Abstract
Background
Evolutionary studies have been conducted that have investigated the chromosomal variance in the genus of Chlamydia. However, no all-encompassing genus-wide comparison has been performed on the plasmid. Therefore, there is a gap in the current knowledge on Chlamydia plasmid diversity.
Aims
This project is aimed to investigate and establish the nature and extent of diversity across the entire genus of Chlamydia, by comparing the sequences of all currently available plasmid carrying strains.
Methods
The PUBMED database was used to identify plasmid sequences from all available strains that met the set quality criteria for their inclusion in the study. Alignments were performed on the 51 strains that fulfilled the criteria using MEGA X software. Following that Maximum Likelihood estimation was used to construct 11 phylogenetic trees of the whole plasmid sequence, the individual 8 coding sequences, the iteron and a chromosomal gene ompA as a comparator.
Results
The genus-wide plasmid phylogeny produced three distinct lineages labelled as alpha, beta and gamma. Nineteen genotypes were found in the initial whole plasmid analysis. Their distribution was allocated as six C. pecorum, two C. pneumoniae, one C. gallinacea, one C. avium, one C. caviae, one C. felis, two C. psittaci, one C. trachomatis, one C. muridarum, and two C. suis. The chromosomal comparative gene ompA supported this distribution, with the same number of primary clades with the same species distribution. However, ompA sequence comparison resulted in fewer genotypes due to a reduced amount of available sequences (33 out of 51). All results were statistically significant.
Conclusion
The results of this study indicate that the common bacterial ancestor of all the species had a plasmid, which has diverged over time. Moreover, it suggests that there is a strong evolutionary selection towards these species retaining their plasmids due to its high level of conservation across the genus, with the notable exception of C. pneumoniae. Furthermore, the evolutionary analysis showed that the plasmid and the chromosome have co-evolved.
Introduction
The Chlamydia are a distinct genus of pathogenic bacteria that can cause infections in humans and animals. Within this genus there are currently eleven recognised species [1] namely: Chlamydia abortus, C. avium, C. caviae, C. felis, C. gallinacea, C. muridarum, C. pecorum, C. pneumoniae, C. psittaci, C. suis, and C. trachomatis. C. trachomatis and C. pneumoniae are the most common chlamydial infections in humans, but zoonotic infections of C. psittaci and C. abortus [1] also occur. C. abortus is endemic among ruminants such as cows and sheep, but can also cause abortion in humans and other mammals. C. pneumoniae is a major cause of community-acquired pneumonia in humans, while C. psittaci is the cause of parrot fever (psittacosis), which may present as atypical pneumoniae that can mimic typhoid fever, but is often asymptomatic [2]. C. trachomatis is the most common bacterial sexually transmitted infection (STI) in humans [3], and it is the leading cause of preventable blindness in developing countries (trachoma) [4]. Less common syndromes and diseases caused by C. trachomatis include lymphogranuloma venereum [5] and reactive arthritis [6]. The remaining species have not been shown to cause disease in humans, but they can infect birds (C. avium and C. gallinacea), cats (C. felis), koalas (C. pecorum), rodents (C. muridarum) and swine (C. suis and C. pecorum).
All Chlamydia have a chromosome of around 1 Mbp. This is supplemented by a roughly 7.5 kbp circular plasmid in all species apart from C. abortus [7]. In the majority of microbial organisms, plasmids have a role in genetic variation and encode environment specific information. This can include antibiotic resistance which can be horizontally transferred and distributed among strains. Currently, there is no evidence of antibiotic resistance located on chlamydial plasmids. Plasmids are present in nine non-trachomatis species of Chlamydia (C. avium [8], C. caviae [9], C. felis [10], C. gallinacea [8], C. muridarum [11], C. pecorum [12], C. pneumoniae [13–15], C. psittaci [16, 17], and C. suis [18–21] and the human pathogen C. trachomatis [1]. Most chlamydial plasmids are 7.5 kbp in length with some exceptions [15, 22, 23]. They contain non-coding RNA with unknown functionality and 8 coding sequences (CDS) [24].
The origin of replication in chlamydial plasmids is thought to be regulated by iterons. Iterons are short repeated DNA sequences occurring between 3–5 iterations that play a crucial role in promoting replication of the plasmid [25]. Most Chlamydia species have four tandem repeat sequences, with each iteration comprising 22 base pairs [23, 26]. In Chlamydia, these repeat sequences have been described as mainly AT rich and have shown to be the most conserved regions by phylogenetic studies (reference?). This supports the notion that they play an important and conserved role in the regulation of plasmid replication.
Plasmid diversity within the genus Chlamydia was first investigated in twelve C. psittaci strains and with the prototype C. trachomatis L2 strain. Homology was recognised among the strains of C. psittaci [23]. However, he plasmid of C. trachomatis was the first characterised and consequently the most studied plasmid from the genus Chlamydia. Further studies established that these plasmids are present in most strains of C. trachomatis. The plasmid is highly conserved, with an intraspecific variation of around 3% [27]. Large-scale deletions or rearrangements of the plasmid are therefore rare [27, 28], but in 2006 a new strain was described that evaded PCR detection due to a single large deletion in CDS1 of its plasmid [29]. In 2018 157 whole genome sequences of C. trachomatis (including plasmid) isolates were deposited into the pubMLST database, where 902 genes were identified.[30] This was followed by the development of the MLST plasmid scheme that permitted the investigation of allelic variance. The results identified 6 plasmid clusters along with 4 chromosomal clusters. A close relationship was noted between the chromosomal genome, plasmid type and the diseases that they caused. They concluded that this would point towards co-evolution of the chromosome of C. trachomatis and their plasmids [31, 32].
Comparison of C. psittaci plasmid pCpA1 (avian), C. pneumoniae pCpnE1 (N16, equine) and C. trachomatis pMoPn (the agent of mouse pneumonia–this has been re-categorised into C. muridarum) and pLGV440 (human) further expanded the field [26]. This study showed maintenance of plasmid size at 7.5 kbp, although there was a single 200 nucleotide deletion in CDS1 of pCpnE1, which split it into two almost equal halves labelled ORF1A and ORF1B (CDS1A and CDS1B). This 200-nucleotide deletion proved to be the largest point of divergence from the other 3 strains. Furthermore, the analysis established the conservation of start codons for the 8 CDS’s between the plasmids: ATG (Methionine) for 1, 3, 4, 5 and 6, and GTG (Valine) for CDS 7 and 8. However, there is still debate over the actual start sequences of CDS 7 and 8 [26]. This highlights the ambiguity in the variance among these plasmids. The phylogenetic analysis concluded that MoPn was more closely related to C. trachomatis L1 (80%) than either the avian or equine strains.
