Abstract
NOD-like receptor 12 (NLRP12) is a member of the nucleotide-binding domain and leucine-rich repeat containing receptor inflammasome family that plays a central role in innate immunity. We previously showed that DNA damage upregulated NLRP12 in hematopoietic stem cells (HSCs) of mice deficient in the DNA repair gene Fanca. However, the role of NLRP12 in HSC maintenance is not known. Here, we show that persistent DNA damage–induced NLRP12 improves HSC function in both mouse and human models of DNA repair deficiency and aging. Specifically, treatment of Fanca–/– mice with the DNA cross-linker mitomycin C or ionizing radiation induces NLRP12 upregulation in phenotypic HSCs. NLRP12 expression is specifically induced by persistent DNA damage. Functionally, knockdown of NLRP12 exacerbates the repopulation defect of Fanca–/– HSCs. Persistent DNA damage–induced NLRP12 was also observed in the HSCs from aged mice, and depletion of NLRP12 in these aged HSCs compromised their self-renewal and hematopoietic recovery. Consistently, overexpression of NLRP12 substantially improved the long-term repopulating function of Fanca–/– and aged HSCs. Finally, persistent DNA damage–induced NLRP12 maintains the function of HSCs from patients with FA or aged donors. These results reveal a potentially novel role of NLRP12 in HSC maintenance and suggest that NLRP12 targeting has therapeutic potential in DNA repair disorders and aging.
Keywords: Aging, Hematology
Keywords: DNA repair, Hematopoietic stem cells
Induction of NLRP12 in response to persistent DNA damage helps maintain hematopoietic stem cell function in the context of Fanconi anemia and age.
Introduction
Hematopoietic stem cells (HSCs) ensure a balanced production of all blood cells throughout life. Under cellular stress conditions, such as DNA damage, or during aging, HSCs gradually lose their self-renewal and regenerative potential, leading to a variety of hematological diseases, including bone marrow (BM) failure and leukemia (1, 2). The most irreversible cause of HSC aging relates to the accumulation of DNA damage. It has been shown that aged HSCs accumulate signatures of widespread DNA damage, including γ-H2AX foci. Recent studies using DNA repair–deficient mouse models indicate that DNA damage response (DDR) could intrinsically and extrinsically regulate HSC maintenance and play important roles in tissue homeostasis of the hematopoietic system (3). Defects in DDR pathways progressively impair the fitness of HSCs and are linked to premature aging (4–8). In fact, it has been shown that mice and patients with mutations in genes encoding for proteins involved in DDR pathways display many aspects of premature stem cell aging (5, 9, 10).
Fanconi anemia (FA) is a rare inherited disease with 22 complementation groups identified thus far (11–13). Among them, the genes encoding the groups A–W (FANCA–FANCW) have been cloned (14–21). At the molecular level, a DNA damage repair–based FA pathway has been established, which consequently influences important cellular processes, such as DNA replication, cell cycle control, and DDR/repair (22, 23). The 2 most important hematological hallmarks of FA are BM failure and progression to leukemia caused by HSC depletion and malignant transformation. We previously demonstrated that preleukemic HSC expansion and subsequent leukemogenic initiation in FA Lin–Sca-1+c-Kit+ (LSK; enriched for HSCs) compartment involves transcriptional alterations not only of genes in DDR/repair pathways but also of those involved in DDR/repair-unrelated cell signaling pathways, such as genes in cell migration, myeloid proliferation, and immune response pathways (24). These studies raise a new perspective on the immunological/DDR/repair interface between HSC defect and malignant transformation. However, the underlying mechanisms remain to be elucidated.
The nucleotide-binding domain and leucine-rich repeat containing receptor (NLR) family of proteins is a group of intracellular receptors that are important for sensing both pathogen-associated and danger-associated molecular patterns (25, 26). NOD-like receptor 12 (NLRP12) is a member of the NLR inflammasome family that plays a central role in innate immunity (25, 26). Unlike other proinflammatory NLR receptors, NLRP12 possesses an antiinflammatory function (27) and has been considered a negative regulator of the canonical and noncanonical pathways of NF-κB (28). In fact, NLRP12-mediated NF-κB suppression was implicated in colonic inflammation and tumorigenesis and osteoclast differentiation (29). Studies using the Nlrp12-knockout mouse model reveal a critical role for NLRP12 in maintaining intestinal homeostasis and providing protection against colorectal tumorigenesis (30). Our previous study showed that chronic DNA damage upregulates NLRP12 in hematopoietic stem progenitor cells (HSPCs) of mice deficient in the DNA repair gene Fanca (24). However, the role of NLRP12 in HSC maintenance is not known.
In the present study, we have investigated the role of persistent DNA damage–induced NLRP12 in maintaining HSC function in mice. We demonstrate that persistent DNA damage–induced NLRP12 improves HSC function in both mouse and human models of DNA repair deficiency (Fanca–/– mice) and aging. Our results indicate that NLRP12 plays a critical role in HSC maintenance under conditions of chronic DNA damage and aging.
Results
Persistent DNA damage induces NLRP12 expression in HSCs deficient in Fanca.
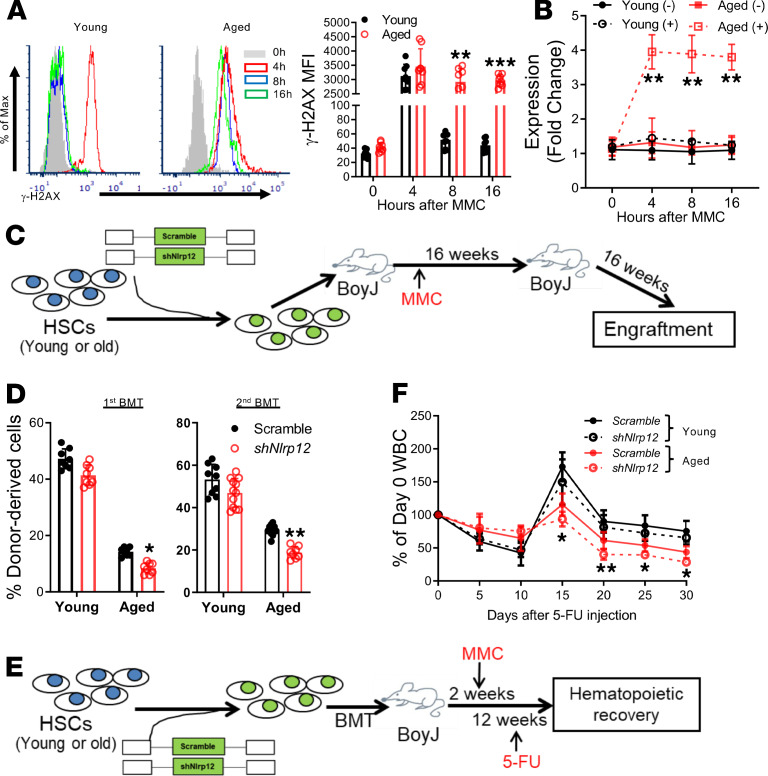
Our previous study showed that DNA damage induces Nlrp12 expression in HSPCs of mice deficient for the DNA repair gene Fanca (24). To further examine the role of the upregulated Nlrp12 in HSC maintenance under chronic DNA damage, we injected WT and Fanca–/– mice with the DNA cross-linking agent mitomycin C (MMC), which generates double-stranded breaks (DSBs). WT LSK (enriched for HSPCs; Figure 1A) cells efficiently repaired DSBs, as evidenced by a progressive decline of γ-H2AX (an established marker of DSBs; ref. 31) within 16 hours after MMC treatment (Figure 1B). In contrast, LSK cells from MMC-treated Fanca–/– mice retained high levels of γ-H2AX throughout this 16-hour period (Figure 1B), indicative of persistent DNA damage. We also performed the Comet assay (32) as a complementary method to measure DNA damage. We found that DNA damage, defined as comet tail moments, was persistently elevated in Fanca–/– cells during the 16-hour period after MMC treatment, whereas DNA damage peaked at 4 hours and returned to baseline at 16 hours after MMC treatment in WT cells (Figure 1C).
Figure 1. Persistent DNA damage induces Nlrp12 upregulation in Fanca–/– HSCs.
