Abstract
PURPOSE
Allele-specific copy number alteration (CNA) analysis is essential to study the functional impact of single-nucleotide variants (SNVs) and the process of tumorigenesis. However, controversy over whether it can be performed with sufficient accuracy in data without matched normal profiles and a lack of open-source implementations have limited its application in clinical research and diagnosis.
METHODS
We benchmark allele-specific CNA analysis performance of whole-exome sequencing (WES) data against gold standard whole-genome SNP6 microarray data and against WES data sets with matched normal samples. We provide a workflow based on the open-source PureCN R/Bioconductor package in conjunction with widely used variant-calling and copy number segmentation algorithms for allele-specific CNA analysis from WES without matched normals. This workflow further classifies SNVs by somatic status and then uses this information to infer somatic mutational signatures and tumor mutational burden (TMB).
RESULTS
Application of our workflow to tumor-only WES data produces tumor purity and ploidy estimates that are highly concordant with estimates from SNP6 microarray data and matched normal WES data. The presence of cancer type–specific somatic mutational signatures was inferred with high accuracy. We also demonstrate high concordance of TMB between our tumor-only workflow and matched normal pipelines.
CONCLUSION
The proposed workflow provides, to our knowledge, the only open-source option with demonstrated high accuracy for comprehensive allele-specific CNA analysis and SNV classification of tumor-only WES. An implementation of the workflow is available on the Terra Cloud platform of the Broad Institute (Cambridge, MA).
INTRODUCTION
Copy number alterations (CNAs) are typically measured by the ratio of tumor to normal DNA abundance. However, tumor purity and ploidy affect this ratio and must be incorporated to infer absolute copy numbers.1,2 Information from germline single-nucleotide polymorphisms (SNPs) further allows deconvolution of absolute copy number into the 2 parental copy numbers. This parental or allele-specific copy number provides a direct readout of loss of heterozygosity (LOH; when either the maternal or paternal copy is lost), which can indicate the complete loss of wild-type function when a somatic mutation in a putative tumor suppressor is identified.3 Inferring allele-specific copy number is further crucial to understanding mutagenesis, allowing determination of clonality and timing of copy number changes at the same locus.2,4,5
Whole-exome sequencing (WES) and targeted panel sequencing have become routine applications in the clinic, providing comprehensive data while saving cost and scarce tumor tissue by eliminating the need for multiple single-analyte assays. Therefore, such comprehensive tests may aid treatment decision making by increasing the detection of actionable alterations, which includes point mutations and amplifications of oncogenes in targeted therapies, microsatellite instability (MSI), and tumor mutational burden (TMB) in immunotherapy.6,7
CONTEXT
Key Objective
The current study explores the feasibility of tumor-only sequencing to determine various complex biomarkers beyond known driver mutations and provides open-source implementations.
Knowledge Generated
We demonstrate that sophisticated algorithms can, in many cases, minimize the need for sequencing matched normal specimens. Our workflow is available for download and in the Terra Cloud platform.
Relevance
Clinical tumor-only sequencing reduces time and cost over matched tumor and normal sequencing and enables analyses of the large number of specimens for which blood samples are unavailable.
Sequencing both tumor and matched normal specimens provides certain benefits over tumor-only sequencing, even in diagnostic settings where alterations of uncertain significance are usually ignored. For example, high-depth sequencing of blood samples can more reliably identify clonal hematopoiesis, hotspot mutations that arose in heme rather than in tumor cells.8-10 Matched normal samples are also commonly required for existing algorithms to detect complex biomarkers such as MSI, TMB, or LOH. Obtaining comprehensive information from clinical tumor-only sequencing data could reduce time and cost, while enabling analyses of the large number of archived specimens for which blood samples are unavailable. However, the reliability of tumor-only sequencing is not well assessed,11,12 and validated open-source analysis tools are lacking.
Without matched normal samples, it is necessary to distinguish algorithmically between somatic mutations and germline variants. Existing approaches commonly involve machine learning using public germline and somatic databases, in silico predictions of the functional impact of mutations, and allelic fractions (the ratios of nonreference to total sequencing reads) of mutations and their neighboring SNPs.13,14 Recently developed tools additionally use allele-specific copy number, allowing the calculation of accurate posterior probabilities for all possible somatic and germline genotypes.15-17 However, in the absence of complete workflows and thorough benchmarking, controversy has persisted over the reliability of tumor-only sequencing.12
We present a complete workflow, along with a Cloud-based implementation, for tumor-only hybrid-capture data. The workflow is based on our previously published tool PureCN.15 We benchmark an improved version against gold standard data sets of matched normal WES and Affymetrix SNP6 microarrays (Affymetrix, Santa Clara, CA) and compare it to alternative recently published methods.17,18 Using the ovarian carcinoma (OV) and lung adenocarcinoma (LUAD) data sets of The Cancer Genome Atlas (TCGA), which represent opposing extremes with respect to tumor purity, copy number heterogeneity, and TMB, we demonstrate high reliability of tumor-only analyses for inference of allele-specific copy number, identification of functional mutations, LOH, mutational signatures, and TMB.
METHODS
Data Download
BAM files were downloaded through the GDC Data Transfer Tool using manifest files built by the GenomicDataCommons R/Bioconductor package.19 The TCGAutils R/Bioconductor package20 was used to annotate the manifest file: TCGAutils::UUIDtoBarcode for transferring universally unique identifiers to TCGA barcodes and TCGAutils::TCGAbiospec for extracting biospecimen data from TCGA barcodes. Capture kit information was obtained via the GDC API. BAM files mapping to multiple capture kits were excluded. BED files containing the locations of baits based on hg19 were lifted over to GRCh38 using hg19ToHg38 liftover chain file downloaded from the University of California, Santa Cruz Genome Browser.21 None of the data analyzed in this study were used to develop or tune the algorithm or parameters and thus represent true validation sets.
Data Processing
ABSOLUTE analysis of TCGA SNP6 microarray data has been described previously.2,22,23 The manually curated ABSOLUTE output was obtained from Synapse24 and lifted over to GRCh38. In addition to the PureCN-based15 workflow described in detail in the Appendix, we applied the FACETS 0.5.618 copy number tool to all samples. Tumor and normal BAM file pairs were processed by snp-pileup with the parameters -g -q15 -Q20 -P100 -r25,0, and the outputs from which were imported using readSnpMatrix and further processed by preProcSample, procSample with cval = 150, and emcncf.
Single-nucleotide variants were called with Mutect 1.1.725 (Appendix). SGZ17 in version 1.0.0 was used to classify the mutation calls into somatic versus germline (Appendix). Variants labeled “germline,” “probable germline,” “somatic,” “probable somatic,” or “somatic subclonal” by SGZ were considered called, and all others were considered uncalled. Finally, we applied deconstructSigs26 to identify the 30 mutational signatures27 curated by the Wellcome Trust Sanger Institute28 (Appendix).
