Abstract
BACKGROUND
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a novel viral pneumonia (COVID-19), which is rapidly spreading throughout the world. The positive result of nucleic acid test is a golden criterion to confirm SARS-CoV-2 infection, but the detection features remain unclear.
METHODS
We performed a retrospective analysis in 5630 high-risk individuals receiving SARS-CoV-2 nucleic acid tests in Wuhan, China, and investigated their characteristics and diagnosis rates.
RESULTS
The overall diagnosis rate was 34.7% (1952/5630). Male (P = 0.025) and older populations (P = 2.525 × 10–39) were at significantly higher risk of SARS-CoV-2 infection. People were generally susceptible, and most cases concentrated in people of 30–79 years. Furthermore, we investigated the association between diagnosis rate and the amount of testing in 501 subjects. Results revealed a 1.27-fold improvement (from 27.9% to 35.5%) of diagnosis rate from testing once to twice (P = 5.847 × 10–9) and a 1.43-fold improvement (from 27.9% to 39.9%) from testing once to 3 times (P = 7.797 × 10–14). More than 3 testing administrations was not helpful for further improvement. However, this improvement was not observed in subjects with pneumonia (P = 0.097).
CONCLUSION
All populations are susceptible to SARS-CoV-2 infection, and male and older-aged populations are at significantly higher risk. Increasing the amount of testing could significantly improve diagnosis rates, except for subjects with pneumonia. It is recommended to test twice in those high-risk individuals whose results are negative the first time, and performing 3 tests is better, if possible.
FUNDING
This work was supported by National Mega Project on Major Infectious Disease Prevention (no. 2017ZX10103005-007) and National Key Research and Development Program of China (no. 2018YFE0204500).
Keywords: COVID-19, Infectious disease
Keywords: Molecular diagnosis
All people are generally susceptible to SARS-CoV-2 infection; however, being male and older increases infection risk, and repeated testing could markedly improve diagnosis rates.
Introduction
An ongoing outbreak of a novel viral pneumonia, formally called the coronavirus disease 2019 (COVID-19), has rapidly spread throughout the world and raised urgent global concerns (1–3). The causative pathogen is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), isolated by Chinese scientists on January 7, 2020 (4, 5). By April 7, 2020, there are 83,095 cumulative confirmed cases in China and more than 1.33 million cumulative confirmed cases in the world (data were available at https://ncov.dxy.cn/ncovh5/view/pneumonia).
Accumulated evidence has confirmed that SARS-CoV-2 infection shows sustained human-to-human transmission (6–8). Although SARS-CoV-2 causes a lower death rate than SARS, its transmission between individuals is much more prevalent. Besides typically infected patients, asymptomatic and mildly symptomatic infectors are becoming important infectious sources, which are not reported in SARS previously (9–12). More notably, the average incubation period of SARS-CoV-2 infection was 5.2 days, but some individuals may have a long incubation period of more than 14 days (13–15). These features determine that rapid and accurate diagnosis of SARS-CoV-2 in high-risk populations such as suspected cases and close contacts is crucial to control the pandemic in the community and hospitals. The Chinese center for disease and prevention recommends a specific real-time reverse transcription PCR (RT-PCR) method for diagnosis of SARS-CoV-2, by detecting its 2 target genes, open reading frame 1ab (ORF1ab) and nucleocapsid protein (N).
Several factors possibly affect detection results of SARS-CoV-2, including patient condition, viral load, sampling quality, specimen delivery time, and the testing process. Delineating features of SARS-CoV-2 in a large population is necessary to provide valuable information for clinical treatment and pandemic control. Here, we performed a large retrospective study to investigate the characteristics and diagnosis rate of 5630 subjects receiving SARS-CoV-2 nucleic acid tests from Wuhan, China.
Results
Characteristics of included subjects and confirmed cases of SARS-CoV-2 infection.