(A) Gating strategy for FACS for LSK (Lin–Sca-1+c-Kit+), SLAM (Lin–Sca-1+c-Kit+CD150+CD48–), MPP (multipotent progenitor; Lin–Sca-1+c-Kit+CD150–CD48–), and HPC (hematopoietic progenitor cell; Lin–c-Kit+Sca-1+CD150–CD48+ and Lin–c-Kit+Sca-1+CD150+CD48+) cell fractions isolated from Fanca–/– mice and their WT littermates. (B and C) Persistent DNA damage in Fanca–/– HSCs. Fanca–/– mice or their WT littermates were i.p. injected with a single dose of MMC (0.75 mg/kg) followed by flow cytometry analysis for γ-H2AX and Comet assay for DNA strand breaks at different time points. Representative flow plots (B, upper) and MFI kinetics (B, lower) and comet images at 8 hours post–MMC treatment (C, left) and olive tail moment (right) are shown. Original magnification, ×100. 0h, untreated control. (D) Kinetics of DNA damage–induced Nlrp12 expression in HSCs. Fanca–/– mice or their WT littermates were i.p. injected with a single dose of MMC (0.75 mg/kg) followed by cell sorting for SLAM cells at different time points. RNAs were then extracted from such cells followed by qPCR analysis for Nlrp12 expression using primers listed in Supplemental Table 1. Samples were normalized to the level of GAPDH mRNA (n = 6–9 per group). (E) Persistent DNA damage induces Nlrp12 upregulation specifically in Fanca–/– HSCs. Whole BM cells (WBMCs) from mice described in D at 0 hours (–MMC) and 16 hours (+MMC) were subjected to cell sorting for SLAM (LSK CD150+CD48–), MPP (LSK CD150–CD48–), or HPC (LSK CD150–CD48+ and LSK CD150+CD48+) cell fractions. RNAs were then extracted from such cells followed by qPCR analysis for Nlrp12 expression. Results are shown as means ± SD of 3 independent experiments (n = 6–9 per group). (F) Increased NLRP12 proteins in Fanca–/– HSCs. The cell fractions of HSCs, MPPs, and HPCs described in E were subjected to intracellular NLRP12 staining and flow cytometry analysis. Representative histogram (left) and quantification of MFI (right) are shown. (n = 6–9 per group). *P < 0.05; **P < 0.01; ***P < 0.001 MMC versus untreated control (0h). Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
Next, we employed ionizing radiation (IR), another effective and commonly used method to induce DNA damage (33–35). We exposed WT and Fanca–/– mice to 500 cGy total body irradiation (TBI, ref. 35) to assess IR-induced DNA damage in HSCs. We observed a significant increase in the formation of γ-H2AX foci in both WT and Fanca–/– SLAM cells at 4 hours following IR treatment (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133365DS1). However, the level of γ-H2AX foci peaked at 4 hours, then went on a progressive decline and returned to baseline at 16 hours in WT cells, whereas the level of γ-H2AX foci remained high in Fanca–/– cells throughout the 16-hour period after IR treatment (Supplemental Figure 1A). Importantly, the persistent accumulation of DNA damage was associated with a persistent activation of DDR in Fanca–/– SLAM cells, as evidenced by persistently high levels of phosphorylation of ATM-S1981, Chk2-T68, and p53-S15 (Supplemental Figure 1B), all of which are established signatures of DDR (36, 37).
Analysis of the kinetics of NLRP12 expression by quantitative PCR (qPCR) showed that Nlrp12 transcript remained significantly elevated in BM LSK cells isolated from Fanca–/– mice during the 16-hour post-MMC period (Figure 1D). On the other hand, MMC treatment did not significantly increase NLRP12 expression in WT cells at each post-MMC time point compared with untreated controls (Figure 1D). We also monitored the kinetics of Nlrp12 expression in irradiated mice and found that IR treatment persistently elevated Nlrp12 expression in Fanca–/– HSCs (Supplemental Figure 1C). Taken together, these results indicate that NLRP12 upregulation resulting from MMC or IR treatment is induced by persistent DNA damage (occurs only in Fanca–/– cells) but not by transitory DNA damage.
We then further analyzed the levels of Nlrp12 expression in 3 defined HSPC compartments: HSCs (SLAM), MPPs, and restricted HPCs (38) (Figure 1A). Analysis by both qPCR (Figure 1E) and intracellular NLRP12 staining (Figure 1F) revealed that the levels of Nlrp12 were significantly increased in Fanca–/– HSCs but not in less primitive MPPs or HPCs. These results indicate that persistent DNA damage induces Nlrp12 expression selectively in Fanca–/– HSCs.
Nlrp12 expression is specifically induced by persistent DNA damage.
To determine whether the induction of Nlrp12 was specifically in response to persistent DNA damage, we took 2 approaches: (a) genetic correction of FA deficiency by lentiviral transduction of functional FANCA cDNA in Fanca–/– HSCs and (b) knocking down the DDR/repair checkpoint protein, eyes absent phosphatase 2 (Eya2), in WT HSCs. It is known that the phosphorylation status of Tyr142 in H2AX plays a critical role in the regulation of DDR and is considered a rate-limiting step for DDR/repair. Recent studies have identified the eyes absent phosphatases, protein-tyrosine phosphatases of the haloacid dehalogenase superfamily, as being responsible for dephosphorylating the C-terminal tyrosyl residue of histone H2AX (39, 40). We transduced freshly sorted BM LSK cells with lentivirus expressing Venus-FANCA (Supplemental Figure 2) or EGFP-Eya2-knockdown (Eya2-KD) shRNA (Supplemental Figure 3) and transplanted the gene-corrected or Eya2-KD cells into lethally irradiated recipients, followed by MMC treatment at 2 weeks posttransplant. We found that complementation of FA deficiency almost completely abolished Nlrp12 upregulation induced by MMC treatment in Fanca–/– HSCs 16 weeks posttransplant (Figure 2A). Conversely, knocking down of Eya2 led to substantially increased Nlrp12 expression in MMC-treated WT HSCs (Figure 2B), which was correlated with prolonged accumulation of γ-H2AX in Eya2-KD WT HSCs (Figure 2C). Furthermore, persistent DNA damage (Figure 2D) and Nlrp12 upregulation (Figure 2E) were also observed in mouse models deficient in genes functioning in homologous recombination (Brca2–/–; ref. 41), nonhomologous end joining (DNA-PKcs3A/3A; ref. 42), and base excision repair (Parp1–/–; ref. 32) pathways. Thus, persistent DNA damage induces Nlrp12 upregulation in HSCs deficient for these canonical DNA repair genes.
Figure 2. Nlrp12 expression is specifically induced by persistent DNA damage.
