraria since 2015 from Celgene, Celldex, Janssen, Lilly, MedImmune, and Verastem and research funding from Apexigen, FibroGen, Inovio, Janssen, and Lilly. He is an inventor on licensed patents relating to cancer cellular immunotherapy (10286066, 9453199, and 7670781) and receives royalties from Boston Children’s Hospital for a licensed research-only monoclonal antibody.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(10):e137263.https://doi.org/10.1172/jci.insight.137263.
Contributor Information
Austin P. Huffman, Email: ahuffman@pennmedicine.upenn.edu.
Jeffrey H. Lin, Email: jeffrlin@pennmedicine.upenn.edu.
Samuel I. Kim, Email: skim0814@sas.upenn.edu.
Katelyn T. Byrne, Email: byrnek@pennmedicine.upenn.edu.
Robert H. Vonderheide, Email: rhv@upenn.edu.
References
- 1.Veatch JR, et al. Endogenous CD4+ T cells recognize neoantigens in lung cancer patients, including recurrent oncogenic KRAS and ERBB2 (Her2) driver mutations. Cancer Immunol Res. 2019;7(6):910–922. doi: 10.1158/2326-6066.CIR-18-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitzer MH, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168(3):487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmi Y, et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature. 2015;521(7550):99–104. doi: 10.1038/nature14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurwitz AA, Yu TF, Leach DR, Allison JP. CTLA-4 blockade synergizes with tumor-derived granulocyte-macrophage colony-stimulating factor for treatment of an experimental mammary carcinoma. Proc Natl Acad Sci USA. 1998;95(17):10067–10071. doi: 10.1073/pnas.95.17.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie AJ, et al. Dual control of antitumor CD8 T cells through the programmed death-1/programmed death-ligand 1 pathway and immunosuppressive CD4 T cells: regulation and counterregulation. J Immunol. 2009;183(12):7898–7908. doi: 10.4049/jimmunol.0901060. [DOI] [PubMed] [Google Scholar]
- 7.Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18(10):635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 8.Bogen B, Fauskanger M, Haabeth OA, Tveita A. CD4+ T cells indirectly kill tumor cells via induction of cytotoxic macrophages in mouse models. Cancer Immunol Immunother. 2019;68(11):1865–1873. doi: 10.1007/s00262-019-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res. 2014;2(2):91–98. doi: 10.1158/2326-6066.CIR-13-0216. [DOI] [PubMed] [Google Scholar]
- 10.Alspach E, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019;574(7780):696–701. doi: 10.1038/s41586-019-1671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odaka M, et al. Eradication of intraperitoneal and distant tumor by adenovirus-mediated interferon-beta gene therapy is attributable to induction of systemic immunity. Cancer Res. 2001;61(16):6201–6212. [PubMed] [Google Scholar]
- 12.Palomba ML, et al. CD8+ T-cell-dependent immunity following xenogeneic DNA immunization against CD20 in a tumor challenge model of B-cell lymphoma. Clin Cancer Res. 2005;11(1):370–379. [PubMed] [Google Scholar]
- 13.Zamarin D, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6(226):226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang H, et al. Radiation-induced equilibrium is a balance between tumor cell proliferation and T cell-mediated killing. J Immunol. 2013;190(11):5874–5881. doi: 10.4049/jimmunol.1202612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu YC, et al. Treatment of patients with metastatic cancer using a major histocompatibility complex class II-restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. J Clin Oncol. 2017;35(29):3322–3329. doi: 10.1200/JCO.2017.74.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banchereau J, et al. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 18.Morrison AH, Diamond MS, Hay CA, Byrne KT, Vonderheide RH. Sufficiency of CD40 activation and immune checkpoint blockade for T cell priming and tumor immunity. Proc Natl Acad Sci U S A. 2020;117(14):8022–8031. doi: 10.1073/pnas.1918971117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winograd R, et al. Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol Res. 2015;3(4):399–411. doi: 10.1158/2326-6066.CIR-14-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne KT, Vonderheide RH. CD40 stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Rep. 2016;15(12):2719–2732. doi: 10.1016/j.celrep.2016.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rech AJ, et al. Radiotherapy and CD40 activation separately augment immunity to checkpoint blockade in cancer. Cancer Res. 2018;78(15):4282–4291. doi: 10.1158/0008-5472.CAN-17-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vonderheide RH. CD40 agonist antibodies in cancer immunotherapy. Annu Rev Med. 2020;71:47–58. doi: 10.1146/annurev-med-062518-045435. [DOI] [PubMed] [Google Scholar]
