Abstract
PURPOSE
To develop an artificial intelligence (AI)–based model for identifying patients with lymph node (LN) metastasis based on digital evaluation of primary tumors and train the model using cystectomy specimens available from The Cancer Genome Atlas (TCGA) Project; patients from our institution were included for validation of the leave-out test cohort.
METHODS
In all, 307 patients were identified for inclusion in the study (TCGA, n = 294; in-house, n = 13). Deep learning models were trained from image patches at 2.5×, 5×, 10×, and 20× magnifications, and spatially resolved prediction maps were combined with microenvironment (lymphocyte infiltration) features to derive a final patient-level AI score (probability of LN metastasis). Training and validation included 219 patients (training, n = 146; validation, n = 73); 89 patients (TCGA, n = 75; in-house, n = 13) were reserved as an independent testing set. Multivariable logistic regression models for predicting LN status based on clinicopathologic features alone and a combined model with AI score were fit to training and validation sets.
RESULTS
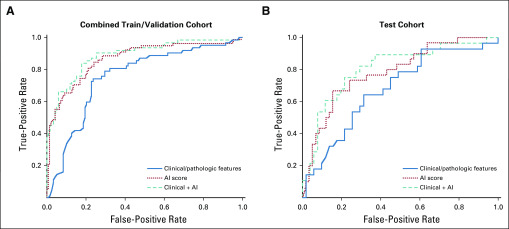
Several patients were determined to have positive LN metastasis in TCGA (n = 105; 35.7%) and in-house (n = 3; 23.1%) cohorts. A clinicopathologic model that considered using factors such as age, T stage, and lymphovascular invasion demonstrated an area under the curve (AUC) of 0.755 (95% CI, 0.680 to 0.831) in the training and validation cohorts compared with the cross validation of the AI score (likelihood of positive LNs), which achieved an AUC of 0.866 (95% CI, 0.812 to 0.920; P = .021). Performance in the test cohort was similar, with a clinicopathologic model AUC of 0.678 (95% CI, 0.554 to 0.802) and an AI score of 0.784 (95% CI, 0.702 to 0.896; P = .21). In addition, the AI score remained significant after adjusting for clinicopathologic variables (P = 1.08 × 10−9), and the combined model significantly outperformed clinicopathologic features alone in the test cohort with an AUC of 0.807 (95% CI, 0.702 to 0.912; P = .047).
CONCLUSION
Patients who are at higher risk of having positive LNs during cystectomy can be identified on primary tumor samples using novel AI-based methodologies applied to digital hematoxylin and eosin–stained slides.
INTRODUCTION
Risk stratification is essential in determining the optimal treatment strategy for patients with bladder cancer. Bladder cancer is broadly divided into two risk groups based on invasion of the muscular wall (muscularis propria) of the bladder: nonmuscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC). In general, NMIBC is associated with a relatively favorable long-term prognosis compared with MIBC, which carries a worse prognosis with up to 50% of patients developing disease recurrence.1,2 For patients diagnosed with localized MIBC, the mainstay of therapy is radical cystectomy (RC) with pelvic lymph node (LN) dissection.3 Although RC achieves negative margins in up to 96% of patients,4 recurrence rates of up to 50% during follow-up suggest that there are likely microscopic extravesical tumor deposits present at the time of surgical resection in a substantial portion of patients.5 To reduce recurrences after surgery, neoadjuvant chemotherapy (NAC) has been administered using a four-drug regimen containing methotrexate, vincristine, doxorubicin, and cisplatin.6 Although there have been attempts to further risk stratify MIBC on the basis of adverse or variant histology and clinical staging,7 there has not yet been widespread adoption of further risk stratification approaches to cluster patients with MIBC into higher and lower risk groups for selection of NAC before surgical intervention. These pathologic features are typically identified on tissue acquired from transurethral resection of bladder tumor (TURBT) procedures and have highly variable concordance with final RC findings in the literature.8 Therefore, it would be ideal if prognostic markers were developed that do not suffer from interobserver variability and reflect the nature of final pathology features at RC.
CONTEXT
Key Objective
To evaluate whether the histologic appearance and features of primary tumors can be used to predict the metastatic potential of disease at the time of surgical intervention using advanced artificial intelligence techniques. Here we investigate whether digital pathology specimens from primary tumors from RC can be used to predict whether the patient will have positive LN findings.
Knowledge Generated
Convolutional neural networks are able to learn distinct features within tumor specimens which, combined with microenvironmental features, are associated with increased likelihood of positive LNs, and these features are independent of known clinicopathologic prognostic features.
Relevance
MIBC suffers from a lack of robust clinical and pathologic variables for stratification of patients with a high risk of recurrence after surgery; the algorithm presented here could serve as an additional prognostic biomarker in this setting.
Traditionally, pathology slides undergo qualitative interpretation and assessment by the pathologist to generate a clinical report from which a small fraction of the information is used for clinical risk evaluation. Advances in artificial intelligence (AI) techniques in computer vision using deep convolutional neural networks (CNNs) have allowed for the development of prognostic models that use digital images of pathology slides that can evaluate the entirety of the available image.9 The theoretical hypothesis of this work is that the histologic appearance of the primary tumor contains information related to the metastatic potential of that tumor. In this study, we tested this hypothesis by developing an AI-based model for identifying patients with LN metastasis based solely on evaluation of primary tumors, and the model was trained by using digitized cystectomy specimens available from The Cancer Genome Atlas (TCGA) Project and validated in a separate test cohort that included patients from our institution.