Statement of Reproducible Research
Analyses presented in this article are reproducible using the code and instructions available through GitHub.29
RESULTS
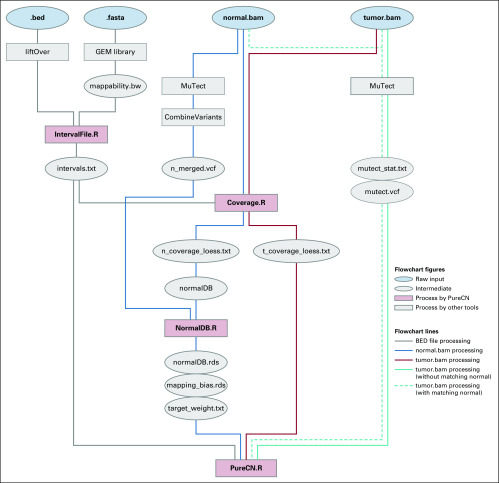
Reliable analysis of clinical tumor-only sequencing data involves multiple nontrivial steps that are distinct from the analysis of matched tumor and normal sequencing. Here, we describe and benchmark a detailed workflow for hybrid-capture tumor-only sequencing data including variant calling, coverage normalization for copy number calling, purity and ploidy inference, and classification of variants by somatic status (Appendix Fig A1).
Tumor Purity and Ploidy Inference
We selected OV and LUAD WES data from TCGA as complementary, representative data sets for our benchmarking study.30,31 Among the TCGA data sets, OV shows the highest tumor purity as a result of the availability of large surgical specimens. High purity complicates somatic versus germline classification because of the overlapping distributions of expected allelic fractions. The LUAD data set, obtained by core needle biopsies, in contrast, ranks among the lowest in tumor purity, presenting a different challenge for copy number calling because of the dilution of signal.15,17 LUAD is additionally challenging because of increased copy number heterogeneity.2,22 Subclonal copy number changes increase the number of copy states, making ploidy inference often ambiguous.2,32
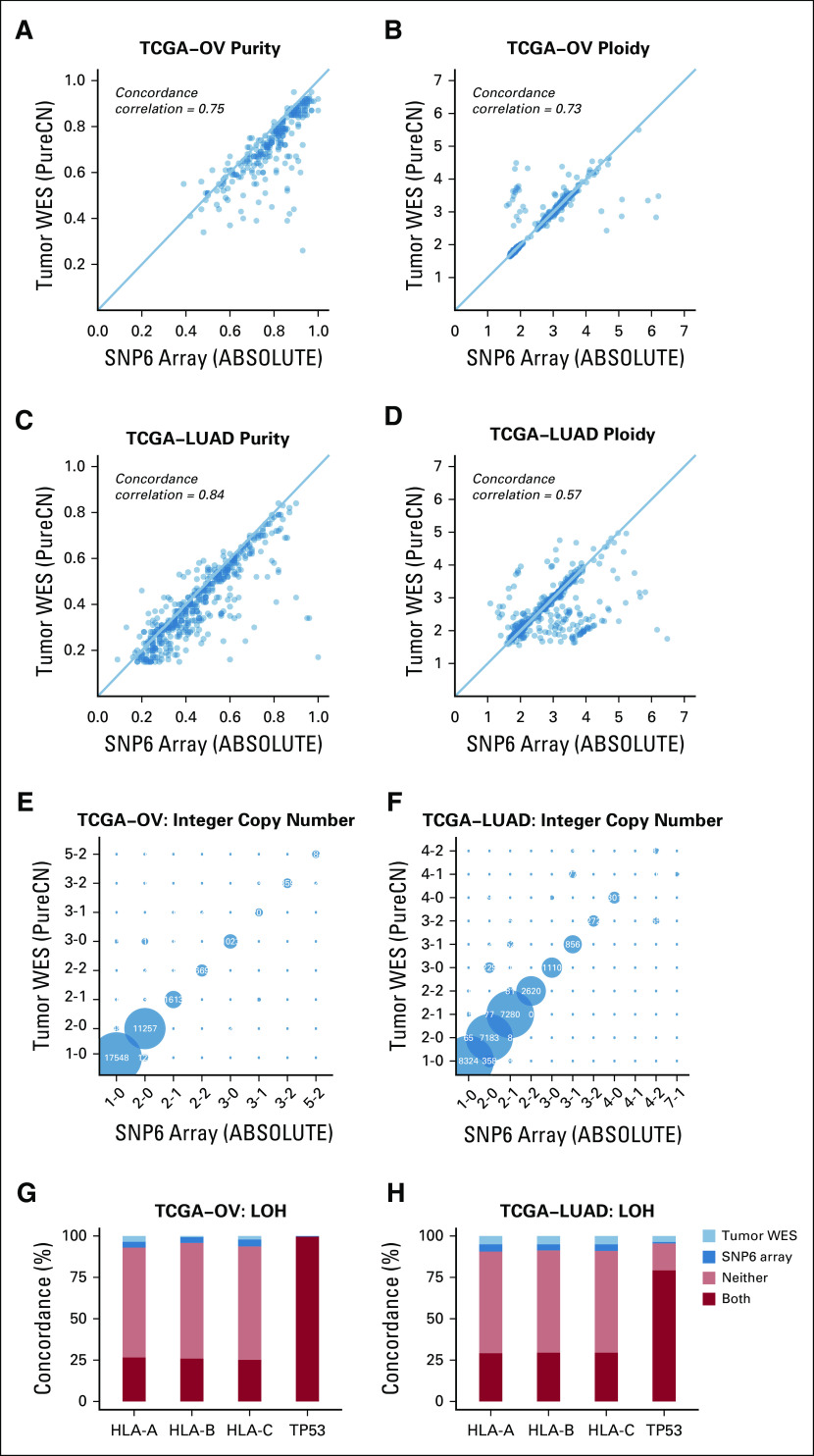
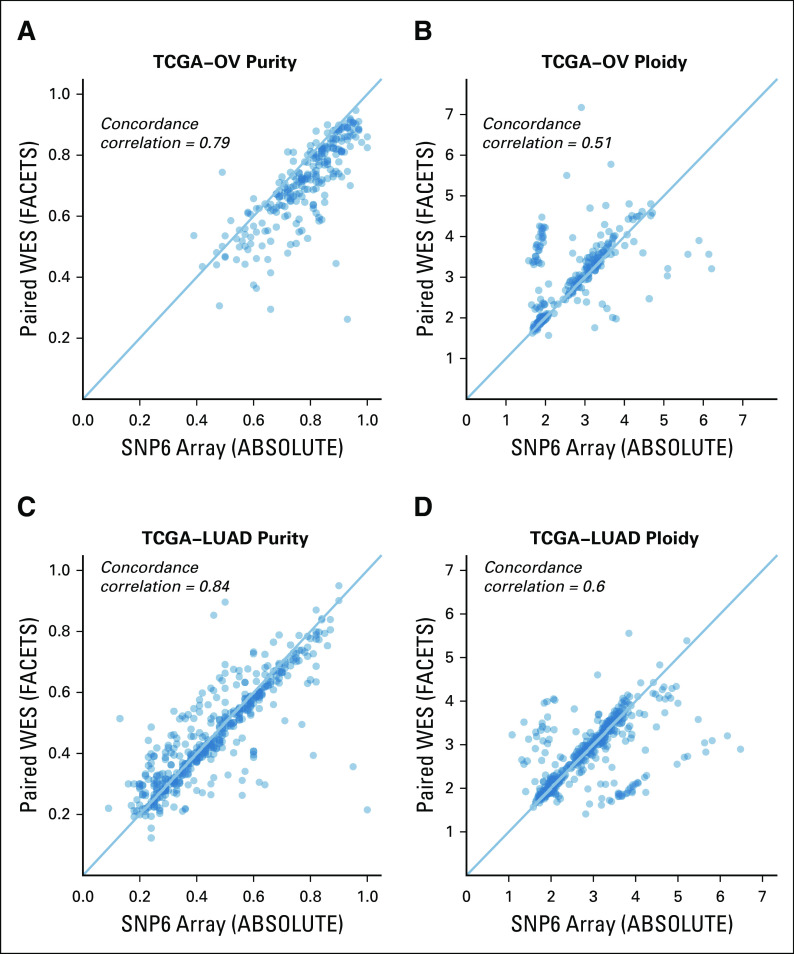
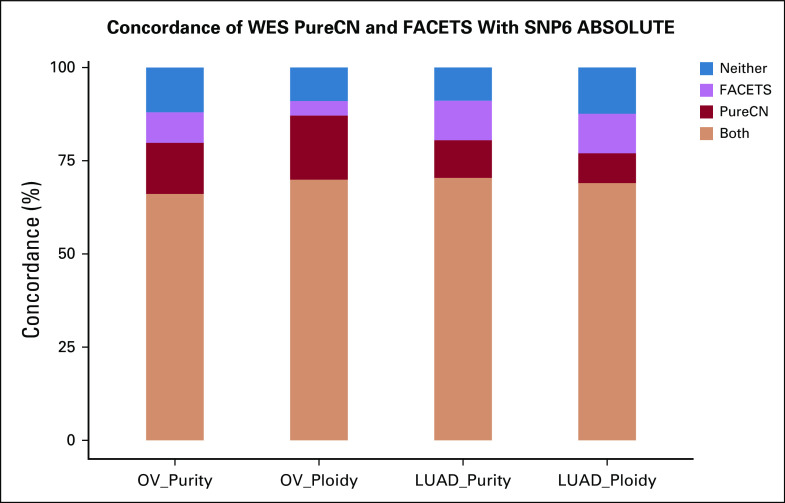
We first compared maximum likelihood purity and ploidy estimates from our workflow using tumor WES with those from manually curated ABSOLUTE SNP6 microarray calls (Figs 1A to 1D, Data Supplement, Appendix). We analyzed 233 OV and 442 LUAD samples and found a high correlation of microarray and WES results for tumor purity (Pearson correlation, r = 0.75 and r = 0.84 for OV and LUAD, respectively) and tumor ploidy for OV (87.1% concordant, defined as ploidy difference < 0.5; Pearson correlation, r = 0.73). Note that since SNP6 and WES data were generated from different tissue slides, a perfect correlation of purity is not expected, whereas ploidy should be in general similar. Ploidy estimates for LUAD were also concordant in the majority of samples (77.1% concordant; Pearson correlation, r = 0.57). In addition, we applied FACETS, a widely used allele-specific CNA analysis tool for tumor and matched normal sequencing,18 to both OV and LUAD paired WES data (Appendix Fig A2). For 68.9% of all samples, all 3 tools generated concordant purity and ploidy calls (Appendix Fig A3). For OV, PureCN showed a higher ploidy concordance with ABSOLUTE than FACETS (87.1% v 73.8%, respectively), whereas for LUAD, its concordance was slightly lower (77.1% v 79.6%, respectively). Samples of discordant ploidy, compared with concordant samples, had lower purity (39.2% v 54.3%, respectively; 2-sided Mann-Whitney, P < .0001) and lower mean coverage (100.4× v 107.2×; 2-sided Mann-Whitney, P = .03).
