Abstract
Background
The crisis of antimicrobial resistance is already here with us, affecting both humans and animals alike and very soon, small cuts and surgeries will become life threatening. This study aimed at determine the whole genome sequences of multi-drug resistant Escherichia coli isolated in a Pastoralist Community of Western Uganda: phylogenomic changes, virulence and resistant genes.
Methods
This was a laboratory based cross sectional study. Bacterial isolates analyzed in this study were 42 multidrug resistant E. coli isolated from stool samples from both humans (n = 30) and cattle (n = 12) in pastoralist communities collected between January 2018-March 2019. Most of the isolates (41/42) were resistant to three or more antibiotics (multi-drug resistant) and 21/42 isolates were ESBL producers; 13/30 from human and 8/12 from cattle. Whole Genome Sequencing (WGS) was carried out at the facilities of Kenya Medical Research Institute-Wellcome trust, Kilifi, to determine the phylogenomic changes, virulence and resistant genes.
Results
At household level, the genomes from both human and animals clustered away from one another except for one instance where two human isolates from the same household clustered together. However, 67% of the E. coli isolated from cattle were closely related to those found in humans. The E. coli isolates were assigned to eight different phylogroups: A, B1, B2, Cladel, D, E, F and G, with a majority being assigned to phylogroup A; while most of the animal isolates were assigned to phylogroup B1. The carriage of multiple AMR genes was higher from the E. coli population from humans than those from cattle. Among these were Beta-lactamase; blaOXA-1: Class D beta-lactamases; blaTEM-1, blaTEM-235: Beta-lactamase; catA1: chloramphenicol acetyl transferase; cmlA1: chloramphenicol efflux transporter; dfrA1, dfrA12, dfrA14, dfrA15, dfrA17, dfrA5, dfrA7, dfrA8: macrolide phosphotransferase; oqxB11: RND efflux pump conferring resistance to fluoroquinolone; qacL, qacEdelta1: quinolone efflux pump; qnrS1: quinolone resistance gene; sul1, sul2, sul3: sulfonamide resistant; tet(A), tet(B): tetracycline efflux pump. A high variation of virulence genes was registered among the E. coli genomes from humans than those of cattle origin.
Conclusion
From the analysis of the core genome and phenotypic resistance, this study has demonstrated that the E. coli of human origin and those of cattle origin may have a common ancestry. Limited sharing of virulence genes presents a challenge to the notion that AMR in humans is as a result of antibiotic use in the farm and distorts the picture of the directionality of transmission of AMR at a human-animal interface and presents a task of exploring alternative routes of transmission of AMR.
Background
The crisis of antimicrobial resistance is already here with us, affecting both humans and animals alike and very soon, small cuts and surgeries will become life threatening. Evidence of non-prescribed use of antimicrobials in livestock to mask ill farming practices and misuse of antibiotics in community pharmacies has been documented globally [1, 2]. Farmers use large amounts of antibiotics in livestock and this is now known as a key driver and recipe to accelerating emergency of antimicrobial resistance [3]. Resistant bacterial clones may spread from animals to humans rendering antibiotics less effective and increases mortality and morbidity in developing nations due to such bacteria [3]. Previous studies in Uganda report abuse of antimicrobials in animal husbandry as a major contributor to antimicrobial resistance emergence among microbes. Additionally, 40% of the persons who visit a health- care facility in Uganda are treated with antibiotics [4]. These antibiotics are mostly obtained without prescription in drug shops and community pharmacies in sub-therapeutic doses. Globalization of trade coupled with the revolutionalization of travel has simplified distribution of resistant bacteria due to easy movement of humans and their goods including livestock across countries making the problem of antimicrobial resistance (AMR) global in nature.
The global spread of multi-drug resistant Enterobacteriaceae especially CTX-M type ESBLs and strains producing carbapenemases such as KPC and NDM warrant a multi-stake holder attention [5]. Resistance against certain antibiotic categories is already high in hospital settings in Uganda but resistance among bacteria from animals is not well documented. Introduction of bacteria with resistant genes into food animals as a result of antibiotic selection in veterinary medicine is now of great concern. Due to the routine consumption of certain antibiotics like tetracyclines in animal husbandry, undesirable shift of resistance towards potent second line and third generation antibiotics is now a reality [6, 7]. Baseline knowledge on the epidemiology and transmission routes of drug resistant microbes in Uganda is needed to drive the necessary prevention, intervention and control measures. While studies have been conducted in health care facilities in mostly urban areas settings [8, 9]. Little effort has been devoted to determining the molecular epidemiology of antimicrobial resistance, including multidrug resistance at a human-animal interface. Pastoralist communities live with their domestic animals inside the park hence a porous interface for microbial and disease transmission which provides a good ground for this study. We aimed to determine the whole genome sequences of multi-drug resistant Escherichia coli isolated in a Pastoralist Community of Western Uganda: phylogenomic changes, virulence and resistant genes.