(A) Genetic correction of Fanca deficiency abolishes Nlrp12 induction under persistent DNA damage. LSK cells from Fanca–/– mice were transduced with lentiviral vector expressing Venus or Venus/FANCA. Two thousand sorted Venus+ cells were transplanted into lethally irradiated BoyJ recipients followed by a single dose of MMC injection at 2 weeks posttransplant. Venus+ SLAM cells were then sorted for qPCR analysis for Nlrp12 expression at the indicated time points. The kinetics of NLRP12 expression is shown. Samples were normalized to the level of GAPDH mRNA (n = 9 per group). (B) Knockdown of Eya2 increases Nlrp12 expression in WT HSCs. LSK cells from WT mice were transduced with lentiviral vector expressing scramble shRNA or shRNA targeting Eya2. Two thousand sorted GFP+ cells were transplanted into lethally irradiated BoyJ recipients followed by single dose of MMC injection. GFP+ SLAM cells were then sorted for qPCR analysis for Nlrp12 at the indicated time points post–MMC treatment. Results are shown as means ± SD of 3 independent experiments (n = 9 per group). CTL, untreated control. (C) Eya2 knockdown induces persistent DNA damage in WT HSCs. WBMCs from the mice described in B were isolated for flow cytometry analysis for γ-H2AX in donor-derived SLAM cells at the indicated time points after MMC injection. (D and E) Persistent DNA damage in HSCs from mice deficient in Brca2, DNAPKcs, and Parp1 genes. Quantification of γ-H2AX levels (D) and Nlrp12 expression (E) in Brca2–/–, DNAPKcs3A/3A, and Parp1–/– HSCs following MMC treatment is shown. (F) Serum TNF-α levels in WT and Fanca–/– mice subjected to DNA damage. WT and Fanca–/– mice were i.p. injected with a single dose of MMC (0.75 mg/kg). The serum of BM was subjected to ELISA 16 hours later for TNF-α levels (n = 5). (G) NF-κB activation in HSCs from WT and Fanca–/– mice subjected to DNA damage. WBMCs from mice described in F were subjected to flow cytometry analysis for phosphorylated p65 (p-p65) in SLAM cells. (H and I) Suppression of inflammatory signaling failed to prevent DNA damage–induced Nlrp12 upregulation in Fanca–/– HSCs. Fanca–/– mice or their WT littermates were injected with a single dose of MMC injection. Anti-TNF antibody (H) or NF-κB inhibitor BAY11-7082 (I) was administered 30 minutes before and after MMC injection. Sixteen hours after MMC treatment, SLAM cells were sorted for qPCR analysis for Nlrp12 expression. Samples were normalized to the level of GAPDH mRNA. Results are shown as means ± SD of 3 independent experiments (n = 6–8 per group). **P < 0.01; ***P < 0.001. Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
Because chronic inflammation also induces sustained DNA damage and DDR (43), and because inflammation has been considered a major damaging stressor in FA hematopoiesis (44–46), we next attempted to determine whether suppression of inflammatory signaling could prevent persistent DNA damage–induced Nlrp12 upregulation in Fanca–/– HSCs. We used 2 approaches to suppress inflammatory signaling: TNF-α neutralization and NF-κB inhibition. As expected, we observed significantly higher levels of TNF-α in the serum (Figure 2F) and phosphorylated p65 in BM SLAM cells (Figure 2G) of Fanca–/– mice, both in steady state and under the DNA-damaging condition, compared with those of WT mice. Treatment with TNF-α–neutralizing antibody was not able to block Nlrp12 upregulation induced by MMC treatment in Fanca–/– HSCs (Figure 2H). Similarly, inhibition of NF-κB by BAY11-7082, an inhibitor of cytokine-induced IκBα phosphorylation (47), also failed to prevent MMC-induced Nlrp12 upregulation in Fanca–/– HSCs (Figure 2I). We confirmed the intended effects of anti–TNF-α neutralization and BAY11-7082 on TNF and NF-κB activity. Specifically, we found that anti–TNF-α neutralization inhibited NF-κB activation (48–50) and that BAY11-7082 suppressed downstream IL-1β and IL-6 expression (ref. 51; Supplemental Figure 4) in cells from mice treated with TNF-α–neutralizing antibody or BAY11-7082, respectively. Taken together, these data indicate that Nlrp12 upregulation is specifically induced by persistent DNA damage in Fanca–/– HSCs.
Knocking down NLRP12 exacerbates the repopulation defect of Fanca–/– HSCs.
We and others have shown that FA HSCs are defective in competitive repopulation (6, 52, 53). To further understand the role of persistent DNA damage–induced NLRP12 in HSC maintenance, we transduced WT and Fanca–/– HSCs with a lentiviral vector expressing scramble shRNA or shRNA targeting Nlrp12 (Supplemental Figure 5, A and C) and transplanted the transduced cells into lethally irradiated BoyJ recipients, followed by MMC treatment at 2 weeks posttransplant (Figure 3A). While transduction with the Nlrp12 shRNA did not affect WT HSCs, knocking down of Nlrp12 exacerbated the repopulation defect of Fanca–/– HSCs in both primary recipients (Figure 3B) and secondary recipients (Figure 3C). It is noteworthy that without MMC treatment, Nlrp12-KD did not have an effect on the repopulating capacity of either Fanca–/– or WT HSCs in both primary recipients and secondary recipients (Figure 3D). These results indicate that DNA damage–induced Nlrp12 expression improves the repopulation function of Fanca–/– HSCs.
Figure 3. Knockdown of Nlrp12 exacerbates the repopulation defect of Fanca–/– HSCs.
(A) Schematic presentation of experimental design. (B) Knockdown of Nlrp12 compromises Fanca–/– HSC repopulating capacity in primary recipients. LSK cells from Fanca–/– mice or their WT littermates were transduced with lentiviral vector expressing scramble shRNA or shRNA targeting Nlrp12. Then, 2000 sorted GFP+ cells, along with 2 × 105 protector cells, were transplanted into lethally irradiated BoyJ recipients followed by a single dose of MMC injection at 2 weeks posttransplant. Donor-derived chimeras were determined by flow cytometry at 16 weeks posttransplant. Representative flow plots (upper) and quantification (lower) are shown (n = 8–9 per group). (C) Knockdown of NLRP12 exacerbates the long-term repopulating defect of Fanca–/– HSCs. WBMCs from the primary recipients described in B were pooled and transplanted into sublethally irradiated secondary BoyJ recipients. Donor-derived chimeras were detected by flow cytometry 16 weeks posttransplant. Representative flow plots (left) and quantification (right) are shown (n = 9 per group). (D) Donor-derived chimera in primary (left) and secondary recipients (right) without MMC treatment. Results are shown as means ± SD of 3 independent experiments (n = 8–9 per group). **P < 0.01. Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
Persistent DNA damage–induced NLRP12 maintains HSC function in aged mice.
Because prolonged DNA damage accumulates due to decreased DNA repair capacity, and because compromised stem cell function plays crucial roles in aging (1–4), we wondered if persistent DNA damage–induced Nlrp12 upregulation also occurred in aged HSCs. Indeed, we found that BM SLAM cells from MMC-injected old mice retained high levels of γ-H2AX for up to 16 hours posttreatment, whereas the level of γ-H2AX in SLAM cells from young mice peaked at 4 hours and returned to the untreated control level 8 hours posttreatment (Figure 4A). Similar to DNA repair–deficient Fanca–/– HSCs, we found that persistent DNA damage increased Nlrp12 expression in aged HSCs compared with HSCs from young mice (Figure 4B). We also observed persistent DNA damage (Supplemental Figure 6A) and upregulated NLRP12 expression (Supplemental Figure 6B) in aged HSCs following IR compared with young HSCs.
Figure 4. Persistent DNA damage–induced Nlrp12 maintains HSC function in aged mice.
(A) MMC induces persistent DNA damage in aged HSCs. Young (6- to 8-week-old) and old mice (20- to 26-month-old) WT mice were i.p. injected with a single dose of MMC (0.75 mg/kg) followed by flow cytometry analysis for γ-H2AX at different time points. Representative flow plots gated on SLAM cells (left) and kinetics (right) are shown. (B) Persistent DNA damage induces Nlrp12 upregulation in aged mice. Young (6- to 8-week-old) and old (20- to 26-month-old) WT mice were subjected to single dose (0.75 mg/kg) of MMC injection. RNA from the sorted SLAM cells was extracted for qPCR analysis for NLRP12. Samples were normalized to the level of GAPDH mRNA. The kinetics of NLRP12 expression is shown. Results are shown as means ± SD of 3 independent experiments (n = 6 per group). (C) Schematic presentation of experimental design. (D) Depletion of Nlrp12 compromises HSC function in aged mice. LSK cells from young or aged mice were transduced with lentiviral vector expressing scramble shRNA or shRNA targeting Nlrp12. Along with 2 × 105 protector cells, 2000 sorted GFP+ cells were transplanted into lethally irradiated BoyJ recipients followed by MMC injection at 2 weeks posttransplant. Donor-derived chimeras in primary recipients (left) or secondary recipients (right) were determined by flow cytometry at 16 weeks posttransplant (n = 9–12). (E) Schematic presentation of experimental design. (F) Knockdown of Nlrp12 compromises hematopoietic recovery in aged mice after 5-FU treatment. LSK cells from young or aged mice were transduced with lentiviral vector expressing scramble shRNA or shRNA targeting Nlrp12. Along with 2 × 105 protector cells, 2000 sorted GFP+ cells were transplanted into lethally irradiated BoyJ recipients followed by MMC injection at 2 weeks posttransplant. Ten weeks later, the mice were administrated with a single dose of 5-FU (150 mg/kg) by i.p. injection. WBC count was monitored over a 30-day period. Results are shown as means ± SD of 3 independent experiments (n = 9 per group). *P < 0.05; **P < 0.01; ***P < 0.001 (shNlrp12 vs. scramble in aged HSCs). Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
To determine whether persistent DNA damage–induced Nlrp12 expression improves the repopulation function of aged HSCs, we performed serial BM transplantation using BM LSK cells from young and old mice transduced with scramble shRNA or shRNA targeting Nlrp12 (Supplemental Figure 5B and Figure 4C). Under the DNA-damaging condition, knocking down of Nlrp12 (Supplemental Figure 5C) did not show an overt effect on the repopulating ability of young HSCs; however, Nlrp12-KD further reduced hematopoietic repopulation of aged HSCs in both primary recipients and secondary recipients (Figure 4D). Interestingly, Nlrp12-KD also reduced the repopulating capacity of unstressed, aged HSCs, albeit not to a statistically significant degree, in both primary recipients and secondary recipients (Supplemental Figure 5D). Thus, persistent DNA damage–induced Nlrp12 expression improves the repopulation function of aged HSCs.
