Abstract
There is limited understanding of the role of host metabolism in the pathophysiology of human tuberculosis (TB). Using high-resolution metabolomics with an unbiased approach to metabolic pathway analysis, we discovered that the tryptophan pathway is highly regulated throughout the spectrum of TB infection and disease. This regulation is characterized by increased catabolism of tryptophan to kynurenine, which was evident not only in active TB disease but also in latent TB infection (LTBI). Further, we found that tryptophan catabolism is reversed with effective treatment of both active TB disease and LTBI in a manner commensurate with bacterial clearance. Persons with active TB and LTBI also exhibited increased expression of indoleamine 2,3-dioxygenase-1 (IDO-1), suggesting IDO-1 mediates observed increases in tryptophan catabolism. Together, these data indicate IDO-1–mediated tryptophan catabolism is highly preserved in the human response to Mycobacterium tuberculosis and could be a target for biomarker development as well as host-directed therapies.
Keywords: Infectious disease, Metabolism
Keywords: Tuberculosis
Tryptophan catabolism is induced in active tuberculosis disease and latent tuberculosis infection and is reversed with effective treatment in a manner commensurate with bacterial clearance.
Introduction
Tuberculosis (TB) is now the leading cause of death due to an infectious disease, resulting in 1.5 million deaths each year (1). A major constraint in the effort to address the global TB epidemic has been limited understanding of TB pathophysiology. The mechanisms used by Mycobacterium tuberculosis (M. tuberculosis) to evade the host immune response and establish latency remain incompletely understood, as do the mechanisms for progression of latent TB infection (LTBI) to active TB disease. A better understanding of the heterogeneity of host-pathogen interactions following infection with M. tuberculosis, particularly those that allow the bacteria to avoid eradication by the host immune response, will be critical to develop effective vaccines, detect persons at greatest risk for progression to active TB disease, and identify potential targets for host-directed therapeutics (2).
Host metabolism is intricately linked to the immune response to infectious pathogens (3). Clinical metabolomics studies to date have shown tryptophan is decreased in the cerebrospinal fluid (CSF) of persons with TB meningitis (4) and gradually declines in the serum and plasma of persons with LTBI progressing to active TB disease (5, 6). Targeted studies of tryptophan in TB disease suggest such declines may reflect induction of indoleamine 2,3-dioxygenase (IDO) (7–10), the rate-limiting host enzyme for catabolism of tryptophan to kynurenine, which is highly upregulated in the lungs of animals with TB disease (11–13). Whereas tryptophan catabolism limits growth of intracellular pathogens such as Toxoplasma gondii (14) and Coxiella burnetii (15), M. tuberculosis is able to synthesize tryptophan itself, allowing it to counter ongoing IDO activity without inhibiting mycobacterial growth (16). In addition to causing local tryptophan depletion, upregulation of IDO-mediated tryptophan catabolism modulates CD4+ T cell responses, limiting inflammation and inducing immune tolerance (17). In pregnancy, induction of IDO in the placenta is necessary to prevent a T cell–mediated immune response against the fetus (18) while induction of IDO in many tumors results in T cell–mediated tolerance and persistence of cancer cells (19). In TB disease, induction of tryptophan catabolism has been put forth as a potential mechanism for M. tuberculosis evasion of T cell responses by inducing tolerance and persistence of the bacilli (12). Thus, observed increases in tryptophan catabolism may not only be associated with active TB disease but also have relevance to disease pathophysiology.
Using an unbiased plasma high-resolution metabolomics (HRM) approach, we aimed to determine whether tryptophan metabolism is among the most regulated metabolic pathways in the host response to TB infection, disease, and treatment response. We sought to comprehensively characterize changes in host tryptophan metabolism in a controlled study of persons with drug-susceptible (DS) pulmonary TB and to validate our findings in a second, demographically distinct population with multidrug-resistant TB (MDR-TB). We further sought to determine whether alterations in tryptophan metabolism observed in persons with active TB disease were also evident in persons with LTBI. Our results show tryptophan metabolism is highly regulated throughout the spectrum of TB infection and disease, characterized by increased catabolism of tryptophan to kynurenine. We show tryptophan catabolism occurs not only in persons with TB disease but also in asymptomatic persons with LTBI and is reversed with treatment of active TB disease and LTBI at a rate commensurate with the rate of bacterial clearance. We further show transcripts of IDO-1 are significantly higher in blood cells of persons with active TB disease and some populations with LTBI versus those not infected with M. tuberculosis, suggesting the observed changes in tryptophan catabolism are primarily mediated through induction of IDO-1. Taken together, these findings indicate that IDO-1–mediated tryptophan catabolism is a highly preserved human response to TB infection and disease and insufficient to restrict M. tuberculosis growth and may therefore be a target for biomarker development as well as host-directed therapies.
Results
Metabolic pathway regulation in human TB disease.
We performed HRM on plasma samples of a well-characterized population of persons with culture-confirmed pulmonary TB disease from Tbilisi, Georgia (n = 89), enrolled as part of a prior randomized controlled trial (20) (Table 1; described in detail in Methods). All persons in the Georgia cohort were HIV negative, had DS-TB, had a positive sputum smear for acid-fast bacilli (AFB) and culture for M. tuberculosis, were enrolled within 7 days of initiation of anti-TB treatment, and were treated with 6 months of directly observed therapy (DOT). Plasma samples were collected for all participants at enrollment and after 1, 2, and 4 months of active TB treatment. Patients with active TB were compared at baseline with control participants (n = 57), which included asymptomatic individuals with LTBI (n = 20) and those without evidence of M. tuberculosis infection (n = 37).
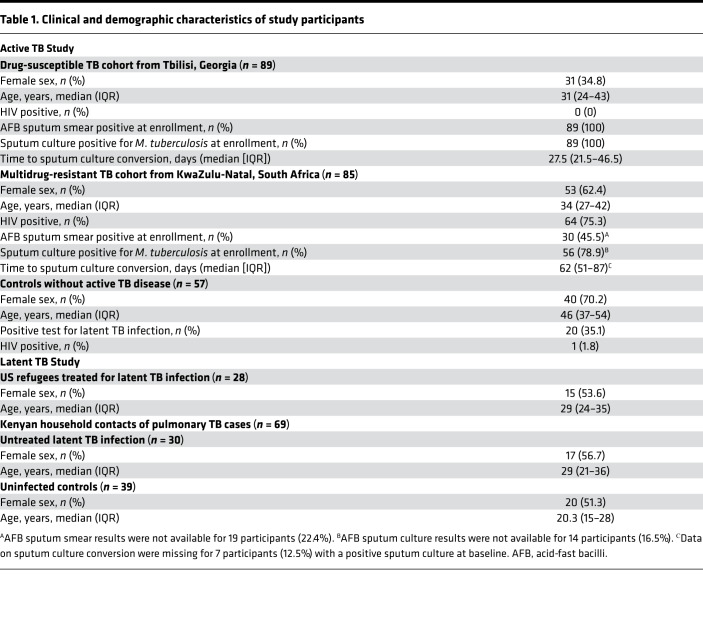
Table 1. Clinical and demographic characteristics of study participants.