Methods
Bacterial strains
Bacterial isolates analyzed in this study were multidrug resistant bacteria isolated from stool samples from both humans (n = 30) and cattle (n = 12) in pastoralist communities of Kasese district between January 2018-March 2019. The cattle were sample because they were the most common animals reared by the pastoralists in Kasese district. The pastoralist communities in Kasese district are settled in and around Queen Elizabeth National Park (QENP). The Kasese side of the National Park has two pastoralist communities in Nyakatonzi and Hima sub-countries. The QEPA lies astride the equator along the latitudes of 0Ê 39' 36" North, 30Ê 16' 30" East. The northern area has been occupied by pastoralists since the 1920s. Specific sites where samples were taken were Bwera-Mpondwe (Bwera) in the east, Hima in the north and Katwe-Kabatoro (Katwe).
Speciation and antibiotic susceptibility testing
Speciation and antibiotic susceptibility of the isolates was done using the Phoenix automated microbiology system (Phoenix 100 ID/DST system) from Becton and Dickson (Franklin Lakes, NJ, USA) and the results interpreted using the CLSI guidelines. Sensitivity testing was carried out using a total of 15 antibiotics which include; ampicillin, amoxicillin-clavulanic acid, cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime, ciprofloxacin, levofloxacin, gentamycin, tetracycline, nitrofurantoin, imipenem, Ertapenem and cotrimoxazole. Multi-drug resistance was defined as one isolate being resistant to three or more classes of antibiotics tested [10].
Extraction of Genomic bacterial DNA
Extraction of Genomic bacterial DNA was done at Molecular Biology Laboratory of Department of Immunology and Molecular Biology of Makerere University using Modified Cetyltrimethylammonium bromide (CTAB) Method as described before [11]. The modified CTAB method uses Enzymes and detergents to lyse cells, release the nucleic acids, and an organic solvent to purify the DNA and absolute alcohol (isopropanol/ethanol) to precipitate out the DNA.
Whole genome-sequencing
Whole Genome Sequencing (WGS) was carried out at the facilities of KEMRI Wellcome Trust Research Programme for the isolates that turned out to be multidrug resistant from house contacts and cattle. Genomic DNA from cultured E. coli was quantified using Qubit and diluted to 0.2ng/μl and library prep performed using the Illumina Nextera XT protocol according to the manufactures instructions. Briefly, gDNA was fragment and tagged with adapter sequences using a transposase enzyme in a process known as tagmentation. Using a limited cycle PCR step, indices were introduced. This helped in demultiplexing after sequencing was complete. A size selection clean-up was done using AMPure beads (AGENCOURT) and normalized to ensure equal representation of all libraries. The normalized libraries were pooled, denatured and loaded on the Miseq platform with an out of 2X200bp [12].
Bioinformatics analysis
Paired-end reads from each E. coli isolate were assembled de novo using spades v 3.11.1 algorithm [13] to generate a draft genome sequence for each isolate and quality assessment of for assemblies was done using QUAST 4.5. Clermont typing method [14, 15] was used to determine phylogroups basing on the genome-clustering tool called Mash [16].
Sequences were analysed using the Nullarbor pipeline (Seeman T, available at: https://github.com/tseemann/nullarbor). In brief, reads were trimmed to remove adaptor sequences and low-quality bases with Trimmomatic, [17] and Kraken (v1.1.1) was used to investigate for contamination [18]. Reads were aligned to the E. coli str. K-12 substr. MG1655 complete genome using the Burrows-Wheeler Aligner MEM (0.7.17-r1188) algorithm [19]. Samples with at least 21x depth of coverage and 76% genome coverage were retained for analysis. SNPs were identified using Freebayes (v1.3.1) with a minimum depth of coverage of 10x and allelic frequency of 0.9 required to confidently call a single nucleotide polymorphism (SNP) [20]. For all isolates, sequence reads covered >76% of the reference genome, A total of 29,525 core SNP sites were identified, with 2,733 genes identified as core genes (present in >99% of isolates).De novo assemblies were also performed using SPAdes (v. 3.13.1), [13] as part of the Nullarbor pipeline, with genes annotated using Prokka (v. 1.14.0, Seemann T, available at: https://github.com/tseemann/prokka).