To further examine the impact of DNA damage–induced Nlrp12 upregulation on stressed hematopoiesis, we treated the MMC-stressed recipients transplanted with Nlrp12-depleted young or aged HSCs with the myeloid-ablating agent fluorouracil (5-FU), after establishing stable donor-derived hematopoiesis, and monitored hematopoietic recovery over a period of 30 days (refs. 52, 54; Figure 4E). We observed a similar drop of white blood cell (WBC) count at the first week after 5-FU injection in the recipients transplanted with young or aged HSCs (Figure 4E). However, WBC recovery in recipients transplanted with young HSCs started as early as 10 days after 5-FU treatment, whereas the recovery of WBC counts in aged HSC–transplanted mice persistently lagged during the next 20-day period, as compared with those of young HSCs (Figure 4F). Knocking down of Nlrp12 further delayed hematopoietic recovery in aged HSC–transplanted recipients (Figure 4F). Together, these results suggest a role of persistent DNA damage–induced NLRP12 in HSC maintenance during aging.
Constitutive overexpression of NLRP12 improves the long-term repopulating function of Fanca–/– and aged HSCs.
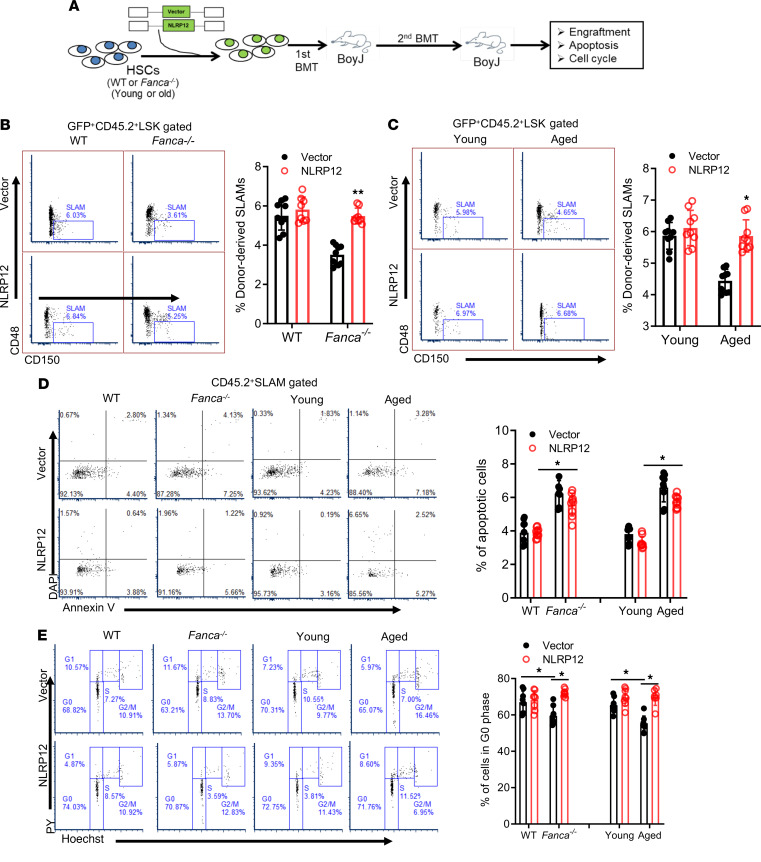
Because we observed a potential link between DNA damage–induced Nlrp12 expression and HSC maintenance, we wondered whether forced expression of Nlrp12 could improve the function of DNA repair–deficient Fanca–/– and aged HSCs. To this end, we transduced BM LSK cells from WT and Fanca–/– mice, or young and aged mice with lentivirus-based EGFP-Nlrp12 expression vector. The GFP+-sorted transduced cells were transplanted into lethally irradiated BoyJ recipients (Figure 5A). Analysis of donor-derived (CD45.2+) SLAM cells from the primary recipients by intracellular NLRP12 staining revealed approximately 5-fold higher level of NLRP12 expression in Fanca–/– or aged HSCs transduced with Nlrp12-expressing lentivirus than those of EGFP-alone control viruses (Supplemental Figure 7). Functional analysis by serial BM transplantation showed that overexpression of Nlrp12 significantly increased the frequencies of both Fanca–/– (Figure 5B) and aged HSCs (Figure 5C) in the secondary transplanted mice. These results suggest that the constitutively overexpressed Nlrp12 may improve the self-renewal ability of Fanca–/– and aged HSCs.
Figure 5. Constitutive overexpression of Nlrp12 improves the long-term repopulating function of Fanca–/– and aged HSCs.
(A) Schematic presentation of experiment design. (B and C) Ectopic expression of Nlrp12 increases HSC frequencies of Fanca–/– and aged HSCs in secondary transplanted mice. LSK cells from WT and Fanca–/– mice (B), or young and old mice (C), were transduced with lentiviral vector expressing EGFP alone (Vector) or EGFP-NLRP12. Two thousand sorted cells, along with 2 × 105 protector cells, were transplanted into lethally irradiated BoyJ recipients. Sixteen weeks posttransplant, WBMCs were pooled for secondary transplantation into sublethally irradiated BoyJ mice. WBMCs from the secondary recipients were subjected to flow cytometry analysis for the frequencies of donor-derived (GFP+CD45.2+) SLAM cells at 16 weeks posttransplant. Representative flow plots (left) and quantification (right) are shown (n = 9 per group). (D) Overexpression of NLRP12 does not affect apoptosis. WBMCs from the secondary recipients described in C and D were subjected to flow cytometry analysis for apoptosis in donor-derived SLAM cells. Representative flow plots (left) and quantification (right) are shown (n = 9 per group). (E) Overexpression of NLRP12 increases quiescence of Fanca–/– and aged HSCs. WBMCs from the recipients described in C and D were subjected to flow cytometry analysis for cell cycle in donor-derived SLAM cells. Representative flow plots (left) and quantification (right) are shown (n = 9 per group). *P < 0.05; **P < 0.01. Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
To investigate the cellular mechanism underlying Nlrp12-mediated improvement of the function of Fanca–/– and aged HSCs, we measured apoptosis of donor-derived HSCs in the secondary recipient mice by annexin V staining. Low levels (<5%) of apoptotic donor-derived HSCs were observed in the recipients transplanted with either WT or young HSCs regardless of NLRP12 expression (Figure 5D). There was a significant increase in apoptosis in donor Fanca–/– and aged HSCs compared with donor WT and young HSCs; this increase was only marginally reduced by constitutive overexpression of Nlrp12 (Figure 5D). We also performed cell cycle analysis to evaluate the effect of NLRP12 overexpression on quiescence of donor Fanca–/– and aged HSCs. We observed a statistically significant reduction of quiescent donor Fanca–/– and aged HSCs compared with donor WT and young HSCs (Figure 5E). Ectopic overexpression of Nlrp12 significantly increased quiescence of donor Fanca–/– and aged HSCs compared with vector-alone controls as determined by Hoechst 33342/pyronin staining (Figure 5E). Similarly, Ki-67/DAPI staining showed a significant increase in cycling Ki-67+ donor-derived SLAM cells in the recipients transplanted with Fanca–/– or aged cells compared with those transplanted with WT or young cells, respectively. Forced expression of Nlrp12 significantly decreased the proportion of cycling HSCs in the recipients compared with those transplanted with vector-alone controls (Supplemental Figure 7). These results suggest that increased quiescence may play a causal role in NLRP12-mediated functional improvement of Fanca–/– and aged HSCs.