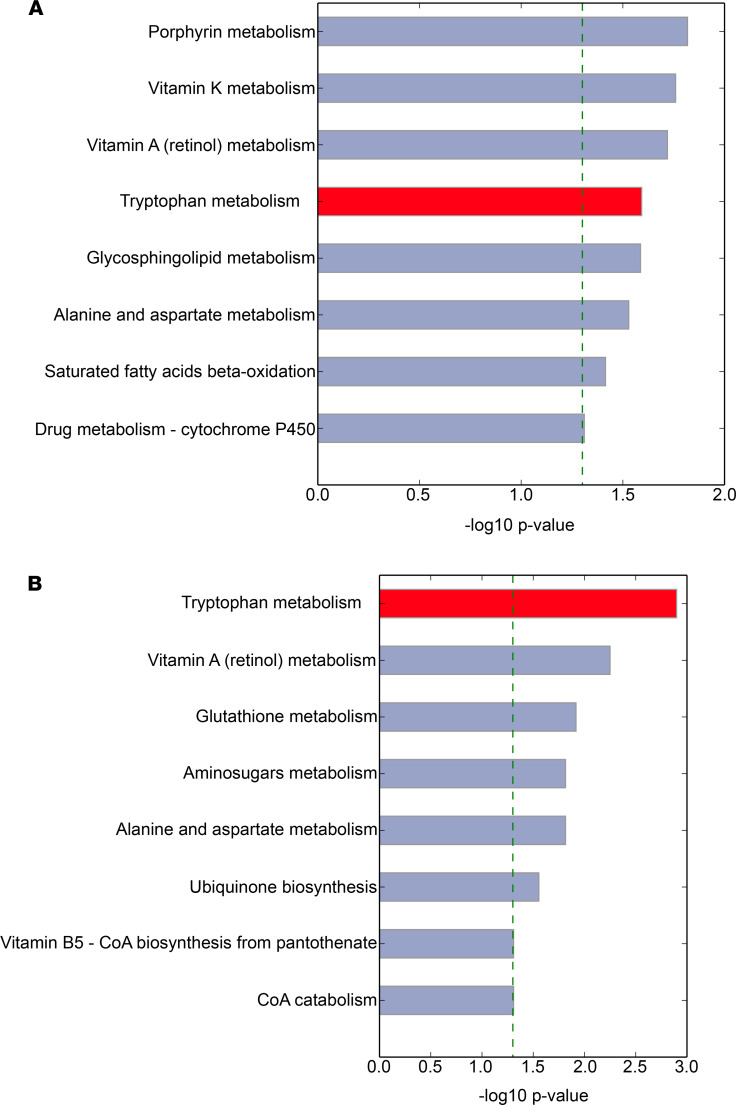
HRM analysis yielded 7498 mass/charge (m/z) features in plasma samples. We first sought to determine which metabolic pathways were significantly regulated in persons with active TB disease versus controls. To accomplish this, we used mummichog, an informatics tool that leverages the organization of metabolic networks to predict functional changes in metabolic pathway activity (21). We found that multiple metabolic pathways were regulated in patients with active TB versus controls, including porphyrin metabolism, vitamin A metabolism, vitamin K metabolism, and tryptophan metabolism (Figure 1A). To evaluate the possibility that observed metabolic differences between groups were due to factors other than TB disease, such as dietary and environmental exposures (22), we also examined metabolic pathway changes over the course of antibiotic therapy. We reasoned that host metabolic differences resulting from TB disease were likely to be reversed with effective TB treatment. Here we found both tryptophan and vitamin A metabolism were also significantly affected by treatment for active TB disease (Figure 1B). Upon further interrogation of these metabolic pathways, we found only the tryptophan pathway contained multiple metabolites that were both significantly different in persons with active TB disease versus controls and significantly changed with anti-TB treatment.
Figure 1. The tryptophan metabolic pathway is highly regulated in the host response to TB disease and chemotherapy-mediated bacterial clearance.
(A) Differences in metabolic pathway activity (21) in persons from the country of Georgia with active TB disease at the time of diagnosis (n = 89) versus controls without active TB disease (n = 57). (B) Changes in metabolic pathway activity in persons from Georgia over 4 months of treatment for drug-susceptible TB (DS-TB) disease with directly observed therapy. The horizontal bars show the magnitude of the –log(P value) for pathway enrichment in each pathway.
Tryptophan catabolism in active TB disease in Georgia.
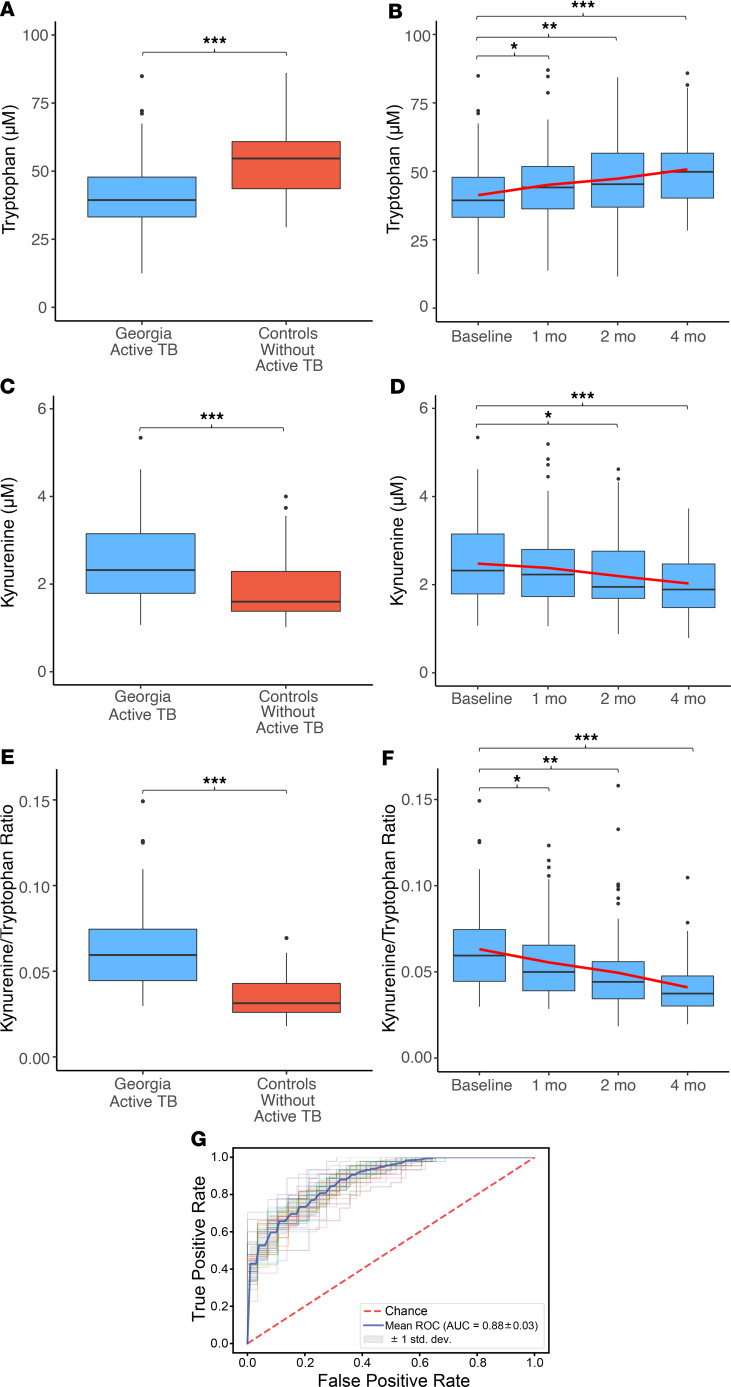
Based on these results, we sought to determine which metabolites in the tryptophan pathway were altered in persons with TB disease and how these metabolites changed with effective TB treatment. We found tryptophan and kynurenine were the most significantly different metabolites in persons with TB disease versus controls (P < 0.001) and that these metabolites were also the most significantly changed with TB treatment (P < 0.001). We therefore quantified plasma concentrations of tryptophan and kynurenine by accurate mass, tandem mass spectrometry (MS/MS), and retention time relative to authentic standards (23–26). We found plasma tryptophan concentrations were significantly decreased in patients with active TB versus controls without active TB but increased in a stepwise fashion with effective TB treatment (Figure 2, A and B). Conversely, we found plasma kynurenine concentrations were significantly elevated in active TB disease and declined in a stepwise fashion with effective TB treatment (Figure 2, C and D).
Figure 2. Tryptophan catabolism in persons with DS pulmonary TB disease in Georgia.
