Abstract
BACKGROUND
The coronavirus disease 2019 (COVID-19), infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a severe outbreak throughout the world. The host immunity of COVID-19 patients is unknown.
METHODS
The routine laboratory tests and host immunity in COVID-19 patients with different severity of illness were compared after patient admission.
RESULTS
A total of 65 SARS-CoV-2–positive patients were classified as having mild (n = 30), severe (n = 20), and extremely severe (n = 15) illness. Many routine laboratory tests, such as ferritin, lactate dehydrogenase, and D-dimer, were increased in severe and extremely severe patients. The absolute numbers of CD4+ T cells, CD8+ T cells, and B cells were gradually decreased with increased severity of illness. The activation markers such as HLA-DR and CD45RO expressed on CD4+ and CD8+ T cells were increased in severe and extremely severe patients compared with mild patients. The costimulatory molecule CD28 had opposite results. The percentage of natural Tregs was decreased in extremely severe patients. The percentage of IFN-γ–producing CD8+ T cells was increased in both severe and extremely severe patients compared with mild patients. The percentage of IFN-γ–producing CD4+ T cells was increased in extremely severe patients. IL-2R, IL-6, and IL-10 were all increased in extremely severe patients. The activation of DC and B cells was decreased in extremely severe patients.
CONCLUSION
The number and function of T cells are inconsistent in COVID-19 patients. The hyperfunction of CD4+ and CD8+ T cells is associated with the pathogenesis of extremely severe SARS-CoV-2 infection.
FUNDING
This work was funded by the National Mega Project on Major Infectious Disease Prevention (2017ZX10103005-007) and the Fundamental Research Funds for the Central Universities (2019kfyRCPY098).
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, Cellular immune response, Innate immunity
The hyperfunction of CD4+ and CD8+ T cells is associated with the pathogenesis of extremely severe SARS-CoV-2 infection
Introduction
In December 2019, a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which can cause severe respiratory infection in humans, was first reported and confirmed in Wuhan, China (1, 2). Very soon, the virus has caused a severe outbreak in China and spread to most regions of the world (3–5). The disease caused by SARS-CoV-2 has been recently named as the coronavirus disease 2019 (COVID-19) by the World Health Organization. Most patients infected with SARS-CoV-2 have mild illness and present common symptoms such as fever, cough, and fatigue (6). A small number of infected patients progress to severe cases with acute respiratory distress syndrome, and some infected patients with severe illness worsen in a short period of time and die of multiple organ failure, especially in elderly patients with comorbidities (7). Up to now, the differences in pathogenesis between mild and severe illness in COVID-19 patients is unknown.
The current studies focus on the genome sequences (1, 8), epidemiology and outbreak investigation (9, 10), detection methods (11, 12), receptor recognition (13), and drug treatment of SARS-CoV-2 (14). There are currently no studies comparing host immunity between patients with mild and severe illness. One previous study showed that C-reactive protein (CRP) and IL-6 were increased in COVID-19 patients, whereas the number of lymphocytes was decreased (7). Another study found that infected patients with severe illness had higher plasma level of cytokines such as IL-2, TNF-α, and IL-10 (6). These studies indicated that the pathogenesis of SARS-CoV-2 infection is associated with the dysregulation of host immunity.
The methods that can be used to predict the development of severe illness are of great value for treatment and prognosis of SARS-CoV-2 infection. The establishment of these methods depends on an understanding of the pathogenesis of the disease. This can also help to design vaccine strategies and find new treatment targets. In this study, we systematically investigated the clinical characteristics, routine laboratory tests, and host immunity of COVID-19 patients with different severity of illness. Our results suggest that the hyperfunction of both CD4+ and CD8+ T cells may lead to the development of extremely severe illness in patients with SARS-CoV-2 infection.
Results
Enrolled patients.