METHODS
Patient Population and Slide Review
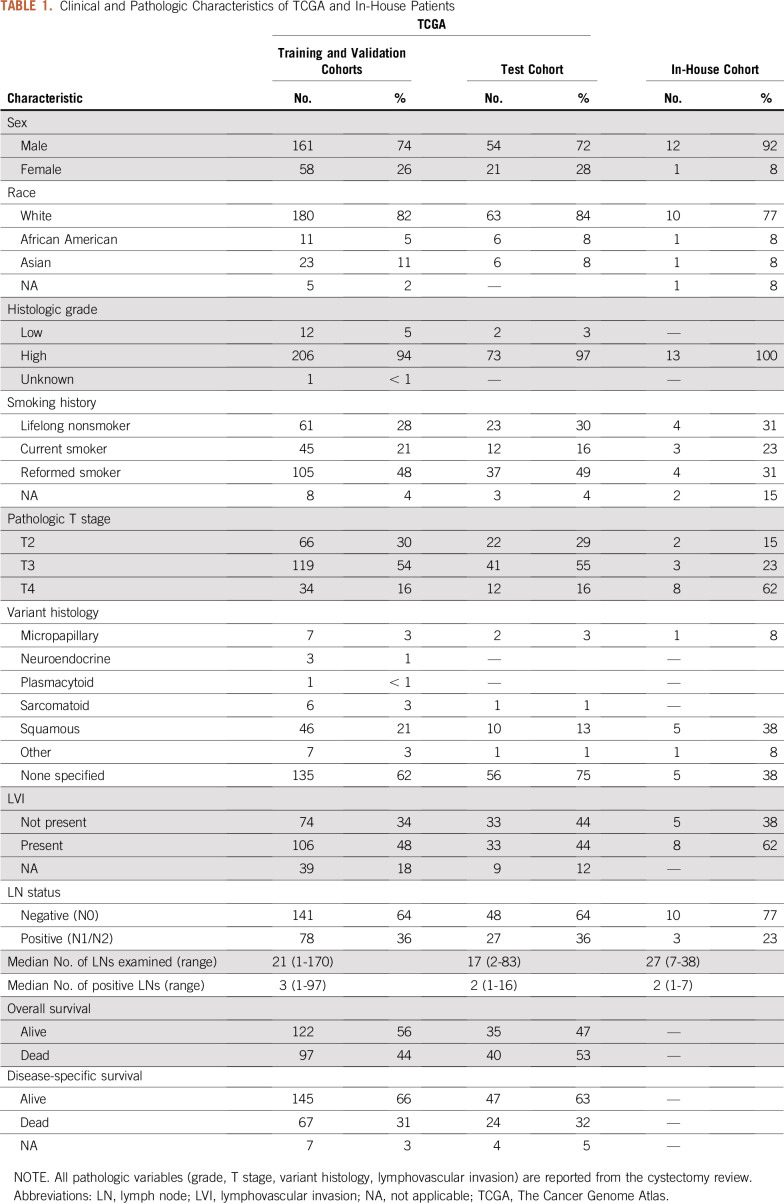
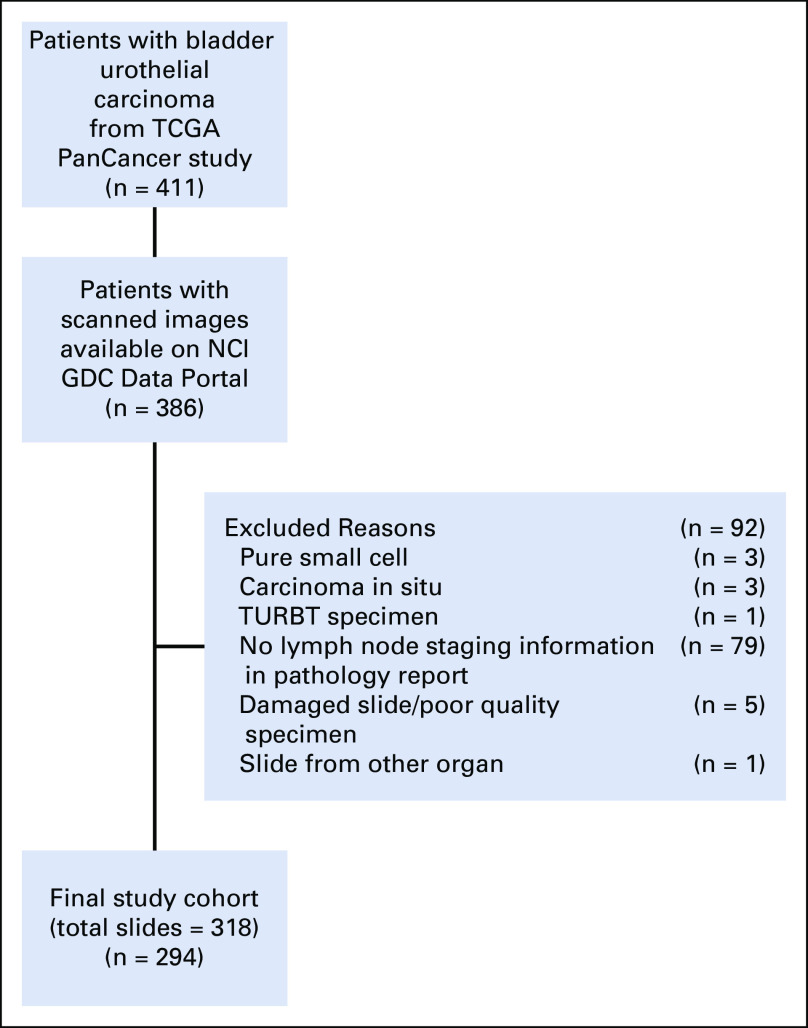
All available digital pathology slides (457 slides from 386 patients) derived from cystectomy specimens that were stained with hematoxylin and eosin (H&E) were downloaded from the National Cancer Institute Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/; accession date: November 5, 2018). The presence of LN metastasis (positive or negative), number of excised LNs, and presence of variant histology on routine H&E examination were collected from surgical cystectomy pathology reports available at the cBioPortal (http://www.cbioportal.org). Clinical, demographic, and patient outcome data were accessioned from publicly available TCGA outcomes resources reported in Liu et al.10 Download instructions for reproducibility are provided in the Appendix. T stages reported in this study refer to pathologic T stage from cystectomy. Of the variant histologies represented in the TCGA cohort (Table 1), the following were classified as high risk: neuroendocrine, micropapillary, and plasmacytoid. A board-certified pathologist (G.T.B.) reviewed all diagnostic slides for the presence and extent of tumor and manually annotated tumor regions within the slide. Ninety-two patients were excluded from this study because of missing or low-quality data (Appendix Fig A1). The number of slides that were publicly available from the TCGA cohort varied; a majority of patients had only one representative slide of the tumor specimen (median, one slide per patient; range, 1-8 slides per patient). At model inference and evaluation, only the first diagnostic slide for each patient (denoted DX1 in the TCGA cohort) was used in analysis, consistent with previous studies using this data set.11
TABLE 1.
Clinical and Pathologic Characteristics of TCGA and In-House Patients
For testing on an external cohort, 43 patients were identified from our local institution who underwent radical cystectomy between 2013 and 2019. Of these patients, 30 were excluded on the basis of the aforementioned exclusion criteria, availability of diagnostic slides for review and digitization, or those undergoing more than two types of neoadjuvant therapy. This study is compliant with local institutional review board and Health Insurance Portability and Accountability Act (HIPAA) guidelines. Although the trained algorithm was applied to all slides from an individual patient, only a single slide with the largest tumor burden was used for statistical analysis to remain consistent with the TCGA data set.