E. coli has been traditionally clustered by phylogroup and have a good association with isolates being a commensal or pathogen [21]. The phylotyping method described by Clermont et al. (2013) [22] was performed in silico. In short, based on the presence or absence of 5 genes: chuA, yjaA, tspE4.C2, arpA and trpA, isolates can be separated into 8 phylogroups (A, B1, B2, D, C, E, F, and cryptic clades).
Each draft genome was screened for the presence of AMR and virulence genes using the software package Abricate (https://github.com/tseemann/abricate), a package for mass screening of contigs for antimicrobial resistance or virulence genes using databases. For antimicrobial resistance screening we used the NCBI Bacterial Antimicrobial Resistance Reference Gene Database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047) and for virulence genes screening VFDB, a reference database for bacterial virulence factors [23]. Statistical analysis and visualisation were performed in R version 3.6.1 (www.r-project.org).
Ethical considerations
The study was approved by the Makerere University School of Biomedical Sciences Higher Degrees and Ethics Committee (SBS-HDREC) and The Uganda National Council of Science and Technology (UNCST). Written informed consent was obtained from all participants. All participant identifying information was kept confidential. Written informed consent from parents/guardians of participants below 18 years was sought and written assent was provided by all minors who were able to read and write who voluntarily participated in this study while those who were not able to read and write provided verbal assent. Verbal assent was witnessed by the parent/guardian.
Results
Antibiotic susceptibility pattern
Antibiotic susceptibility testing was carried out on all the 42 E. coli isolated from both human and animals. A majority of the isolates displayed high resistance to ampicillin, cefazolin, trimethoprim/sulphurmethoxazole, amoxicillin/clavulanic acid and Cefotaxime. Low resistance to imipenem, ciprofloxacin, Levofloxacin Ertapenem were recorded whereas only one isolate was resistant to gentamicin. Most of the isolates 41/42 were resistant to three or more antibiotics (multi-drug resistant) and 21/42 isolates were ESBL producers.
Whole genome sequencing and de novo assembly
Computation of the total number of reads and quality metrics of the showed homogenous results with a good quality profile for all isolates assemblies. Estimated average depth of coverage for 42 E. coli isolates on which WGS was performed was 28× with an average of 197 contigs >500-base pairs after genome assembly using SPAdes.
Phylogenetic analysis of the E. coli Isolated from humans and cattle
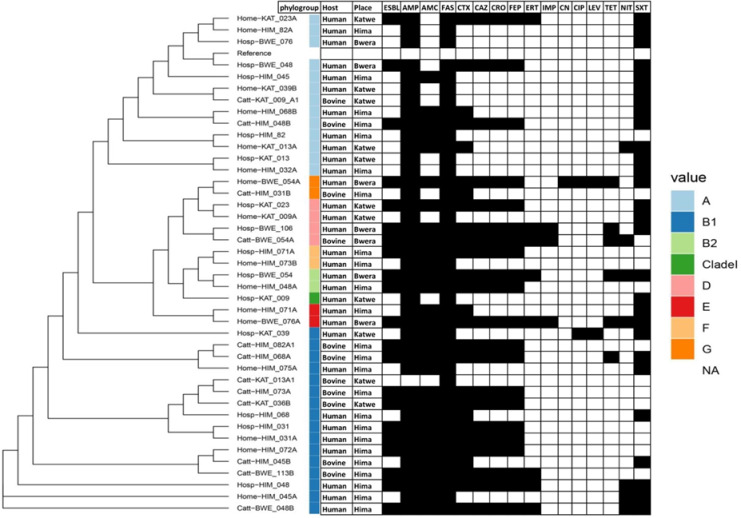
The genomes of E. coli from humans and animals were clustered according to single nucleotide polymorphism in core genes. At household level, the genomes from both human and animals clustered away from one another except for one instance where two human isolates from the same household clustered together. However, 67% of the E. coli isolated from cattle were closely related to those found in humans (Fig 1). All the four major E. coli phylotypes were identified through in silico phylotyping. Most isolates belonged to phylotype B1, which accounted for 38% (n = 16) of E. coli isolates, evenly split between human and cattle samples (Fig 1). Phylotypes A and D accounted for 31% (n = 13) and 9.5% (n = 4) of isolates respectively, with the majority of the phylogroup A isolated from human samples and only two from cattle. The majority of isolates from Hima location are phylotype B1 and A.
Fig 1. Maximum likelihood phylogenetic trees based on SNP differences within the core genomes and antibiotic susceptibility of E. coli isolated from humans and animals in Kasese district.