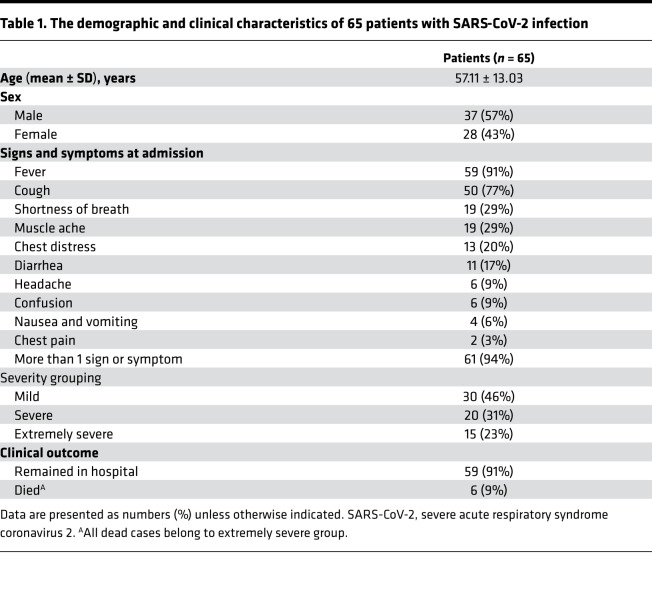
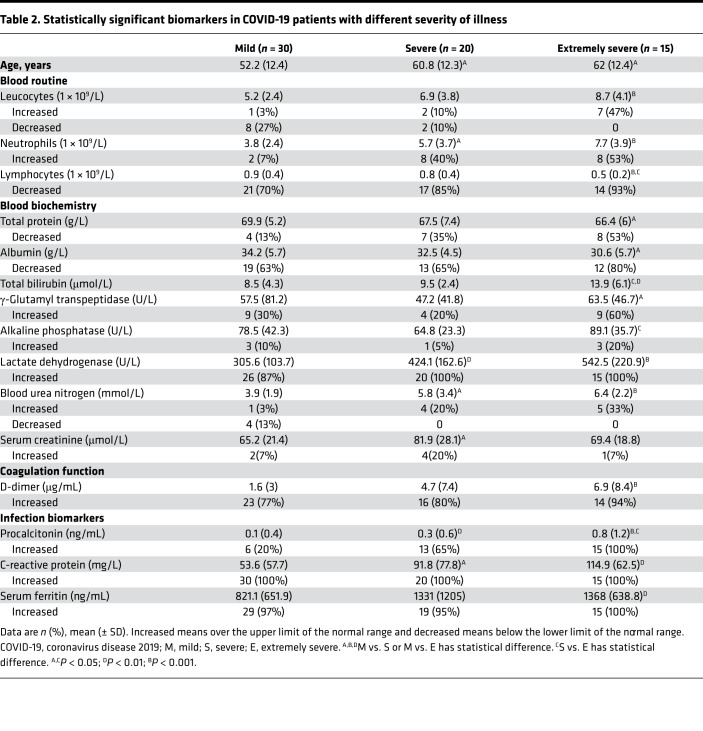
The demographic and clinical characteristics of COVID-19 patients are shown in Table 1. A total of 65 COVID-19 patients with different severity of illness (mild, n = 30; severe, n = 20; extremely severe, n = 15) were recruited. The percentage of males was higher than females. Fever and cough were the most common symptoms, and over 90% of patients had more than 1 symptom. Six patients died and all belonged to the extremely severe group. The ages of severe patients (60.8 ± 12.3) and extremely severe patients (62 ± 12.4) were significantly higher than mild patients (52.2 ± 12.4) (Table 2). The results of blood routine, biochemistry, coagulation function, and infection biomarkers in different groups are shown in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137799DS1). The biomarkers that had significant differences among the 3 groups are shown in Table 2. We observed that many indicators — such as leucocyte number, lymphocyte number, total protein, total bilirubin, lactate dehydrogenase, urea nitrogen, D-dimer, CRP, and ferritin — had significant differences among the 3 groups, especially between the mild and extremely severe groups. Similarly, there were more indicators, such as lymphocyte number, total protein, albumin, urea nitrogen, and D-dimer, in the extremely severe group that were out of normal ranges. The normal ranges of laboratory tests are shown in Supplemental Table 2.
Table 1. The demographic and clinical characteristics of 65 patients with SARS-CoV-2 infection.
Table 2. Statistically significant biomarkers in COVID-19 patients with different severity of illness.
T cell, B cell, and NK cell counts.
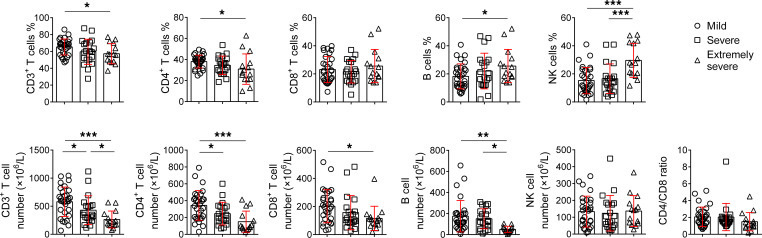
The percentages of CD3+ and CD4+ T cells in extremely severe patients were significantly lower than in mild patients. In contrast, the percentages of B cells and NK cells were relatively increased in extremely severe patients. The absolute numbers of CD3+ T cells, CD4+ T cells, CD8+ T cells, and B cells were all gradually decreased with increased severity of illness, and these cells in extremely severe patients were all significantly lower than in mild patients. The number of B cells in extremely severe patients was also significantly lower than in severe patients (Figure 1).
Figure 1. TBNK lymphocyte counting.
The percentages and absolute numbers of T cells, B cells, and NK cells, and the results of CD4/CD8 ratio in different groups are shown in graphs (mean ± SD). *P < 0.05, **P < 0.01, ***P < 0.001 (1-way ANOVA).