FIG 1.
Accuracy of purity, ploidy, and exome-wide copy number inference. (A-D) Comparison of purity and ploidy estimates from paired SNP6 microarray data (ABSOLUTE2) against those from tumor-only whole-exome sequencing (WES) data (PureCN) in ovarian cancer (OV) and lung adenocarcinoma (LUAD) samples. (E-F) Shown are concordances of the major and minor allele copy numbers of ABSOLUTE copy number alteration (can) calls with the corresponding tumor-only WES PureCN calls for all altered regions where both tools could make a call. Bubbles on the diagonal represent concordant calls. States where the minor copy number is 0 (1-0, 2-0, 3-0, 4-0) are regions in loss of heterozygosity (LOH). (G-H) Concordance of LOH calls between the 2 analyses was further reviewed on HLA-A/B/C and TP53 loci for the cases with sufficient power to detect LOH: LOH observed in both microarray and WES analyses (both, dark red); absent in both analyses (neither, orange); detected only from microarray data (SNP6 array, dark blue); and detected only from WES data (tumor WES, light blue). TCGA, The Cancer Genome Atlas.
Allele-Specific Copy Number and LOH
We further analyzed the accuracy of allele-specific copy number analysis by comparing ABSOLUTE from SNP6 data with the corresponding numbers called by PureCN on WES data. We restricted our comparison to the samples with concordant ploidy calls and tumor purity > 30% and demonstrated a high concordance of copy number calls (Figs 1E and 1F).
In an LOH event, the minor copy number is by definition 0; LOH calling is thus a special case of allele-specific copy number calling. We examined 2 specific loci of main clinical interest, HLA-A/B/C and TP53, in more detail. TP53 is lost most frequently in ovarian cancer, and HLA LOH is the major interest in immunotherapy.33,34 HLA and TP53 loci were investigated in 143 and 223 OV cases, respectively, where both tumor-only WES and SNP6 array made LOH calls (Data Supplement). For LUAD, the same comparison was done in 298 and 332 samples for HLA and TP53 loci, respectively. In OV, the mean agreement in LOH status between tumor-only WES and SNP6 microarray was 94.2% for HLA and 99.6% for TP53 (Fig 1G). In LUAD, it was 91.0% for HLA and 95.5% for TP53 (Fig 1H), with the discordant samples showing low purity (average of 30.9% v 43.3% tumor purity for discordant v concordant samples, respectively; 2-sided Mann-Whitney, P < .0005).