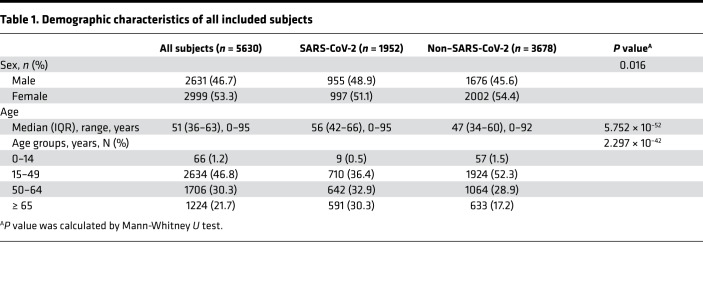
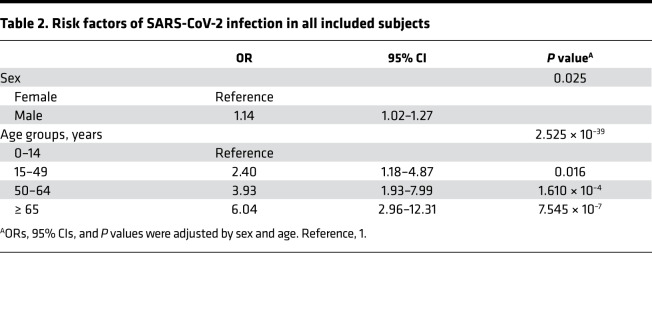
Demographic characteristics of 5630 subjects are shown in Table 1. A total of 34.7% (1952 of 5630) of subjects were positive for SARS-CoV-2 nucleic acid test. More males were observed in the SARS-CoV-2 group than Non–SARS-CoV-2 group (48.9% vs. 45.6%, P = 0.016). The ages of the SARS-CoV-2 group were also older than ages of Non–SARS-CoV-2 group (all P < 0.05). Binary logistic regression analysis showed that males had a 1.14-fold increased risk of SARS-CoV-2 infection than females (OR = 1.14, 95% CI = 1.02–1.27, P = 0.025). Older age was another risk factor, and people older than 65 years had a 6.04-fold increased risk of SARS-CoV-2 infection (OR = 6.04, 95% CI = 2.96–12.31,P = 7.545 × 10–7). More details are shown in Table 2.
Table 1. Demographic characteristics of all included subjects.
Table 2. Risk factors of SARS-CoV-2 infection in all included subjects.
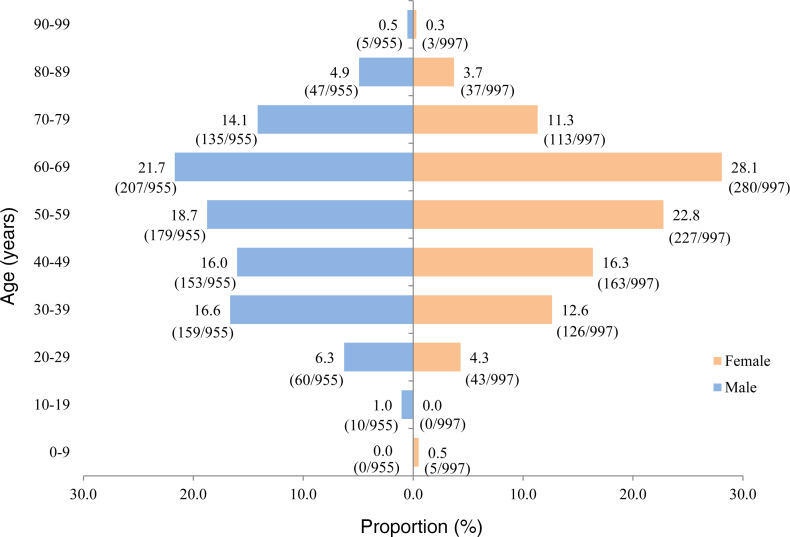
Moreover, we summarize characteristics of subjects of the SARS-CoV-2 group based on sex and age (10 years in each group) in Figure 1. In these 1952 confirmed cases of SARS-CoV-2 infection, the proportion of males (48.9% [955 of 1952]) was close to that of females (51.1% [997 of 1952]). The age of confirmed cases was in approximately normal distribution, regardless of whether subjects were male or female. The cases mainly concentrated in ages of 30–79 years, with a highest proportion in the age group of 50–69 years. These results suggested that all populations were susceptible to SARS-CoV-2 infection, especially in the middle-aged and elderly people.