Image-Based Classification Pipeline
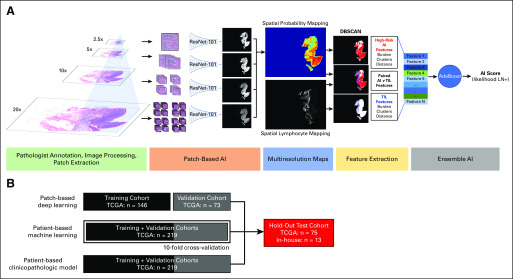
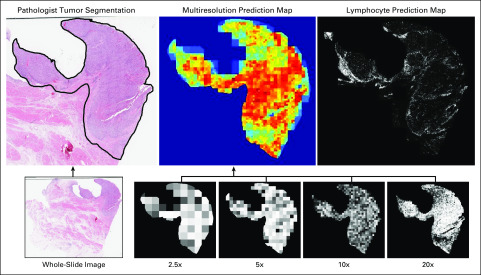
The entire image processing and classification pipeline is shown in Figure 1. TCGA patients were randomly assigned to the training cohort (50% [n = 146]), the validation cohort (25% [n = 73]), or the testing cohort (25% [n = 75]), and an in-house cohort was added to the testing set (Fig 1B). Given the large input size of digital pathology images, only regions containing tumor from the bladder cystectomy specimens were used. These regions were evenly divided into smaller image patches for deep learning-based classification. Multiresolution probability maps were constructed to allow for spatially resolved predictions from all deep learning models and were combined with microenvironmental features (burden and spatial organization of lymphocyte infiltration) to derive a final patient-level AI score, which represents the likelihood of a patient having positive LN status at the time of surgery. These procedures were then applied to the patients in the test cohort. Full details are provided in the Appendix.
FIG 1.
(A) Deep learning workflow. After tumor regions were annotated by a pathologist, patches of equal size were extracted across four resolutions for training the patch-based convolutional neural network application (ResNet-101). Spatial referencing of patch location and relative size (compared with highest resolution) allowed for construction of multiresolution probability maps from convolutional neural network outputs. These probability maps were used to define high-risk regions based on various probability thresholds as input to density-based spatial clustering of applications with noise (DBSCAN) and combined with lymphocyte infiltration maps for input to a machine learning–based classifier. (B) Training, validation, and testing schema. For patch-based deep learning, training and validation cohorts were kept separate. For patient-based machine learning, training and validation sets were combined, and 10-fold cross validation was used in model development. For clinicopathologic-based logistic regression analysis, training and validation cohorts were combined. AI, artificial intelligence; LN, lymph node; TCGA, The Cancer Genome Atlas; TIL, tumor-infiltrating lymphocytes.
Stain normalization and patch extraction.
For each digital slide, images were read in at a 4:1 ratio from apparent 40× magnification digital images to allow for whole-slide image processing.12,13 Image preprocessing included stain normalization, background artifact removal, and stain deconvolution using previously published methods.14,15 Region selection was determined by fitting a minimum number of non-overlapping regions that fully contained pathologist annotations. Within each region, 1, 4, 16, and 64 patches were mapped corresponding to 300 × 300 pixel images at magnifications of 2.5×, 5×, 10×, and 20×, respectively. The same coordinates were used to guide extraction of 576 images of 100 × 100 pixels at an effective 20× resolution from the original H&E images for estimating the presence of lymphocyte infiltration using the algorithm of Saltz et al.11,16
Patch-based classification learners.
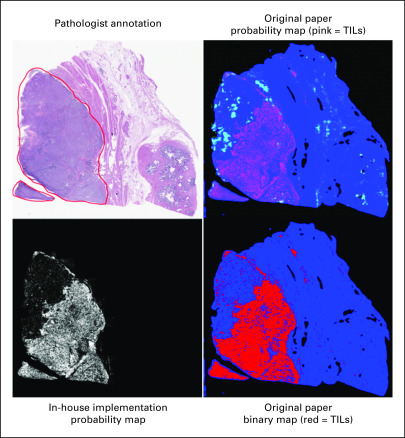
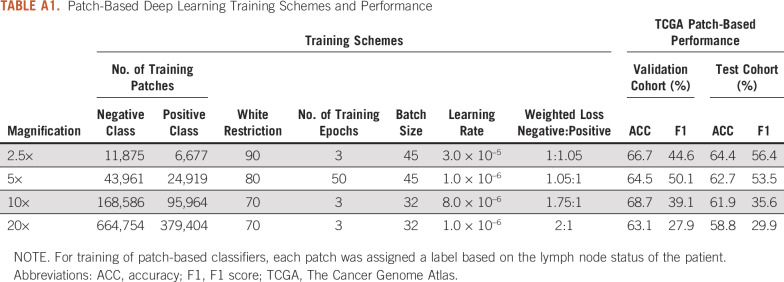
Four different models using ResNet-101 architecture were trained from patches at 2.5×, 5×, 10×, and 20× magnifications, respectively, using fast.ai library (https://github.com/fastai/fastai) with weighted cross-entropy loss. Augmentation techniques that included orientation and intensity were applied randomly during training to reduce overfitting patient-specific and slide-specific characteristics. Final training conditions are included in the Appendix and Appendix Table A1. At deployment, the corresponding output (probability of being LN positive) was saved for each patch image at every resolution. For lymphocyte classification, publicly available code and model weights were downloaded from Saltz et al,11 and consistency in results from our preprocessing methods compared with those in the original study were confirmed (Appendix Fig A2). This model was applied to the slides for all patients, and the probability of containing a lymphocyte was saved for each patch.
Post-processing and patient-based classification learner.