Classification of Variants by Somatic Status
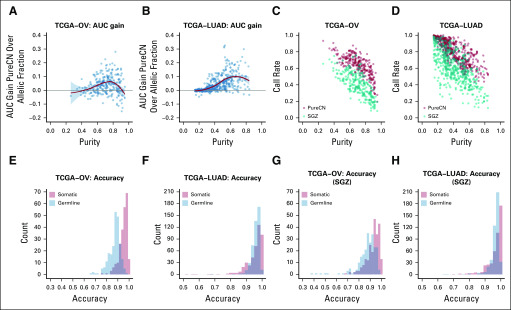
We next evaluated the somatic status predictions of variants not found in public germline databases. We first compared predictions against a simple model that uses only allelic fractions. This essentially compared the performance of commonly used ad hoc allelic fraction filters such as 0.4 against our model that adjusts allelic fractions for allele-specific copy number. We observed a significant improvement over this simple model in tumors with purity > 30% (Figs 2A and 2B, Data Supplement). At tumor purity < 30%, inclusion of copy number does not provide a benefit for classification because of the large difference in expected allelic fractions of germline and somatic variants. A small number of cases were observed in which the simple model performed slightly better in terms of area under the curve; these were mainly cases with small numbers of CNAs. However, the complex model still provides a benefit in that it returns a probability.
FIG 2.
Accuracy of variant classification. (A-B) Gain in area under the curve (AUC) of the somatic status prediction by PureCN over a model that only uses allelic fractions, shown as a function of tumor purity. (C-D) Correlation of tumor purity and call rates in ovarian cancer (OV) and lung adenocarcinoma (LUAD) for PureCN (red) and SGZ (teal).17 (E-H) Histograms of accuracy rates for all samples. These are the fractions of variants correctly called as somatic (orange) or germline (blue). TCGA, The Cancer Genome Atlas.
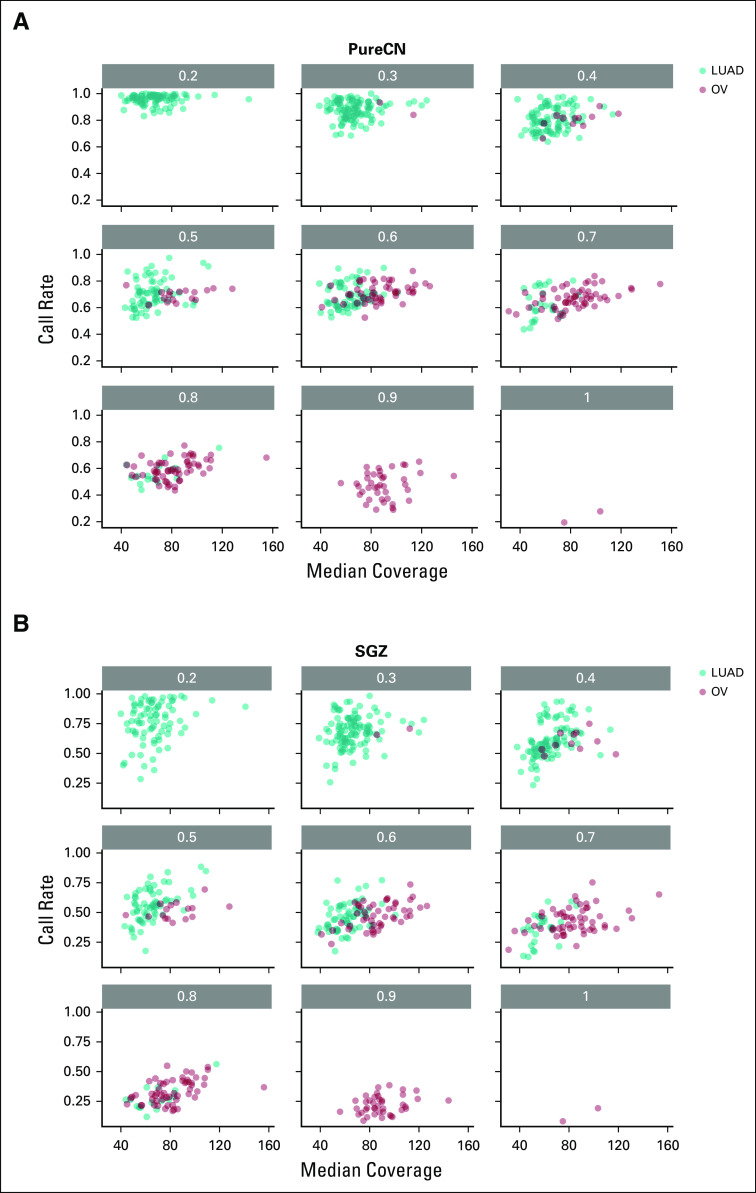
We then examined how many variants can be classified as either germline or somatic with reasonable certainty (Data Supplement). As expected, this call rate was largely a function of tumor purity (Figs 2C and 2D). Increasing sequencing coverage also increased these rates (Appendix Figs A4A and A4B). Somatic variants were classified with higher median accuracy than germline variants (96.1% v 88.1%, respectively, in OV; and 97.2% v 96.6%, respectively, in LUAD; Figs 2E and 2F). This is also expected because the somatic group includes subclonal mutations, which are usually easier to classify than monoclonal mutations because of their lower allelic fractions and therefore higher allelic fraction difference compared with germline. We observed a similar median somatic and germline accuracy using SGZ (94.0% and 88.9%, respectively, in OV; and 98.4% and 97.3%, respectively, in LUAD; Figs 2G and 2H),17 but with lower median call rates (39.5% and 59.5% for OV and LUAD, respectively, for SGZ v 64.4% and 82.2%, respectively, for PureCN).
We further investigated the ability to detect functionally important mutations using a driver detection algorithm.35 Well-defined LUAD driver genes such as TP53, KRAS, KEAP1, and STK11 were called in both tumor-only and paired analyses. We observed a small number of false-positive hits from sequencing artifacts that the matched normals, but not the pool of normals, filtered out (Data Supplement).