Figure 1. Characteristics of SARS-CoV-2 cases based on sex and age.
Association between diagnosis rates and the amount of testing.
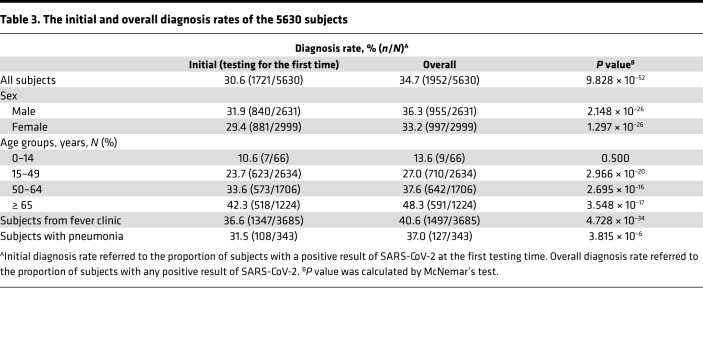
The overall diagnosis rate of 5630 subjects was 34.7% (1952 of 5630). Because some subjects were tested multiple times due to medical considerations (detailed in Methods), we further compared the initial and overall diagnosis rates in all included subjects in order to examine whether increasing the amount of testing improve diagnosis rate. As shown in Table 3, results revealed that the overall diagnosis rate was significantly improved compared with the diagnosis rate of testing only once (34.7% vs. 30.6%, P = 9.828 × 10–52). The improvements were also demonstrated in males, females, different age groups (age ≥ 15 years), subjects from fever clinic, and subjects with pneumonia (all P < 0.05).
Table 3. The initial and overall diagnosis rates of the 5630 subjects.
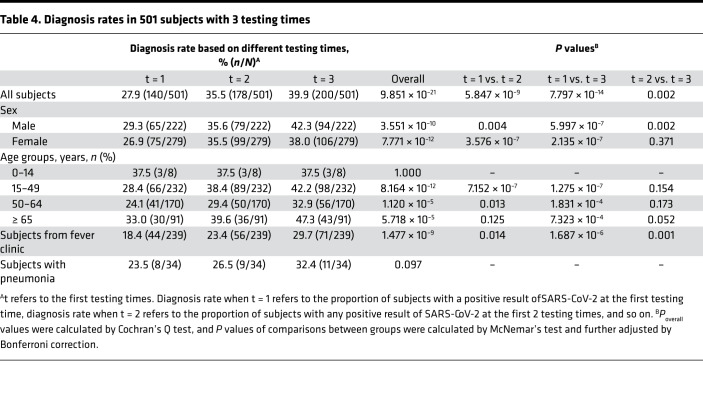
Testing the same population with the same amount of tests could provide an accurate comparison of improvement of diagnosis rates. Due to actual factors, only a small part of subjects were eligible for analysis if they met the following criteria: (a) subjects were suspected cases or close contacts of confirmed cases of SARS-CoV-2; (b) subjects could be continuously followed in our hospital after the first testing; and (c) subjects received 3 or more SARS-CoV-2 nucleic acid tests. A total of 501 subjects fulfilled the inclusion criteria. Table 4 demonstrates the association between diagnosis rate and the amount of testing in these subjects. Results revealed that increasing the amount of testing could significantly improve the overall diagnosis rates (Poverall = 9.851 × 10–21). There was a 1.27-fold improvement from testing once to twice (from 27.9% to 35.5%) and a 1.43-fold improvement from testing once to 3 times (from 27.9% to 39.9%). Multiple between-group comparisons further supported the result (Pt = 1 vs. t = 2 = 5.847 × 10–9, Pt = 1 vs. t = 3 = 7.797 × 10–14, Pt = 2 vs. t = 3= 0.002). Similar trends were also observed in most subgroup analysis, but it was notable that increasing the amount of testing did not improve the diagnosis rate of subjects with pneumonia (Poverall = 0.097). In subjects older than 14 years and without pneumonia, diagnosis rates were significantly improved when the amount of testing increased from 1 to 2, except for subjects older than 65 years, and improved more when the amount of testing increased from 1 to 3 in all subgroups (all Pt = 1 vs. t = 3 < 0.05). However, when the amount of testing increased from 2 to 3, the improvement was not significant in some subgroups. In addition, there were 154 subjects who received 4 administrations of SARS-CoV-2 nucleic acid tests. Thus, we also compared the diagnosis rates when the amount of testing increased from 3 to 4, and we identified that the overall diagnosis rate was not improved (P = 1.000). These results suggest that increasing the amount of testing could significantly improve the overall diagnosis rate of SARS-CoV-2, and 2 or 3 may be suitable.