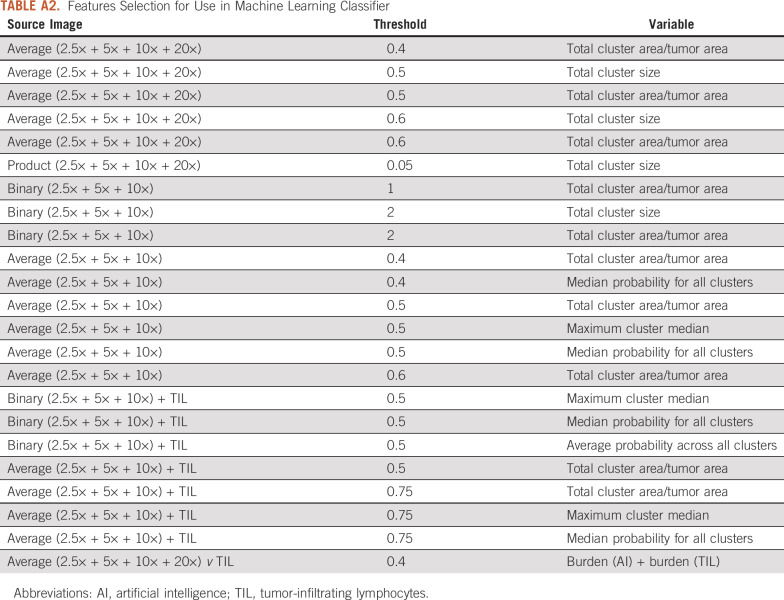
Given that all patches were extracted within known coordinates, the final result is a multiresolution prediction map in which every pixel represents the average probability of LN status across deep learning models. A simplified majority vote score was calculated as the burden of tumor patches with probability greater than 50% for LN positivity across individual models. The final score was derived from high-risk clusters (ie, high probability) that were identified with density-based spatial clustering of applications with noise (DBSCAN) using starting conditions derived from varying probability thresholds and extraction of metrics related to size, number, density, and distance between clusters and lymphocytes (Fig 1A). A full description is presented in the Appendix and Appendix Table A2. An adaptive boosting classification ensemble of decision trees (AdaBoost17) was fit using 10-fold cross validation within combined training and validation cohorts (Fig 1). Variable selection18 and training conditions are detailed in the Appendix. Classification performance was determined by cross-validation out-of-bag estimates for the training and validation cohorts and was then applied to testing cohorts. Classification outputs were saved as binary (yes/no LN positive) and continuous (range, 0-1), which is referred to as the AI score hereafter. All post-processing and AdaBoost classifications were developed in MATLAB (R2018b; www.mathworks.com).18
Statistical Analysis
The accuracy of AI-based models was reported for patch-level and patient-level results in each cohort. The final AI score, reflecting the probability of LN positivity (range, 0-1), was used to derive the area under the curve (AUC). The accuracy, specificity, sensitivity, and positive predictive value reported in the combined training and validation sets are the binary out-of-bag predictions in 10-fold cross validation.
A multivariable logistic regression model for prediction of LN status based on clinicopathologic features was developed after univariable assessment. Variables with known clinical significance and those with significant association with LN status were fit using backward Akaike information criterion. A combined model that included the AI score was added to the multivariable model and fit to the training and validation sets. Both multivariable models were applied to test sets. The classification performance of the logistic regression model was reported by the AUC and evaluated using the pROC package in R.
All reported P values are two-sided, and P < .05 was used to determine statistical significance. Statistical analyses were conducted using R software version 3.4.1 (http://www.r-project.org/). All code used for model development and statistical analysis is publicly available at https://github.com/NIH-MIP/BLCA_LNprediction.git.
RESULTS
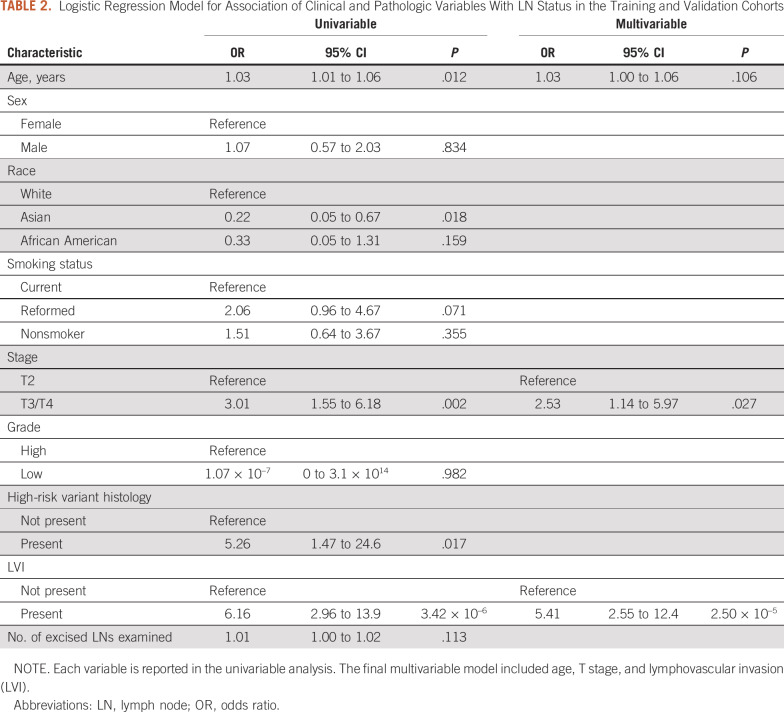
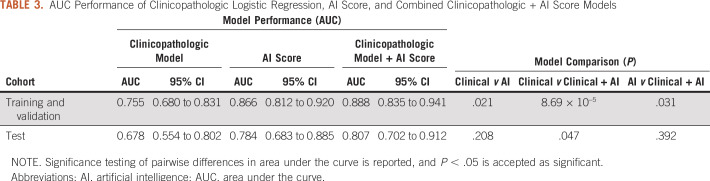
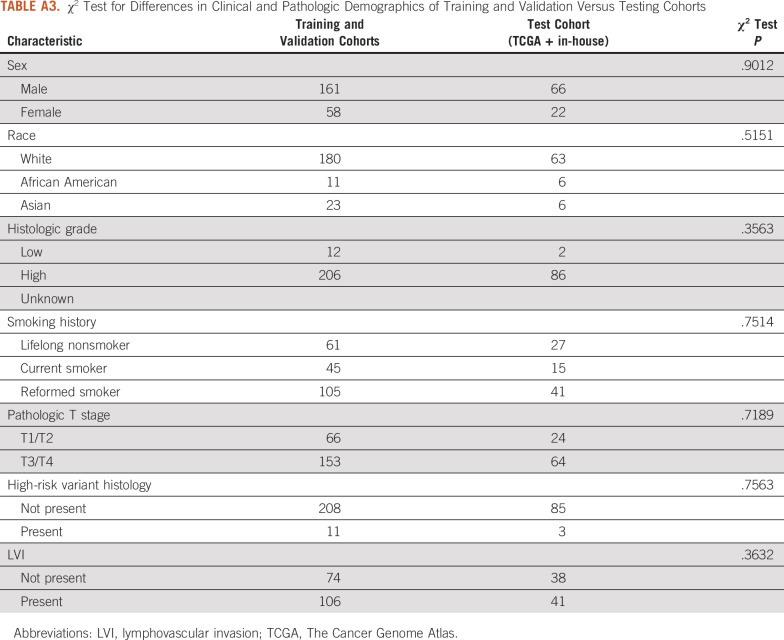
The final cohort consisted of 294 patients from the TCGA cohort and 13 patients from the in-house cohort; 105 patients from the TCGA cohort and 3 patients from the in-house cohort had positive LN metastasis at the time of surgery. Patient demographics are listed in Table 1. There were no statistically significant differences between TCGA patient samples used for the training and validation cohorts compared with those left out in the test cohort (Appendix Table A3). For multivariable analysis, a final model was constructed by using the following input variables: age, disease stage, presence of high-risk histology, lymphovascular invasion (LVI), and number of LNs excised (Table 2). After backward Akaike information criterion selection, the final multivariable model included age, T stage, and the presence of LVI. Using this model for predicting LN status resulted in an AUC of 0.755 (95% CI, 0.680 to 0.831) in the combined training and validation cohorts (Table 3). When applied to the testing cohort, the performance of the multivariable model decreased to an AUC of 0.678 (95% CI, 0.554 to 0.802).
