Abstract
Background/Aim: Chondroid syringoma is a rare benign tumor emanating from sweat glands. Although rearrangements of the pleomorphic adenoma gene 1 (PLAG1) have been reported in such tumors, information on PLAG1 fusion genes is very limited. Materials and Methods: Cytogenetic, fluorescence in situ hybridization, RNA sequencing, array comparative genomic hybridization, reverse transcription polymerase chain reaction, and Sanger sequencing analyses were performed on two chondroid syringoma cases. Results: Both tumors had structural rearrangements of chromosome 8. An NDRG1-PLAG1 transcript was found in the first tumor in which exon 3 of PLAG1 was fused with exon 1 of NDRG1. A TRPS1-PLAG1 chimeric transcript was detected in the second chondroid syringoma in which exon 2 or exon 3 of PLAG1 was fused with exon 1 of TRPS1. Conclusion: The NDRG1-PLAG1 and TRPS1-PLAG1 resemble other PLAG1 fusion genes inasmuch as the expression of PLAG1 comes under the control of the NDRG1 or TRPS1 promoter.
Keywords: Chondroid syringoma, PLAG1, fusion genes, NDRG1- PLAG1, TRPS1-PLAG1, chromosome 8
Chondroid syringoma, also known as mixed tumor of the skin, is a rare benign tumor emanating from sweat glands and able to display a wide array of histological patterns (1-5). It may have histological similarities with another benign exocrine gland tumor, pleomorphic adenoma or mixed tumor of the salivary glands, something that may cause differential diagnostic difficulties (3,5,6). The name chondroid syringoma was coined by Hirsch and Helwing (3) because of the invariable presence in the tumor of sweat gland elements (syringoma) and because cartilage-like material (chondroid) was also present in most tumors they studied (3). Chondroid syringoma most commonly occurs in the head-and-neck region of middle-aged males (3,5,7-9) but can also be found in the axilla and on the anterior chest, trunk, extremities, and scrotum (3,7,10-19).
We studied genetically two chondroid syringomas finding fusion of the pleomorphic adenoma gene 1 (PLAG1) at 8q12.2 in one with the gene N-myc downstream regulated 1 (NDRG1), which maps to chromosome subband 8q24.22, and in the second with the gene transcriptional repressor GATA binding 1 (TRPS1), which maps to 8q23.3.
Materials and Methods
Ethics statement. The study was approved by the regional Ethics Committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no) and written informed consent was obtained from the patients. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
Case description
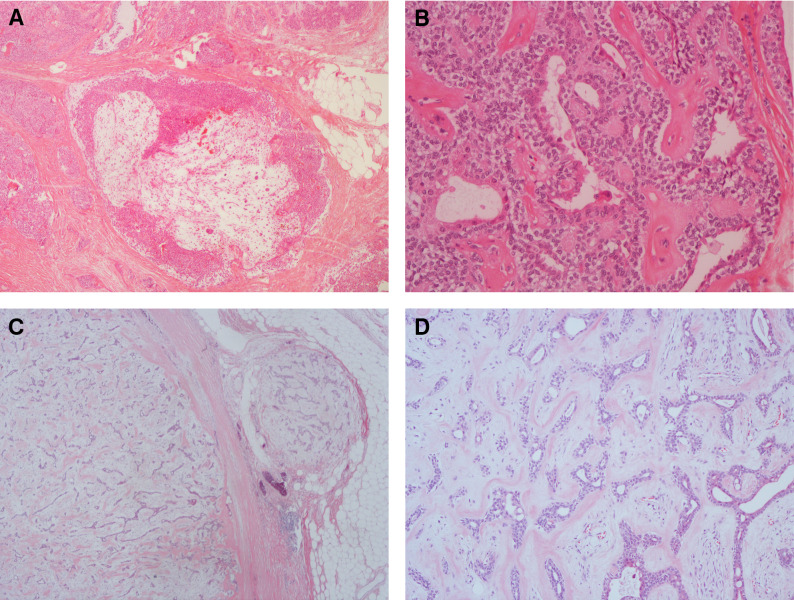
Case 1. A 60-year-old male without any relevant medical previous history was surgically treated for a tumor located at the left ankle. The tumor was found to be consistent with a chondroid syringoma. Surgical margins were doubtful. Six years later re-excision was performed. Macroscopically the tumor was lobulated, but well demarcated. The cut section was soft and gelatinous intermingled with harder areas. Microscopically the tumor was composed of small cystic spaces, areas with myxoid tissue and areas with solid growth of cubic epithelium (Figure 1A and B).
Figure 1. Microscopic examination of chondroid syringoma from case 1 (A and B) and case 2 (C and D). (A) Tumor with myxoid areas, ×40. (B) Ductal structures embedded in solid growth of cubic epithelial cells, intermingled with collagen band, ×200. (C) Partly capsulated tumor with slit like ductal spaces embedded in chondromyxoid stroma, ×20. (D) High magnification of the capsulated tumor (from C), ×100.