TMB
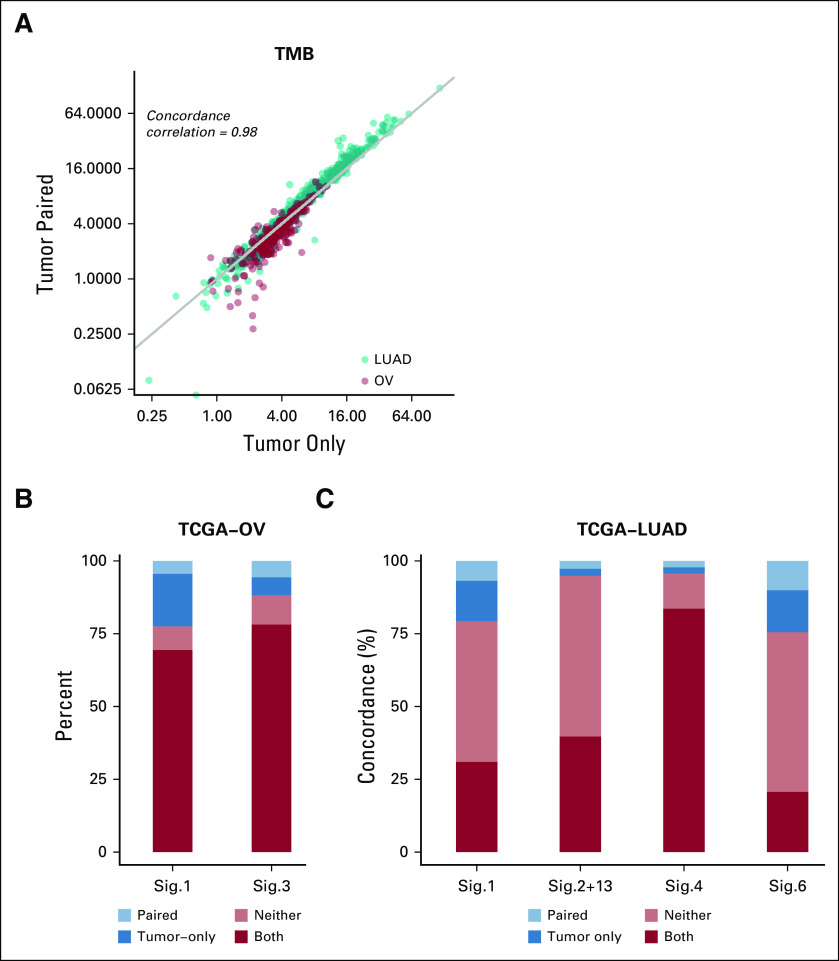
We next sought to investigate the accuracy of the variant classification for determining TMB (Appendix). From the comparison of tumor-only and paired analysis modes, we found a high concordance (Pearson correlation, r = 0.98) and good calibration of somatic mutation rates per megabase in both OV and LUAD (Fig 3A; Data Supplement). The mean absolute difference in somatic rates per megabase of the matched versus tumor-only pipeline was 0.60 Mb for OV and 1.74 Mb for LUAD. A simplified pipeline that removed variants with allelic fraction > 0.4 and was otherwise identical showed differences of 0.9 Mb for OV and 1.80 Mb for LUAD compared with the matched pipeline (Data Supplement).
FIG 3.
Tumor mutational burden (TMB) and mutational signatures. (A) TMB from ovarian cancer (OV; red) and lung adenocarcinoma (LUAD; teal) samples in tumor-only versus paired modes shown on a log scale. (B-C) Concordance of COSMIC mutational signatures (Sig.) between tumor-only and paired analysis modes: mutational signatures observed in both tumor-only and paired modes of analysis (both, red); absent in both analyses (neither, orange); detected only from tumor-only analysis (tumor only, dark blue); or detected only from paired mode of analysis (paired, light blue). TCGA, The Cancer Genome Atlas.
Mutational Signatures
To further evaluate the clinical utility of our workflow, we assessed the accuracy of mutational signature identification36 from tumor WES data with or without matched normal profile. Among 30 validated mutational signatures, we investigated the 2 OV-associated mutational signatures with known etiology in detail (Fig 3B). Signature 1 has been found in all cancer types and is linked to aging. Signature 3 is associated with homologous repair deficiency, a potential biomarker for PARP inhibition in ovarian cancer.37 We obtained a high agreement for mutational signature calls from tumor-only and paired analyses (77.5% for signature 1 and 88.1% for signature 3), confirming that our workflow can detect mutational signatures without matched normal profile even in high-purity samples.
In LUAD data, we again reproduced previously associated signatures of known etiology. In addition to the aging signature 1, we found a significant fraction of samples dominated by the APOBEC (signature 2 and signature 13), tobacco (signature 4), and DNA mismatch repair deficiency (signature 6) signatures (Fig 3C, Data Supplement). We observed high agreement between tumor-only and matched normal data for all these signatures (79.3%, 94.8%, 95.7%, and 75.5% for signatures 1, combined 2 and 13, 4, and 6, respectively).
Terra Pipeline
The described workflow is available as a shareable workspace on Terra (formerly known as FireCloud) of the Broad Institute (Cambridge, MA; Appendix). Users can thus easily test the workflow and apply it to their own data stored in Google Cloud Storage (Google, Mountain View, CA) or to data already hosted by Terra, such as TCGA.
DISCUSSION
We present a complete workflow for reliable analysis of clinical tumor-only WES data without matched normal samples. This workflow is validated on OV and LUAD data from TCGA and benchmarked against a gold standard, manually curated analysis of SNP6 microarray data with matched normals. Our workflow estimates tumor purity, ploidy, LOH, TMB, and mutational signatures with high concordance to established workflows for SNP6 and WES data with paired tumor and normal samples.
TMB is an emerging biomarker for response to immunotherapy,38-41 but the current lack of standards significantly challenges implementing TMB testing in the clinic.42 To our knowledge, this is the first thoroughly validated open-source, tumor-only TMB pipeline. This open-source reference implementation will help establish standards for TMB calling and support its implementation in standard clinical settings where tumor-only WES is performed.
Although high tumor purity challenges somatic status classification (Figs 2C and 2D), the proposed approach to determining clinically relevant biomarkers such as TMB and somatic signatures is surprisingly robust to varying tumor purity (Fig 3). Notably, signatures of clear etiology such as homologous repair deficiency, APOBEC, or smoking had a significantly higher concordance with matched analyses than broader and less certain signatures, such as those associated with aging. In contrast, we also note that high tumor purity is beneficial for LOH and copy number calling. Still, all parts of the workflow achieved high accuracy in tumors of 40%-60% purity, the range in which most clinical tissue specimens fall.
Increasing sequencing coverage increases the accuracy of somatic status classification16 and ploidy inference. The results presented in this study are based on relatively low-coverage WES sequencing to an average of 100×. The substantial improvements in sequencing costs and runtimes of current-generation instruments such as Illumina NovaSeq (Illumina, San Diego, CA) make much deeper sequencing of WES feasible. Therefore, we expect accuracies reported here to be pessimistic estimates for assays implemented in the clinic.
The average runtime of a WES sample was approximately 3 hours and required 3.5 GB of RAM on an Intel Xeon E5-2680 v4 cluster node (Intel, Santa Clara, CA). Parallelization could reduce the runtime to approximately 30 minutes per sample, making application in high-throughput clinical settings feasible. This is an order of magnitude more than matched tumor and normal allele-specific CNA callers.18 These tools usually average coverage and SNP allelic fractions across segments in their likelihood models to reduce data points dramatically. Because without matched normal the germline status of variants is not available, PureCN in contrast includes this uncertainty in the likelihood model, resulting in the longer runtime.