(A) Plasma tryptophan concentrations were significantly lower in persons with DS pulmonary TB disease from Georgia (light blue; n = 89) versus controls without active TB disease (red; n = 57) and (B) significantly increased after 1, 2, and 4 months of DOT compared with baseline. The red line indicates the trend of the mean over time. (C) Plasma kynurenine concentrations were significantly higher in Georgian patients with DS-TB versus controls and (D) significantly declined after 1, 2, and 4 months of active TB treatment. (E) The plasma kynurenine/tryptophan (K/T) ratio was also significantly higher in patients with active TB versus controls and (F) declined with antibiotic therapy in a stepwise fashion. (G) The receiver operator characteristic curve (ROC) for the plasma K/T ratio demonstrated excellent classification accuracy for identification of pulmonary TB. Active TB cases from Georgia were compared with controls using a Wilcoxon rank-sum test. Changes in tryptophan, kynurenine, and the K/T ratio relative to baseline were compared using a Wilcoxon signed-rank test (*P ≤ 0.05, **P < 0.01, and ***P < 0.001). The AUC for the ROC curve was calculated using logistic regression with 2-fold crossvalidation. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
We investigated whether the observed changes in plasma tryptophan and kynurenine concentrations were the result of alterations in IDO activity. Upregulation of IDO in the lungs of rhesus macaques with TB disease results in increased pulmonary tryptophan catabolism, which is reflected by plasma measurements of the kynurenine-to-tryptophan (K/T) ratio (12). We therefore examined the plasma K/T ratio as a marker of IDO-mediated tryptophan catabolism (6–10, 12). We found the plasma K/T ratio more clearly separated persons with active TB disease from controls without active TB than the plasma tryptophan or kynurenine concentration alone (Figure 2E) with excellent classification accuracy for active TB disease (AUC 0.88; Figure 2G). Further, the plasma K/T ratio was significantly lower after 1 month of effective treatment in patients with DS-TB, with continued stepwise declines after 2 and 4 months of treatment (Figure 2F).
We also examined all other metabolites in the tryptophan metabolic pathway detected by our HRM platform. We found the kynurenine metabolite 2-aminophenol and the tryptophan catabolism metabolite indole-3-ethanol were significantly elevated in persons with active TB versus controls but did not change with TB treatment (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137131DS1). Concentrations of 5-hydroxyindoleacetate increased with TB treatment while anthranilate and 3-hydroxyanthranilate declined with TB treatment. However, these metabolites were not significantly different between persons with active TB versus controls at baseline, and their role in the human metabolic response to TB disease remains unclear (Supplemental Table 1).
Tryptophan catabolism in active TB disease in South Africa.
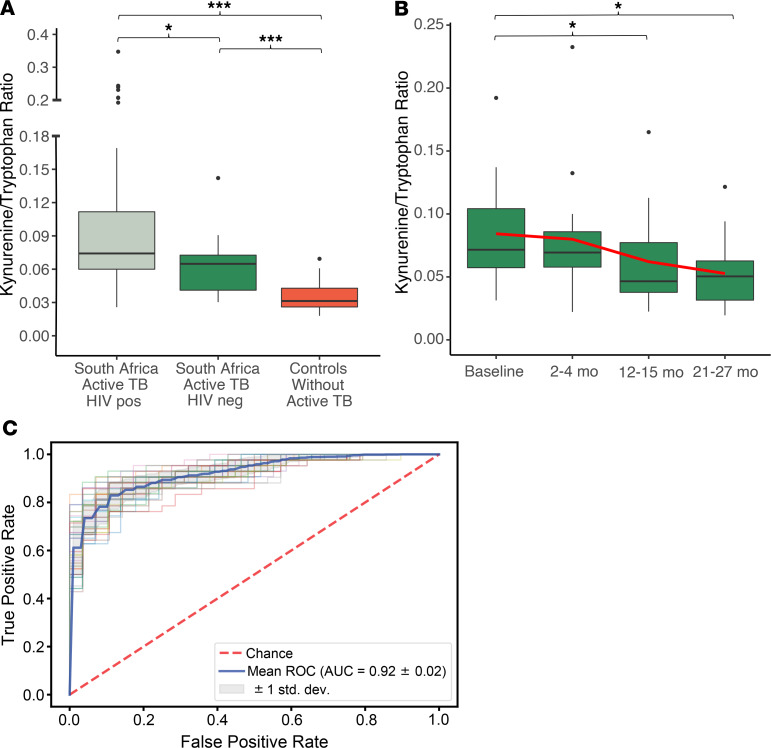
Given that the observed plasma K/T ratio differentiated patients with active TB in the country of Georgia from controls without active TB and declined with effective TB treatment in a manner related to the bacterial burden, we sought to validate our results in a separate, demographically distinct population. Thus, we examined tryptophan catabolism in persons with MDR-TB from KwaZulu-Natal province in South Africa (n = 85) with a high prevalence of HIV coinfection (75%) (27). All South African participants had a positive sputum culture for M. tuberculosis and were enrolled after phenotypic drug susceptibility testing (DST) demonstrated MDR-TB (2–3 months after pulmonary TB was first diagnosed). Most South African participants had an additional sputum specimen collected at study enrollment (83%), among whom 46% had a positive AFB sputum smear and 79% remained culture positive for M. tuberculosis. HRM was used to compare plasma samples from South African patients with pulmonary MDR-TB at baseline to the same group of control participants (n = 57) described above. The plasma K/T ratio was significantly higher in South African MDR-TB patients both with and without HIV coinfection versus controls (Figure 3A). Persons with TB disease coinfected with HIV had a higher plasma K/T ratio versus persons with TB disease alone (P = 0.01), consistent with prior observations that HIV also induces tryptophan catabolism (28, 29). The plasma K/T ratio again demonstrated excellent classification accuracy for active TB disease (AUC = 0.92; Figure 3C), and a cutoff of 0.03 had a sensitivity of 99% in both cohorts. Although specificity at this cutoff was low (42%), it shows the K/T ratio could have potential as a rule out test for TB disease.
Figure 3. Tryptophan catabolism in persons with multidrug-resistant pulmonary TB disease in South Africa.
(A) The plasma K/T ratio was significantly higher in South African multidrug-resistant TB (MDR-TB) patients with (n = 64) and without (n = 21) HIV coinfection versus controls (n = 57). (B) In MDR-TB patients with plasma samples available for the duration of the 2-year treatment period (12 HIV positive, 5 HIV negative), the plasma K/T ratio significantly declined at both 12- to 15-month and 21- to 27-month time points relative to baseline. The red line indicates the trend of the mean over time. (C) The ROC curve for the plasma K/T ratio demonstrated excellent classification accuracy for identification of pulmonary TB in South Africa. The plasma K/T ratio in active TB cases was compared with controls using a Wilcoxon rank-sum test, and changes relative to baseline were compared using a Wilcoxon signed-rank test (*P ≤ 0.05, and ***P < 0.001). The AUC for the ROC curve was calculated using logistic regression with 2-fold crossvalidation. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
For 37 South Africans with MDR-TB, plasma samples were available at baseline and after 2–4 months of MDR-TB treatment. Over this time there was no significant change in the plasma K/T ratio (Supplemental Figure 1). This is consistent with the slower rate of clearance of viable bacteria observed with second-line therapy used to treat MDR-TB at the time of the study (30). However, in the 17 participants with serial plasma samples collected over the duration of the 2-year study period, the plasma K/T ratio was significantly lower at both 12- to 15-month and 21- to 27-month time points relative to baseline (Figure 3B).
Tryptophan catabolism and disease severity.