TABLE 2.
Logistic Regression Model for Association of Clinical and Pathologic Variables With LN Status in the Training and Validation Cohorts
TABLE 3.
AUC Performance of Clinicopathologic Logistic Regression, AI Score, and Combined Clinicopathologic + AI Score Models
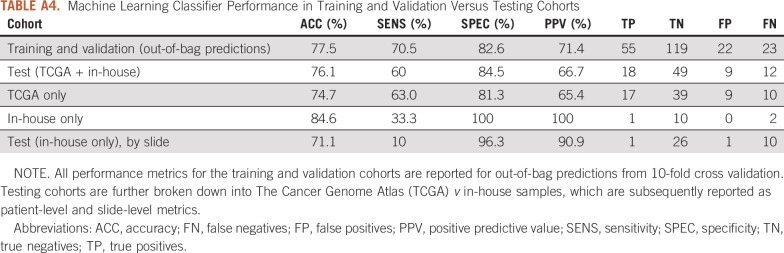
By combining features extracted from both multiresolution prediction maps and lymphocyte infiltration maps (Fig 1A), the AI score (likelihood of LN positivity) achieved an AUC of 0.866 (95% CI, 0.812 to 0.920) in the training and validation cohorts and 0.784 (95% CI, 0.702 to 0.896) in the test cohort (Fig 2; Table 3). The AI score remained significant when it was included in the multivariable logistic regression model with clinicopathologic variables (odds ratio, 6.36; 95% CI, 3.66 to 12.1; P = 1.08 × 10−9), which significantly outperformed clinicopathologic features alone in the hold-out test cohort with an AUC of 0.807 versus 0.678 (P = .047; Table 3). On its own, the AI model achieved 82.6% specificity with 71.4% positive predictive value for the combined training and validation cohorts (cross-validated out-of-bag predictions) and 84.5% specificity with 66.7% positive predictive value in the test cohort (Appendix Table A4). Specifically, accuracy was 84.6% on the patient level and accuracy was 71.1% on the slide level within the in-house test cohort.
FIG 2.
Receiver operating characteristic curves for (A) combined training and validation cohorts and (B) test cohort. In the training and validation cohorts, artificial intelligence (AI) score is reported for out-of-bag predictions from 10-fold cross validation. In both cases, the clinicopathologic + AI model significantly outperformed the clinicopathologic model alone.
Slide images from a representative true-positive patient from the test cohort are shown in Figure 3. In assessing the relationship of the AI score to known adverse pathologic features, binary classification by AI score was significantly associated with the presence of high-risk variant histology in both training and testing cohorts (χ2 test P = .019 and P = .044, respectively). In the training set, the AI score binary classification was significantly associated with LVI (χ2 test P = .0005), likely reflecting the correlation of each to LN status. In the test set, no association between AI score and LVI was observed (χ2 test P = 1).
FIG 3.
Representative example slides from a true-positive patient from an independent test set. Tumor regions from the whole slide image annotated by the pathologist were fed into each pretrained convolutional neural network for 2.5×, 5×, 10×, and 20× magnifications. All were averaged for a multiresolution prediction map and combined with a lymphocyte prediction map for machine learning–based classification.
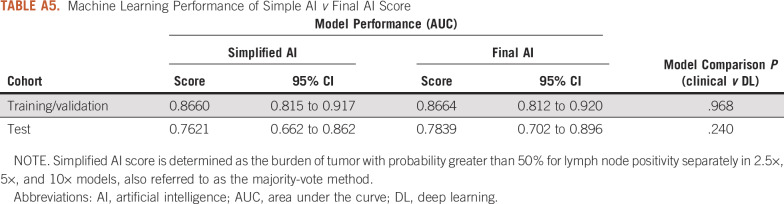
Results pertaining to patch-based training and performance at the individual magnification level and at combined magnification levels are presented in the Appendix. The overall AI score generalized better in the test cohort compared with the simplified majority vote score in which AUC was 0.762 (95% CI, 0.662 to 0.862; P = .24) in the test cohort (Appendix Table A5). Post-processing of multiresolution prediction maps and lymphocyte infiltration maps revealed that the fraction of lymphocyte-positive regions in the tumor area that were positive by the majority vote method was significantly lower than the fraction of lymphocyte-negative regions in tumor area that were negative by the majority vote method (Wilcoxon signed-rank P = 3.66 × 10−8).
DISCUSSION
Recurrence rates remain high for patients undergoing radical cystectomy after diagnosis of localized MIBC, in which current prognostication and treatment selection remains highly dependent on tumor staging, grading, and the presence of variant histologic features.19 These features are often limited or inconsistently reported.20 Thus, there is an urgent need for improved prognostication for patients with bladder cancer. In this study, we demonstrated the ability of deep learning algorithms to identify pathologic features within primary tumors associated with metastatic spread to LNs. Within this novel approach, we demonstrated that the combination of deep learning predictions with microenvironmental features results in a robust AI-based method that significantly improves prognostication compared with clinicopathologic features. This AI approach is not subject to observer variability, which demonstrates consistent mapping to the risk of LN metastasis that generalized well to a leave-out test cohort of 88 patients from 22 centers.