Table 4. Diagnosis rates in 501 subjects with 3 testing times.
Discussion
Speeding up the diagnosis of suspected cases and close contacts is the key to win the battle of the SARS-CoV-2 pandemic. In this retrospective study, we included 5630 subjects receiving a SARS-CoV-2 nucleic acid test from Wuhan, China. By analyzing the big data, we answered several crucial issues as follows: (a) the overall diagnosis rate of high-risk populations was 34.7%, and being male and of an older age were risk factors of SARS-CoV-2 infection; (b) all populations were susceptible to SARS-CoV-2 infection, especially people 50–69 years old; and (c) increasing the amount of testing could significantly improve the overall diagnosis rate of SARS-CoV-2, and 2 or 3 times may be suitable.
The positive result of a nucleic acid test is a golden criterion to confirm SARS-CoV-2 infection. Although reverse transcription PCR (RT-PCR) is an excellent recommended method for SARS-CoV-2 detection, the results are in risk of bias by viral load, specimen type, sample collection, and quality control of the testing process (16). Our study had distinct advantages to ensure the accuracy of testing results, including specimen collection by experienced clinicians, quick specimen transportation maintained in viral-transport medium, and timely detection under strict quality control in a laboratory certificated by College of American Pathologists (CAP).
Sex differences are quite common in human diseases. We identified that being male was a significant risk factor of SARS-CoV-2 infection, which could be supported by previous evidence. Sex differences were confirmed and considered a feature of SARS-CoV infection (17, 18). Recently, a systematic review and meta-analysis from arXiv preprint also found that males were more susceptible and vulnerable to SARS-CoV-2 than females (19). The mechanism of sex difference in SARS-CoV-2 is unclear, but potential explanations include smoking condition, different levels of ACE2 expression, and distinct immune responses between males and females (20–22). Aging is another crucial risk factor of human diseases. Results from our study also found that older people were more likely to be infected by SARS-CoV-2. In addition, we observed that SARS-CoV-2–infected cases occurred in all ages and presented in an approximately normal distribution. This finding was consistent with previous studies (14, 23). The smaller numbers of SARS-CoV-2 cases from children and quite older people are possibly due to less outdoor activities and, thus, less exposure to this virus.
Moreover, we identified that increasing testing times significantly improved diagnosis rate, with a 1.27-fold improvement from testing once to twice (from 27.9% to 35.5%) and a 1.43-fold improvement from testing once to 3 times (from 27.9% to 39.9%). More than 3 testing times seemed not to be helpful for further improvement. Increasing the amount of testing could effectively reduce biases from several actual factors, such as sample collection, viral load of different disease process, and patients’ conditions. However, increasing testing times did not help improve diagnosis rate in people with pneumonia. A probable explanation is that SARS-CoV-2 cases with pneumonia have a high viral load and could be detected at the first testing. Based on these results, we recommend that 2 testing times should be performed in those high-risk people when the testing result is negative for the first testing, and 3 testing times is better, if available.
A main limitation of this study was that only throat swabs were evaluated. Compared with specimens from the lower respiratory tract, including bronchoalveolar lavage fluid and sputum, nasopharyngeal swabs and throat swabs are more convenient and suitable for universal screening of SARS-CoV-2 cases in high-risk populations, such as suspected cases or those with close contact with suspected cases. A recent study found that positive rate of nasal swabs (5 of 8) was higher than that of throat swabs (126 of 398), but the result was possibly biased by the quite smaller number of nasal swabs (24). Another study reported a diagnosis rate of 38.25% in 4818 subjects for nasopharyngeal swabs (25). It seemed a litter higher than our diagnosis rate of 34.7% in 5630 subjects. In spite of this, biases from sampling, testing, or patients’ conditions could be also reduced by increasing the amount of testing. This strategy is theoretically suitable to improve diagnosis rate of nasopharyngeal swabs.