The most extensive (non-trachomatis) phylogenetic study has been performed on C. pecorum plasmids [26]. In total 21 strains were used to characterise and compare the structure of the plasmids. The results showed a highly conserved plasmid with a low G+C content (31.6%). They varied in either 7,547 or 7,548 bp in length. The alignment of the 21 strains resulted in 12 unique plasmid genotypes labelled alphabetically from A to L. The 11 koala strains were identified into five genotypes, with SAK09Ure, SAK84Ure, and VicR6UGT Genotype A; NoHerEyes, PMHaUre, TedHUre and DbDeUG Genotype B; HazBoEye and HazBoUgt Genotype C; finally, Marsbar and IpTale Genotypes D and E, respectively. The three porcine strains were all Genotype L (R106, L1 and 1886). The four ovine strains were allocated into 3 plasmid genotypes, with pCpecs CurE11Rec and CurE19Rec labelled as Genotype F, while remaining strains (W73 and IPA) were allocated into Genotypes G and H. The three bovine strains (WAB31Ileal, 66P130, and LW623) were of a unique genotypes, labelled I, J and K, respectively [12, 26].
Most recently, an epidemiological study of C. gallinacea compared a recently isolated C. gallinacea strain Jx-1 with type strain 08-1274/3 [33]. Jx-1 was found to contain a plasmid which was sequenced and deposited into GenBank. pJx-1 shared 99.9% identity to p1274 with only two point mutations at position 6573 (C to T) and 7170 (C to A) in PGP 6 reflecting the close relatedness of these two strains.
Phylogenic studies have been performed both within and between species, however, at the present time no all-encompassing genus-wide comparison has been performed. Advances in sequencing technology means there is now an abundance of plasmid sequences available for all known species in public databases. It is time to draw all this information together. Therefore, the aim of this study was to investigate the nature and extent of diversity across the non-trachomatis chlamydial species, by analysing all currently published plasmid-carrying strains.
Materials and methods
Plasmid sequence data
Whole genome sequences of non-trachomatis species that were available in PubMed’s Nucleotide database were included for analysis. Furthermore, due to the large deletion in the only available peer-reviewed WGS of C. suis strain MD56, 3 directly submitted (unpublished) C. suis strains were used to provide greater insight into the significance of the deletions present on MD56. In total 50 non-trachomatis Chlamydia strains have been identified to have a plasmid (Table 1). The C. trachomatis LGV440 (L2) was included for comparative reasons, as it has been widely studied.
Table 1. List of all the Chlamydial species that are being used for the study.
Relevant information on length of plasmid, accession number, PUBMED ID and source provided.
| Genus and Strain | Length in BP | PUBMED | Acession |
|---|---|---|---|
| C. avium | |||
| 10DC88 | 7099 | 24461712 | CP006572.1 |
| C. caviae | |||
| GPIC (pCpGP1) | 7966 | 12682364 | AE015926.1 |
| C. felis | |||
| C-56 (pCFe1) | 7552 | 16766509 | AP006862.1 |
| C. gallinacae | |||
| Jx-1 | 7492 | 29212448 | CP019793.1 |
| 08-1274/3 | 7619 | 24461712 | CP015841.1 |
| C. muridarum | |||
| Nigg (pMoPn) | 7501 | 10684935 | AE002162.1 |
| C. pecorum | |||
| 66P130 | 7548 | 26870613 | KT223766 |
| 1886 | 7548 | 26870613 | KT223767 |
| CurE11Rec | 7547 | 26870613 | KT223768 |
| CurE19Rec | 7547 | 26870613 | KT223769 |
| DbDeUG | 7547 | 26870613 | KT223770 |
| IPA | 7547 | 26870613 | KT223771 |
| IpTaLe | 7547 | 26870613 | KT223772 |
| LW623 | 7547 | 26870613 | KT223774 |
| SaK09Ure | 7547 | 26870613 | KT223777 |
| SaK84Ure | 7547 | 26870613 | KT223778 |
| VicR6UGT | 7547 | 26870613 | KT223779 |
| L1 | 7548 | 26870613 | KT223773 |
| R106 | 7548 | 26870613 | KT223776 |
| Marsbar | 7547 | 26870613 | KT223775 |
| W73 | 7547 | 26870613 | KT223780 |
| HazBoEye | 7547 | 26870613 | KT352920 |
| HazBoUGT | 7547 | 26870613 | KT352921 |
| NoHeEye | 7547 | 26870613 | KT352922 |
| PMHaUre | 7547 | 26870613 | KT352923 |
| TedHUre | 7547 | 26870613 | KT352924 |
| WaB31Ileal | 7548 | 26870613 | KT223781 |
| C. pneumoniae | |||
| N16 (pCpnE1) | 7368 | 9202459 | X82078.1 |
| B21 | 7533 | 24503994 | AZNB01000165.1 |
| LPCoLN | 7530 | 19749045 | CP001714.1 |
| C. psittaci | |||
| 84/55 | 7487 | 23209198 | CP003812 |
| CB7 | 7577 | 24903864 | JMBZ01000059.1 |
| HJ | 5258 | 25744990 | JPIH02000081.1 |
| 6BC | 7553 | 21441521 | CP002550.1 |
| Cal10 | 7553 | 21622741 | AEZD01000004.1 |
| CP3 | 7552 | 23209198 | AFVN01000005.1 |
| M56 | 7553 | 23209198 | CP003814.1 |
| NJ1 | 7552 | 23209198 | AFVK01000004.1 |
| WC | 7553 | 23209198 | CP003818.1 |
| VS225 | 7553 | 23209198 | CP003817.1 |
| MN | 7491 | 23209198 | CP003815.1 |
| WSRT30 | 7553 | 23209198 | CP003819.1 |
| DD34 | 7553 | 25887617 | AFVL01000005.1 |
| Frances | 7553 | 25887617 | AFVM01000003.1 |
| UGA | 7553 | 25887617 | AWXQ01000007.1 |
| RD1 | 7553 | 21183672 | FQ482150.1 |
| C. suis | |||
| MD56 | 5976 | 24812227 | AYKJ01000001.1 |
| SWA-2 | 7494 | Direct submission | LT821324.1 |
| SWA-14 | 7494 | Direct submission | LT860208.1 |
| SWA-86 | 7494 | Direct submission | LT860208.1 |
| C. trachomatis | |||
| LGV440 | 7500 | 2836808 | X06707.3 |
For each plasmid, their corresponding chromosomal major outer membrane protein (ompA) gene was selected for comparative phylogenetic analysis. This gene was chosen as it has been extensively studied with established variance present across interspecies analyses [31]. Thirty-three of the 51 plasmids sequences were identified with a corresponding ompA gene in the database.