Classification of tumors into risk groups based on histologic characterization is a key part of clinical risk stratification in multiple types of cancers such as Gleason scoring in prostate cancer or Fuhrman grading in clear cell renal cell carcinoma.21,22 Here, expert pathologists used experience-based knowledge to define specific morphologic features to create risk groups, which are often subsequently retrospectively validated against patient outcomes. With our AI approach, CNNs learn the imaging characteristics of a tumor that are most strongly associated with LN spread. Current literature supports the notion that CNNs could have the capacity to learn unique features at each magnification level of digital pathology slides.23,24 Here, we trained separate models at multiple resolutions and spatially aggregated them to generate a tumor-level probability map. We found that the CNNs learned features known to be associated with poor prognosis, such as high-risk variant histologies. Given the improved prognostic performance of the AI approach over using standard clinicopathologic variables, we suggest that the AI approach learned unique features associated with the potential for LN spread not reported or described in traditional clinical workflow. Furthermore, this approach achieved similar performance results when applied to an independent albeit small in-house cohort.
A unique aspect of this study was the evaluation of tumor-level prognosis in the setting of substantial intratumor heterogeneity.25 The multiresolution probability maps created from CNN outputs demonstrated heterogeneity in the distribution of high-risk features defined as high probability areas. Furthermore, the information learned directly from the H&E slides was more predictive when combined with information from the tumor microenvironment (tumor-infiltrating lymphocytes). Of the twenty-six highly predictive features selected for use in the machine learning-based ensemble classifier, eight accounted for the presence and burden of infiltrating lymphocytes. The spatial distribution of tumor-infiltrating lymphocytes on H&E images was shown to have prognostic importance in multiple tumor types in the original publication of the algorithm used in this study.11 Independent studies that evaluated certain populations of lymphocytes by immunohistochemistry have already demonstrated more favorable prognosis in patients with high lymphocyte density at cystectomy.26,27 Taken together, our data demonstrate that the presence of any lymphocyte on H&E staining (ie, not distinguished by phenotype) significantly improved discrimination of patient samples with high versus low metastatic potential when combined with output from deep learning–based algorithms, indicating that the complex effect of microenvironmental features on prognosis can be learned from AI-based systems.
The clinical application of this work may have an impact when applied to TURBT specimens for selecting patients most likely to benefit from NAC. A prospective trial of NAC + RC versus RC alone demonstrated improved survival for patients treated with NAC + RC.6 However, meta-analysis of NAC showed that the overall survival benefit for NAC was only 5%.19 These trials were performed without substratifying patients with MIBC into risk groups; thus, the results of these trials may be diminished by the inclusion of low-risk patients who would not have developed recurrence with observation alone. To address the need for further stratification, previous studies have used clinical and pathologic variables to substratify MIBC into low-risk and high-risk groups and have proposed that low-risk patients may forego NAC.28 This strategy is based on the association between adverse outcomes and advanced clinical T stage, hydronephrosis, and adverse variant pathologic features (LVI, micropapillary features, or neuroendocrine features).28 However, clinical T stage is frequently discordant with pathologic T stage,29,30 and the presence of variant features varies widely (7% to 81%) among reported series.8 Validation of the approach used in this study in TURBT specimens may motivate physicians to use it as a prognostic marker in efforts to stratify patients before surgery.
This study has several important limitations. Although this methodology was developed in a heterogeneous data set, all of the specimens were cystectomy specimens. An independent test set was created to model evaluation, but it consisted mainly of TCGA patients because of the limited availability of patients from our own institution (n = 13). Within this in-house subset, accuracy remained high with 11 of 13 correctly classified. Deep learning algorithms are highly prone to overfitting, especially when trained on small data sets. Although our algorithms were trained on only 146 patients, each model was trained from thousands of patches per resolution (Appendix Table A1). We used techniques such as pretrained weights, stain normalization, and image augmentation to avoid overfitting. However, overall evaluation was still limited to patient-level analysis and therefore requires further validation in external populations.
Because of the variation in the number of available slides on TCGA, validation of the AI-based model in the TCGA testing cohort used only a single slide labeled DX1. In addition, some pathology slides had a substantial number of artifacts such as ink and dust, so they required preprocessing. Tumor heterogeneity could lead to incorrect classification of samples when evaluated across all tumor slides. Slide-level performance in the in-house cohort tracked similarly with patient-level performance. There was also a variable number of LNs sampled, which diminished the quality of the ground truth labels. Variability in TCGA clinical data led to missing data for LVI in 16% of the patients. Given the clinical importance of this pathologic feature, it is possible that the power of the clinical model may differ when compared with a complete set with nonmissing values.
In conclusion, we demonstrated a novel methodology for identifying patients who are at higher risk of having positive LNs during cystectomy based on digital H&E slides from the primary tumor. This methodology is not subject to intra- or interobserver variation and can be generalized to multiple centers. All code used in developing this methodology is publicly available. This methodology may be useful for identifying patients most likely to benefit from additional therapy beyond surgical resection.
Appendix
DATA SOURCE AND DOWNLOAD PROCEDURES FOR STUDY REPRODUCIBILITY
Digital Pathology Images
Central access site: portal.gdc.cancer.gov
Select “repository”
Under “File” Filter options select:
Data Type → Slide Image
Experimental Strategy → Diagnostic Slide
Under “Cases” Filter options select Primary Site → Bladder
Download manifest file
Follow instructions from NCI GDC on downloading and using the GDC Transfer Tool: https://docs.gdc.cancer.gov/Data_Transfer_Tool/Users_Guide/Getting_Started/
Pathology Reports
Central access site: https://www.cbioportal.org/
Query → “Bladder/Urinary Tract” → “Bladder Cancer (TCGA, PanCancer Atlas)”
Direct link to study: https://www.cbioportal.org/study/clinicalData?id=blca_tcga_pan_can_atlas_2018
Individual patients can be searched/selected within “Clinical Data” Tab
Clinical End Point Data
Central access site: https://gdc.cancer.gov/about-data/publications/pancanatlas
Download file under “TCGA-Clinical Data Resource (CDR) Outcome”
Corresponding publication: https://www.cell.com/cell/fulltext/S0092-8674(18)30229-0
Patch Extraction and Stain Normalization
For digital slides, images were read in at a 4:1 ratio from apparent 40× magnification (effective 10× magnification) digital images to allow for whole-slide image processing.13,14 Staining was normalized to an independent reference specimen not included in the data cohort using the Macenko method.7 Background artifacts and ink markings were removed and the background intensity was set before deconvolution of the 3-channel image using hematoxylin and eosin stains and the Ruifrok and Johnson method.8 All stain normalization and color deconvolution was implemented by using the Stain Normalization Toolbox for MATLAB (https://warwick.ac.uk/fac/sci/dcs/research/tia/software/sntoolbox/).