This study has several limitations. First, we focused on benchmarking our tumor-only workflow where it differs from standard matched tumor and normal analyses. A systematic evaluation of accuracy for the variant calling steps upstream of this workflow is beyond the scope of this study.43,44 Second, our workflow is currently not designed for whole-genome sequencing (WGS) data. In contrast to gold standard WGS tools, PureCN was designed for high-coverage data (> 100×) and currently does not use information largely unavailable in hybrid-capture data such as split reads or SNP phasing. These would be straightforward additions once high-coverage diagnostic WGS becomes common in oncology. However, support for WGS would likely require the implementation of additional heuristics to achieve acceptable runtimes, for example by averaging information in noncoding regions. Third, as with allele-specific CNA calling in matched tumor and normal data, purity and ploidy inference can be ambiguous in a minority of cases of low purity or of high heterogeneity. Therefore, our pipeline provides tools that allow manual correction of results by trained curators, described in the documentation of the PureCN package. Importantly, the accuracy of TMB calling was robust even to inaccuracies in ploidy, partly because different ploidy solutions can be equivalent for variant classification.17
Fourth, all samples in this study originated from high-quality fresh frozen samples from only 2 cancer types, and only limited benchmarking on formalin-fixed paraffin-embedded samples was previously done.15 Cancer types that have proven to be difficult to analyze with ABSOLUTE (eg, chromosomally stable samples from patients with myeloproliferative disease2) are expected to be similarly challenging with PureCN. Finally, reliable labeling of clonal hematopoiesis from tumor-only or low-coverage matched normal sequencing remains a shortcoming but is an area of research we are currently pursuing.
As a result of the high concordance with matched tumor and normal sequencing, the proposed workflow supports the clinical application of tumor-only sequencing, especially in diagnostic settings. Furthermore, implementation of the workflow on Terra will facilitate users, even those with no coding experience, to process their own data in the Cloud.
APPENDIX
Installation
PureCN can be obtained and installed under the Artistic 2.0 license from Bioconductor (https://doi.org/doi:10.18129/B9.bioc.PureCN), Bioconda (https://bioconda.github.io/recipes/bioconductor-purecn/README.html), or GitHub (https://github.com/lima1/PureCN). Unless otherwise specified, R scripts referred to in this Appendix are part of the PureCN package.
Dependencies
Genome reference FASTA files were downloaded from National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/001/405/GCA_000001405.15_GRCh38/seqs_for_alignment_pipelines.ucsc_ids/GCA_000001405.15_GRCh38_no_alt_analysis_set.fna.gz). dbSNP version 135 was downloaded from https://software.broadinstitute.org/gatk/download/bundle, and COSMIC version 77 was downloaded from https://cancer.sanger.ac.uk/cosmic. As an alternative to dbSNP, PureCN also supports population allele frequencies provided by databases such as GnomAD or ExAC provided as POPAF or POP_AF info field in VCFs. By default, in this case, a population allele frequency of 0.001 or higher defines known germline variants.
The workflow described in this article assumes that germline single-nucleotide polymorphisms (SNPs) and somatic mutations were identified by MuTect 1.1.7,25 which requires Java 1.7. Other commonly used variant callers with tumor-only mode can be used instead, but the resulting VCFs need to be filtered for common artifacts before subjecting them to this workflow.
This workflow further requires a read mappability file for the reference FASTA file. The mappability score mi is defined as 1/(No. of alignments) for a k-mer starting at position i in the reference genome. For hg19, these scores can be downloaded as precomputed for various k-mer sizes from the University of California, Santa Cruz (UCSC) Genome Browser (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/encodeDCC/wgEncodeMapability/). For other reference genomes, we require the GEM library version 1.315 (Derrien T, et al: PLoS One 7:e30377, 2012).
Finally, GATK3 CallableLoci was used to collect callable regions with sufficient coverage, mappability, and sequence quality. VCFs were combined into a single multisample VCF with GATK3 CombineVariants.
Variant Calling
MuTect was run separately on both tumor and normal BAM files (Appendix Fig A1) using arguments –dbSNP Homo_sapiens_assembly38.dbsnp.vcf and –cosmic CosmicCodingMuts.vcf. For benchmarking purposes, MuTect was also run in matched normal mode by providing both tumor and normal BAM files and otherwise identical parameters (Appendix Fig A1, dashed line). Capture kit intervals were not provided to include all SNPs with sufficient coverage in the flanking regions of baits. Normal samples were run in artifact detection mode (–artifact_detection_mode argument) and then combined into a single multisample VCF (referred as normal.panel.vcf.file in the following) using GATK3 CombineVariants with argument –minimumN 5. The latter specifies the minimum number of normal VCF files containing the variant call to be included in the normal database. This was set to a high value to ensure that all individual-specific germline variants were ignored, which would otherwise indirectly provide matched normal information for some tumors.
Reference Files Generation
The first step of the workflow (Appendix Fig A1) is the generation of reference files for each capture kit. To exclude regions of low read mappability, bigWig files were generated from the GRCh38 reference genome assembly without ALT contigs using the GEM library (Derrien T, et al: PLoS One 7:e30377, 2012) and the UCSC wigToBigWig tool. The k-mer size in gem-mappability was set to the read lengths of the studies, and the maximum number of mismatches and edit distances was set to 2 (–m and –e arguments, respectively), matching the settings used by ENCODE.
Next, the script IntervalFile.R was used with default arguments to annotate the regions defined by the baits BED file with mean GC-content, mean mappability, and gene symbols. IntervalFile.R further splits on- and off-target regions into bins of maximum 400 bp and 200 kbp, respectively (Appendix Fig A1, black line), as previously described (Talevich E, et al: PLOS Comput Biol 12:e1004873, 2016).
Only capture kits that were used for > 100 samples were considered, and separate normal databases were built for each kit (Appendix Fig A1, blue lines). With these criteria, 233 ovarian carcinoma (OV) tumors and their matched normal samples were processed from 2 different capture kits (Custom V2 Exome Bait, 48 RXN X 16 tubes; and SureSelect Human All Exon 38 Mb v2). For lung adenocarcinoma (LUAD), 442 tumors and matched normal samples were processed (Custom V2 Exome Bait, 48 RXN X 16 tubes kit).
Coverage files were generated for all normal samples using Coverage.R (Appendix Fig A1, blue lines) with default arguments. This script normalizes on- and off-target coverages independently for GC-content.
The normal coverage databases for the LUAD and the 2 OV capture kits (output file normalDB.rds) were then generated with the NormalDB.R script with default arguments. In brief, outlier normal samples with very high or very low coverage were excluded (median coverage, > 4× or < 0.25×, respectively). Furthermore, intervals with no read count in > 3% of samples and average coverage < 25% of the chromosome median were removed. In total, 157 and 176 process-matched normals from 2 OV capture kits and 250 normals from LUAD were used to build the 3 normal databases.
For each interval, NormalDB.R then calculates the inverse of the log2–copy number ratio standard deviation across all normal samples and creates the interval_weights.txt output file, later used by the segmentation function to downweight intervals with high variance in normal controls.