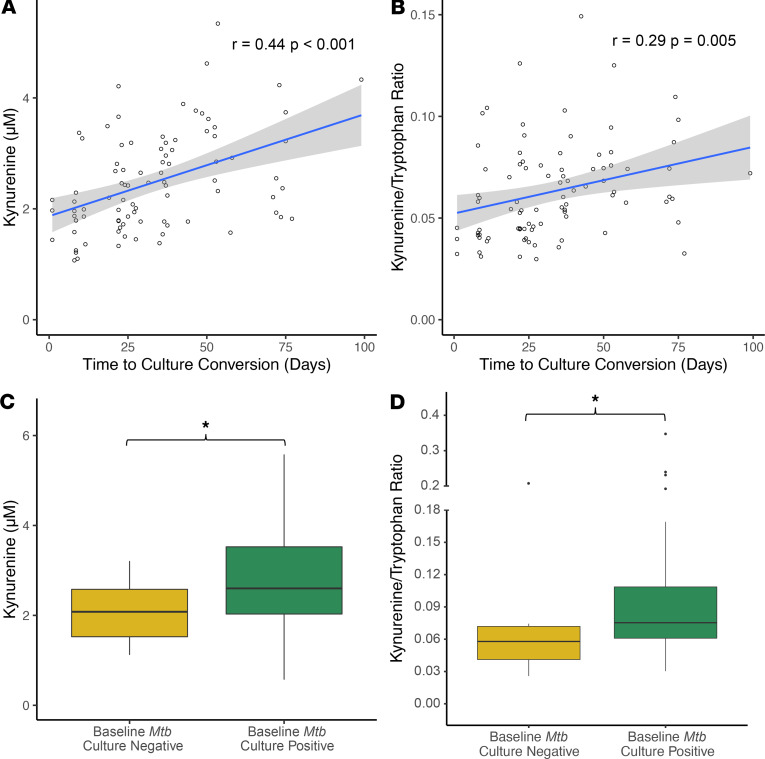
To determine whether the plasma K/T ratio was associated with disease severity, we examined the relationship between baseline plasma tryptophan and kynurenine concentrations and sputum culture results. In the Georgia DS-TB cohort, all participants were sputum culture positive for M. tuberculosis at baseline, with a sputum culture repeated every 2 weeks during the first 8 weeks of treatment and then repeated monthly for the remainder of the 16-week study period. We found both the baseline plasma kynurenine concentration and plasma K/T ratio were significantly and positively correlated with subsequent time to sputum culture conversion (r = 0.44, P < 0.001; and r = 0.29, P = 0.005 respectively; Figure 4, A and B).
Figure 4. The baseline plasma K/T ratio is associated with time to sputum culture conversion.
In persons from Georgia with active TB disease (n = 89), (A) the baseline plasma kynurenine concentration and (B) the baseline plasma K/T ratio were significantly and positively correlated with subsequent time to sputum culture conversion. In persons from South Africa with active TB disease and baseline sputum culture results (n = 71), (C) the baseline plasma kynurenine concentration and (D) the baseline plasma K/T ratio were significantly higher in persons who remained sputum culture positive for M. tuberculosis versus those who did not. For correlation analyses r indicates the Pearson correlation coefficient. Persons from South Africa who remained sputum culture positive for M. tuberculosis versus those who were not were compared using a Wilcoxon rank-sum test (*P ≤ 0.05). The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
In the South Africa MDR-TB cohort, participants were enrolled following referral to an MDR-TB treatment clinic and had initially received first-line DS-TB treatment before establishing a diagnosis of MDR-TB (27). Repeat sputum cultures were collected at enrollment (time of MDR-TB diagnosis) and then monthly for participants who remained culture positive. The plasma kynurenine concentration and the K/T ratio were both significantly higher in persons who remained sputum culture positive for M. tuberculosis at enrollment versus those who did not (P = 0.02, and P = 0.01, respectively; Figure 4, C and D). However, for those known to be culture positive at enrollment, there was no correlation between baseline plasma kynurenine or the K/T ratio and time to sputum culture conversion (P = 0.42, and P = 0.19, respectively).
Tryptophan catabolism in LTBI.
The observation that the plasma K/T ratio declined with successful treatment of active TB in a stepwise manner that mirrored the time course of treatment for DS-TB and MDR-TB suggests that the K/T ratio is a function of the burden of live bacteria in TB. To test this hypothesis, we examined subjects with LTBI, generally believed to be a paucibacillary state (31, 32). We enrolled 2 cohorts of individuals with LTBI: one from a refugee health clinic in the United States (DeKalb County Board of Health, metro Atlanta, Georgia; n = 28), presumed to have been infected in their country of origin, and another from a group of household contacts of active pulmonary TB cases in Kenya (n = 30). We compared the plasma K/T ratio at the time of LTBI diagnosis in both cohorts to a group of Kenyan household contacts without evidence of infection with M. tuberculosis (n = 39) as characterized by a negative QuantiFERON-TB Gold (QFT). Both US refugees and Kenyan household contacts with LTBI had a significantly higher plasma K/T ratio compared with QFT-negative controls (Figure 5A).
Figure 5. The plasma K/T ratio is increased in persons with LTBI and normalizes with latent TB treatment.
(A) Kenyan household contacts (n = 30) and US refugees (n = 28) with LTBI had a significantly higher plasma K/T ratio compared with Kenyan household contacts testing negative for latent TB (n = 39). (B) In persons treated for latent TB with 3 months of weekly isoniazid and rifapentine (3HP; n = 28), the plasma K/T ratio decreased in a stepwise fashion 3 and 6 months after treatment start and remained significantly lower after 9 and 12 months compared with baseline. The red line indicates the trend of the mean over time. (C) Although the plasma K/T ratio was similar at baseline for Kenyan contacts and US refugees with LTBI, it was significantly lower 6 and 12 months after starting 3HP treatment in the US refugee cohort. Cohorts with LTBI were compared with each other and to uninfected controls using a Wilcoxon rank-sum test. For the latent TB treatment cohort, each treatment time point was compared with baseline using a Wilcoxon signed-rank test (*P ≤ 0.05, and **P < 0.01). The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
After enrollment, the cohort of refugees was treated for LTBI with 12 weekly doses of high-dose isoniazid and rifapentine (3HP) given by DOT, and serial plasma samples were obtained every 3 months for 1 year after treatment initiation. HRM demonstrated that the plasma K/T ratio declined in a stepwise fashion 3 and 6 months after treatment start and remained significantly lower than baseline at 9 and 12 months (Figure 5B). Plasma tryptophan concentrations also significantly increased with LTBI treatment and were maximal at 6 months (Supplemental Table 1). Although kynurenine was not significantly affected by treatment, there was a significant decrease in the downstream metabolite indole-3-acetaldehyde over time, which may reflect altered metabolic flux through the tryptophan catabolic pathway (33) or the impact of antibiotics on the intestinal microbiome (34). Kenyan household contacts with LTBI did not receive antibiotic treatment and were followed every 6 months for 1 year. The plasma K/T ratio remained unchanged in the Kenyan household contacts, while it was significantly lower in the refugee cohort 6 and 12 months after initiating LTBI treatment with 3HP (Figure 5C).
It has been hypothesized that some individuals infected with M. tuberculosis are able to clear the organism immunologically (31). If this were the case, we would expect some individuals to demonstrate a lower plasma K/T ratio at baseline that does not change with LTBI therapy. Indeed, we found 4 of the 28 individuals undergoing LTBI treatment exhibited such a pattern, though the small number of subjects precludes definitive conclusions.
IDO transcription in active TB disease and LTBI.
We thought it was likely that observed changes in tryptophan catabolism were mediated by induction of the host enzyme IDO-1, which is highly expressed in pulmonary epithelial and endothelial cells in mice with TB disease (11) and at the periphery of the TB granuloma in rhesus macaques with TB disease (12, 35). However, increased IDO-1 expression in humans with active TB and LTBI has not been previously reported. Furthermore, it is unknown whether increased tryptophan catabolism in human TB disease is solely due to increased IDO-1 expression or whether IDO-2, which is expressed at lower levels in the lung and in myeloid dendritic cells (36, 37), may also play a role. To examine this question, we interrogated publicly available whole blood transcriptomics data sets to determine whether there was evidence of increased IDO-1 and IDO-2 expression in persons with active TB and LTBI with and without HIV coinfection relative to persons without evidence of infection with M. tuberculosis.