All patches were derived in reference to 1,200 × 1,200 pixel regions at 10× magnification, with the number of regions determined by fitting the minimum number of non-overlapping regions that fully contained the pathologist’s annotations. Within each of these reference regions, patch images were extracted at four separate image resolutions. The reference 1,200 × 1,200 region was resampled to a single 300 × 300 image (1/4 original size) corresponding to effective 2.5× magnification, four 300 × 300 images (1/2 original size) corresponding to effective 5× magnification, and sixteen 300 × 300 images (no resampling) corresponding to 10× magnification. Coordinates of the reference region at 10× magnification were mapped to 40× magnification coordinates (4,800 × 4,800 pixels) to allow for extraction of sixty-four 300 × 300 images at 20× effective magnification, for which background artifacts and stain deconvolution were applied in reference to the corresponding region. The same coordinates were used to guide extraction of 576 images of 100×100 pixels at effective 20× resolution from the original (non-normalized, non-deconvolved) hematoxylin and eosin image for estimation of lymphocyte infiltration using the methods of Saltz et al.9,10
Patch-Based Classification
Four different models using ResNet-101 architecture were trained from patches at 2.5×, 5×, 10×, and 20× magnifications. All patches were at an effective 300 × 300 image size from extraction and were labeled according to patient lymph node (LN) status. Patches containing less than 90%, 80%, 70%, and 70% of white space (determined by proportion of normalized grayscale image > 0.8) for 2.5×, 5×, 10×, and 20× magnifications, respectively, were considered for input. A weighted cross-entropy loss was applied at the time of patch-level training to offset bias from the observed class imbalance at the patient level. Data augmentation techniques, including image rotation, flipping, warping, brightness, and contrast were applied randomly at training time to reduce overfitting to patient-specific and slide-specific characteristics. The final training parameters, including number of epochs, learning rate, imbalance technique, and data augmentation scheme, are included in Appendix Table A1. All models were trained using fast.ai library (https://github.com/fastai/fastai). After training was completed, all models were deployed to all patches within training, validation, and testing cohorts and the probability of being LN positive was saved for each patch image at every resolution. For lymphocyte classification, publicly available code and model weights were downloaded from Saltz et al.9 These models were applied to all 100 × 100 pixel images from all patient samples extracted during preprocessing, and the probability of containing lymphocytes was saved for each patch. Consistency in lymphocyte maps derived from the preprocessing methods in this study versus the original study were confirmed (Appendix Fig A2).
Post-Processing and Patient-Based Classification
Given that patches at all resolutions were derived spatially within known coordinates from the original image, a multiresolution prediction map was derived from all LN status classification and lymphocyte classification results. The final prediction maps were created at 1/200 the size of original 40× apparent magnification image, such that each lymphocyte prediction corresponded to a single pixel, each effective 20× prediction corresponded to 3 × 3 pixel area, each 10× prediction corresponded to a 6 × 6 pixel area, each 5× prediction corresponded to a 12 × 12 pixel area, and each 2.5× prediction corresponded to a 24 × 24 pixel area. Lymphocyte prediction maps were derived separately from LN classification maps. For each LN classification, maps of each resolution and per-pixel average prediction across all resolutions were created. The final result is an image in which every pixel represents the regional probability of LN status.
From the resultant pixel-based probability maps, high-risk clusters (ie, high probability of LN positivity by classification algorithms) were identified by using density-based spatial clustering of applications with noise (DBSCAN) implementation in MATLAB. Twelve different starting conditions derived from varying threshold probability cutoffs within various combinations of multiresolution probability maps. For each condition, metrics related to the size, number, density, and distance between clusters were extracted. In addition, distance between each resultant cluster and lymphocyte-rich pixels was tabulated. Finally, overall average probability of each resolution and total average across all resolutions were calculated for each patient. In total, 716 elements were extracted from representative slides for each patient in all cohorts. A full description of probability map derivation, DBSCAN parameters, and extracted features is provided in Appendix Table A2.
For image-based classification, an adaptive boosting classification ensemble of decision trees (AdaBoost) was fit using 10-fold cross validation within combined training and validation cohorts. Feature selection was determined during each partition on the basis of predictor importance estimates from decision trees using all variables. Within each partition, the top 20 important features were selected, resulting in 40 unique variables selected across 10-folds. Any variables selected as a top feature in more than one partition were considered for final model inclusion (n = 26), in which the final model was trained by repeating 10-fold cross validation selecting 15 features using an interaction test for variable selection.11 The final model was trained by using a maximum of three splits, a learning rate of 0.001, 4,000 learning cycles, and a 1.382 misclassification cost for false positives. Classification performance was determined by out-of-bag estimates during cross validation. The final model was then applied to testing cohorts. Classification outputs were saved as binary (yes/no for LN positive) and continuous (classification fit: range, 0-1 probability of LN positivity), which is referred to as the artificial intelligence (AI) score. The image-based classifier was developed in MATLAB (R2018b, www.mathworks.com) using Statistics and Machine Learning Toolbox.
FIG A1.
Final study selection. GDC, Genomic Data Commons; NCI, National Cancer Institute; TCGA, The Cancer Genome Atlas; TURBT, transurethral resection of bladder tumor.
FIG A2.
Representative lymphocyte map from in-house implementation v the original publication. TILs, tumor-infiltrating lymphocytes.
TABLE A1.
Patch-Based Deep Learning Training Schemes and Performance
TABLE A2.
Features Selection for Use in Machine Learning Classifier
TABLE A3.
χ2 Test for Differences in Clinical and Pathologic Demographics of Training and Validation Versus Testing Cohorts
TABLE A4.
Machine Learning Classifier Performance in Training and Validation Versus Testing Cohorts
TABLE A5.
Machine Learning Performance of Simple AI v Final AI Score
SUPPORT
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
AUTHOR CONTRIBUTIONS
Conception and design: Stephanie A. Harmon, Thomas H. Sanford, Sherif Mehralivand, Peter L. Choyke, Baris Turkbey
Provision of study materials or patients: Stephanie A. Harmon, Thomas H. Sanford, Piyush K. Agarwal
Collection and assembly of data: Stephanie A. Harmon, Thomas H. Sanford, Chris Yang, Vladimir A. Valera, Piyush K. Agarwal, Baris Turkbey
Data analysis and interpretation: Stephanie A. Harmon, Thomas H. Sanford, G. Thomas Brown, Joseph M. Jacob, Vladimir A. Valera, Joanna H. Shih, Peter L. Choyke, Baris Turkbey
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stephanie A. Harmon
Patents, Royalties, Other Intellectual Property: Patent: System and Method for Evaluation of Disease Burden
Sherif Mehralivand
Stock and Other Ownership Interests: AbbVie
Piyush K. Agarwal
Employment: Pfizer (I)
Travel, Accommodations, Expenses: AstraZeneca/MedImmune
Peter L. Choyke
Patents, Royalties, Other Intellectual Property: Patent holder for magnetic resonance imaging-ultrasound fusion technology licensed to InVivo which markets it as UroNav. However, as a government employee I personally receive no financial benefit from this patent.