The black shading indicates resistance. ESBL = Extended Spectrum β-lactamase, AMP = ampicillin, AMC = amoxicillin/clavulanic acid, FAS = cefazolin, CTX = Cefotaxime, CAZ = ceftazidime, CRO = ceftriaxone, FEP = cefepime, ERT = ertapenem, IMP = imipenem, CN = gentamicin CIP = ciprofloxacin, LEV = levofloxacin, TET = tetracycline, NIT = nitrofurantoin, SXT = trimethoprim/sulphurmethoxazole.
Antibiotic susceptibility pattern
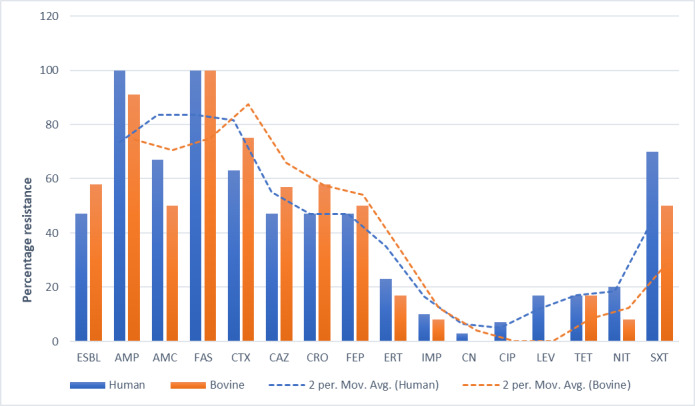
Antibiotic susceptibility testing was carried out on all the 42 E. coli isolated from both human and cattle. Generally, a majority of the isolates displayed high resistance to cefazolin (100%), ampicillin (98%), Cefotaxime (67%), trimethoprim/sulphurmethoxazole (64%) and amoxicillin/clavulanic acid (62%). Low resistance to ciprofloxacin (5%), imipenem (10%), and Levofloxacin (12%) were recorded whereas only one isolate was resistant to gentamicin (2%). Most of the isolates 41/42 were resistant to three or more antibiotics (multi-drug resistant) and 21/42 isolates were ESBL producers (Fig 1). All the isolates assigned to phylogroup A were resistant to ampicillin, cefazolin and trimethoprim/sulfamethoxazole but sensitive to imipenem, gentamicin, ciprofloxacin, levofloxacin and tetracycline. All phylogroup B1 isolates were resistant to cefazolin but sensitive to imipenem, gentamicin and tetracycline. There was also high resistance against ampicillin (94%), amoxicillin/clavulanic acid (94%).
Generally, there was a similar pattern of antibiotic resistance among the E. coli isolated from human and that isolated from cattle with the trends in cattle appearing higher than those in humans (Fig 2). All the isolates from cattle and humans were resistant to cefazolin. All the human isolates were resistant to ampicillin and highly resistant to trimethoprim/sulfamethoxazole 21(70%), amoxicillin/clavulanic acid, 20(67%) and Cefotaxime, 19(63%). Low resistance against gentamicin 1(3%), ciprofloxacin, 2(7%) and imipenem, 3(10%) was detected among the isolates from humans. The isolates from cattle were highly resistant to ampicillin, 11(91%) and Cefotaxime, 9(75%) and no resistance was detected against gentamicin, ciprofloxacin and levofloxacin. There was no significant difference in resistance to particular drugs between the human and cattle isolates except for Cotrimoxazole (p<0.05).
Fig 2. Phenotypic antibiotic resistance patterns in humans and cattle.
ESBL = Extended Spectrum β-lactamase, AMP = ampicillin, AMC = amoxicillin/clavulanic acid, FAS = cefazolin, CTX = Cefotaxime, CAZ = ceftazidime, CRO = ceftriaxone, FEP = cefepime, ERT = ertapenem, IMP = imipenem, CN = gentamicin CIP = ciprofloxacin, LEV = levofloxacin, TET = tetracycline, NIT = nitrofurantoin, SXT = trimethoprim/sulphurmethoxazole.