Persistent DNA damage–induced NLRP12 maintains the function of HSCs from FA patients or aged donors.
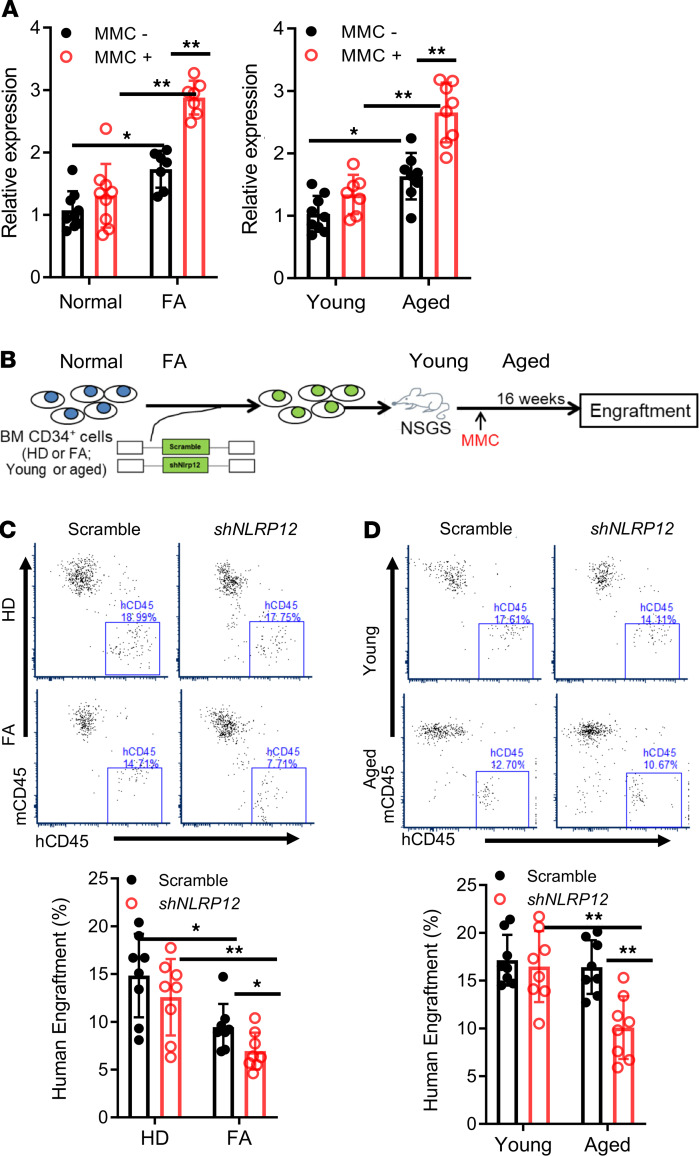
To evaluate whether our findings in the mouse models were extendable to humans, we next assessed the role of DNA damage–induced NLRP12 in HSCs from FA patients, which are DNA repair deficient (11–13), and aged donors. qPCR analysis showed that NLRP12 expression was significantly increased in BM CD34+ cells from FA patients under the DNA damage condition in comparison with healthy donors (Figure 6A). Similarly, HSPCs from aged donors also exhibited higher levels of DNA damage–induced NLRP12 expression compared with those from young donors (Figure 6A). To assess the in vivo effect of persistent DNA damage–induced NLRP12 expression on HSCs of FA patients and old people, we transduced BM CD34+ cells from FA patients and aged donors with scramble shRNA or shRNA targeting NLRP12 (Supplemental Figure 8) and performed human xenotransplantation assay using the humanized NOD/SCID-γ IL-2γ–/– SGM3 (NSGS) mice as recipients (refs. 55, 56; Figure 6B). We found that knocking down of NLRP12 further reduced the repopulating capacity of HSCs from patients with FA (Figure 6C). Similarly, NLRP12-KD also compromised hematopoietic repopulation of HSCs from aged donors (Figure 6D). Together, these results indicate that persistent DNA damage–induced NLRP12 has a role in the maintenance of HSCs from patients with FA or aged donors.
Figure 6. Persistent DNA damage–induced NLRP12 maintains the function of HSCs from patients with FA and aged donors.
(A) Increased NLRP12 expression in HSPCs from patients with FA or aged donors under DNA damage. Real-time qPCR mRNA measurement of NLRP12 in BM CD34+ cells from healthy donors (Normal) and patients with FA (upper) or young and aged donors (lower) after 16-hour culture in the presence or absence of MMC (10 nM). Samples were normalized to the level of GAPDH mRNA. Results are shown as means ± SD of 3 independent experiments (n = 6–9 per group). (B) Schematic presentation of experimental design. (C and D) Persistent DNA damage–induced NLRP12 maintains the function of HSCs from patients with FA or aged donors. BM CD34+ cells from the indicated donors or patients were transduced with scramble shRNA or shRNA targeting NLRP12. Two thousand sorted GFP+ cells were transplanted into sublethally irradiated NSGS recipients followed by MMC injection at 2 weeks posttransplant. Human engraftment was analyzed 16 weeks after transplant by flow cytometry using anti-human CD45 antibody. Results are shown as means ± SD of 3 independent experiments (n = 9 per group). *P < 0.05; **P < 0.01; ***P < 0.001. Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups.
Discussion
In this study, we have investigated the relationship between persistent DNA damage–induced NLRP12 and HSC function using the mouse models deficient for the core complex of the FA DNA repair pathway (Fanca–/–) and aging. Our study identifies a functional link between persistent DNA damage–induced NLRP12 upregulation and HSC maintenance. There are several findings that highlight the significance of upregulated NLRP12 in maintaining HSC function: (a) persistent DNA damage induced NLRP12 upregulation in DNA damage repair–deficient Fanca–/– and aged HSCs, (b) NLRP12 expression was specifically induced by persistent DNA damage but not by inflammation, (c) knocking down of NLRP12 exacerbated the repopulation defect of Fanca–/– and aged HSCs, (d) constitutive overexpression of NLRP12 markedly improved long-term repopulating function of HSCs of DNA repair–deficient Fanca–/– mice and aged mice, (e) overexpression of Nlrp12 substantially increased quiescence of donor Fanca–/– and aged HSCs in transplanted recipients, and (f) persistent DNA damage–induced NLRP12 improved the function of HSCs from patients with FA or aged donors.
The innate immune system is an important part of host defense from infection by other organisms (57, 58). Mounting evidence suggests that HSPCs are not simply a source of leukocytes in the BM but are active players in the innate immune response to local and systemic insults, including DNA damage (59). Recent studies have also suggested the mobilization of HSCs as a result of innate immunity-mediated sterile inflammation in the BM microenvironment (58). However, how innate immune response regulates HSC function remains elusive. It has also been shown that different types of DNA damage lead to induction of immune responses. While transient DNA damage is associated with protection from immune responses to systemic stress, immune responses induced by persistent DNA damage are linked to degenerative disorders and organ decline in aging (60). Although the mechanistic link between persistent DNA damage and induction of immune response is still a subject of intense debate, our results raise key questions, such as why the immune system responds to persistent DNA damage and how it senses such damage. We propose that persistent DNA damage is seen by the immune system as signals eliciting alerts of danger and systemic protection. Therefore, it is logical to anticipate beneficial effects of persistent DNA damage recognition by the immune system. In this context, our study indicates for the first time to our knowledge that the interplay between persistent DNA damage and the innate immune receptor NLRP12 plays a crucial role in maintaining HSC function. Protection appears to be accomplished through a mechanism involving upregulating NLRP12 in response to persistent DNA damage.