T cell subset analysis.
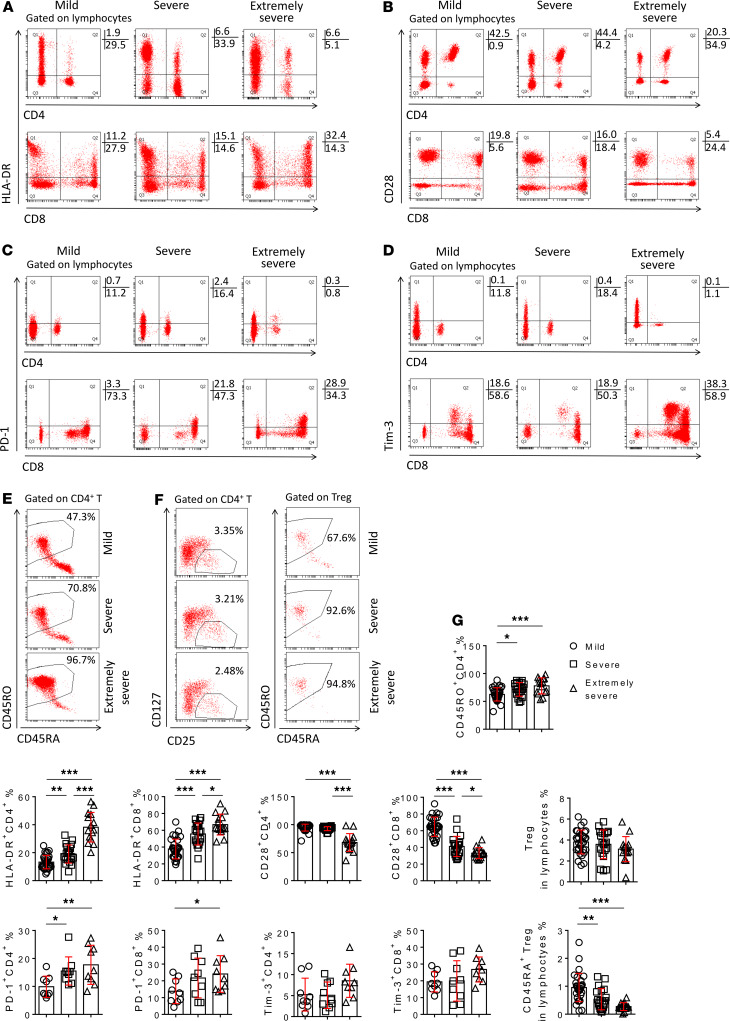
The expression of activation marker HLA-DR on both CD4+ and CD8+ T cells was significantly increased in severe and extremely severe patients compared with mild patients, especially the increase of that on CD4+ T cells in extremely severe patients (Figure 2, A and G). The expression of CD45RO on CD4+ T cells had a similar trend as HLA-DR (Figure 2, E and G). In contrast, the costimulatory molecule CD28 expressed on CD4+ and CD8+ T cells was decreased in severe and extremely severe patients. CD28 expression on CD4+ T cells in extremely severe patients was significantly lower than in severe patients (Figure 2, B and G). Furthermore, the expression of PD-1 on both CD4+ and CD8+ T cells was significantly increased in extremely severe patients compared with mild patients (Figure 2, C and G). The expression of Tim-3 on CD4+ and CD8+ T cells also had an increased trend in extremely severe patients, but no significant difference was found among the 3 groups (Figure 2, D and G). Although the percentage of Tregs had no difference among the 3 groups, the percentage of natural Tregs (CD45RA+ Tregs) was significantly decreased in severe patients, especially in those with extremely severe illness, when compared with mild patients (Figure 2, F and G).
Figure 2. The phenotypes and subsets of T cells.
(A–E) FACS dot plots showing the expression of HLA-DR (A), CD28 (B), PD-1 (C), Tim-3 (D) on CD4+ and CD8+ T cells, and the expression of CD45RA and CD45RO (E) on CD4+ T cells in representative patients with different severity of illness. (F) FACS dot plots showing the gating of Tregs, and the expression of CD45RA and CD45RO on Tregs in representative patients with different severity of illness. (G) The percentages of HLA-DR+, CD28+, PD-1+, and Tim-3+ cells in CD4+ or CD8+ T cells, the percentage of CD45RO+ cells in CD4+ T cells, the percentage of Tregs in lymphocytes, and the percentage of CD45RA+ Tregs in lymphocytes in patients with different groups are shown in graphs (mean ± SD). *P < 0.05, **P < 0.01, ***P < 0.001 (1-way ANOVA).
Lymphocyte function analysis.