Phylogenetic analysis
MEGA X (version 10.0.5) was used to execute alignments. Alignment were performed with ClustalW, following which a Maximum Likelihood tree was constructed for each CDS. The Maximum Likelihood model selects trees and branches based on the highest probability of them forming. The overall probability for each tree is presented in a log-likelihood function. General likelihood values are small, therefore applying them in log function will present a more distinguishable data set. However, these results will be negative due to the values being less than one in the function. This method assists in the reconstruction of the evolutionary history of these plasmids and might reveal possible recombination. The eight coding sequences (CDS) were individually compared, which made it possible to identify individual gene recombination events as branch swaps on the constructed phylogenies. The 51 whole plasmid sequences were then aligned and a ML phylogenetic tree was constructed. Finally, the same phylogenetic method was used to establish the diversity within the iteron across the extracted plasmids.
SnapGene (version 4.2.7) was used to extract each CDS from whole plasmid sequence data. An assumption was made that most of these genes would be labelled uniformly. The following species had the correct labelling C. caviae, C. felis, C. muridarum, and C. pneumoniae. The rest had errors ranging from incorrectly labelling CDS1 as CDS2 (all the C. pecorum strains), to missing labels which required the use of a diagram based on C. pneumoniae LPCoLN to establish the correct CDS (Table 1).
The phylogeny of all the alignments was deduced using the Maximum Likelihood method with the Tamura-Nei nucleotide substitution model with uniform rates. The phylogeny test was statistically supported by 1000 bootstrap replications to provide the probability of each branch's formation. Bootstrapping is a method of re-sampling each branch to provide a confidence to each outcome [34]. Essentially it illustrates the probability of each branch being recovered if the taxa were sampled multiple times. As recommended [34], the parameters were set for 1000 repetitions and the cut off value of x ≥70% was used, due to studies [35] showing that it corresponds to a 95% confidence that the clade is real. Thus, any value above 70% (0.7) would suggest a statistically significant result. For tree inference options the initial tree was automatically formed, and nearest-neighbour-interchange was applied for the heuristic method. No outgroup was selected for the analysis of the divergence.
Mega X was further used to perform the p-distance analysis on nucleotide level, with 1000 bootstrap repetitions to determine the standard error (SE). Gaps were considered as complete deletions. The model to determine substitutions was set as the p-distance method, with substitutions type selected as nucleotide and data to include transitions, transversions, and deletions.
Results
Structural composition of plasmids
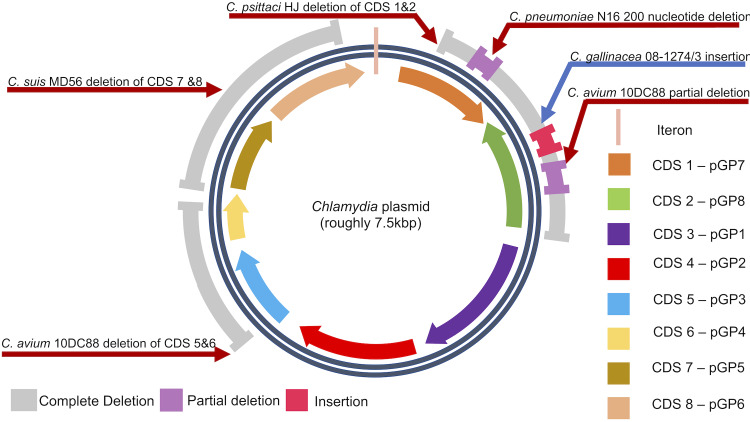
The alignments of the plasmid sequences provided the initial results on the organisational differences between strains. Structurally, most strains adhere to the currently accepted model for chlamydial plasmids (Fig 2). With a few exceptions, they all contain eight coding sequences (CDS) that can be found at very similar spatial locations. The direction of transcription for each gene is conserved with CDS2 being the only gene that is inversely transcribed.
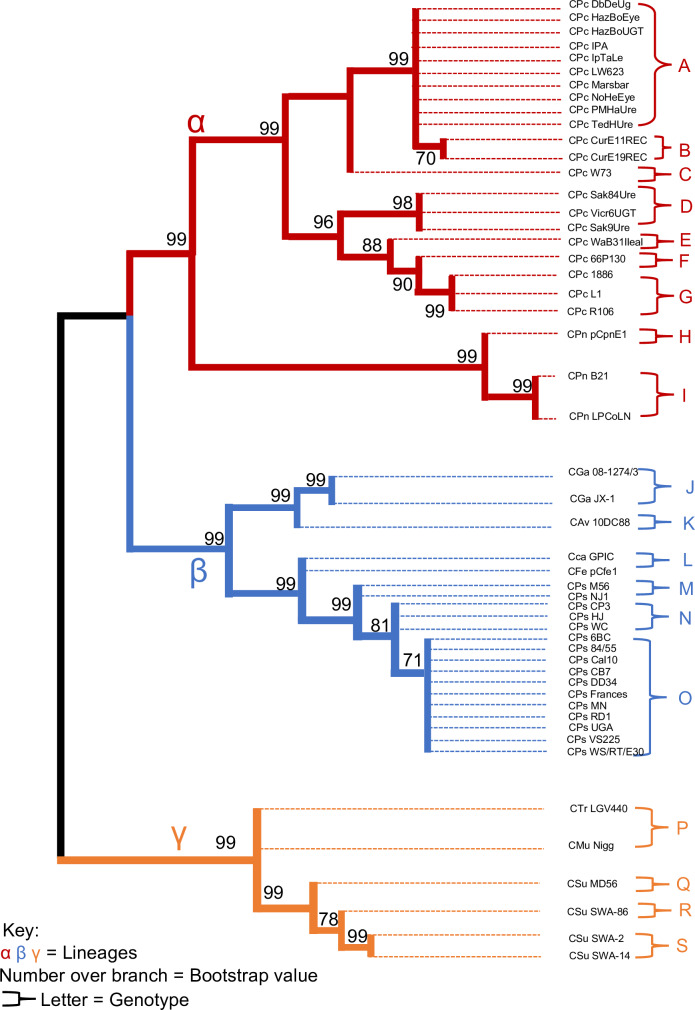
Fig 2. Maximum Likelihood estimate phylogeny for 51 Chlamydial plasmids with a log likelihood of log(-60175.44).