NLRP12, a member of the NOD-like receptor family, is expressed in myeloid and BM cells and was implicated as a checkpoint regulator of inflammatory cytokines, as well as an inflammasome activator (27). NLRP12 has been shown to play important roles in innate immunity (61). Our previous study showed that preleukemic HSC expansion and subsequent leukemogenic initiation in the FA HSPC compartment involves transcriptional alterations not only of genes in DDR/repair but also of those involved in DDR/repair-unrelated cell signaling pathways, including many genes in the cell migration, myeloid proliferation, and immune response pathways (24). These studies raised a new perspective on the immunological/DDR interface between HSC defect and malignant transformation. In the current study, we further investigate the role of persistent DNA damage–induced NLRP12 expression in HSC maintenance. Our results suggest that persistent DNA damage–induced NLRP12 expression plays a crucial role in functional improvement of DNA repair–deficient Fanca–/– and aged HSCs. However, whether persistent DNA damage–induced innate immune response, that is, NLRP12 upregulation in the current study, plays a role in leukemogenesis in DNA repair disorders and during aging requires further investigation.
One intriguing finding of our current study is the observation that NLRP12 expression is specifically induced by persistent DDR but not inflammation. NLRP12 belongs to a superfamily of innate immune receptors involved in inflammatory response. However, unlike other proinflammatory NLR receptors, NLRP12 is believed to possess antiinflammatory function (27). In the present study, we show that suppression of persistent DNA damage by genetic correction of Fanca–/– HSCs abolished NLRP12 upregulation (Figure 2A). Conversely, induction of persistent DNA damage by knocking down the DDR checkpoint protein Eya2 in WT HSCs upregulated NLRP12 (Figure 2B). However, suppression of inflammatory signaling by TNF neutralization or NF-κB inhibition failed to prevent NLRP12 upregulation in Fanca–/– HSCs (Figure 2, C and D). These results suggest that the mechanisms by which NLRP12 (and probable other innate immune receptors) regulates HSC function under chronic DNA damage and inflammation may be different.
Another notable observation of the current study is that constitutive overexpression of NLRP12 improves the function of aged HSCs. During aging, HSCs gradually lose their self-renewal and regenerative potential and accumulate DNA damage (1–3). Mice or patients deficient in DNA damage repair genes exhibit a premature stem cell aging phenotype (62). Our present data show an aging-related persistent DNA damage–induced NLRP12 expression in HSCs, which plays an important role in maintaining the function of the aged HSCs (Figure 4). Mechanistically, this regulation is, at least in part, through improving HSC quiescence (Figure 5).
In summary, the current study used both mouse and human models of DNA repair deficiency and aging to demonstrate that persistent DNA damage–induced NLRP12 improves the function of DNA repair–deficient or aged HSCs. These findings reveal a positive role of a special innate immune response induced by persistent DNA damage in HSC maintenance and suggest that genetic or pharmacological activation of NLRP12 may have therapeutic value in DNA repair disorders and aging.
Methods
Mice and treatments.
Fanca+/– mice were provided by Madeleine Carreau (Laval University, Quebec City, Quebec, Canada) (63) and used to generate Fanca–/– mice and their WT littermates. Parp1–/– mice were generated by backcrossing 129SParp1tm1zqw/J (The Jackson Laboratory) with WT C57BL/6 mice (32). DNAPKcs3A/3A mice (a gift from Benjamin P.C. Chen, University of Texas Southwestern Medical Center at Dallas, Dallas, Texas, USA) were generated by interbreeding heterozygous DNAPKcs+/3A mice (41). Brca2fl/fl Vav1Cre mice were generated by Cre-mediated deletion of floxed alleles by crossing Brca2tm1/Brn (National Cancer Institute; ref. 42) with the Vav1Cre strain (64). Young (10- to 12-week-old) or aged (20- to 26-month-old) C57BL/6 mice (65) were used for the aging study. All the animals, including BoyJ (C57BL/6: B6, CD45.1+) recipient mice, were maintained in the animal facility at West Virginia University (WVU) Health Sciences Center. Nonobese diabetic severe combined immunodeficiency (NOD/SCID IL2γ–/– SGM3) (NSGS) mice, which express transgenic cDNAs encoding human stem cell factor (also termed KITLG), granulocyte-macrophage colony stimulating factor, and interleukin 3 (55) were purchased from The Jackson Laboratory and housed in a transgenic animal facility at WVU. NSGS mice at the age of 6–8 weeks were used as recipients for BM transplants (54). All experimental procedures conducted in this study were approved by the Institutional Animal Care and Use Committee of WVU.
For in vivo MMC (MilliporeSigma) treatment, experimental mice were i.p. injected with a single dose of MMC (0.75 mg/kg; MilliporeSigma) (66). For anti–TNF-α antibody and NF-κB inhibition treatments, mice were i.p. injected with 20 μg of neutralizing mouse anti–TNF-α antibody (R&D Systems, Bio-Techne) (67) or BAY11-7082 (20 mg/kg; Calbiochem) (47) 30 minutes before and after MMC injection. Mice were then sacrificed for BM cell isolation 16 hours after the final infection.
5-FU treatment was adapted from previous studies (52, 54). Briefly, a single dose (150 mg/kg) of 5-FU (MilliporeSigma) was administrated i.p. into the experimental mice. Kinetics of hematopoietic recovery was monitored.
For assessment of IR-induced DNA damage and Nlrp12 expression, mice were irradiated with 500 cGy TBI using a Shepherd cesium137 irradiator (35). At the indicated time points after TBI, mice were euthanized, and BM cells were isolated for further analyses.
Flow analysis and cell sorting.
Femurs and tibias were flushed to dissociate the BM fraction. Cells were resuspended in 5 mL PBS/0.5% BSA and filtered through a 70-μm filter (BD Biosciences, 3523350). The mononuclear cells were isolated by Ficoll-Paque (GE Healthcare Life Sciences, 95040-394) gradient centrifugation. For LSK staining, cells were labeled by the biotin-conjugated anti-lineage antibody cocktail (BioLegend, 133307) followed by staining with a secondary PerCP-Cy5.5 anti-streptavidin antibody (BioLegend, 405214), PE-Cy7 anti–Sca-1 antibody (BD Biosciences, 558162), and APC-Cy7 anti–c-Kit antibody (BD Biosciences, 553356). To access the long-term HSC subpopulation, cells were stained with LSK antibodies in addition to CD45.2-eFluor 450 (eBioscience, Thermo Fisher Scientific, 48-0454-82). Flow cytometry was performed on an LSRFortessa (BD Biosciences), and analysis was done with FCS Express 6 software (De Novo Software).
For the cell sorting, lineage-negative cells were enriched using lineage depletion reagents (Miltenyi Biotec, 130-090-858) according to the manufacturer’s instructions. The HSC/SLAM (Lin–Sca-1+c-Kit+CD150+CD48–), MPP (Lin–Sca-1+c-Kit+CD150–CD48–) or HPC (Lin–c-Kit+Sca-1+CD150–CD48+ and Lin–c-Kit+Sca-1+CD150+CD48+) cell fractions were acquired by using the FACSAria II sorter (BD Biosciences).
For donor-derived chimera analysis, peripheral blood from the recipient mice were subjected to staining using PE–anti-CD45.1 and APC–anti-CD45.2 (both from BD Biosciences, 553776 and 558702) antibodies followed by flow cytometry analysis.
For cell cycle analysis, surface marker–stained cells were fixed and permeabilized using Cytofix/Cytoperm buffer (BD Pharmingen, 554722) followed by intensive wash using Perm/Wash Buffer (BD Pharmingen, 554723). Cells were then labeled with Hoechst 33342 and Pyronin Y staining buffer (10 mg/mL Hoechst 33342 and 150 ng/mL Pyronin Y in Perm/Wash buffer; MilliporeSigma, 92-32-0) at 37˚C for 1 hour or anti–mouse Ki-67 antibody (BD Pharmingen, 550609) and DAPI (MilliporeSigma) at room temperature for 30 minutes followed by flow cytometry analysis on CD45.2+ SLAM–gated population.
For intracellular staining, surface marker–stained cells were fixed and permeabilized using Cytofix/Cytoperm buffer followed by intensive wash using Perm/Wash Buffer. Cells were then incubated with antibodies against NLRP12 (BioLegend, 675202), γ-H2AX (biotin conjugated, MilliporeSigma, 16-193), or phosphorylated p65 (Cell Signaling Technology, 3033S) at 4°C for 30 minutes. After washing, cells were incubated with secondary antibody and analyzed by flow cytometry analysis.
Serum TNF-α level measurement.