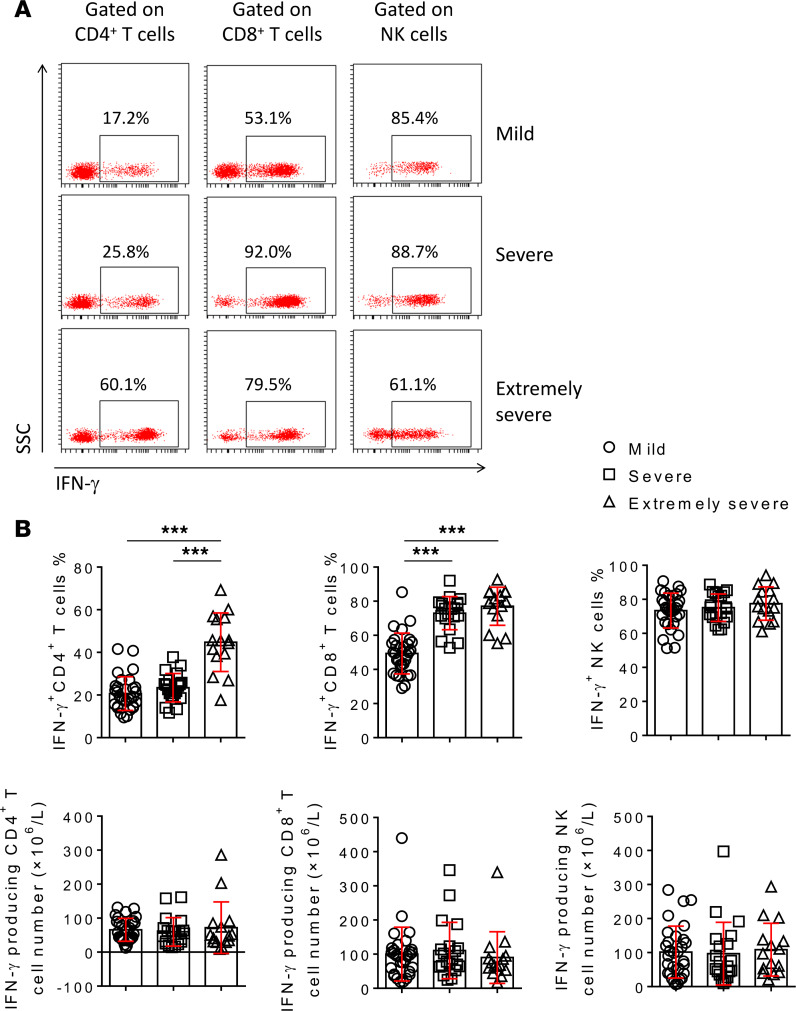
The results of PMA/ionomycin-stimulated lymphocyte function were approximately consistent with the results of phenotype analysis. In particularly, the percentage of IFN-γ–producing CD4+ T cells in extremely severe patients was much higher than in both mild and severe patients. The percentage of IFN-γ–producing CD8+ T cells in both severe and extremely severe patients was significantly higher than in mild patients, but the percentage of IFN-γ–producing CD8+ T cells had no difference between severe and extremely severe group. The percentage of IFN-γ–producing NK cells had no difference among the 3 groups (Figure 3, A and B). Although the percentages of IFN-γ–producing CD4+ and CD8+ T cells in severe and extremely severe patients were increased, the absolute numbers of them had no difference among these 3 groups (Figure 3B). These data suggest that the increased IFN-γ–producing ability of T cells, especially the CD4+ T cells, is associated with the pathogenesis of extremely severe SARS-CoV-2 infection.
Figure 3. The IFN-γ–producing ability of CD4+ T cells, CD8+ T cells, and NK cells.
PMA/ionomycin-stimulated lymphocyte function assay was performed in patients with different severity of illness. (A) FACS dot plots showing the producing of IFN-γ in CD4+ T cells, CD8+ T cells, and NK cells in representative patients with different severity of illness after stimulation. (B) The percentages of IFN-γ+ cells in CD4+ T cells, CD8+ T cells, and NK cells, and the absolute numbers of IFN-γ producing CD4+ T cells, CD8+ T cells, and NK cells are shown in graphs (mean ± SD). ***P < 0.001 (1-way ANOVA).
Cytokine profiles.
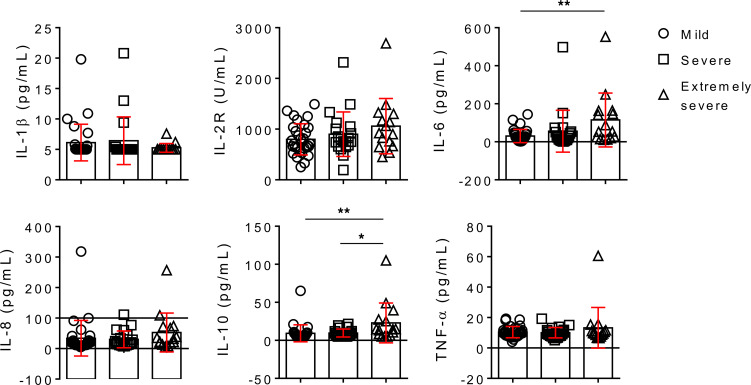
There are no differences of IL-1β, IL-8, and TNF-α in COVID-19 patients with different severity of illness. The level of IL-2 receptor (IL-2R) was gradually increased with increased severity of illness but had no significant difference among the 3 groups. The levels of IL-6 and IL-10 were also gradually increased with increased severity of illness, and the levels of IL-6 and IL-10 in extremely severe patients were significantly higher than in mild patients (Figure 4).