Reads harboring nonreference alleles have a lower chance of passing filters, thus resulting in average allelic fractions of heterozygous SNPs below the expected 0.5. Therefore, NormalDB.R next computes a position-specific nonreference mapping bias (output file mapping_bias.rds) for all variants in the normal.panel.vcf.file, provided through the –normal_panel argument. Mapping bias is defined as the ratio of the sum of all alt reads over all samples versus the total number of reads of heterozygous SNPs (allelic fraction > 0.05 and < 0.9). This procedure further uses an empirical Bayes approach that adds the average number of nonreference and total reads per SNP across all samples to this ratio, thus forcing the mapping bias of rare or low-coverage SNPs closer to the average mapping bias.
Whole-Exome Copy Number Calling
Tumor coverages were calculated and GC-normalized using the Coverage.R script with default arguments, analogous to the normal coverages (Appendix Fig A1, red line). The PureCN.R script was then used for the main copy number calling step that includes tumor purity and ploidy inference as well as classification of somatic status and clonality for all variants.15 The –postoptimize flag as well as all previously mentioned reference files were provided. Variants in the UCSC simple repeat track were excluded (–snpblacklist argument). Otherwise, default parameters were used.
In brief, tumor versus normal log2–copy number ratio was first calculated and denoised using tangent normalization (Beroukhim R, et al: Nature 463:899-905, 2010), again independently for on- and off-target regions. This was changed from older PureCN versions (< 1.8) described previously,15 in which the n most similar normals were used for normalization of the tumor coverage. Since PureCN v1.10, mapping bias of variants not found in the normal database is imputed by averaging the mapping bias of the 5 neighbors on both sides, weighting each of the 10 SNPs by corresponding number of samples in the database. Then DNAcopy (Venkatraman ES, et al: Bioinformatics 23:657-663, 2007) was used for the segmentation of merged on- and off-target log2 ratios. Reliable germline SNPs present in the normal database without major mapping bias were used to improve the segmentation by Ward clustering and identification of copy-neutral loss of heterozygosity (LOH). Candidate purity and ploidy combinations for the segmented log2 ratios were identified in a 2D-grid search and subsequently optimized using simulated annealing. Allelic variants were finally fitted to all local optima, calculating somatic posterior probabilities for all variants.
The likelihood model of PureCN has been described previously.15 PureCN versions > 1.8.0 differ in 2 minor details. First, the uncertainty of copy number log2 ratio standard deviation is now included in the optimization. This is an advantage in high-quality samples where shifts in log2 ratio across chromosomes can sometimes exceed the average noise within segments. Second, the observed sample ploidy can differ from the true ploidy, especially in smaller gene panels that cover only small fractions of the genome. Previously described PureCN versions modeled this potential deviation as a function of the sample noise, but this was changed to a function of tumor purity.
Classification of Variants by Somatic Status
Variants with a somatic posterior probability ≥ 0.8 were classified as somatic, whereas those with a probability of ≤ 0.2 were classified as germline. Although this cutoff may seem arbitrary and liberal, the assumption is that such a classification of specific variants is mostly of interest when additional information strongly suggests functional significance, such as determined by in silico functional prediction tools or as a result of location in hotspot domains of relevant genes. All variants found in germline databases with small prior probability of being somatic were excluded from benchmarking.
SGZ17 in version 1.0.0 was applied to all WES data. SGZ is methodologically similar to PureCN but does not include the uncertainty of allele-specific copy number in the posterior probability calculation and is not correcting allelic fractions for nonreference mapping bias. Because SGZ does not ship with a copy number tool, allele-specific copy number data as generated by the PureCN callLOH function were provided. Variants flagged by PureCN for recurrent presence in the pool of normals or for high imputed mapping bias were excluded. The same set of variants was thus used for both tools. Parameters of both tools including classification cutoffs were specified before data analysis.
Tumor Mutational Burden
To call tumor mutational burden (TMB), defined as the number of somatic mutations per megabase, Dx.R was run with the –callable and –snpblacklist flags and otherwise default arguments, confining the regions of interest to bases reliably callable by MuTect and excluding simple repeats. Callable regions were obtained by GATK3 CallableLoci with a minimum read depth of 30 (–minDepth argument) and otherwise default parameters. Noncoding regions were excluded from the CallableLoci output using FilterCallableLoci.R. Mutations with a posterior probability > 0.5 for being somatic, and that were also not included in germline databases and not flagged by PureCN, were included in the TMB calculation. In the matched tumor and normal TMB pipeline, somatic variants were assigned a prior somatic probability of 0.999 and germline SNPs a prior of 0.0001; otherwise, identical parameters were used. Note that for TMB, we use a different posterior probability cutoff of 0.5 instead of 0.8. The assumption here is that assignment errors in the 0.2 to 0.8 range will roughly cancel each other out. The inclusion of all variants is also important to avoid a bias in which TMB is underestimated in higher purity samples where variant classification is more difficult. Importantly, PureCN reports both somatic and private germline rates (number of SNPs per megabase not found in the used germline database); clear outlier cases in private germline rates, which should not show dramatic differences across individuals, indicate that many of the uncertain variants were misclassified. In our experience, this is rare even in high-purity samples.
Mutational Signatures
Dx.R was run with the –signature argument that internally calls the deconstructSigs R package.26 Somatic variant filtering was identical to the TMB step, with the exception that noncoding regions were kept to increase the number of mutations. Samples with ≤ 50 somatic mutations were excluded as recommended,26 leaving 160 OV and 368 LUAD samples for analysis.
Terra Workspace
To make our copy number variant (CNV) analysis method easy to use on new data, we built a shareable workspace on Terra (formerly known as FireCloud) of the Broad Institute (Cambridge, MA). Currently, our Tumor_Only_CNV workspace consists of 7 workflows, written in workflow description language, and 5 Jupyter Notebooks, written in R. Workflows cover the whole analysis process from the raw input files to final outputs, with Notebooks describing the accessory data processing and downstream analysis steps. The workspace (https://app.terra.bio/#workspaces/waldronlab-sehyun/Tumor_Only_CNV) can be accessed by logging into Terra with a Gmail account.
For public accessibility, our workspace is built on synthetic example data, consisting of 92 normals and 8 tumors labeled as “neutral” and “case,” respectively. Users can apply the Terra workspace to their own data stored in Google Cloud Storage (Google, Mountain View, CA) or to data already hosted by Terra, such as The Cancer Genome Atlas (TCGA).
The 7 workflows are named based on the order of the processes and the major tool used in each workflow. Three workflows with the prefix “1_” can be run at the same time because they do not require any output from other workflows as their input. Briefly, 1_PureCN_IntervalFile generates an interval file from a BED file. 1_MuTect1_Variants_Calling on tumor samples identifies germline SNPs and somatic mutations. 1_MuTect1_PON is applied to a pool of process-matched normals and builds a single VCF containing variants present in more than a user-defined number of samples, which is later used to filter nonreference read mapping biases. 2_PureCN_Coverage runs separately on tumor and process-matched normal samples to calculate GC-normalized coverage. 3_PureCN_normalDB builds a normal database for coverage normalization. 4_PureCN_PureCN takes the assay-specific files created from the previous workflows to normalize, segment, and determine purity and ploidy of tumor samples. Finally, 5_PureCN_Dx extracts copy number and mutation metrics from 4_PureCN_PureCN output.