Comparison of virulence genes identified in E. coli genomes, isolated and sequenced from human and cattle samples
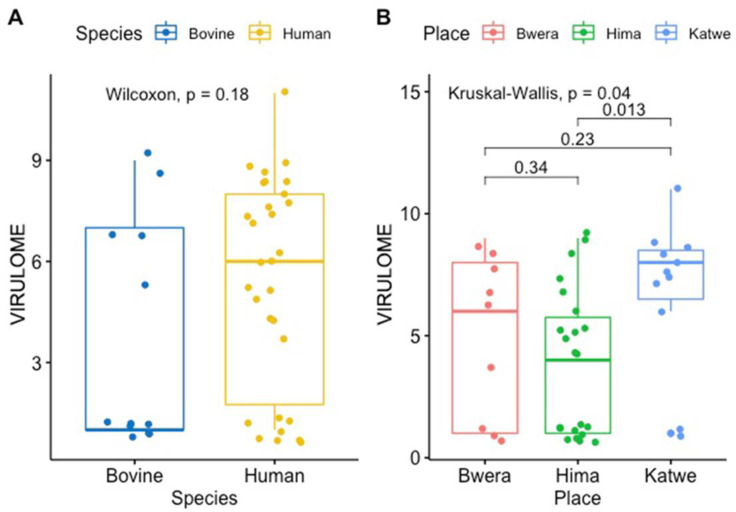
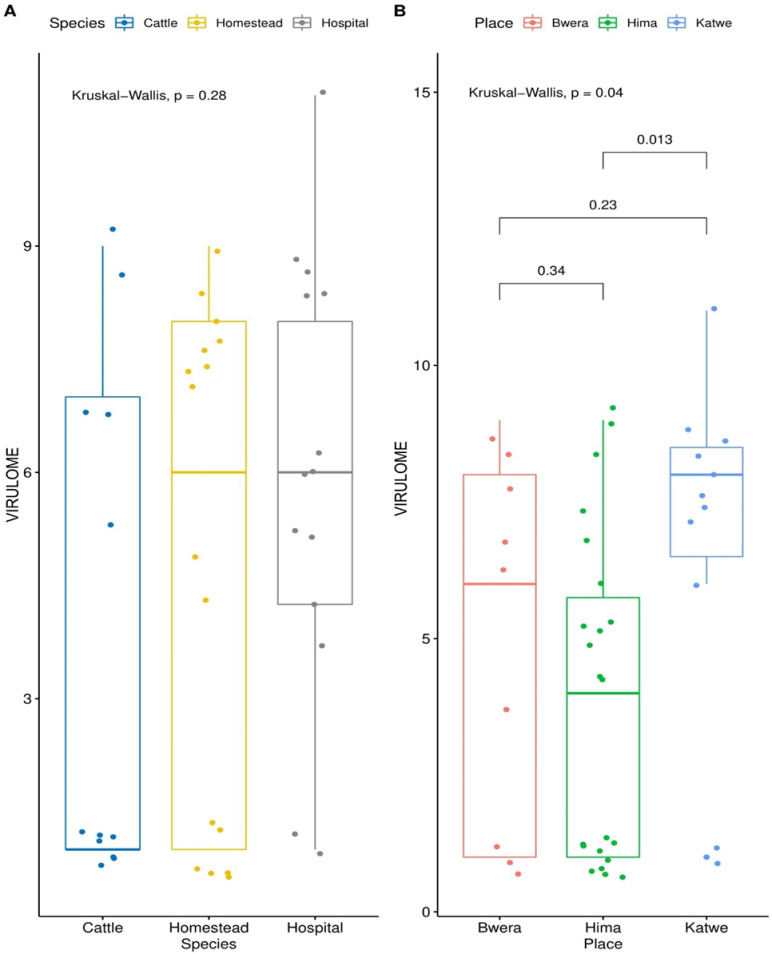
Antimicrobial resistance genes were identified in all 30 E. coli genomes from human and the 12 E. coli genomes from cattle. The carriage of virulence genes was significantly higher among the E. coli isolates from humans compared to those of cattle origin (Fig 3) and this rule out the possibility of clonal relationship between the isolates from the two hosts (p = 0.18). Isolates from Katwe location had significantly higher number of virulence genes compared to the other two sites, Bwera and Hima (p = 0.04). The human samples have a significantly higher number of virulence genes compared to bovine samples. However, there was no difference in the median of the virulence genes from the E. coli isolated from the hospital and that isolated from the community (Fig 4).
Fig 3. Box plots showing the median values for the number of virulence genes in the two groups along with the interquartile range.
The human samples had a significantly higher number of virulence genes compared to bovine samples. The E. coli strains from Katwe had significantly higher number of virulence genes compared to Bwera and Hima.
Fig 4. Box plots comparing the variation in virulence of the E. coli from hospital and the community.
Homestead = community isolated E. coli, species = human/cattle.