Figure 4. Cytokine profile analysis.
The serum levels of IL-1β, IL-2R, IL-6, IL-8, IL-10, and TNF-α in different groups are shown in graphs (mean ± SD). *P < 0.05, **P < 0.01 (1-way ANOVA).
DC and B cell subset analysis.
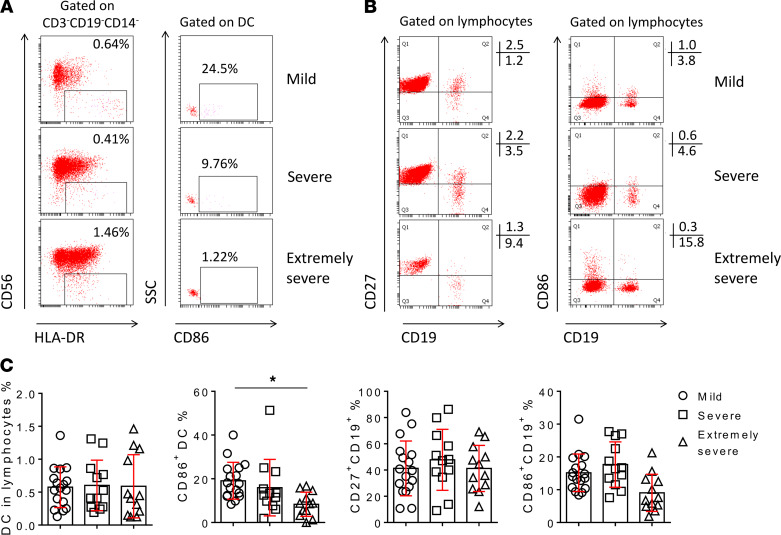
Although the percentage of DC in lymphocytes had no difference in patients with different severity of illness, the expression of CD86 on DC was significantly decreased in extremely severe patients compared with mild patients (Figure 5, A and C). Similarly, the expression of CD86 on B cells had a decreased trend in extremely severe patients, but no significant difference was found among these groups. The percentage of memory B cells (CD27+CD19+) had no difference among the 3 groups (Figure 5, B and C).
Figure 5. DC and B cell subsets.
(A) FACS dot plots showing the gating of DC and the expression of CD86 on DC in representative patients with different severity of illness. (B) FACS dot plots showing the expression of CD27 and CD86 on CD19+ B cells in representative patients with different severity of illness. (C) The percentages of DC in lymphocytes, CD86+ cells in DC, and CD27+ and CD86+ cells in B cells in different groups are shown in graphs (mean ± SD). *P < 0.05 (1-way ANOVA).
Discussion
SARS-CoV-2 infection has become an urgent public health challenge in China and throughout the world. Most infected patients have mild illness and are recovered after 2–3 weeks. However, once infected patients progress to severe illness with acute respiratory distress syndrome, over 10% of them worsen in a short period of time and die of multiple organ failure (7). As the pathogenesis of SARS-CoV-2 infection is unknown, there is no standard treatment and most patients receive symptomatic treatment. Thus, a further understanding of the pathogenesis of the disease is urgently needed. The most important finding of this study is that the hyperfunction of CD4+ and CD8+ T cells is associated with the pathogenesis of extremely severe SARS-CoV-2 infection.
Previous study has found that some routine laboratory biomarkers are out of reference ranges and are higher in more severe disease (6). In accordance with this study, we also found that some biomarkers such as lactate dehydrogenase and D-dimer were higher in severe illness, but these biomarkers were not all specific to SARS-CoV-2. Similar to other viral infection, neutrophils and procalcitonin had no obvious change in mild patients, while inflammatory biomarkers such as CRP and erythrocyte sedimentation rate were increased. These data suggest that the pathogenesis of extremely severe SARS-CoV-2 infection is involved with the disorder of inflammatory response.
T lymphocytes, especially CD8+ cytotoxic T cells, are the most important immune cells to protect against viral infection (15–17). One of the typical characteristics of SARS-CoV-2 infection is decreased number of lymphocytes, which is in accordance with previous findings showing that the number of lymphocytes could be used as a reference index in the diagnosis of SARS-CoV-2 infection in the clinic (7). We further found that both CD4+ and CD8+ T cells were gradually decreased with increased severity of illness. The previous concept is that a substantial decrease in the total number of lymphocytes indicates that SARS-CoV consumes immune cells and inhibits the body’s cellular immune function, and damage to T lymphocytes might be an important factor leading to exacerbations of patients (18).