- 24.Beatty GL, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331(6024):1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajor DL, et al. Long-term outcomes of a phase I study of agonist CD40 antibody and CTLA-4 blockade in patients with metastatic melanoma. Oncoimmunology. 2018;7(10):e1468956. doi: 10.1080/2162402X.2018.1468956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Hara MH, et al. Abstract CT004: A Phase Ib study of CD40 agonistic monoclonal antibody APX005M together with gemcitabine (Gem) and nab-paclitaxel (NP) with or without nivolumab (Nivo) in untreated metstatic ductal pancreatic adenocarcinoma (PDAC) patients. Cancer Res. 2019;79(13 suppl):CT004 [Google Scholar]
- 27.Nowak AK, Robinson BW, Lake RA. Synergy between chemotherapy and immunotherapy in the treatment of established murine solid tumors. Cancer Res. 2003;63(15):4490–4496. [PubMed] [Google Scholar]
- 28.Vonderheide RH, et al. Clinical activity and immune modulation in cancer patients treated with CP-870,893, a novel CD40 agonist monoclonal antibody. J Clin Oncol. 2007;25(7):876–883. doi: 10.1200/JCO.2006.08.3311. [DOI] [PubMed] [Google Scholar]
- 29.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335):eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 33.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD, Wherry EJ. A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog. 2011;7(7):e1002098. doi: 10.1371/journal.ppat.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014;2014:292376. doi: 10.1155/2014/292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhong M, Wang C, Xu Y, Gao WQ, Zhang Y. CCL5-deficiency enhances intratumoral infiltration of CD8+ T cells in colorectal cancer. Cell Death Dis. 2018;9(7):766. doi: 10.1038/s41419-018-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dangaj D, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35(6):885–900.e10. doi: 10.1016/j.ccell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3+Treg cells in pancreatic ductal adenocarcinoma. Oncogene. 2017;36(21):3048–3058. doi: 10.1038/onc.2016.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aldinucci D, Casagrande N. Inhibition of the CCL5/CCR5 axis against the progression of gastric cancer. Int J Mol Sci. 2018;19(5):E1477. doi: 10.3390/ijms19051477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makino Y, et al. Impaired T cell function in RANTES-deficient mice. Clin Immunol. 2002;102(3):302–309. doi: 10.1006/clim.2001.5178. [DOI] [PubMed] [Google Scholar]
- 40.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4(12):2171–2185. doi: 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long KB, Gladney WL, Tooker GM, Graham K, Fraietta JA, Beatty GL. IFNγ and CCL2 cooperate to redirect tumor-infiltrating monocytes to degrade fibrosis and enhance chemotherapy efficacy in pancreatic carcinoma. Cancer Discov. 2016;6(4):400–413. doi: 10.1158/2159-8290.CD-15-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vonderheide RH. The immune revolution: a case for priming, not checkpoint. Cancer Cell. 2018;33(4):563–569. doi: 10.1016/j.ccell.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cambien B, et al. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRβ in colorectal carcinoma. PLoS One. 2011;6(12):e28842. doi: 10.1371/journal.pone.0028842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willenbrock F, et al. Abstract B40: High circulating CCL5 is associated with poor prognosis in locally advanced pancreatic cancer (LAPC): biomarker analysis from the randomized phase II SCALOP trial. Cancer Res. 2019;79(24 suppl):B40 [Google Scholar]
- 45.Jiao X, et al. Recent advances targeting CCR5 for cancer and its role in immuno-oncology. Cancer Res. 2019;79(19):4801–4807. doi: 10.1158/0008-5472.CAN-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pervaiz A, Zepp M, Mahmood S, Ali DM, Berger MR, Adwan H. CCR5 blockage by maraviroc: a potential therapeutic option for metastatic breast cancer. Cell Oncol (Dordr) 2019;42(1):93–106. doi: 10.1007/s13402-018-0415-3. [DOI] [PubMed] [Google Scholar]
- 47.Reshef R, et al. Blockade of lymphocyte chemotaxis in visceral graft-versus-host disease. N Engl J Med. 2012;367(2):135–145. doi: 10.1056/NEJMoa1201248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reshef R, et al. High graft CD8 cell dose predicts improved survival and enables better donor selection in allogeneic stem-cell transplantation with reduced-intensity conditioning. J Clin Oncol. 2015;33(21):2392–2398. doi: 10.1200/JCO.2014.60.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moy RH, et al. Clinical and immunologic impact of CCR5 blockade in graft-versus-host disease prophylaxis. Blood. 2017;129(7):906–916. doi: 10.1182/blood-2016-08-735076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huffman AP, et al. Pharmacodynamic monitoring predicts outcomes of CCR5 blockade as graft-versus-host disease prophylaxis. Biol Blood Marrow Transplant. 2018;24(3):594–599. doi: 10.1016/j.bbmt.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Lo A, et al. Tumor-promoting desmoplasia is disrupted by depleting fap-expressing stromal cells. Cancer Res. 2015;75(14):2800–2810. doi: 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus database (GSE150176).