Other Relationship: Aspyrian Therapeutics, Philips Healthcare, GE Health Care
Baris Turkbey
Patents, Royalties, Other Intellectual Property: Royalties from US government patents for magnetic resonance imaging/ultrasound fusion biopsy computer-aided diagnosis software.
Other Relationship: NVIDIA, Philips Healthcare
No other potential conflicts of interest were reported.
REFERENCES
- 1.Millán-Rodríguez F, Chéchile-Toniolo G, Salvador-Bayarri J, et al. Primary superficial bladder cancer risk groups according to progression, mortality and recurrence. J Urol. 2000;164:680–684. doi: 10.1016/s0022-5347(05)67280-1. [DOI] [PubMed] [Google Scholar]
- 2.Mari A, Campi R, Tellini R, et al. Patterns and predictors of recurrence after open radical cystectomy for bladder cancer: A comprehensive review of the literature. World J Urol. 2018;36:157–170. doi: 10.1007/s00345-017-2115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gakis G, Efstathiou J, Lerner SP, et al. ICUD-EAU International Consultation on Bladder Cancer 2012: Radical cystectomy and bladder preservation for muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2013;63:45–57. doi: 10.1016/j.eururo.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Dotan ZA, Kavanagh K, Yossepowitch O, et al. Positive surgical margins in soft tissue following radical cystectomy for bladder cancer and cancer specific survival. J Urol. 2007;178:2308–2312. doi: 10.1016/j.juro.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Bassi P, Ferrante GD, Piazza N, et al. Prognostic factors of outcome after radical cystectomy for bladder cancer: A retrospective study of a homogeneous patient cohort. J Urol. 1999;161:1494–1497. [PubMed] [Google Scholar]
- 6.Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349:859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 7.Kluth LA, Black PC, Bochner BH, et al. Prognostic and prediction tools in bladder cancer: A comprehensive review of the literature. Eur Urol. 2015;68:238–253. doi: 10.1016/j.eururo.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 8.Chalasani V, Chin JL, Izawa JI. Histologic variants of urothelial bladder cancer and nonurothelial histology in bladder cancer. Can Urol Assoc J. 2009;3:S193–S198. doi: 10.5489/cuaj.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology: New tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linkert M, Rueden CT, Allan C, et al. Metadata matters: Access to image data in the real world. J Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg IG, Allan C, Burel JM, et al. The open microscopy environment (OME) data model and XML file: Open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macenko M, Niethammer M, Marron JS, et al. A method for normalizing histology slides for quantitative analysis. 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Boston, MA. June 28-July 1, 2009; pp. 1107–1110. [Google Scholar]
- 15.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 16.Hou L, Nguyen V, Kanevsky AB, et al. Sparse autoencoder for unsupervised nucleus detection and representation in histopathology images. Pattern Recognit. 2019;86:188–200. doi: 10.1016/j.patcog.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh WY. Regression trees with unbiased variable selection and interaction detection. Stat Sin. 2002;12:361–386. http://pages.stat.wisc.edu/~loh/treeprogs/guide/guide02.pdf [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: A statistical view of boosting. Ann Stat. 2000;28:337–407. [Google Scholar]
- 19.Veskimäe E, Espinos EL, Bruins HM, et al. What is the prognostic and clinical importance of urothelial and nonurothelial histological variants of bladder cancer in predicting oncological outcomes in patients with muscle-invasive and metastatic bladder cancer? A European Association of Urology Muscle Invasive and Metastatic Bladder Cancer Guidelines Panel Systematic Review. Eur Urol Oncol. 2019;2:625–642. doi: 10.1016/j.euo.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Wu B, Zha Z, et al. Clinicopathological factors in bladder cancer for cancer-specific survival outcomes following radical cystectomy: A systematic review and meta-analysis. BMC Cancer. 2019;19:716. doi: 10.1186/s12885-019-5924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein JI, Allsbrook WC, Jr, Amin MB, et al. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 22.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 23.BenTaieb A, Li-Chang H, Huntsman D, et al. A structured latent model for ovarian carcinoma subtyping from histopathology slides. Med Image Anal. 2017;39:194–205. doi: 10.1016/j.media.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Sarma KV, Chung Ho K, et al. A multi-scale U-Net for semantic segmentation of histological images from radical prostatectomies. AMIA Annu Symp Proc. 2018;2017:1140–1148. [PMC free article] [PubMed] [Google Scholar]
- 25.Warrick JI, Sjödahl G, Kaag M, et al. Intratumoral heterogeneity of bladder cancer by molecular subtypes and histologic variants. Eur Urol. 2019;75:18–22. doi: 10.1016/j.eururo.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Sjödahl G, Lövgren K, Lauss M, et al. Infiltration of CD3+ and CD68+ cells in bladder cancer is subtype specific and affects the outcome of patients with muscle-invasive tumors. Urol Oncol. 2014;32:791–797. doi: 10.1016/j.urolonc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Yu A, Mansure JJ, Solanki S, et al. Presence of lymphocytic infiltrate cytotoxic T lymphocyte CD3+, CD8+, and immunoscore as prognostic marker in patients after radical cystectomy. PLoS One. 2018;13:e0205746. doi: 10.1371/journal.pone.0205746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culp SH, Dickstein RJ, Grossman HB, et al. Refining patient selection for neoadjuvant chemotherapy before radical cystectomy. J Urol. 2014;191:40–47. doi: 10.1016/j.juro.2013.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Rundstedt FC, Mata DA, Kryvenko ON, et al. Utility of clinical risk stratification in the selection of muscle-invasive bladder cancer patients for neoadjuvant chemotherapy: A retrospective cohort study. Bladder Cancer. 2017;3:35–44. doi: 10.3233/BLC-160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherif A. The risk of oversimplification in risk-stratification of neoadjuvant chemotherapy-responses in muscle invasive bladder cancer. Transl Androl Urol. 2019;8:S337–S340. doi: 10.21037/tau.2019.05.03. [DOI] [PMC free article] [PubMed] [Google Scholar]