We also believe the function of lymphocytes in the final stage of COVID-19 patients is in the status of inhibition, as exuberant inflammatory responses can promote T cell apoptosis. However, before this happened, we speculate that lymphocytes are in the status of hyperfunction. In the early stages of disease, the migration of lymphocytes from blood to the lung may be the cause of lymphocyte deficiency in peripheral blood, which is mediated by antigenic stimulation. The phenotype and function results of T cells support the speculation. We found that activation marker HLA-DR was increased and that costimulatory molecule CD28 was decreased in patients with severe illness, which suggests the activation of T cells in severe patients. The expressions of PD-1 and Tim-3 on CD8+ T cells were also increased in extremely severe patients, which is in accordance with previous studies showing that PD-1 and Tim-3 can also be used as markers of acute T cell activation (19–21). The lymphocyte function assay further demonstrated that the IFN-γ–producing ability of CD8+ T cells was increased in severe patients. If the activation of CD8+ T cells cannot eliminate the virus, CD4+ T cells will be activated to further increase immune responses. Thus, we found that the IFN-γ–producing ability of CD4+ T cells was also remarkably increased in extremely severe patients. We speculate that the hyperfunction of CD4+ T cells will initiate macrophage activation syndrome, which leads to cytokine storm in extremely severe patients.
The increased level of IL-2R supports the hyperfunction of CD4+ T cells. The loss of natural Tregs and increased level of IL-10 also support the hyperfunction of host immunity in extremely severe patients. Regarding why older people have higher mortality after infection, one possible reason is that they have low storage of lymphocytes, especially the CD8+ T cells (22). As insufficient numbers of lymphocytes migrate to infection sites, the host might compensatorily increase the function of CD4+ T cells, which finally induces the occurrence of cytokine storm. It also emphasizes that evaluation of host immunity plays an important role in the treatment of immune-related diseases.
We hypothesize that the host immunity in severe COVID-19 patients might have the following characteristics. First, antigen presenting cells may be in the state of consumption. The number of B cells is decreased, and the activation of DC and B cells is decreased. Second, the number and function of T cells may be inconsistent. Although the numbers of CD4+ and CD8+ T cells are decreased, the IFN-γ–producing ability of both them is increased, especially the increased ability of CD4+ T cells. The increase of IL-2R supports the hyperfunction of CD4+ T cells in the pathogenesis of extremely severe illness. Third, negative immune regulation may be activated. The number of Tregs, especially the natural Tregs, is consumed. The increase of IL-10 in extremely severe patients may support the activation of Tregs. However, the characteristics of host immunity in extremely severe patients have not always been like this. Given that the continuous and overwhelming inflammatory responses will finally induce the apoptosis of lymphocytes, lymphocyte anergy will occur in the later stages of infection. Thus, the function of lymphocytes can be completely different in different stages of infection. This could be the reason why the results of lymphocyte function in our research are different from previous published results (17, 18).
Three limitations should be mentioned. First, the number of patients in this study is relatively small. The results should be validated in another prospective study. Second, we only analyzed peripheral blood lymphocytes. A further analysis of lymphocytes from alveolar lavage fluid is needed. Third, the expression of some immunological markers, such as CD28 and CD45RO, is different in different age stages (23). Therefore, continuously monitoring host immunity in the same patients is needed. However, there was no significant difference in age between severe and extremely severe patients.
This study suggests that the hyperfunction of CD4+ and CD8+ T cells is associated with the pathogenesis of extremely severe SARS-CoV-2 infection.
Methods
Patients.
This study was carried out in January 2020 at Tongji Hospital (Wuhan, China). The suspected patients who had typical clinical symptoms (fever, cough, or shortness of breath) and radiological characteristics (unilateral pneumonia, bilateral pneumonia, or ground-glass opacity) were admitted to the fever ward of Tongji Hospital. Then, the patients with positive SARS-CoV-2 real-time RT-PCR results aged 18–80 years were continuously recruited.
The routine laboratory tests including blood routine, biochemistry, coagulation function, and infection biomarkers (procalcitonin, erythrocyte sedimentation rate, ferritin, CRP) were performed in each patient after patient admission. The host immunity was simultaneously evaluated by analysis of TBNK lymphocyte number; PMA/ionomycin-stimulated lymphocyte function; T cell, B cell, and DC subsets; and cytokine profiles, as described in Methods. The treatment of patients was mainly based on symptomatic treatment, as there is currently no effective antiviral therapy for SARS-CoV-2. The demographic and clinical information, laboratory results, and outcome data were finally collected from electronic medical records.
Grouping criteria.