In summary, the overall diagnosis rate of SARS-CoV-2 was 34.7% in 5630 subjects from Wuhan, China. All populations were susceptible, and being male and of an older age were risk factors of SARS-CoV-2 infection. Increasing the amount of testing could significantly improve the overall diagnosis rate of SARS-CoV-2, except for suspected cases with pneumonia. We recommend that 2 testing times should be performed in those high-risk people when the testing result is negative for the first testing, and 3 testing times is better, if available. To the best of our knowledge, this is the first study to delineate characteristics and testing strategy of SARS-CoV-2 in a large population. This work facilitates a better understanding of SARS-CoV-2 detection and provides directive and feasible information to combat the SARS-CoV-2 pandemic.
Methods
Data collection.
Data were collected from January 22 to February 18, 2020, at Tongji Hospital in Wuhan, China. A total of 5630 subjects who were at high risk of SARS-CoV-2 infection were included. These subjects met one of the following criteria: (a) subjects presented fever and/or typical respiratory symptoms; (b) subjects presented radiological characteristics, including bilateral pneumonia, unilateral pneumonia, or ground-glass opacity; and (c) subjects had potential epidemiological exposure history of COVID-19. Throat swabs were collected from each subject by clinicians; they were maintained in viral-transport medium and transferred to the Department of Laboratory Medicine (Tongji Hospital) for SARS-CoV-2 testing as soon as possible. Testing results were reported within 24 hours of sample collection. Characteristics of subjects were obtained from medical records.
Detection of SARS-CoV-2.
Viral RNA extraction was conducted using a Magnet Viral Nucleic Acid Extraction Kit on PAN9600 Automated Nucleic Acid Extraction System (Tianlong). The SARS-CoV-2 nucleic acid detection kit (Shanghai Huirui Biotechnology Co. Ltd) was approved by National Health Commission of China, which was provided by Wuhan Center for Disease Control and Prevention. In the early outbreak of SARS-CoV-2, Shanghai Huirui Biotechnology Co. Ltd was one of the first recommended companies providing SARS-CoV-2 nucleic acid detection kits by the National Health Commission of China. Two target genes, ORF1ab and N, were simultaneously amplified using the real-time RT-PCR assay. If the Ct value was lower than 35, the result was considered as positive. If the Ct value was greater than or equal to 39.2, the result was considered as negative. If the Ct value was between 35 and 39.2, the result required retesting for confirmation. Three negative controls and 1 positive control were set in each run of the experiment to ensure the quality-of-detection process. All procedures were performed according to the manufacturer’s protocol.
SARS-CoV-2 is a new virus leading to a rapid outbreak of novel viral pneumonia, and many features of this disease were unclear during its early outbreak in Wuhan. To better admit SARS-CoV-2 patients, some doctors suggested testing subjects more than once based on their medical condition. Usually, a subject was tested multiple times due to the following reasons: (a) a suspected case presented typical clinical symptoms (e.g., fever, cough) and/or abnormal imaging of chest radiography/CT, but the result of nucleic acid test of SARS-CoV-2 was negative or inconclusive; (b) a positive case did not show typical clinical symptoms, but the result of the nucleic acid test of SARS-CoV-2 was close to cutoff value of the diagnostic kit; and (c) a positive case had an improved condition after receiving clinical treatments for several days and was then checked for the status of SARS-CoV-2. When testing for SARS-CoV-2 multiple times, the time interval between 2 tests was at least 2 days.
Statistics.
Continuous variables are expressed as median and interquartile range (IQR). Categorical variables are expressed as frequency or percentage. Mann-Whitney U test was used to compare the difference of independent samples between 2 groups. McNemar’s test or Cochran’s Q test was used to examine the difference of related samples between 2 or multiple groups, respectively. Binary logistic regression analysis was performed to estimate the ORs and 95% CI of factors associated with the diagnosis rate of SARS-CoV-2 infection. P values of multiple between-group comparisons were adjusted by Bonferroni correction. Statistical analyses were performed using IBM SPSS Statistics version 20.0 and GraphPad Prism 8.0.2. A 2-sided P value below 0·05 was considered as statistically significant.