In persons with or without HIV coinfection, IDO-1 transcripts were significantly increased in active TB relative to controls with diseases other than TB and no evidence of LTBI (Figure 6) (38–43). In HIV-negative persons with active TB disease, IDO-1 transcripts were also increased relative to healthy controls. Studies demonstrated substantial heterogeneity with regard to IDO-1 expression in persons with LTBI. In children, levels of IDO-1 expression were similar in LTBI and active TB, and both children with LTBI and those with active TB demonstrated significantly higher IDO-1 expression versus controls with diseases other than TB and no evidence of LTBI (42). The one study that enrolled HIV-positive adults with LTBI as well as those with diseases other than TB and negative LTBI testing also showed a significant increase in IDO-1 transcripts in LTBI (43). In studies of HIV-negative adults, however, IDO-1 expression in persons with LTBI was closer to that of controls without evidence of infection with M. tuberculosis and significantly lower than persons with active TB (41, 43).
Figure 6. Whole blood transcriptomics demonstrates increased IDO-1 transcription in HIV-negative and HIV-positive persons with active TB disease and LTBI.
The bubble plots represent whole blood measurement of transcriptional changes in the tryptophan catabolic enzyme IDO-1 in HIV-negative persons (left box) and HIV-positive persons (right box). Study accession numbers from the publicly available National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) data repository are shown on the x axis, and each comparison is shown on the y axis. The color scale for each dot represents the log2 fold change in expression for persons with active TB disease (ATB) and LTBI relative to healthy controls (HC) and person with diseases other than TB and no evidence of infection with M. tuberculosis (OD). The dot size represents the –log P value for each comparison. ‡studies that were performed in children.
Although we did find evidence for increased IDO-2 transcripts in persons with active TB disease, particularly in HIV-positive persons with active TB and LTBI versus uninfected controls, increases in transcripts were smaller relative to IDO-1 (Supplemental Figure 2 and Supplemental Table 2). In HIV-negative individuals, most studies demonstrated a trend toward increased IDO-2 transcripts in persons with active TB disease relative to persons with LTBI and uninfected controls, though the increase generally did not meet statistical significance. These data indicate that increased tryptophan catabolism in active TB disease and LTBI is primarily mediated through induction of IDO-1, while IDO-2 may also play a role in active TB disease.
Discussion
In this multicohort study, we used HRM with an unbiased approach to metabolic pathway analysis to discover that the tryptophan pathway is highly regulated in the host response to TB infection and disease, as well as chemotherapy-mediated bacterial clearance. Observed alterations in tryptophan metabolism were primarily driven by increased catabolism of tryptophan to kynurenine, which is evident not only in active TB disease but also in asymptomatic persons with LTBI. We further show that increases in tryptophan catabolism are reversed with effective treatment of active TB disease and LTBI at a rate that closely mirrors the clinical response to therapy. Persons with active TB disease and some populations with LTBI also demonstrate significant increases in IDO-1 transcripts, suggesting increases in tryptophan catabolism are mediated by induction of IDO-1. Together, these data indicate IDO-1–mediated tryptophan catabolism is a highly preserved human response to M. tuberculosis that occurs throughout the spectrum of TB infection and disease in a bacterial burden–dependent manner yet is not sufficient to control M. tuberculosis replication.
The finding that increased tryptophan catabolism is evident across diverse human populations infected with M. tuberculosis and tightly connected to the burden of bacilli suggests it is likely to be of great benefit to either the host or the pathogen. The immunomodulatory effects associated with induction of IDO are well documented (17, 18, 28, 44) and have clear potential to benefit M. tuberculosis. Ongoing IDO-mediated tryptophan catabolism results in decreased proliferation of CD4+ T cells (17, 44). In malignancy and chronic HIV infection, induction of IDO activity also shifts CD4+ T cell differentiation toward regulatory T cells and away from Th17 cells, resulting in tolerance of tumor cells and antimicrobial products, respectively (17, 28). Similarly, upregulation of IDO in the granulomas of macaques with pulmonary TB is associated with decreased proliferation of M. tuberculosis antigen-specific T cells (12, 35) while increased IDO signaling in mice with pulmonary TB has been associated with inhibition of Th17 cells (11). Induction of IDO-mediated tryptophan catabolism may therefore allow M. tuberculosis to survive at the site of infection by modulating CD4+ T cell responses to induce immune tolerance and bacterial persistence (12, 28). Inhibition of IDO enzymatic activity using the drug 1-methyl-tryptophan in macaques with TB disease improved differentiation of CD4+ T cells, improved T cell trafficking in the granuloma, and attenuated disease (12). This suggests that induction of tryptophan catabolism primarily is pathogen beneficial, and the net effect of therapeutic IDO inhibition would be host beneficial.
Increased tryptophan catabolism could also protect the host from excessive inflammation. Mice lacking an IFN-γ receptor on nonhematopoietic cells demonstrate pathologically robust inflammatory responses to M. tuberculosis, which are associated with an absence of IDO expression (11). However, IDO-1–knockout mice show similar levels of pulmonary inflammation in response to M. tuberculosis versus WT mice (13), indicating therapeutic inhibition of IDO would not lead to a pathological inflammatory response. Further study of the impact of tryptophan catabolism on the activation and differentiation of CD4+ T cells in TB, as well as the potential value of IDO inhibitors and kynureninases as host-directed therapies, is warranted.
Increases in the plasma K/T ratio and IDO-1 transcripts in active TB and LTBI observed in our study may be at least partially the result of IFN-γ signaling, which is known to induce IDO expression (11). During progression from LTBI to active TB disease, this may lead to a positive feedback loop, whereby IFN-γ–mediated induction of IDO-1 results in further alteration of antigen-specific T cell responses (45), allowing for increased pathogen replication and further stimulation of IFN-γ production. Such a cascade would explain why we observed marked increases in tryptophan catabolism in all active TB cohorts at the time of diagnosis, which was associated with prolonged time to sputum culture clearance. However, effective chemotherapy appears to interrupt this cascade, with declines in the plasma K/T ratio observed in patients with DS-TB after 1 month of effective treatment, which continued through month 4. These declines mirrored the response to DS-TB treatment, which results in cure after 6 months of therapy in the vast majority of cases (46). In those with MDR-TB, declines in the plasma K/T ratio were more gradual, in keeping with the 2-year treatment course that was standard for MDR-TB at the time of the study (27).
Data from other human studies support a role for tryptophan catabolism in pulmonary TB disease (5–10). A recent study also found tryptophan concentrations were significantly decreased in the CSF of persons with TB meningitis versus controls (4). However, whereas we found a higher plasma K/T ratio was associated with longer times to sputum culture clearance in pulmonary TB, low tryptophan was protective in TB meningitis (4). If tryptophan catabolism modulates T cell responses to create tolerance, such immune modulation may be protective in the central nervous system, where robust inflammatory responses are generally detrimental (47). In conditions such as HIV and pregnancy, which similarly induce IDO-mediated tryptophan catabolism (29, 44), further study is needed to determine whether this may contribute to the elevated TB progression risk observed in these populations (48–50).
Our study findings also indicate the plasma K/T ratio may have utility as a biomarker of disease activity following infection with M. tuberculosis. Prior studies have shown tryptophan catabolism is increased in persons with TB disease (4–10), and we similarly found the plasma K/T ratio differentiated persons with pulmonary TB disease from controls with excellent classification accuracy. We additionally found the plasma K/T ratio was increased in persons with LTBI and declined with treatment of LTBI or active TB disease in a stepwise fashion that paralleled the clinical response to treatment. These findings suggest the plasma K/T ratio closely reflects disease activity across the spectrum of TB infection and disease. Further evaluation of plasma kynurenine, tryptophan, and the K/T ratio as markers of treatment response, M. tuberculosis eradication, and risk of relapse after treatment is warranted.