This conforms to the idea that these 8 CDSs play a significant role in the function of the bacteria, due to their preservation across all species. However, the current study indicates significant variation in the following strains: C. avium 10DC88, C. gallinacea 08-1274/3, C. pneumoniae N16, C. psittaci HJ, and C. suis MD56 (Fig 2). Complete gene deletion occurs in C. avium at CDS5 and CDS6.
It has been hypothesised that the plasmid has a role in virulence, with genes CDS5 and CDS6 being the most likely candidates, making them essential for infection and propagation in vivo [36] However, both CDS5 and CDS6 have been proven to be redundant in the survival of a plasmid, illustrated by the ability of C. avium 10DC88 to lose both of these genes. Furthermore, both CDS5 and CDS6 are dispensable for Chlamydia propagation in in vitro studies [37]. Moreover, C. avium 10DC88 has an early stop codon that partitions CDS2 into two separate genes, identified as 2A and 2B. Due to the absence of more examples of C. avium plasmid sequences, it is not possible to (a) check the validity of the data and (b) determine the origin of this mutation that gives rise to this early stop codon.
However, the whole plasmid phylogeny (Fig 1) provides us with the tool to investigate the nature of this deletion by examining the relatedness of C. avium’s sequences to those of C. gallinacea. When compared to C. gallinacea JX-1, there is a single nucleotide deletion that caused a frameshift and the codon for lysine is replaced with a stop codon. C. gallinacea 08-1274/03 also has an early stop codon in CDS 2, caused by a 62-nucleotide insertion into the middle of the gene. As in C. avium, the consequence of this is the formation of two separate genes labelled 2A and 2B. C. psittaci HJ has the largest deletion among all strains, resulting in the loss of CDS 1 and CDS 2, which makes it the smallest plasmid. Furthermore, C. suis MD56 has a complete deletion of CDS 7 and CDS 8, a feature which is absent in the directly submitted C. suis samples. This would suggest that this is not a fixed mutation in C. suis, and is instead unique to MD56. CDS 7 translates into a protein that is involved in the partitioning of the plasmid, whilst CDS 8 has a function in the control of replication. The apparent importance of these genes to plasmid maintenance and the absence of these mutations in the directly submitted sequences may lead us to question the authenticity of the C. suis MD56 sequence. However, in the event that the deletions in MD56 are genuine, they would indicate that those genes are redundant and that there may be failsafe mechanisms to substitute for those mutations in the plasmid. Finally, C. pneumoniae plasmid pCpnE1 contains a 200-nucleotide deletion in CDS 1 that has resulted in the production of two distinct CDSs labelled 1A and 1B. This was previously identified in 1997 [15], and the validity of this mutation is corroborated by a similar deletion in the Swedish New Variant C. trachomatis strain, supporting the notion that CDS2 compensates for loss of CDS1 activity [29].
Fig 1. General structure of chlamydial plasmids with annotated mutations and their strain names.
Whole plasmid phylogeny
The results for each phylogenetic tree are listed with their highest log likelihood, average p-distance with SE, and number of alleles constructed (Table 2). Each tree has been condensed to show only values of and above 70 (0.7). Therefore, all the clades that have been generated are statistically significant [35].
Table 2. Annotation of each element of the plasmid.
This includes the putative function and length of each element. The table includes the results of the average p-distance, alleles present and log likelihood of tree formation.
| Annotation | Hypothetical function | Base pair length range | Mean P-distance | SE | Alleles | Phylogeny log(x) |
|---|---|---|---|---|---|---|
| CDS1/pgp7 | Integrase | 252–1017 | 0.248 | 0.007 | 14 | -7012.49 |
| CDS2/pgp8 | Integrase | 450–1068 | 0.254 | 0.008 | 12 | -5960.31 |
| CDS3/pgp1 | Replicative DNA helicase | 1298–1422 | 0.26 | 0.006 | 14 | -10908.87 |
| CDS4/pgp2 | Virulence plasmid protein | 924–1065 | 0.172 | 0.007 | 14 | -6799.12 |
| CDS5/pgp3 | Virulence plasmid protein | 795–825 | 0.278 | 0.008 | 14 | -6281.03 |
| CDS6/pgp4 | Virulence plasmid protein | 306–209 | 0.181 | 0.012 | 8 | -1728.8 |
| CDS7/pgp5 | Plasmid partitioning protein | 627–810 | 0.254 | 0.01 | 15 | -5880.2 |
| CDS8/pgp6 | Plasmid replication protein | 252–1017 | 0.274 | 0.009 | 14 | -6252.29 |
| Repeat sequence | Iteron | 88bp(22x4 tandem repeat) | 0.017 | 0.005 | 4*(1 incomplete) | -215.66 |
| Whole Plasmid | - | 5258–7966 | 0.5 | 0.001 | 19 | -60175.44 |
| Chromosomal ompA | Membrane transporter | 1062–1209 | 0.24 | 0.008 | 15 | -11947.12 |
The entire plasmid phylogeny produced three distinct lineages that are labelled as alpha, beta and gamma (Fig 2). For the total number of 51 plasmids, the corresponding 19 genotypes were found in the initial whole plasmid analysis (Table 2). Their distribution was allocated as follows: seven C. pecorum (A, B, C, D, E, F and G), two C. pneumoniae (H and I), one C. gallinacea (J), one C. avium (K), one C. Caviae (L), one C. felis (M), two C. psittaci (N and O), one C. trachomatis (P), one C. muridarum (Q), and two C. suis (R and S). The chosen chromosomal comparator gene ompA had the same number of primary clades. However, it resulted in fewer genotypes, the total number being 15 for the 33 sequences that were available.