Study approval.
This retrospective study was approved by the Ethics Committee of Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology (TJ-IRB20200201). Because of the rapid emergence of this infectious disease, and retrospective and anonymous analysis, written informed consent was waived.
Author contributions
YL, LC, and ZS designed the study and supervised the research. NS, YZ, XW, and JP performed the assays and collected data. NS, WL, and FW performed statistical analyses. All authors contributed to the writing and editing of the manuscript and approved the manuscript
Acknowledgments
This work was supported by National Mega Project on Major Infectious Disease Prevention (no. 2017ZX10103005-007) and National Key Research and Development Program of China (no. 2018YFE0204500). We thank all the doctors, nurses, laboratory technicians, and other people who have fought bravely against SARS-CoV-2 in the front line. We also thank those who have given selfless support to the fight against the pandemic
Version 1. 04/30/2020
In-Press Preview
Version 2. 05/21/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(10):e137662.https://doi.org/10.1172/jci.insight.137662.
Contributor Information
Na Shen, Email: shenna@tjh.tjmu.edu.cn.
Yaowu Zhu, Email: yaowu_zhu@163.com.
Xiong Wang, Email: wangxiong@tjh.tjmu.edu.cn.
Jing Peng, Email: ppjj6@126.com.
Weiyong Liu, Email: wyliu@hust.edu.cn.
Feng Wang, Email: xijiange_1@163.com.
Yanjun Lu, Email: junyanlu_2000@163.com.
Liming Cheng, Email: chengliming2015@163.com.
Ziyong Sun, Email: zysun@tjh.tjmu.edu.cn.
References
- 1.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phelan AL, Katz R, Gostin LO. The Novel Coronavirus Originating in Wuhan, China: Challenges for Global Health Governance. JAMA. 2020;323(8):709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 7.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ki M, Task Force for 2019-nCoV Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol Health. 2020;42:e2020007. doi: 10.4178/epih.e2020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothe C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahase E. China coronavirus: mild but infectious cases may make it hard to control outbreak, report warns. BMJ. 2020;368:m325. doi: 10.1136/bmj.m325. [DOI] [PubMed] [Google Scholar]
- 12.Tong ZD, et al. Potential Presymptomatic Transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerging Infect Dis. 2020;26(5):1052–1054. doi: 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. doi: 10.7326/M20-0504. Lauer SA, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application [published online March 10, 2020]. Ann Intern Med . [DOI] [PMC free article] [PubMed]
- 14.Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine Association. [An update on the epidemiological characteristics of novel coronavirus pneumonia COVID-19] Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):139–144. doi: 10.3760/cma.j.issn.0254-6450.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Guan WJ, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 17.Leung GM, et al. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann Intern Med. 2004;141(9):662–673. doi: 10.7326/0003-4819-141-9-200411020-00006. [DOI] [PubMed] [Google Scholar]
- 18.Channappanavar R, Fett C, Mack M, Ten Eyck PP, Meyerholz DK, Perlman S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J Immunol. 2017;198(10):4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei X, et al. Sex Differences in Severity and Mortality Among Patients With COVID-19: Evidence from Pooled Literature Analysis and Insights from Integrated Bioinformatic Analysis. arXiv. https://arxiv.org/abs/2003.13547 Published March 30, 2020. Accessed May 1, 2020.
- 20.Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8(4):e20. doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel SK, Velkoska E, Burrell LM. Emerging markers in cardiovascular disease: where does angiotensin-converting enzyme 2 fit in? Clin Exp Pharmacol Physiol. 2013;40(8):551–559. doi: 10.1111/1440-1681.12069. [DOI] [PubMed] [Google Scholar]
- 22.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.1001/jama.2020.3786. Wang W, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens [published ahead of print March 11, 2020]. JAMA . [DOI] [PMC free article] [PubMed]
- 25.Liu R, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]