This study is subject to several limitations. Because this study was conducted in human subjects, host metabolic changes were measured in plasma rather than the site of disease. Because increases in tryptophan catabolism in TB are likely occurring predominantly in the lungs, tissue-level alterations in tryptophan and kynurenines may be much greater than those observed in the plasma, where the substrate and products are diluted in the plasma volume. The original studies that provided the plasma samples for this metabolomics study were not designed to examine the plasma K/T ratio as a biomarker of active TB disease, and there were no non-TB pulmonary disease control groups included. Though we showed a strong association between the K/T ratio and the burden of TB, the observational nature of the study precludes us from demonstrating that induction of tryptophan catabolism is causally linked with TB progression and treatment response. An effect of other interventions in the parent studies on the plasma K/T ratio, including receipt of differing antibiotic regimens, high-dose vitamin D in the Georgia cohort and antiretroviral therapy (ART) in the South Africa cohort, cannot be excluded. Participants in the South African cohort received DS-TB treatment before MDR-TB diagnosis, and some were culture negative at the time of study enrollment, limiting our ability to evaluate the correlation of the baseline plasma K/T ratio and time to culture conversion in this population. The metabolomics method used in this study is not optimized for detection of more hydrophobic host lipids, and it is possible that metabolically relevant changes in host lipid metabolism were not captured. Albumin, which binds tryptophan and could affect plasma tryptophan concentrations (51), was not measured.
In summary, the plasma K/T ratio and transcription of IDO-1 are significantly increased across multiple human populations at varying points on the spectrum of TB infection and disease in a bacterial burden–dependent manner, indicating induction of tryptophan catabolism is a highly preserved host response that reflects TB disease activity. Induction of tryptophan catabolism was even evident in asymptomatic persons with LTBI and was attenuated with effective treatment in those with DS-TB, MDR-TB, and LTBI. Collectively, these data provide evidence for an important role of IDO-1–mediated tryptophan catabolism in the pathophysiology of TB infection and disease in humans that could be a target for biomarker development and host-directed therapies.
Methods
Study participants.
For all cohorts, blood was collected in ethylenediaminetetraacetic acid–containing tubes and centrifuged; isolated plasma was immediately frozen and stored at –80°C. Samples collected outside of the United States were subsequently shipped on dry ice to Emory University, Atlanta, Georgia, USA. All samples remained frozen during transit and were kept at –80°C before metabolomics analysis.
Active TB study with Georgia pulmonary TB cohort.
Persons with pulmonary TB were selected from a randomized, double-blind controlled trial of adjunctive high-dose cholecalciferol (vitamin D3) for TB treatment conducted in the country of Georgia (ClinicalTrials.gov identifier NCT00918086) (20). All persons included in this metabolomics substudy were HIV negative and had DS-TB. Inclusion criteria for patients included age at least 18 years and newly diagnosed active TB disease, suggested by a positive AFB sputum smear and confirmed by positive sputum culture for M. tuberculosis. Baseline plasma samples for HRM were obtained from eligible subjects within 7 days of initiating therapy with conventional dosing of first-line anti-TB drugs (isoniazid, rifampin, pyrazinamide, and ethambutol) (20). Phenotypic DST was performed on M. tuberculosis isolates recovered from all persons with pulmonary TB using the absolute concentration method (52). All patients were treated for DS-TB with 6 months of DOT per WHO guidelines (46); 2 months of isoniazid, rifampin, pyrazinamide, and ethambutol followed by 4 months of isoniazid and rifampin. Plasma samples were also collected at baseline and after 4, 8, and 16 weeks of active TB treatment. Sputum cultures were collected at baseline and every 2 weeks to determine each participant’s time to culture conversion. As outlined in the parent project, adjunctive high-dose vitamin D3 had no impact on culture clearance of M. tuberculosis in DS-TB (20).
Active TB study with South Africa pulmonary TB cohort.
Persons with pulmonary TB from KwaZulu-Natal province, South Africa, were enrolled as part of a study of MDR-TB and TB/HIV coinfection (27). All persons in the South African cohort had MDR-TB as demonstrated by a positive sputum culture for M. tuberculosis and phenotypic DST indicating resistance to at least both isoniazid and rifampin. Baseline plasma samples were collected within 7 days of starting conventional treatment for MDR-TB. Persons with HIV coinfection were continued on conventional ART, and those not previously on ART were started on treatment. All patients were referred to a dedicated MDR-TB treatment center and treated with a standardized drug regimen that included kanamycin (15 mg/kg, maximum 1 g daily), moxifloxacin (400 mg daily), ethionamide (15–20 mg/kg, maximum 750 mg daily), terizidone (15–20 mg/ kg, maximum 750 mg daily), ethambutol (15–20 mg/kg, maximum 1200 mg daily), and pyrazinamide (20–30 mg/kg, maximum 1600 mg daily). All persons who completed the study were treated for a period of 2 years, and serial plasma samples were obtained 2–4 months, 12–15 months, and 21–27 months after treatment initiation.
Active TB study with controls without active TB disease.
Plasma from persons with and without LTBI was analyzed for cross-sectional comparison with pulmonary TB cases. Persons with LTBI were enrolled from the DeKalb County Board of Health in DeKalb County, Georgia, USA. All persons with LTBI had positive test results from at least 2 FDA-approved tests for LTBI (QFT, TSPOT.TB, and tuberculin skin test [TST]). All tests were interpreted according to the guidelines from the Centers for Disease Control and Prevention (53, 54). Controls without LTBI were United States–born adults at low risk for M. tuberculosis exposure and infection, who had at least 1 negative TST within the year before plasma collection documented in medical records (55).
Latent TB study with US refugee cohort.
Persons in the LTBI treatment cohort were enrolled from the Refugee Health Clinic at the DeKalb County Board of Health (in metropolitan Atlanta, Georgia, USA). All persons were at epidemiologically high risk for TB exposure (born in a country of medium or high TB incidence), recently arrived in the United States (within the prior year), and had a positive QFT. Active TB was ruled out with a negative symptom screen and a negative chest radiograph, as well as an AFB sputum culture, if indicated. All persons in the LTBI treatment cohort were treated with 3HP via DOT (56). Baseline plasma samples were obtained before treatment start, with serial plasma samples obtained 3, 6, 9, and 12 months after treatment initiation.
Latent TB study with Kenyan household contact cohort.
HIV-negative close contacts were enrolled in Kisumu, Kenya, if they were living in the same household as a person diagnosed with active pulmonary TB. All household contacts underwent testing for LTBI with QFT. In those with a positive QFT, active TB disease was ruled out with a symptom screen, chest radiograph, and AFB sputum culture, if indicated. Those with a negative QFT were used as uninfected controls. Persons with LTBI (positive QFT, negative chest radiograph, no symptoms) had additional plasma samples obtained 6 and 12 months after enrollment.
Plasma metabolomics analysis.
Deidentified samples were randomized by a computer-generated list into blocks of 40 samples before transfer to the analytical laboratory, where personnel were blinded to clinical and demographic data. Thawed plasma (65 μL) was treated with 130 μL acetonitrile (2:1, v/v) containing an internal isotopic standard mixture (3.5 μL/sample), as previously described (57). The internal standard mix for quality control consisted of 14 stable isotopic chemicals covering a broad range of small molecules (57). Samples were mixed and placed on ice for 30 minutes before centrifugation to remove protein. The resulting supernatant was transferred to low-volume autosampler vials maintained at 4°C and analyzed in triplicate. The active TB study data were analyzed using an Orbitrap Fusion Mass Spectrometer, and the latent TB study data were analyzed using a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, San Jose, California, USA). In both studies, hydrophilic interaction chromatography was used (Higgins Analytical, Targa, Mountain View, California, USA, 2.1 × 10 cm) with a formic acid/acetonitrile gradient. The high-resolution mass spectrometer was operated in positive electrospray ionization mode over scan range of 85 to 1275 m/z and stored as .Raw files (23). Data were extracted and aligned using apLCMS (58) and xMSanalyzer (59) with each feature defined by specific m/z value, retention time, and integrated ion intensity (23). Three technical replicates were performed for each plasma sample and median summarized (60).
Metabolite identification and reference standardization.