In terms of intraspecies analysis, the 21 individual C. pecorum strains were allocated into 7 distinct genotypes. C. pecorum porcine plasmids 1886, L1, and R106 were identified in the same clade and were annotated as genotype G with a 0.99 bootstrap value. The largest plasmid group, labelled genotype A, contained the following 10 strains; DbDeUG, HazBoEye, HazBoUGT, IPA, IpTaLe, LW623, Marsbar, NoHeEye, PMHaUre, and TedHUre. The remaining three Koala strains Sak09Ure, Sak84Ure, and Vicr6UGT were identified in the same genotype labelled D. A subclade was formed for the two ovine strains CurE11REC and CurE19REC, identified as genotype B. The remaining 3 strains (W73, WaB31Ileal, and 66P130) were identified as distinct plasmid types (C, E and F respectively).
The ompA phylogeny (Fig 3) resulted in the equal number of primary clusters (labelled alpha, beta, and gamma), which were all allocated with the same species as the whole plasmid phylogeny.
Fig 3. The Maximum Likelihood estimate for 33 ompA strains with a log likelihood of log(-11947.22).
Coding sequence phylogenies
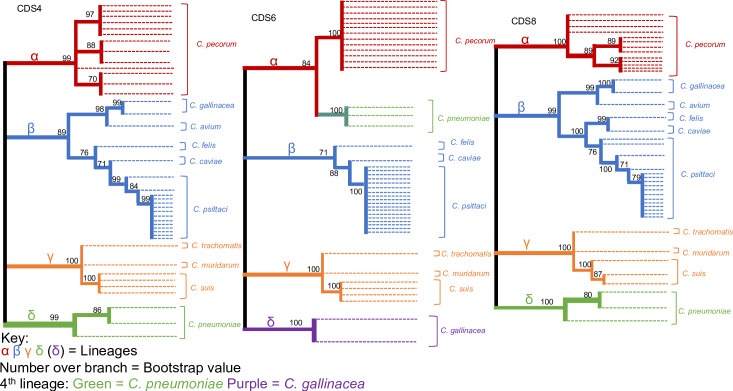
All 8 coding sequences were further analysed to obtain a higher resolution of the extent of divergence. The ML tree for the 8 genes was subcategorised into two groups (A&B) according to their number of lineages. Group A (Fig 4) accommodated CDS1, CDS2, CDS3, CDS5, and CDS7 (pgp 7, 8, 1, 3, and 5). All 5 genes supported 3 lineages, with CDS1, CDS3, and CDS5 mirroring the distribution of species seen in the whole plasmid phylogeny. The remaining two genes each had a major differentiating point that distinguished themselves as unique from the rest of the group and one another. CDS2 had the largest point of divergence with C. pneumoniae forming the third gamma lineage by itself, supported by a 1.00 bootstrap value. This led to the migration of the cluster that contains C. muridarum, C. suis, and C. trachomatis to the alpha lineage (0.87 bootstrap value), hence suggesting a closer relationship with C. pecorum species than the others. CDS7 had a less dramatic change to its structure with C. pneumoniae crossing over to the Beta cluster (1.00 bootstrap support), thus leaving C. pecorum on its own in the alpha cluster. Within group A, CDS2 had the lowest number of genotypes at 12. Conversely, CDS7 was identified to have the most genotypes with 15. The remaining three CDS’ (CDS1, CDS3 and CDS5) generated 14 unique genotypes.
Fig 4. The Maximum Likelihood estimate for gene group A with the following five coding sequences being present; CDS1, CDS2, CDS3, CDS5, and CDS7.
CDS4, CDS6 and CDS8 were allocated into Group B (Fig 5), which is categorised by the emergence of a 4th primary cluster, labelled delta. CDS 4 and 8 have C. pneumoniae as their 4th delta lineage, with a bootstrap value of 0.99 and 1.00 respectively. Considering the previously discussed branch swap in CDS7, and the unique cluster formation in CDS2, it could be possible that C. pneumoniae is a common ancestor from which other species have diverged. However, it is also possible that recombination has occurred with some of its genes.
Fig 5. The Maximum Likelihood estimate for gene group B with the following 3 coding sequences being present; CDS4, CDS6, and CDS8.
Both CDS 4 & 8 had 14 genotypes with each species having its own unique sequence. Analysis of CDS4 identified C. gallinacea as a 4th lineage with a 1.00 bootstrap value. C. pneumoniae was located on the alpha cluster, conforming to the structure of the whole plasmid phylogeny. CDS6 was divided into 8 sequence types, which was the lowest across all CDSs.
Iteron phylogeny
The iteron was shown to be the most highly conserved sequence of the plasmid across the species. 43 of the 51 plasmids were identified as identical with a 0.88 bootstrap value (Fig 6). These 43 strains consist of all the C. pecorum, C. psittaci, C. gallinacea, C. avium, C. caviae, C. felis plasmids and one C. pneumoniae strain pCpnE1 (N16) plasmid. The plasmids from the remaining C. pneumoniae strains formed a sub-clade with a 0.96 bootstrap value. C. suis formed its own clade with the strains SWA-2, SWA-14, and SWA-84. Due to a large deletion leaving it with a 2-tandem repeat, C. suis strain MD556 was identified within its own cluster. Finally, C. trachomatis and C. muridarum were shown to have a similar iteron. Despite the formation of 4 clades in the phylogenetic tree, the differences between these sequences are extremely minor as shown by the average p-distance of 0.017 (SE = 0.008) (Table 2), which is the lowest across all elements of the plasmid. The high degree of conservation of the iteron among chlamydial species strongly suggests its importance in the regulation of replication of Chlamydial plasmids.