Four of the 5 notebooks cover preprocessing of input files. 1_Annotate_Manifest demonstrates how to build sample manifest files when using TCGA data, using GenomicDataCommons and TCGAutils R/Bioconductor packages. 2_Build_Data_Table illustrates how to custom-subset data in a proper format for the Terra data model. 3_Format_BED creates a properly formatted BED file before it is used in the 1_PureCN_IntervalFile workflow. 4_Download_SNP_Blacklist shows how to directly download UCSC simple repeats, an input for the snpblacklist variable of PureCN and Dx workflows. The final notebook, 5_Downstream_Analysis, demonstrates how to extract major results from the analysis, such as purity and ploidy estimates, LOH, and TMB. As a result of lack of gene annotation in the synthetic example data, some outputs from 4_PureCN_PureCN and 5_PureCN_Dx are not available with synthetic data sets. Instead, we provide a list of default outputs from one of the ovarian cancer samples analyzed in this article.
FIG A1.
Copy number alterations analysis workflow. Raw input data files and the intermediate/processed data files are depicted as blue and gray oval shapes, respectively. R scripts provided by PureCN are depicted by rose squares, and third-party tools are depicted by gray squares. Gray solid lines indicate how the target region information is processed. Blue and red solid lines describe how normal and tumor BAM files are processed, respectively. Dashed and solid teal lines show how germline single-nucleotide polymorphisms and somatic mutations were prepared with or without matched normal, respectively.
FIG A2.
Purity and ploidy estimates using an alternative tool. Purity and ploidy estimates from paired whole-exome sequencing (WES) data were obtained using FACETS. As in Figure 1, 233 ovarian cancer (OV) and 442 lung adenocarcinoma (LUAD) samples were analyzed and compared with ABSOLUTE calls. (A) Purity and (B) ploidy estimates of OV. (C) Purity (436 cases are plotted because FACETS did not return a purity estimate for 6 of the LUAD samples as a result of insufficient information) and (D) ploidy estimates of LUAD. TCGA, The Cancer Genome Atlas.
FIG A3.
Concordance of PureCN and FACETS with ABSOLUTE. From 233 ovarian cancer (OV) and 436 lung adenocarcinoma (LUAD) cases, concordance was calculated of whole-exome sequencing (WES)–based estimates from PureCN and FACETS with SNP6 array-based ABSOLUTE calls. Concordance was defined as a purity difference < 0.1 and a ploidy difference < 0.5. Estimates agreed by all 3 methods (both, orange); agreed by ABSOLUTE and PureCN only (PureCN, red); or agreed by ABSOLUTE and FACETS only (FACETS, purple); or neither PureCN nor FACETS agreed with ABSOLUTE (neither, blue).
FIG A4.
Correlation of call rates and median sequencing coverage. Median coverage is plotted against call rate for different purity ranges. LUAD, lung adenocarcinoma; OV, ovarian cancer.
SUPPORT
Supported by National Human Genome Research Institute Grant No. U24-HG010263 and Division of Cancer Epidemiology and Genetics, Informatics Technology for Cancer Research program National Cancer Institute Grant No. U24-CA180996.
Preprint version available on bioRxiv.
AUTHOR CONTRIBUTIONS
Data analysis and interpretation; Sehyun Oh, Ludwig Geistlinger, Markus Riester
Collection and assembly of data; Sehyun Oh, Marcel Ramos
Conception and design: Sehyun Oh, Martin Morgan, Levi Waldron, Markus Riester
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to: www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Markus Riester
Employment: Novartis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Van Loo P, Nordgard SH, Lingjærde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21:374–382.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi W, Ng CKY, Lim RS, et al. Reliability of whole-exome sequencing for assessing intratumor genetic heterogeneity. Cell Rep. 2018;25:1446–1457. doi: 10.1016/j.celrep.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KS, Yadav VK, Pei S, et al. SomVarIUS: Somatic variant identification from unpaired tissue samples. Bioinformatics. 2016;32:808–813. doi: 10.1093/bioinformatics/btv685. [DOI] [PubMed] [Google Scholar]
- 14.Kalatskaya I, Trinh QM, Spears M, et al. ISOWN: Accurate somatic mutation identification in the absence of normal tissue controls. Genome Med. 2017;9:59. doi: 10.1186/s13073-017-0446-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riester M, Singh AP, Brannon AR, et al. PureCN: Copy number calling and SNV classification using targeted short read sequencing. Source Code Biol Med. 2016;11:13. doi: 10.1186/s13029-016-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halperin RF, Carpten JD, Manojlovic Z, et al. A method to reduce ancestry related germline false positives in tumor only somatic variant calling. BMC Med Genomics. 2017;10:61. doi: 10.1186/s12920-017-0296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun JX, He Y, Sanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLOS Comput Biol. 2018;14:e1005965. doi: 10.1371/journal.pcbi.1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen R, Seshan VE. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan M, Davis SR: GenomicDataCommons: A bioconductor interface to the NCI Genomic Data Commons. https://www.biorxiv.org/content/early/2017/04/04/117200.
- 20. Ramos M, Schiffer L, Waldron L: TCGAutils: TCGA utility functions for data management version 1.2.1 from Bioconductor. https://rdrr.io/bioc/TCGAutils/
- 21. University of California, Santa Cruz: Genome Browser. http://hgdownload.soe.ucsc.edu/goldenPath/hg19/liftOver/hg19ToHg38.over.chain.gz.
- 22.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor AM, Shih J, Ha G, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Synapse. https://www.synapse.org/#!Synapse:syn7416143.
- 25.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wellcome Trust Sanger Institute: Mutational Signatures (v2 - March 2015). http://cancer.sanger.ac.uk/cosmic/signatures_v2.
- 29. GitHub: CNV analysis workflow code for the manuscript. https://github.com/shbrief/CNVWorkflow_Code.
- 30.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha G, Roth A, Khattra J, et al. TITAN: Inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014;24:1881–1893. doi: 10.1101/gr.180281.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGranahan N, Rosenthal R, Hiley CT, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martincorena I, Raine KM, Gerstung M, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 38.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Büttner R, Longshore JW, López-Ríos F, et al. Implementing TMB measurement in clinical practice: Considerations on assay requirements. ESMO Open. 2019;4:e000442. doi: 10.1136/esmoopen-2018-000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krøigård AB, Thomassen M, Lænkholm A-V, et al. Evaluation of nine somatic variant callers for detection of somatic mutations in exome and targeted deep sequencing data. PLoS One. 2016;11:e0151664. doi: 10.1371/journal.pone.0151664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput Struct Biotechnol J. 2018;16:15–24. doi: 10.1016/j.csbj.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]