Identities of metabolites of interest were confirmed using ion dissociation methods (MS/MS). Fragmentation spectra were generated using a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer with parallel reaction monitoring mode using a targeted inclusion list. The MS/MS spectra were compared with authentic reference standards to confirm the chemical identity of the metabolites of interest. Tryptophan and kynurenine were confirmed and quantified by accurate mass, MS/MS, and retention time relative to authentic standards (Supplemental Figure 4 and refs. 23–26).
Transcriptomics analysis.
Whole blood gene expression data sets publicly available in the GEO database were selected for further analysis if they enrolled persons with active TB disease and at least one of the following groups: persons with LTBI, healthy controls, or controls with diseases other than TB (38–43, 63, 64). To ensure that control patients did not also have LTBI, only controls enrolled from countries with low TB transmission (annual TB incidence of less than 20/100,000) or with negative testing for LTBI (QFT and/or TST) were considered for this analysis. Moreover, only studies of persons with known HIV status were included.
Raw gene expression data and metadata associated with the studies fulfilling the abovementioned criteria were acquired from the GEO database. Lumi and oligo packages from Bioconductor were used for reading and preprocessing of Illumina and Affymetrix arrays, respectively (65, 66). Each probe was annotated using its corresponding Human Genome Organisation Gene Nomenclature Committee gene symbol. The differential expression of genes between different experimental groups was computed using the Linear Models for Microarray Data (limma) package in R (61). A P value less than or equal to 0.05 was considered statistically significant.
Statistics.
Statistical comparisons of metabolite intensity values and concentrations were performed in R version 3.5.0. For the untargeted metabolomics analysis, metabolite intensity values were log2-transformed and compared between groups and over time using limma and repeated measures limma, respectively (61). The limma package provides a statistical framework for differential expression analysis of high-dimensional data sets (61). Metabolic pathway enrichment analysis was performed using mummichog, a Python-based informatics tool that leverages the organization of metabolic networks to predict functional changes in metabolic pathway activity (3, 21, 62).
Cross-sectional comparisons of plasma tryptophan concentration, kynurenine concentration, and the K/T ratio were tested using the Wilcoxon rank-sum test. Changes relative to baseline during treatment of active TB and LTBI were tested using a Wilcoxon signed-rank test. A P value less than or equal to 0.05 was considered statistically significant. ROC curves for the K/T ratio were constructed for both active TB populations relative to controls using logistic regression with 2-fold cross-validation as well as cross-cohort validation (Supplemental Figure 3).
Study approval.
All studies were approved by the Institutional Review Board (IRB) of Emory University (Atlanta, Georgia, USA) and by the individual IRBs associated with the original cohort studies: the Ethics Committee of the National Center for Tuberculosis and Lung Diseases of Georgia (Tbilisi, Georgia), the University of KwaZulu-Natal IRB (Durban, South Africa), the Georgia Department of Public Health IRB (Atlanta, Georgia, USA), and the Kenya Medical Research Institute Scientific and Ethics Review Unit (Nairobi, Kenya), respectively, depending on the site of participant enrollment. All subjects provided written informed consent.
Author contributions
JMC, TRZ, HMB, DPJ, JDE, RRK, NRG, JCMB, CLD, and JR conceptualized and designed the study. JMC, TRZ, HMB, DPJ, JDE, NRG, JCMB, KL, AN, NSS, NI, SGO, NT, CLD, and JR were responsible for sample collection, sample processing, and data acquisition, analysis, and interpretation. JMC, AS, and SL were responsible for evaluation of classification models, whole blood transcriptomics analysis, and data visualization. JMC wrote the original draft. All authors reviewed and contributed to the final draft.
Supplementary Material
Acknowledgments
We thank all the study teams and study sites from the multiple cohorts leveraged by this metabolomics study, including the DeKalb County Board of Health, Kenya Medical Research Institute, Georgia National Center for Tuberculosis and Lung Disease, and University of KwaZulu-Natal. We also thank A. Crampin, M. Kaforou, and the ILULU consortium for providing additional data on latent TB testing for our transcriptomics analysis (Figure 6). This work was supported in part by grants from the NIH, including R21 AI130918 (to TRZ, NRG, JCMB, DPJ, RRK, HMB), the TBRU ASTRa Study Group U19 AI111211 (to HMB, JDE, JMC, NRG, CLD, JR, AN), R01 AI087465 (to NRG, NSS, JCMB), T32 AI074492 (to JMC), K23 AI144040 (to JMC), K24 AI114444 (to NRG);by Georgia Clinical and Translational Science Alliance UL1 TR002378 (to TRZ, HMB, JMC); as well as by grants from the Emory University Global Health Institute (to TRZ, HMB, NT, RRK) and the Emory Medical Care Foundation (to RRK, JMC, HMB, TRZ).
Version 1. 05/05/2020
In-Press Preview
Version 2. 05/21/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(10):e137131.https://doi.org/10.1172/jci.insight.137131.
Contributor Information
Jeffrey M. Collins, Email: jmcoll4@emory.edu.
Amnah Siddiqa, Email: amnahsiddiqa@gmail.com.
Dean P. Jones, Email: dpjones@emory.edu.
Ken Liu, Email: ken.liu@emory.edu.
Russell R. Kempker, Email: rkempke@emory.edu.
Azhar Nizam, Email: anizam@emory.edu.
N. Sarita Shah, Email: saritashah2@gmail.com.
Nazir Ismail, Email: Naziri@nicd.ac.za.
Samuel G. Ouma, Email: souma@kemricdc.org.
Nestani Tukvadze, Email: marikushane@yahoo.com.
Shuzhao Li, Email: shuzhao.li@gmail.com.
Cheryl L. Day, Email: cheryl.day@emory.edu.
Jyothi Rengarajan, Email: jrengar@emory.edu.
James C.M. Brust, Email: jcmbrust@gmail.com.
Neel R. Gandhi, Email: neel.r.gandhi@emory.edu.
Joel D. Ernst, Email: Joel.Ernst@ucsf.edu.
Henry M. Blumberg, Email: hblumbe@emory.edu.
Thomas R. Ziegler, Email: tzieg01@emory.edu.
References
- 1. WHO. World Health Organization Global Tuberculosis Report 2019. WHO. https://www.who.int/tb/publications/global_report/en/ Published October 17, 2019. Accessed April 30, 2020.