The COVID-19 patients with different severity of illness were classified into 3 groups according to the guideline of diagnosis and treatment for SARS-CoV-2 pneumonia made by the Chinese National Health Commission as follows: (a) mild illness, in which the patients had typical symptoms and radiological findings; (b) severe illness, in which the patients met one of the following criteria: respiratory distress (respiration rate ≥ 30 times/min), oxygen saturation (SpO2) ≤ 93% in the resting state, and arterial partial pressure of O2 and the fraction of inspired oxygen (PaO2/FiO2) ratio ≤ 300 mmHg; and (c) extremely severe illness, in which the patients met 1 of the following criteria: respiratory failure requiring mechanical ventilation, shock, and organ failure needing intensive care unit (ICU) treatment. The classification of patients was performed by 2 clinicians: one was from Department of Respiratory and Critical Care Medicine, and the other was from ICU.
Real time RT-PCR.
Throat-swab specimens from the upper respiratory tract that were obtained from patients at admission were maintained in viral-transport medium. SARS-CoV-2 was confirmed by using TaqMan One-Step RT-PCR Kits from Shanghai Huirui Biotechnology Co. Ltd and Shanghai BioGerm Medical Biotechnology Co. Ltd, both of which have been approved by the China Food and Drug Administration. Briefly, RNA was extracted from clinical samples. A total of 5 μL of RNA was used for real-time RT-PCR, which targeted the ORF1ab and N gene. Real-time RT-PCR was performed using the following conditions: 50°C for 15 minutes and 95°C for 5 minutes, 45 cycles of amplification at 95°C for 10 seconds, and 55°C for 45 seconds. The positive SARS-CoV-2 real-time RT-PCR result was defined if both ORF1ab and nucleoprotein cycle thresholds were < 35.
Flow cytometry analysis.
Heparinized peripheral blood was collected from study participants. Fluorescence-labeled mAbs were added to label surface markers on stimulated or unstimulated cells. For labeling intracellular antigens, the cells were fixed and permeabilized with Fixation/Permeabilization Buffer (BD Biosciences) and then stained with mAbs. Isotype controls with irrelevant specificities were included as negative controls. The cell pellets were finally analyzed with FACSCanto flow cytometer (BD Biosciences).
TBNK lymphocyte counting.
The percentages and absolute numbers of CD4+ T cells, CD8+ T cells, B cells, and NK cells were determined by using TruCOUNT tubes and BD Multitest 6-color TBNK Reagent Kit (BD Biosciences) according to the manufacturer’s instructions. In brief, 50 μL of whole blood was labeled with 6-color TBNK Ab cocktail for 15 minutes in room temperature. After adding 450 μL of FACS lysing solution, samples were analyzed with FACSCanto flow cytometer using FACSCanto clinical software (BD Biosciences).
T cell subsets.
The following mAbs were added to 100 μL of whole blood: anti-CD45 (catalog 652803), anti-CD3 (catalog 663490), anti-CD4 (catalog 560345), anti-CD8 (catalog 335822), anti-CD28 (catalog 662797), anti–HLA-DR (catalog 652809), anti-CD45RA (catalog 662840), anti-CD45RO (catalog 663530), anti-CD25 (catalog 662525), anti-CD127 (catalog 560822), anti–PD-1 (catalog 557860), and anti–Tim-3 (catalog 565558) (all from BD Biosciences). Different T cell subsets were defined as following: costimulatory molecule (CD28+CD4+ or CD28+CD8+ T cells), activated T cells (HLA-DR+CD4+ or HLA-DR+CD8+ T cells), naive/memory CD4+ T cells (CD45RA+/CD45RO+ CD4+ T cells), Tregs (CD25hiCD127loCD4+ T cells), and natural Tregs (CD45RA+ Tregs). PD-1 and Tim-3 were also used as activation markers of T cells. Gating strategies of T cell subsets are shown in Supplemental Figure 1, A, B, and F.
B cell subsets.
The following mAbs were added to 100 μL of whole blood: anti-CD45 (catalog 662912), anti-CD19 (catalog 341113), anti-CD27 (catalog 662839), and anti-CD86 (catalog 555660) (all from BD Biosciences). Different B cell subsets were defined as following: memory B cells (CD27+CD19+ cells) and activated B cells (CD86+CD19+ cells). Gating strategies of B cell subsets are shown in Supplemental Figure 1E.
DC subsets.
The following mAbs were added to 100 μL of whole blood: anti-CD45 (catalog 652803), anti-CD3 (catalog 663490), anti-CD19 (catalog 349209), anti-CD14 (catalog 564054), anti-CD56 (catalog 663487), anti–HLA-DR (catalog 652827), and anti-CD86 (catalog 555660) (all from BD Biosciences). The percentage of CD3–CD19–CD14–CD56–HLA-DR+ cells in CD45+ lymphocytes was defined as the percentage of DC. The expression of CD86 on DC was defined as activated DC. Gating strategies of DC subsets are shown in Supplemental Figure 1D.