Detection of virulence factors genes
A total of 174 different virulence genes were detected from the E. coli genomes. The virulence genes; cheY, csgG, entC, entF, entS, fepB, fepC, fepD and flgG were detected in all the E. coli isolates. Other virulent genes that were commonly prevalent include; csgB, csgD, csgE, entA, entB, fdaC, fimA, fim B, fimC, fimD, fimE, fimF, fimG, fimH, fimI, flgG, flhA, fliG, fliM, flip, gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, gspM, ompA, yagV/ecpE, yagW/ecpD, yagX/ecpC, yagY/ecpB, yagZ/ecpA and ykgk/ecpB. The proportion of virulence genes from the two hosts was analyzed and presented in box plots (Fig 3).
Discussion
Whole genome sequencing was used to analyse non–duplicate E. coli isolated from both human and cattle in pastoralist communities of Kasese district. Phylogenetic analysis showed that the genomes of the human E. coli generally clustered together and away from those of cattle origin and likewise the genomes from cattle tended to cluster together and away from those of the cattle origin. The human isolates were mainly assigned to phylogroup A and B1 whereas the cattle isolates were mostly assigned to phylogroup B1. This finding agrees with the Zambian study which reported that most of the cattle isolates of E. coli were assigned to phylogroup B1 which is commonly associated with commensal existence [24]. Phylogroups B2, Clade l, E and F; were associated with only the E. coli isolated from humans, three human isolates were assigned to phylogroup D and only one cattle isolate was assigned to group D. The presence of phylogroups B2 and D among human isolates presents a risk for infection due to E. coli in humans. Phylogroups B2 and D have been reported to be pathogenic and responsible for extra-intestinal infections by a number of studies [25–28].
Phenotypic resistance testing revealed a similar resistance pattern among the human isolates than the cattle isolates for the 15 antibiotics tested in this study. A similar study in Lusaka-Zambia demonstrated significantly higher levels of resistance among the human E. coli isolates when the same antibiotics were tested [24]. Every year, humans produce, use, and consume approximately 175,000 tons of antibiotics [29] which makes emergency of AMR in humans, animals and environment inevitable. The resistance genes were also compared in E. coli isolates originating from the two hosts. The carriage of multiple AMR genes among the E. coli population from humans was higher than those from cattle (Fig 5). There have been several debates trying to link the emergency of AMR in humans to antibiotic use in food animals [30–32]. Our study, like the study in Zambia [24], showed that there was much more diversity in the resistance genotypes present in human isolates than in the cattle isolates. The deference in the diversity of the resistance genotype could indicate the existence of independent AMR selection pressures in human and animals. Whereas other studies have demonstrated the predominance of blaCTX-M-15 allele among human and animal isolates of E. coli, from the community [33, 34], this study identified arsB-mob (80%) as the most predominant allele (Fig 2). This discrepancy could be due to the different methods used and undetermined geographical factors. Another study in livestock in Uganda indicated that blaCTX-M and carbapenems resistant genes were not harboured by E. coli from livestock [35]. The prevalence of cephalosporin resistance genes in the E. coli isolated from cattle in this study presents a risk of transmission of these resistant genes from animals to humans. Similar observations have been made by Okubo et al., in Uganda [35]. In this study, the prevalence of resistance genes was high both in human and cattle isolates.
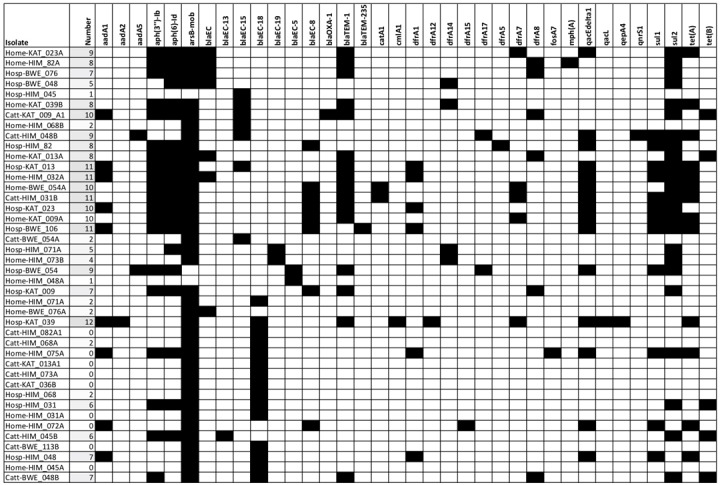
Fig 5. Heat map showing AMR genes found in the genomes of sequenced E. coli isolates.