Serum levels of TNF-α were measured using specific capture Quantikine ELISA kits (R&D Systems, Bio-Techne). Briefly, BM from the indicated mice were collected and allowed to clot for 2 hours at room temperate before centrifuging for 20 minutes at 2000 g. Serum was removed and assayed immediately following the manufacturer’s instructions.
qPCR analysis.
Total RNA was extracted using RNeasy Mini Kit (QIAGEN) following the manufacturer’s procedure. Reverse transcription was carried out at 42°C for 60 minutes and stopped at 95°C for 5 minutes using random hexamers and Superscript II RT (Invitrogen, Thermo Fisher Scientific, 18064014). First-strand cDNA was used for real-time PCR analysis using primers listed in Supplemental Table 1. Samples were normalized to the level of GAPDH mRNA.
Molecular subcloning and materials.
For lentiviral vector construction, full-length FANCA cDNA was subcloned into pRRL-SIN-cPPT-MNDU3-MCS-IVW (TMND-IRES-Venus) vector (24, 68). Hairpin sequence for control (CTTACGCTGAGTACTTCGA), Eya2 (GTGTTTCAGAGACAATCAT-3′), or Nlrp12 (GCTAATGGAAGACCGCAATGC) were subcloned into SFLV-EGFP-shRNA vector (69). To generate lentiviral expression vectors for Nlrp12, the Nlrp12 cDNA (purchased from Origene) was subcloned into the pLVX-IRES-GFP vector (Takara Bio, formerly Clontech Laboratories).
Lentivirus was produced in HEK293T cells (ATCC) after transfection of 20 μg plasmid, 15 μg pCMVΔ8.91 helper plasmid, and 6 μg pMD.G (70) using standard calcium phosphate transfection procedures. Fresh medium change was performed 12 hours after transfection. Supernatants from the cell culture were collected 48 hours after transfection, filtered through 0.45-μm-pore-size filters, and concentrated at 4000 g for 2.5 hours at 4°C to harvest viral particles. The virus pellet was resuspended in sterile PBS and stored at –80°C.
Lentiviral transduction.
As previously described (71), sorted BM LSK cells were prestimulated 5–10 hours in a 24-well dish in serum-free medium. The lentiviral media were added to the cells and spinoculated for 90 minutes at 270 g in the presence of 8 μg/mL polybrene (MilliporeSigma). This process was repeated 24 hours later with a fresh batch of lentiviral media.
BM transplantation.
Two thousand freshly isolated LSK cells or lentivirus-transduced cells (CD45.2+), along with 2 × 105 protector cells from congenic BoyJ mice (CD45.1+), were transplanted into lethally irradiated (11.75 Gy) BoyJ mice. Recipients were subjected to MMC or 5-FU injection at the indicated time points posttransplant.
For serial BM transplantation, 1~3 million WBMCs from primary recipients were pooled and injected into sublethally irradiated (7.0 Gy) BoyJ recipients. Donor-derived chimera were detected by flow cytometry at 16 weeks posttransplant using antibodies against CD45.1 and CD45.2.
For human xenotransplantation, 2000 sorted GFP+ cells were transplanted into sublethally irradiated (250 cGy) NSGS recipients (55, 56). Human engraftment was determined by flow cytometry using anti-human CD45 antibody 16 weeks after BM transplantation.
Human BM cells.
The BM samples from patients with FA and control healthy donors were received with informed consent through the Fanconi Anemia Cell Repository at Cincinnati Children’s Hospital Medical Center. Mononuclear cells were purified by Ficoll-Paque density centrifugation (GE Healthcare Life Sciences) and ammonium chloride red cell lysis. Density-separated cells were magnetically sorted for CD34 via the StemSep system (STEMCELL Technologies) according to the manufacturer’s instructions. The BM samples for the aging study were purchased from BioIVT.
Online supplemental material.
Supplemental Methods and Supplemental Table 1 are presented first. Supplemental Figure 1 shows persistent DNA damage induced by IR in Fanca–/– cells. Supplemental Figure 2 shows transduction efficiency and FCS of lentiviral vector expressing Venus or Venus/FANCA. Supplemental Figure 3 shows transduction efficiency and FCS of lentiviral vector expressing scramble shRNA or shRNA targeting Eya2. Supplemental Figure 4 shows the abolishment of downstream pathways after anti–TNF-α or BAY11-7082 treatment. Supplemental Figure 5 shows transduction efficiency and FCS of lentiviral vector expressing scramble shRNA or shRNA targeting Nlrp12. Supplemental Figure 6 shows persistent DNA damage induces Nlrp12 upregulation in aged HSCs. Supplemental Figure 7 shows ectopic expression of Nlrp12 in transduced cells. Supplemental Figure 8 shows overexpression of NLRP12 increases quiescence of Fanca–/– and aged HSCs. Supplemental Figure 9 shows efficient NLRP12 knockdown in human samples.
Statistics.
Paired or unpaired 2-tailed Student’s t test was used for 2-group comparison and 1-way ANOVA for comparison of more than 2 groups. P values less than 0.05 were considered statistically significant. Results are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
Study approval.
All experimental procedures conducted in this study were approved by the Institutional Animal Care and Use Committee of WVU.
Author contributions
QL performed the research and analyzed the data; LW, ZM, FAC, and HHM performed some of the research and assisted with data analysis. WD designed the research, analyzed the data, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Madeleine Carreau (Laval University) for Fanca+/– mice and Benjamin P.C. Chen (University of Texas Southwestern Medical Center at Dallas) for DNAPKcs3A/3A mice. WD is supported by an NIH/National Heart, Lung, and Blood Institute grant (R01HL151390); NIH Tumor Microenvironment Center of Biomedical Excellence Award (P20GM121322); WVU Health Sciences Center and School of Pharmacy start-up funds; a Leukemia Research Foundation Award; and an American Cancer Society Institutional Research Grant. The BM samples from FA patients and control healthy donors were received through the Fanconi Anemia Cell Repository at Cincinnati Children’s Hospital Medical Center.
Version 1. 05/21/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(10):e133365.https://doi.org/10.1172/jci.insight.133365.
Contributor Information
Qiqi Lin, Email: qiqi.lin@hsc.wvu.edu.
Limei Wu, Email: limei.wu@hsc.wvu.edu.
Zhilin Ma, Email: zhilin.ma@hsc.wvu.edu.
Wei Du, Email: wei.du@hsc.wvu.edu.