- 2.Ernst JD. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe. 2018;24(1):34–42. doi: 10.1016/j.chom.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, et al. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169(5):862–877.e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Laarhoven A, et al. Cerebral tryptophan metabolism and outcome of tuberculous meningitis: an observational cohort study. Lancet Infect Dis. 2018;18(5):526–535. doi: 10.1016/S1473-3099(18)30053-7. [DOI] [PubMed] [Google Scholar]
- 5.Weiner J, et al. Metabolite changes in blood predict the onset of tuberculosis. Nat Commun. 2018;9(1):5208. doi: 10.1038/s41467-018-07635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vrieling F, et al. Plasma metabolomics in tuberculosis patients with and without concurrent type 2 diabetes at diagnosis and during antibiotic treatment. Sci Rep. 2019;9(1):18669. doi: 10.1038/s41598-019-54983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y, et al. Serum indoleamine 2,3-dioxygenase activity predicts prognosis of pulmonary tuberculosis. Clin Vaccine Immunol. 2012;19(3):436–442. doi: 10.1128/CVI.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki Y, et al. Indoleamine 2,3-dioxygenase in the pathogenesis of tuberculous pleurisy. Int J Tuberc Lung Dis. 2013;17(11):1501–1506. doi: 10.5588/ijtld.13.0082. [DOI] [PubMed] [Google Scholar]
- 9.Adu-Gyamfi CG, et al. Plasma indoleamine 2, 3-dioxygenase, a biomarker for tuberculosis in human immunodeficiency virus-infected patients. Clin Infect Dis. 2017;65(8):1356–1358. doi: 10.1093/cid/cix550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi W, et al. Plasma indoleamine 2,3-dioxygenase activity as a potential biomarker for early diagnosis of multidrug-resistant tuberculosis in tuberculosis patients. Infect Drug Resist. 2019;12:1265–1276. doi: 10.2147/IDR.S202369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvignes L, Ernst JD. IFNγ-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31(6):974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautam US, et al. In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2018;115(1):E62–E71. doi: 10.1073/pnas.1711373114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenthal A, et al. M. tuberculosis induces potent activation of IDO-1, but this is not essential for the immunological control of infection. PLoS One. 2012;7(5):e37314. doi: 10.1371/journal.pone.0037314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Däubener W, et al. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect Immun. 2001;69(10):6527–6531. doi: 10.1128/IAI.69.10.6527-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesan S, Roy CR. Host cell depletion of tryptophan by IFNγ-induced Indoleamine 2,3-dioxygenase 1 (IDO1) inhibits lysosomal replication of Coxiella burnetii. PLoS Pathog. 2019;15(8):e1007955. doi: 10.1371/journal.ppat.1007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YJ, et al. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell. 2013;155(6):1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson TS, Munn DH. Host indoleamine 2,3-dioxygenase: contribution to systemic acquired tumor tolerance. Immunol Invest. 2012;41(6-7):765–797. doi: 10.3109/08820139.2012.689405. [DOI] [PubMed] [Google Scholar]
- 18.Munn DH, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281(5380):1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 19.Brochez L, Chevolet I, Kruse V. The rationale of indoleamine 2,3-dioxygenase inhibition for cancer therapy. Eur J Cancer. 2017;76:167–182. doi: 10.1016/j.ejca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Tukvadze N, et al. High-dose vitamin D3 in adults with pulmonary tuberculosis: a double-blind randomized controlled trial. Am J Clin Nutr. 2015;102(5):1059–1069. doi: 10.3945/ajcn.115.113886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DP. Sequencing the exposome: a call to action. Toxicol Rep. 2016;3:29–45. doi: 10.1016/j.toxrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go YM, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol Sci. 2015;148(2):531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Go YM, et al. Metabolic characterization of the common marmoset (Callithrix jacchus) PLoS One. 2015;10(11):e0142916. doi: 10.1371/journal.pone.0142916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker DI, et al. Metabolomic assessment of exposure to near-highway ultrafine particles. J Expo Sci Environ Epidemiol. 2019;29(4):469–483. doi: 10.1038/s41370-018-0102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Accardi CJ, et al. High-resolution metabolomics for nutrition and health assessment of armed forces personnel. J Occup Environ Med. 2016;58(8 suppl 1):S80–S88. doi: 10.1097/JOM.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 27.Brust JCM, et al. Improved survival and cure rates with concurrent treatment for multidrug-resistant tuberculosis-human immunodeficiency virus coinfection in South Africa. Clin Infect Dis. 2018;66(8):1246–1253. doi: 10.1093/cid/cix1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Favre D, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2(32):32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, et al. Plasma indoleamine 2,3-dioxygenase activity is associated with the size of the human immunodeficiency virus reservoir in patients receiving antiretroviral therapy. Clin Infect Dis. 2019;68(8):1274–1281. doi: 10.1093/cid/ciy676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuen CM, et al. Association between regimen composition and treatment response in patients with multidrug-resistant tuberculosis: a prospective cohort study. PLoS Med. 2015;12(12):e1001932. doi: 10.1371/journal.pmed.1001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai M, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 32.Dutta NK, Karakousis PC. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev. 2014;78(3):343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18(5):379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehra S, et al. Granuloma correlates of protection against tuberculosis and mechanisms of immune modulation by Mycobacterium tuberculosis. J Infect Dis. 2013;207(7):1115–1127. doi: 10.1093/infdis/jis778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 37.Papatheodorou I, et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloom CI, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8(8):e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blankley S, et al. The transcriptional signature of active tuberculosis reflects symptom status in extra-pulmonary and pulmonary tuberculosis. PLoS One. 2016;11(10):e0162220. doi: 10.1371/journal.pone.0162220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhagen LM, et al. A predictive signature gene set for discriminating active from latent tuberculosis in Warao Amerindian children. BMC Genomics. 2013;14:74. doi: 10.1186/1471-2164-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson ST, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370(18):1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaforou M, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10(10):e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellor AL, et al. Prevention of T cell-driven complement activation and inflammation by tryptophan catabolism during pregnancy. Nat Immunol. 2001;2(1):64–68. doi: 10.1038/83183. [DOI] [PubMed] [Google Scholar]
- 45.Munn DH, et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22(5):633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 46. WHO. Guidelines for Treatment of Drug-Susceptible Tuberculosis and Patient Care - 2017 Update. WHO. https://www.who.int/tb/publications/2017/dstb_guidance_2017/en/ 2017. Accessed April 30, 2020.
- 47.Mook-Kanamori BB, Geldhoff M, van der Poll T, van de Beek D. Pathogenesis and pathophysiology of pneumococcal meningitis. Clin Microbiol Rev. 2011;24(3):557–591. doi: 10.1128/CMR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlowski A, Jansson M, Sköld M, Rottenberg ME, Källenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8(2):e1002464. doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta A, et al. Postpartum tuberculosis incidence and mortality among HIV-infected women and their infants in Pune, India, 2002-2005. Clin Infect Dis. 2007;45(2):241–249. doi: 10.1086/518974. [DOI] [PubMed] [Google Scholar]
- 50.Zenner D, Kruijshaar ME, Andrews N, Abubakar I. Risk of tuberculosis in pregnancy: a national, primary care-based cohort and self-controlled case series study. Am J Respir Crit Care Med. 2012;185(7):779–784. doi: 10.1164/rccm.201106-1083OC. [DOI] [PubMed] [Google Scholar]
- 51.Fuller RW, Roush BW. Binding of tryptophan to plasma proteins in several species. Comp Biochem Physiol, B. 1973;46(2):273–276. doi: 10.1016/0305-0491(73)90318-0. [DOI] [PubMed] [Google Scholar]
- 52.Lomtadze N, et al. Prevalence and risk factors for multidrug-resistant tuberculosis in the Republic of Georgia: a population-based study. Int J Tuberc Lung Dis. 2009;13(1):68–73. [PMC free article] [PubMed] [Google Scholar]
- 53.American Thoracic Society, Centers for Disease Control Prevention, Infectious Diseases Society of America American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: controlling tuberculosis in the United States. Am J Respir Crit Care Med. 2005;172(9):1169–1227. doi: 10.1164/rccm.2508001. [DOI] [PubMed] [Google Scholar]
- 54.Mazurek GH, et al. Updated guidelines for using Interferon Gamma Release Assays to detect Mycobacterium tuberculosis infection - United States, 2010. MMWR Recomm Rep. 2010;59(RR-5):1–25. [PubMed] [Google Scholar]
- 55.Al Mheid I, et al. Effects of a health-partner intervention on cardiovascular risk. J Am Heart Assoc. 2016;5(10):e004217. doi: 10.1161/JAHA.116.004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Centers for Disease Control Prevention Recommendations for use of an isoniazid-rifapentine regimen with direct observation to treat latent Mycobacterium tuberculosis infection. MMWR Morb Mortal Wkly Rep. 2011;60(48):1650–1653. [PubMed] [Google Scholar]
- 57.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9(1 suppl):S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25(15):1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uppal K, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins JM, Kempker RR, Ziegler TR, Blumberg HM, Jones DP. Metabolomics and mycobacterial disease: don’t forget the bioinformatics. Ann Am Thorac Soc. 2016;13(1):141–142. doi: 10.1513/AnnalsATS.201510-676LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagan T, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178(6):1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter ND, et al. Blood transcriptional biomarkers for active tuberculosis among patients in the United States: a case-control study with systematic cross-classifier evaluation. J Clin Microbiol. 2016;54(2):274–282. doi: 10.1128/JCM.01990-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esmail H, et al. Complement pathway gene activation and rising circulating immune complexes characterize early disease in HIV-associated tuberculosis. Proc Natl Acad Sci U S A. 2018;115(5):E964–E973. doi: 10.1073/pnas.1711853115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 66.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.