Fig 6. The maximum likelihood estimate for 51 iteron plasmid strains with a log likelihood of log (-215.66).
Pairwise matrices
Pairwise matrix computation was performed on each CDS to determine the most and least conserved genes (Table 2). The average p-distance, with standard error (SE) in brackets (x), were the following (starting with CDS1 and ending with CDS8 in numerical order: 0.248 (0.007), 0.254 (0.008), 0.260 (0.006), 0.172 (0.007), 0.278 (0.008), 0.181 (0.012), 0.254 (0.01), and 0.274 (0.009). These results identified CDS4 as the most conserved and CDS5 as the least conserved gene across the sampled species. Due to mutations in CDS2, the test was recalculated with the elimination of the mutated strains to test the difference they make across the species. The results showed a very minor difference, with a p-distance of 0.246 and an unchanged SE.
Discussion
Advances in genome sequencing technology have provided significantly more data to enhance our understanding of the evolution and phylogeny of the chlamydial plasmid. The availability of open databases has provided us with seamless access to all currently sequenced strains.
The structural distribution of the genes in the plasmid, and their direction of transcription was conserved. This is a clear indicator that the plasmid is essential for the survival of the organism in plasmid-bearing species. However, the presence of mutations indicates that some parts of the plasmid are potentially redundant. The pairwise distance matrices analysis provides necessary information about which CDSs and genetic elements are the most conserved. This identified CDS4 as the most conserved (p-distance of 0.181 with SE = 0.012) and CDS5 (p-distance of 0.278 with SE = 0.008) as the least conserved gene across the genus (Table 2). This analysis conforms with the previously studied hypothesis that identified CDS5 (virulence associated protein) as the most polymorphic gene within C. trachomatis with a p-distance of 0.008 [30]. However, the same study identified CDS2 as the most conserved and least diverse gene on the plasmid with only 11 allelic variations within the C. trachomatis species [30], whilst a more recent study identified CDS6 as the most conserved plasmid gene in C. trachomatis across a much larger sample set (but CDS2 was the most conserved at the amino acid level) [27] Our calculations gave a p-distance of 0.254 and SE = 0.008 for CDS2, which was considerably higher compared to CDS4 with a p-distance of 0.181. One might argue that this difference could be due to two of the present species (C. avium 10DC88 and C. gallinacea 08-1274/3) possessing mutations. However, with their elimination, the mean distance only decreased to 0.246. The virulence associated CDS4 is the most conserved plasmid gene on a genus-wide spectrum. This means that CDS4 has an integral role in chlamydial survival and/or pathogenesis as nature selects the retention of this gene with little diversity.
Lineages
The phylogenetic tree for the whole plasmid sequences clearly illustrates three distinct and well-supported lineages, labelled as alpha, beta, and gamma (Fig 1). The alpha lineage consists of C. pecorum and C. pneumoniae. Our study was able to categorise the 21 strains of C. pecorum into 8 unique plasmid genotypes (alphabetically from A to L). This is different from a previous study [26], which predicted 12 plasmid types. This could be attributed to the differences in phylogenetic techniques used, as Bayesian inference can produce different results compared to Maximum likelihood estimations [34].
The koala LPCoLN and bandicoot B21 C. pneumoniae strains were clustered in the alpha clade (I), while equine pCpnE1 (N16) strain formed its own genotype H. From an evolutionary point of view, the alpha lineage is strongly rooted in domesticated mammals, with strains infecting bovine, equine, ovine and porcine hosts. Koalas have been infected with both C. pecorum and C. pneumoniae, despite being undomesticated wild animals. This supports the hypothesis that the koala habitat overlaps with farmland and transmission between domesticated (farm) animals and koalas is occurring [38].
The beta lineage contains C. avium, C. caviae, C. gallinacea, C felis, and C. psittaci. Within this group, C. avium and C. gallinacea were most closely related with a 0.99 bootstrap value. However, they ended up forming separate plasmid types, seen as J for C. gallinacea and K for C. avium. Phylogeny pointed towards three clades (M, N and O) within C. psittaci, of which clade O had the following 11 strains: pCps 6BC, 84/55, Cal10, CB7, DD34, Frances, MN, RD1, UGA, VS225, and WS/RT/E30. Clade M contained the pigeon strain HJ and muskrat strain M56. The remaining strains belonged to genotype N. Finally, both C. caviae and C. felis were shown to have a closer relationship with C. psittaci than the clade formed under C. avium and C. gallinacea, hence they were classified into clade L. This whole cluster mainly infects avian species such as poultry and psittacine birds. However, they also appear to be more related to cat and guinea pig specific Chlamydia. A deeper evolutionary history, in the form of more strains, is required to improve our understanding of how C. caviae and C. felis are related to avian strains.
The final lineage gamma was comprised of the following species; C. muridarum, C. suis, and C. trachomatis. C. muridarum and C. trachomatis were allocated into clade P. C. suis was shown to have 3 different plasmid types (Q, R and S). pCsu MD56 would obviously be presented as a unique plasmid (Q) due to the ≈2kbp deletion of CDS 7 and CDS 8. Of the remaining strains, SWA-2 and SWA-14 were predicted as identical (hence identified as plasmid S), and SWA-84 as a unique plasmid R. The relatedness of these three chlamydial species can be attributed to the habitat of their host species. As humans have progressed from their more tribal history, their contact with Muridae and Porcine species has increased. However, this study is limited in the ability to suggest the direction that evolution has taken. This can be improved by changing the technique used for the construction of the phylogenetic trees. Rather than using the bootstrap method to estimate the probabilistic significance of the results, average p-distance can be used with a rooted group. The rooted group would have to be an assumed or proven ancestor, which would provide information about the direction that natural selection has taken.
When compared to the ompA phylogeny (Fig 3) the maximum likelihood estimation resulted in a perfectly mirrored image of the whole plasmid, suggesting that the chromosome and plasmid have co-evolved. This is consistent with earlier studies that have investigated the diversity within the C. trachomatis species[32]. However, a caveat is that the smaller sample size of the ompA genes is a limiting factor in this analysis. Due to the presence of only 33 sequences out of the possible 51 (at the time of this study), it would be a challenge to determine how well the genotypes of ompA sequences compare to that of the plasmid. For future work, sequencing of the ompA of the 18 missing strains would help form a more accurate representation of the similarity between the chromosome and plasmid diversity.