References
- 1.de Haan G, Lazare SS. Aging of hematopoietic stem cells. Blood. 2018;131(5):479–487. doi: 10.1182/blood-2017-06-746412. [DOI] [PubMed] [Google Scholar]
- 2.Young K, Borikar S, Bell R, Kuffler L, Philip V, Trowbridge JJ. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J Exp Med. 2016;213(11):2259–2267. doi: 10.1084/jem.20160168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Zhou ZW, Ju Z, Wang ZQ. DNA damage response in hematopoietic stem cell ageing. Genomics Proteomics Bioinformatics. 2016;14(3):147–154. doi: 10.1016/j.gpb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13(5):376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 5.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 6.Parmar K, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28(7):1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasher JM, et al. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1–/– mice. EMBO J. 2005;24(4):861–871. doi: 10.1038/sj.emboj.7600542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132(4):681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Ju Z, et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat Med. 2007;13(6):742–747. doi: 10.1038/nm1578. [DOI] [PubMed] [Google Scholar]
- 10.Alter BP, Rosenberg PS, Giri N, Baerlocher GM, Lansdorp PM, Savage SA. Telomere length is associated with disease severity and declines with age in dyskeratosis congenita. Haematologica. 2012;97(3):353–359. doi: 10.3324/haematol.2011.055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagby GC. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10(1):68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Green AM, Kupfer GM. Fanconi anemia. Hematol Oncol Clin North Am. 2009;23(2):193–214. doi: 10.1016/j.hoc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 14.Bogliolo M, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92(5):800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123(1):26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 16.Hodskinson MR, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54(3):472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer SL, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5(2):135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virts EL, et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum Mol Genet. 2015;24(18):5093–5108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V. Update of the human and mouse Fanconi anemia genes. Hum Genomics. 2015;9:32. doi: 10.1186/s40246-015-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JY, et al. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet. 2016;53(10):672–680. doi: 10.1136/jmedgenet-2016-103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knies K, et al. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest. 2017;127(8):3013–3027. doi: 10.1172/JCI92069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11(7):467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu J. Fanconi’s anemia. In: Young NS, ed. Bone Marrow Failure Syndromes. Philadelphia, Pennsylvania, USA: WB Saunders; 2000:47–68. [Google Scholar]
- 24.Du W, Amarachintha S, Wilson A, Pang Q. The immune receptor Trem1 cooperates with diminished DNA damage response to induce preleukemic stem cell expansion. Leukemia. 2017;31(2):423–433. doi: 10.1038/leu.2016.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ataide MA, et al. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog. 2014;10(1):e1003885. doi: 10.1371/journal.ppat.1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vladimer GI, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37(1):96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linz BM, et al. Innate immune cell recovery is positively regulated by NLRP12 during emergency hematopoiesis. J Immunol. 2017;198(6):2426–2433. doi: 10.4049/jimmunol.1601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jéru I, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A. 2008;105(5):1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity. 2012;36(5):742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaki MH, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20(5):649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114(3):371–383. doi: 10.1016/S0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Sipple J, Pang Q, Du W. Salidroside stimulates DNA repair enzyme Parp-1 activity in mouse HSC maintenance. Blood. 2012;119(18):4162–4173. doi: 10.1182/blood-2011-10-387332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox Signal. 2014;21(2):251–259. doi: 10.1089/ars.2013.5668. [DOI] [PubMed] [Google Scholar]
- 34.Borrego-Soto G, Ortiz-López R, Rojas-Martínez A. Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet Mol Biol. 2015;38(4):420–432. doi: 10.1590/S1415-475738420150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Himburg HA, et al. Distinct bone marrow sources of pleiotrophin control hematopoietic stem cell maintenance and regeneration. Cell Stem Cell. 2018;23(3):370–381.e5. doi: 10.1016/j.stem.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally JP, et al. Manipulating DNA damage-response signaling for the treatment of immune-mediated diseases. Proc Natl Acad Sci U S A. 2017;114(24):E4782–E4791. doi: 10.1073/pnas.1703683114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs KM, et al. Unique epigenetic influence of H2AX phosphorylation and H3K56 acetylation on normal stem cell radioresponses. Mol Biol Cell. 2016;27(8):1332–1345. doi: 10.1091/mbc.E16-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Jakob B, et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39(15):6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnan N, et al. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284(24):16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31(6):955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, et al. Spontaneous tumor development in bone marrow-rescued DNA-PKcs(3A/3A) mice due to dysfunction of telomere leading strand deprotection. Oncogene. 2016;35(30):3909–3918. doi: 10.1038/onc.2015.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding N, Maiuri AR, O’Hagan HM. The emerging role of epigenetic modifiers in repair of DNA damage associated with chronic inflammatory diseases. Mutat Res. 2019;780:69–81. doi: 10.1016/j.mrrev.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 45.Hu L, Huang W, Hjort E, Eklund EA. Increased Fanconi C expression contributes to the emergency granulopoiesis response. J Clin Invest. 2013;123(9):3952–3966. doi: 10.1172/JCI69032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagby G. Recent advances in understanding hematopoiesis in Fanconi Anemia. F1000Res. 2018;7:105. doi: 10.12688/f1000research.13213.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dewan MZ, et al. Rapid tumor formation of human T-cell leukemia virus type 1-infected cell lines in novel NOD-SCID/gammac(null) mice: suppression by an inhibitor against NF-kappaB. J Virol. 2003;77(9):5286–5294. doi: 10.1128/JVI.77.9.5286-5294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukla R, et al. Proinflammatory cytokine TNF-α increases the stability of hepatitis B virus X protein through NF-κB signaling. Carcinogenesis. 2011;32(7):978–985. doi: 10.1093/carcin/bgr057. [DOI] [PubMed] [Google Scholar]
- 49.Maguire O, O’Loughlin K, Minderman H. Simultaneous assessment of NF-κB/p65 phosphorylation and nuclear localization using imaging flow cytometry. J Immunol Methods. 2015;423:3–11. doi: 10.1016/j.jim.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon HJ, et al. Stepwise phosphorylation of p65 promotes NF-κB activation and NK cell responses during target cell recognition. Nat Commun. 2016;7:11686. doi: 10.1038/ncomms11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheleschi S, et al. MicroRNA mediate visfatin and resistin induction of oxidative stress in human osteoarthritic synovial fibroblasts via NF-κB pathway. Int J Mol Sci. 2019;20(20):E5200. doi: 10.3390/ijms20205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du W, et al. Fancb deficiency impairs hematopoietic stem cell function. Sci Rep. 2015;5:18127. doi: 10.1038/srep18127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang QS, et al. Fancd2–/– mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. 2010;116(24):5140–5148. doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi M, Srour EF. Regulation of murine hematopoietic stem cell quiescence by Dmtf1. Blood. 2011;118(25):6562–6571. doi: 10.1182/blood-2011-05-349084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wunderlich M, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du W, Li XE, Sipple J, Pang Q. Overexpression of IL-3Rα on CD34+CD38- stem cells defines leukemia-initiating cells in Fanconi anemia AML. Blood. 2011;117(16):4243–4252. doi: 10.1182/blood-2010-09-309179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adamiak M, Ratajczak MZ. Innate immunity and mobilization of hematopoietic stem cells. Curr Stem Cell Rep. 2017;3(3):172–180. doi: 10.1007/s40778-017-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratajczak MZ, Adamiak M, Plonka M, Abdel-Latif A, Ratajczak J. Mobilization of hematopoietic stem cells as a result of innate immunity-mediated sterile inflammation in the bone marrow microenvironment-the involvement of extracellular nucleotides and purinergic signaling. Leukemia. 2018;32(5):1116–1123. doi: 10.1038/s41375-018-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Granick JL, Simon SI, Borjesson DL. Hematopoietic stem and progenitor cells as effectors in innate immunity. Bone Marrow Res. 2012;2012:165107. doi: 10.1155/2012/165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poliezhaieva T, Ermolaeva MA. DNA damage in protective and adverse inflammatory responses: friend of foe? Mech Ageing Dev. 2017;165(pt A):47–53. doi: 10.1016/j.mad.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Lupfer C, Kanneganti TD. Unsolved mysteries in NLR biology. Front Immunol. 2013;4:285. doi: 10.3389/fimmu.2013.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geiger H, Denkinger M, Schirmbeck R. Hematopoietic stem cell aging. Curr Opin Immunol. 2014;29:86–92. doi: 10.1016/j.coi.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Wong JC, Alon N, Mckerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12(16):2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 64. doi: 10.1002/stem.3169. Ma Z, et al. Hes1 deficiency causes hematopoietic stem cell exhaustion [published online March 4, 2020]. Stem Cells . [DOI] [PMC free article] [PubMed]
- 65.Florian MC, et al. Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 2018;16(9):e2003389. doi: 10.1371/journal.pbio.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carreau M, et al. Bone marrow failure in the Fanconi anemia group C mouse model after DNA damage. Blood. 1998;91(8):2737–2744. doi: 10.1182/blood.V91.8.2737.2737_2737_2744. [DOI] [PubMed] [Google Scholar]
- 67.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120(pt 9):1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Erden O, Li L, Ye Q, Wilson A, Du W. Binding to WGR domain by salidroside activates PARP1 and protects hematopoietic stem cells from oxidative stress. Antioxid Redox Signal. 2014;20(12):1853–1865. doi: 10.1089/ars.2013.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sertorio M, Du W, Amarachintha S, Wilson A, Pang Q. In vivo RNAi screen unveils PPARγ as a regulator of hematopoietic stem cell homeostasis. Stem Cell Reports. 2017;8(5):1242–1255. doi: 10.1016/j.stemcr.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schambach A, et al. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp Hematol. 2006;34(5):588–592. doi: 10.1016/j.exphem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Zhao H, Pestina TI, Nasimuzzaman M, Mehta P, Hargrove PW, Persons DA. Amelioration of murine beta-thalassemia through drug selection of hematopoietic stem cells transduced with a lentiviral vector encoding both gamma-globin and the MGMT drug-resistance gene. Blood. 2009;113(23):5747–5756. doi: 10.1182/blood-2008-10-186684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.