Lymphocyte function.
PMA/ionomycin-stimulated lymphocyte function assay was performed as described in our previous studies (22, 24). Briefly, 100 μL of whole blood was diluted with 400 μL of IMDM medium and stimulated with Leukocyte Activation Cocktail (containing PMA, ionomycin and brefeldin A, BD GolgiPlug) for 4 hours. The cells were labeled with mAbs (anti-CD45, anti-CD3, anti-CD4, anti-CD8, and anti-CD56) and then were fixed and permeabilized. The cells were stained with intracellular anti–IFN-γ (catalog 554702) Ab and were analyzed with flow cytometry. Gating strategies of lymphocyte function assay are shown in Supplemental Figure 1C.
Cytokine profile analysis.
Serum samples were collected from study participants. The levels of IL-1β, IL-2R, IL-8, IL-10, and TNF-α in serum were measured according to an automatic procedure of a solid-phase 2-site chemiluminescent immunometric assay via IMMULITE 1000 Analyzer (Siemens). The level of IL-6 was measured by the electrochemiluminescence method (Roche Diagnostics).
Statistics.
The results are presented as mean ± SD. Continuous variables were compared with 1-way ANOVA. Statistical significance was determined as P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001). Statistical analyses were performed using SPSS version 19.0.
Study approval.
This study was approved by the ethical committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. All patients gave written informed consent.
Author contributions
FW, HH, and ZS designed the study. FW, HH, YL, GT, SW, MH, WL, YZ, QL, and LM performed experiments. MF and HZ collected clinical information and classified patients. FW, HH, and ZS performed statistical analysis and drafted the manuscript. All authors contributed to data acquisition, data interpretation, and reviewed and approved the final version of this manuscript.
Supplementary Material
Acknowledgments
This study was funded by the National Mega Project on Major Infectious Disease Prevention (2017ZX10103005-007) and the Fundamental Research Funds for the Central Universities (2019kfyRCPY098).
Version 1. 04/23/2020
In-Press Preview
Version 2. 05/21/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(10):e137799.https://doi.org/10.1172/jci.insight.137799.
Contributor Information
Feng Wang, Email: fengwang@tjh.tjmu.edu.cn.
Hongyan Hou, Email: 815739195@qq.com.
Ying Luo, Email: luoying_2017@163.com.
Guoxing Tang, Email: 15971449367@163.com.
Shiji Wu, Email: wilson547@163.com.
Min Huang, Email: huangminry2013@163.com.
Weiyong Liu, Email: weiyongliu@gmail.com.
Yaowu Zhu, Email: yaowu_zhu@163.com.
Qun Lin, Email: 278231949@qq.com.
Liyan Mao, Email: 862200862@qq.com.
Minghao Fang, Email: fangmh@tjh.tjmu.edu.cn.
Huilan Zhang, Email: huilanz_76@163.com.
Ziyong Sun, Email: zysun@tjh.tjmu.edu.cn.
References
- 1.Zhu N, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JF, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Lancet Emerging understandings of 2019-nCoV. Lancet. 2020;395(10221):311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassetti M, Vena A, Giacobbe DR. The novel Chinese coronavirus (2019-nCoV) infections: Challenges for fighting the storm. Eur J Clin Invest. 2020;50(3):e13209. doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu A, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao S, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei J, Li J, Li X, Qi X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology. 2020;295(1):18. doi: 10.1148/radiol.2020200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94(7):e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt ME, Varga SM. The CD8 T Cell Response to Respiratory Virus Infections. Front Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baazim H, et al. CD8+ T cells induce cachexia during chronic viral infection. Nat Immunol. 2019;20(6):701–710. doi: 10.1038/s41590-019-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeidi A, et al. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Front Immunol. 2018;9:2569. doi: 10.3389/fimmu.2018.02569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu WJ, et al. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau J, et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat Commun. 2017;8:14572. doi: 10.1038/ncomms14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA. 2002;99(19):12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amancha PK, Hong JJ, Ansari AA, Villinger F. Up-regulation of Tim-3 on T cells during acute simian immunodeficiency virus infection and on antigen specific responders. AIDS. 2015;29(5):531–536. doi: 10.1097/QAD.0000000000000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, et al. Combination of lymphocyte number and function in evaluating host immunity. Aging (Albany NY) 2019;11(24):12685–12707. doi: 10.18632/aging.102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin L, et al. Aging of immune system: Immune signature from peripheral blood lymphocyte subsets in 1068 healthy adults. Aging (Albany NY) 2016;8(5):848–859. doi: 10.18632/aging.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou H, et al. Establishment of the Reference Intervals of Lymphocyte Function in Healthy Adults Based on IFN-γ Secretion Assay upon Phorbol-12-Myristate-13-Acetate/Ionomycin Stimulation. Front Immunol. 2018;9:172. doi: 10.3389/fimmu.2018.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.