Irrespective of lineage, all Chlamydia species carrying a plasmid have consistently had a unique plasmid genotype compared to other species. This further supports the notion that Chlamydial plasmids are highly conserved on the level of the entire plasmid and that they have co-evolved with the chromosome[32]. However, this type of preservation is not unanimously present when analysing single genes. The individual CDS ML estimations provided us with two distinct phylogeny types that were allocated as Group A (Fig 4) and Group B (Fig 5). These two phylogeny types were determined by the number of primary clusters they generated. Group A closely resembled the structure of the whole plasmid with 3 lineages, while Group B has an extra 4th cluster. The main discussion point for both groups is the nature of C. pneumoniae’s branch swap, and whether it is a main ancestor and point of evolutionary direction. This study is aimed at identifying points of divergence, therefore rooting of another hypothesised common ancestor of Chlamydia would be required to answer this question. However, these results demonstrate that the C. pneumoniae plasmid is unique compared to the others as it can be located in different lineages dependant on the gene.
Plasmid loss and acquisition
There is no evidence for acquisition of a specific plasmid from another chlamydial species based on the current animal chlamydial plasmids in the databases, this may be because of the relatively small sample size and that such events are rare and have yet to be uncovered; indeed, sequencing of a much larger sample of C. trachomatis genomes revealed occasional evidence for plasmid transfer between clades [39]. In the most well studied biological situation, we know that C. trachomatis plasmids cannot by themselves replicate in the closely related C. muridarum, the barrier is at the level of plasmid replication and the C. trachomatis plasmid must acquire the CDS2 from the C. muridarum plasmid to replicate in the C. muridarum host [40]. C. abortus is the only species that universally does not carry a plasmid. A phylogenetic tree rooted on C. psittaci showed the genomes of all the C. abortus isolates so far sequenced fall into two long branches [41]. This analysis also postulates that C. abortus evolved from a plasmid-bearing ancestor of C. psittaci. In this work, ‘dating’ the origins of the C.abortus species within the genomic phylogeny was based on estimated mutation rates, suggesting the origins of C. abortus can be traced back in millennia and thus could coincide/approximate to the domestication of sheep, some 9–10,000 years ago. The complete absence of plasmids and limited relative diversity in C. abortus genomes (in comparison to other chlamydial species) indicates that the transmission of the C. abortus ancestor to domesticated sheep probably happened once (or a few times), and was co-incident with plasmid loss. A genetic explanation for the continued pathogenicity of C. abortus in the face of plasmid-loss still eludes evolutionary studies.
A further point of interest is that plasmids have not been found in any human isolates of C. pneumoniae sequenced so far [42]. Human C. pneumoniae genomes are extremely homologous suggesting a more recent movement of the pathogen from animals to humans than the emergence of C. abortus from C. psittaci. It has been proposed that humans were infected by an animal isolate of C. pneumoniae which then adapted to humans by gene decay and plasmid loss [43]. It seems likely that this ‘jump’ to humans occurred possibly only once and was accompanied by the loss of the ancestral C. pneumoniae plasmid, then the strain spread through the human population. Recently we developed a vector-based genetic transformation system for C. pneumoniae using the plasmid that came from the equine isolate of C. pneumoniae N16 [44]. This recombinant equine plasmid can replicate efficiently in human C. pneumoniae isolates and whilst human C. pneumoniae have undergone gene decay, they have clearly not lost the ability to support the replication of an animal C. pneumoniae plasmid. This equine plasmid presumably originates from a C. pneumoniae that was a common ancestor for current human and animal C. pneumoniae. Intriguingly it was also shown that the recombinant equine C. pneumoniae plasmid could replicate in several strains of C. felis [44]. Whilst the chromosomes and plasmids of chlamydial species have co-evolved to the extent that the plasmids are species-specific/exclusive C. felis has yet to reach that level of divergence and so can support the replication of plasmids from other Chlamydia without the need for acquisition of accessory plasmid sequences from the recipient host plasmid.
Conclusions
This phylogenetic study provides a framework for investigating the biological properties of the chlamydial plasmid. The phylogeny divided the genus into three distinct lineages designated alpha, beta and gamma. Our results indicate that the lineages diverged from a common ancestor of all the chlamydial species that carried a plasmid. Moreover, there is evidently a strong evolutionary selection towards these species retaining their plasmids since almost all species within the Chlamydiaceae carry a plasmid. No evidence was found for interspecies plasmid transfer.