AMR genes are annotated with the NCBI Bacterial Antimicrobial Resistance Reference Gene Database with >95% coverage. The names of the strains indicated on the y axis are presented in the same order as in Fig 2. aadA1, aadAd2, aadAD5: Streptomycin 3''-adenylyltransferase; aph(3'')-Ib, aph(6)-Id: Streptomycin phosphotransferase; arsB-mob: Arylsulfatase B; blaEC, blaEC-13, blaEC-15, blaEC-18, blaEC-19, blaEC-5, blaEC-8: Beta-lactamase; blaOXA-1: Class D beta-lactamases; blaTEM-1, blaTEM-235: Beta-lactamase; catA1: chloramphenicol acetyl transferase; cmlA1: chloramphenicol efflux transporter; dfrA1, dfrA12, dfrA14, dfrA15, dfrA17, dfrA5, dfrA7, dfrA8: trimethoprim resistant dihydrofolate reductase; fosA: fosfomycin thiol transferase; mph(A): macrolide phosphotransferase; oqxB11: RND efflux pump conferring resistance to fluoroquinolone; qacL, qacEdelta1: Small multidrug resistance gene; qepA4: quinolone efflux pump; qnrS1: quinolone resistance gene; sul1, sul2, sul3: sulfonamide resistant; tet(A), tet(B): tetracycline efflux pump (Fig 5).
A high variation of virulence genes was registered among the E. coli genome from humans than those of cattle origin further supports the hypothesis that the E. coli isolated from the two hosts have a common ancestral origin. However, some studies have found clonal commonality between E.coli isolated from the hospital, the community and animals [36]. The reason for this discrepancy is mainly due to the different methods used. Also, there was no difference in median of virulence genes between the E. coli isolated from hospital from that isolated from the community and this may imply that the E. coli isolated from the two hosts have similar evolutionary pressures.
Conclusions
From the analysis of the core genome and phenotypic resistance, this study has demonstrated that the E. coli of human origin and those of cattle origin may have a common ancestry. Limited sharing of virulence genes presents a challenge to the notion that AMR in humans is as a result of antibiotic use in the farm and distorts the picture of the directionality of transmission of AMR at a human-animal interface and presents a task of exploring alternative routes of transmission of AMR. A major limitation of this study is the small number of isolates sequenced and only one species of animals studied, therefore, we recommend a larger study for better generalization of results.
Acknowledgments
We gratefully acknowledge our participants for making this study possible. Heartfelt thanks to Stallone Kisembo and Zaydah De Laurent for the great work they did as research assistants.
Abbreviation
- ESBLs
Extended spectrum β-lactamases
- CTAB
Cetyltrimethylammonium bromide
- DNA
Deoxyribonucleic acid
- CLSI
Clinical and Laboratory Standards Institute guidelines
- WGS
Whole Genome Sequencing
- AMR
antimicrobial resistance
- QEPA
Queen Elizabeth National Park
- UNCST
Uganda National Council for Science and Technology
Data Availability
Data is available in a public repository; DOI 10.17605/OSF.IO/KPHRD.
Funding Statement
This work was supported by the DELTAS Africa Initiative [grant# 107743/Z/15/Z]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant #107743/Z/15/Z] and the UK government. The views expressed in this manuscript are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
References
- 1.Elliot K. Antibiotics on the farm: Agriculture's role in drug resistance. Center for Global Development. 2015. [Google Scholar]
- 2.O'Neill J. Antimicrobials in agriculture and the environment: Reducing unnecessary use and waste: the review on antimicrobial resistance. 2015. [Google Scholar]
- 3.Maria Angeles Argudín Ariane Deplano, Meghraoui Alaeddine, Magali Dodémont Amelie Heinrichs, Denis Olivier, et al. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes Antibiotics. Antibiotics. 2017;6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. Journal of Multidisciplinary Healthcare. 2013;6:303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P, Naas T, P L.. Global spread of carbapenemase -producing Enterobacteriaceae. Emerg infect Dis. 2011; 17:1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byarugaba D.K, Roger Kisameand, S Olet. Multi-drug resistance in commensal bacteria of food animal origin in Uganda. Africa Journal of microbiology Research. 2011;5(12):1539–48. [Google Scholar]
- 7.Walters MS RJ, Mikoleit M, Kadivane S, Ouma C, et al. Shifts in Geographic Distribution and Antimicrobial Resistance during a Prolonged Typhoid Fever Outbreak—Bundibugyo and Kasese Districts, Uganda, 2009–2011. PLoS Negl Trop Dis. 2014;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omulo S, Thumbi SM, Njenga MK, Call DR. A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrobial Resistance and Infection Control. 2015;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.P Katete D., Kabugo U, Baluku H, Nyakarahuka, L Kyobe. Prevalence and Antimicrobial Susceptibility Patters of Bacteria from Milkmen and cows with clinical Mastitis in and around Kampala-Uganda. PLoS One. 2013;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallen A.J, Yi Mu, Sandra Bulens, Arthur R, Susan Petit, G Ken., et al. Health care -associated invasive MRSA INFECTIONS, 2005–2008; For the active core surveilence (ABCs) MRSA investigators of the Emerging infectious program. JAMA. 2010;304(6):641–7. [DOI] [PubMed] [Google Scholar]