Supporting information
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Acknowledgments
This study was performed as part of the medical degree (BM5) from the University of Southampton. The work was supported by the faculty associated research team headed by Prof Ian Clarke as the project supervisor.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Sachse K, Bavoil PM, Kaltenboeck B, Stephens RS, Kuo C-C, Rosselló-Móra R, et al. Emendation of the family Chlamydiaceae: Proposal of a single genus, Chlamydia, to include all currently recognized species. Systematic and Applied Microbiology. 2015;38(2):99–103. [DOI] [PubMed] [Google Scholar]
- 2.Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am. 2010;24(1):7–25. [DOI] [PubMed] [Google Scholar]
- 3.Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M. Genital Chlamydia trachomatis: An update. Indian J Med Res. 138 India2013. p. 303–16. [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon AW, Peeling RW, Foster A, Mabey DCW. Diagnosis and Assessment of Trachoma. Clin Microbiol Rev. 172004. p. 982–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabey D, Peeling RW. Lymphogranuloma venereum. Sex Transm Infect. 2002;78(2):90–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villareal C, Whittum-Hudson JA, Hudson AP. Persistent Chlamydiae and chronic arthritis. Arthritis Res. 4 London2002. p. 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rockey DD. Unraveling the basic biology and clinical significance of the chlamydial plasmid. J Exp Med. 2011;208(11):2159–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Systematic and Applied Microbiology. 2014;37(2):79–88. [DOI] [PubMed] [Google Scholar]
- 9.Read TD, Myers GSA, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Research. 2003;31(8):2134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Lent S, Piet JR, Beeckman D, van der Ende A, Van Nieuwerburgh F, Bavoil P, et al. Full genome sequences of all nine Chlamydia psittaci genotype reference strains. J Bacteriol. 2012;194(24):6930–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, et al. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J Bacteriol. 2014;196(5):989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelocnik M, Bachmann NL, Kaltenboeck B, Waugh C, Woolford L, Speight KN, et al. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genomics. 2015;16:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusher M, Storey CC, Richmond SJ. Plasmid diversity within the genus Chlamydia. Journal of General Microbiology. 1989;135(5):1145–51. [DOI] [PubMed] [Google Scholar]
- 14.Storey C, Lusher M, Yates P, Richmond S. Evidence for Chlamydia pneumoniae of non-human origin. Journal of General Microbiology. 1993;139(11):2621–6. [DOI] [PubMed] [Google Scholar]
- 15.Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology-Sgm. 1997;143:1847–54. [DOI] [PubMed] [Google Scholar]
- 16.Grinblat-Huse V, Drabek EF, Creasy HH, Daugherty SC, Jones KM, Santana-Cruz I, et al. Genome sequences of the zoonotic pathogens Chlamydia psittaci 6BC and Cal10. J Bacteriol. 2011;193(15):4039–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth-Smith HM, Harris SR, Rance R, West AP, Severin JA, Ossewaarde JM, et al. Genome sequence of the zoonotic pathogen Chlamydophila psittaci. J Bacteriol. 2011;193(5):1282–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dugan J, Rockey DD, Jones L, Andersen AA. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother. 2004;48(10):3989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dugan J, Andersen AA, Rockey DD. Functional characterization of IScs605, an insertion element carried by tetracycline-resistant Chlamydia suis. Microbiology. 2007;153(Pt 1):71–9. [DOI] [PubMed] [Google Scholar]
- 20.Binet R, Maurelli AT. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U S A. 2009;106(1):292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donati M, Huot-Creasy H, Humphrys M, Di Paolo M, Di Francesco A, Myers GS. Genome Sequence of Chlamydia suis MD56, Isolated from the Conjunctiva of a Weaned Piglet. Genome Announc. 2014;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986;16(1):52–62. [DOI] [PubMed] [Google Scholar]
- 23.Hugall A, Timms P, Girjes AA, Lavin MF. Conserved DNA sequences in Chlamydial plasmids. Plasmid. 1989;22(2):91–8. [DOI] [PubMed] [Google Scholar]
- 24.Pawlikowska-Warych M, Sliwa-Dominiak J, Deptulan W. Chlamydial plasmids and bacteriophages. Acta Biochimica Polonica. 2015;62(1):1–6. [DOI] [PubMed] [Google Scholar]
- 25.Konieczny I, Bury K, Wawrzycka A, Wegrzyn K. Iteron Plasmids. Microbiology spectrum. 2014;2(6). [DOI] [PubMed] [Google Scholar]
- 26.Jelocnik M, Bachmann NL, Seth-Smith H, Thomson NR, Timms P, Polkinghorne AM. Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution. PeerJ. 2016;4:e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones CA, Hadfield J, Thomson NR, Cleary DW, Marsh P, Clarke IN, et al. The Nature and Extent of Plasmid Variation in Chlamydia trachomatis. Microorganisms. 2020;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadfield J, Harris SR, Seth-Smith HMB, Parmar S, Andersson P, Giffard PM, et al. Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Genome Res. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann B. A new genetic variant of Chlamydia trachomatis. Sex Transm Infect. 83 England2007. p. 253–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Versteeg B, Bruisten SM, Pannekoek Y, Jolley KA, Maiden MCJ, van der Ende A, et al. Genomic analyses of the Chlamydia trachomatis core genome show an association between chromosomal genome, plasmid type and disease. BMC Genomics. 19 London2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris SR, Clarke IN, Seth-Smith HM, Solomon AW, Cutcliffe LT, Marsh P, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44(4):413–9, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W, Jelocnik M, Li J, Sachse K, Polkinghorne A, Pannekoek Y, et al. From genomes to genotypes: molecular epidemiological analysis of Chlamydia gallinacea reveals a high level of genetic diversity for this newly emerging chlamydial pathogen. BMC Genomics. 2017;18(1):949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holder M, Lewis PO. Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet. 2003;4(4):275–84. [DOI] [PubMed] [Google Scholar]
- 35.Hillis DM, Bull JJ. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Systematic Biology. 1993;42(2):182–92. [Google Scholar]
- 36.Ramsey KH, Schripsema JH, Smith BJ, Wang Y, Jham BC, O'Hagan KP, et al. Plasmid CDS5 influences infectivity and virulence in a mouse model of Chlamydia trachomatis urogenital infection. Infect Immun. 2014;82(8):3341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, et al. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One. 2013;8(3):e59195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polkinghorne A, Hanger J, Timms P. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet Microbiol. 2013;165(3–4):214–23. [DOI] [PubMed] [Google Scholar]
- 39.Hadfield J, Harris SR, Seth-Smith HMB, Parmar S, Andersson P, Giffard PM, et al. Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Genome Res. 2017;27(7):1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Cutcliffe LT, Skilton RJ, Ramsey KH, Thomson NR, Clarke IN. The genetic basis of plasmid tropism between Chlamydia trachomatis and Chlamydia muridarum. Pathogens and disease. 2014;72(1):19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seth-Smith HMB, Buso LS, Livingstone M, Sait M, Harris SR, Aitchison KD, et al. European Chlamydia abortus livestock isolate genomes reveal unusual stability and limited diversity, reflected in geographical signatures. BMC Genomics. 2017;18(1):344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinmaier T, Hoser J, Eck S, Kaufhold I, Shima K, Strom TM, et al. Genomic factors related to tissue tropism in Chlamydia pneumoniae infection. BMC Genomics. 2015;16:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers GS, Mathews SA, Eppinger M, Mitchell C, O'Brien KK, White OR, et al. Evidence that human Chlamydia pneumoniae was zoonotically acquired. J Bacteriol. 2009;191(23):7225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shima K, Wanker M, Skilton RJ, Cutcliffe LT, Schnee C, Kohl TA, et al. The Genetic Transformation of Chlamydia pneumoniae. mSphere. 2018;3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Raw unannotated MEGA X data.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.