- 11.Kateete DP, Nakanjako R, Okee M, Joloba ML, Najjuka CF. Genotypic diversity among multidrug resistant Pseudomonas aeruginosa and Acinetobacter species at Mulago Hospital in Kampala, Uganda. BMC Research Notes. 2017;10(1):284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irenge LM, Ambroise J, Bearzatto B, Durant J-F, Chirimwami RB, Gala J-L. Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: characterization of phylogenomic changes, virulence and resistance genes. BMC Infect Dis. 2019;19(1):137–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology: a journal of computational molecular cell biology. 2012;19(5):455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom. 2018;4(7):e000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clermont O, Bonacorsi S, Bingen E. Rapid and Simple Determination of the<em>Escherichia coli</em> Phylogenetic Group. Applied and Environmental Microbiology. 2000;66(10):4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biology. 2016;17(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology. 2014;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L H. Aligning sequence reads, clone sequences and assembly contigs with BWAMEM. arXiv 2013; 1303.3997v1302. 2013. [Google Scholar]
- 20.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012; 1207.3907v1202. 2012. [Google Scholar]
- 21.Clermont O, Gordon D, and Denamur E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015;161(Pt 5):980–8. [DOI] [PubMed] [Google Scholar]
- 22.Clermont O, Christenson JK, Denamur E, and Gordon DM. The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Zheng DD, Jin Q, LH C, aY J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–D692. 2019;47(D1):D687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mainda G, Lupolova N, Sikakwa L, Richardson E, Bessell PR, Malama SK, et al. Whole Genome Sequence Analysis Reveals Lower Diversity and Frequency of Acquired Antimicrobial Resistance (AMR) Genes in E. coli From Dairy Herds Compared With Human Isolates From the Same Region of Central Zambia. Frontiers in microbiology. 2019;10:1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duriez Patrick, Clermont Oliver, Bonacorsi Stephane, Bingen Edouardo, Chaventre Andre, Elion Jacques, et al. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations Microbiology. 2001;147(6):1671–6. [DOI] [PubMed] [Google Scholar]
- 26.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Molecular Biology and Evolution. 1998;15(12):1685–95. [DOI] [PubMed] [Google Scholar]
- 27.Moazen M., Atefi M., Ebrahimi A., Razmjoo and P., Dastjerdi Mv. Evaluation of Chemicnal and Microbiological quality in 21 brands ofg Iranian bottled drinking water in 2012: A comparison study on label and real contents. Journal of Environmental and Public Health. 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, et al. Predominance of CTX-M-15 among ESBL Producers from Environment and Fish Gut from the Shores of Lake Victoria in Mwanza, Tanzania. Frontiers in microbiology. 2016;7(1862). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. The Lancet Infectious diseases. 2013;13(12):1057–98. [DOI] [PubMed] [Google Scholar]
- 30.Walsh F. The multiple roles of antibiotics and antibiotic resistance in nature. Frontiers in microbiology. 2013;4(255). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillings M. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in microbiology. 2013;4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mark Woolhouse, Melissa Ward, Jeremy VBaF. Antimicrobial resistance in humans, livestock and the wider environment. Phill Trans R Soc B.370:20140083 - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, et al. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC infectious diseases. 2016;16:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, et al. Multiple ESBL-Producing Escherichia coli Sequence Types Carrying Quinolone and Aminoglycoside Resistance Genes Circulating in Companion and Domestic Farm Animals in Mwanza, Tanzania, Harbor Commonly Occurring Plasmids. Frontiers in microbiology. 2016;7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okubo T, Yossapol M, Maruyama F, Wampande EM, Kakooza S, Ohya K, et al. Phenotypic and genotypic analyses of antimicrobial resistant bacteria in livestock in Uganda. Transboundary and Emerging Diseases. 2019;66(1):317–26. [DOI] [PubMed] [Google Scholar]
- 36.Tyrrell JM, Wootton M, Toleman MA, Howe RA, Woodward M, Walsh TR. Genetic & virulence profiling of ESBL-positive E. coli from nosocomial & veterinary sources. Veterinary microbiology. 2016;186:37–43. [DOI] [PubMed] [Google Scholar]