Case 2. A 59-year-old male was surgically treated for a tumor on the left thigh. The tumor was located in the subcutaneous adipose tissue, measuring 18 mm in greatest diameter. Macroscopically the tumor was white and with rubbery turgor. Microscopically the tumor was solid with cribriform growth of cubic cells separated by collagen bundles and small areas with myxoid material, intermingled with loose arranged spindle cells (Figure 1C and D).
G-banding and karyotyping. Fresh tissue from a representative area of the tumors was analyzed cytogenetically as previously described (20).
Fluorescence in situ hybridization (FISH) analysis. In order to characterize the ring and the der(15) chromosome of case 1 (see below), FISH was performed on metaphase spreads using whole chromosome painting probes for chromosomes 8 and 15 (Cytocell, Oxford Gene Technology, Begbroke, Oxfordshire, UK).
The BAC probes were purchased from BACPAC Resource Center located at the Children’s Hospital Oakland Research Institute (Oakland, CA) (https://bacpacresources.org/) (Table I). Detailed information on the FISH procedure was given elsewhere (21).
Table I. BAC probes used for FISH experiments.
DNA was extracted and probes were labelled and hybridized using Abbottʼs nick (Abbott Molecular, Des Plaines, IL, USA) translation kit according to the manufacturer’s recommendations. The PLAG1 probe was labelled with Texas Red-5-dCTP (PerkinElmer, Boston, MA, USA) in order to obtain a red signal. The probes for NDRG1 and TRPS1 were labelled with fluorescein-12-dCTP (PerkinElmer, Boston, MA, USA) in order to obtain green signals. FISH mapping of the probes on normal controls was performed to confirm their chromosomal location. Chromosome preparations were counterstained with 0.2 μg/ml DAPI and overlaid with a 24×50 mm2 coverslip. Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK).
Array comparative genomic hybridization (aCGH) analysis. Genomic DNA was extracted using the Maxwell RSC Instrument and the Maxwell RSC Tissue DNA Kit (Promega, Madison, USA). The concentration was measured using the Quantus Fluorometer and the QuantiFluor ONE dsDNA System (Promega, Madison, WI, USA). Promegaʼs human genomic female DNA was used as reference DNA. For aCGH, CytoSure array products were used (Oxford Gene Technology) according to the company’s protocols. Thus, the CytoSure Genomic DNA Labelling Kit was used for labelling of 1 μg of each patient and reference DNAs and the CytoSure Cancer +SNP array for hybridization. The slides were scanned in an Agilent scanner using the Agilent Feature Extraction Software (version 10.7.3.1). Data were analysed with the CytoSure Interpret analysis software (version 4.9.40). The genomic imbalances were identified using the Circular Binary Segmentation (CBS) algorithm (22) and added a custom-made aberration filter defining a copy number aberration (CNA) as a region with a minimum five probes gained/lost. Annotations are based on human genome build 19.
RNA sequencing. Total RNA was extracted from frozen (–80˚C) tumor tissue adjacent to that used for cytogenetic analysis and histologic examination using miRNeasy Mini Kit (Qiagen, Hilden, Germany). For case 1, one μg of total RNA from the tumor was sent to the Genomics Core Facility at the Norwegian Radium Hospital, Oslo University Hospital (http://genomics.no/oslo/) for high-throughput paired-end RNA-sequencing according to the Illumina TruSeq Stranded mRNA protocol. The softwares FusionCatcher, deFuse, TopHat-Fusion, and FuSeq were used to find fusion transcripts (23-28). In addition, the “grep” command was used to search the fastq files of the sequence data for PLAG1 sequence. The principle of this approach has been described elsewhere (29,30). The search term was the 20-nucleotide-sequence (nt) “ATTGGCCAAAATGGGAAGGA” which corresponds to the first 20 nt in exon 3 of PLAG1 or nt 286-305 in the PLAG1 reference sequence (accession number: NM_002655.2).
Reverse transcription (RT) PCR and Sanger sequencing analyses. The primers used for PCR amplifications and Sanger sequencing analyses are shown in Table II. One μg of total RNA was reverse-transcribed in a 20 μl reaction volume using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA).
Table II. Primers used for PCR amplification and Sanger sequencing analyses.
For amplification of the NDRG1-PLAG1 fusion transcript, the primer combinations were NDRG1-1F1/PLAG1-498R1 and NDRG1-23F1/PLAG1-458R1. For amplification of the TRPS1-PLAG1 fusion transcript, the primer combinations were TRPS1-200F1/PLAG1-498R1 and TRPS1-318F1/PLAG1-458R1. All PCR amplifications were performed in 25 μl reaction volume which contained 12.5 μl Premix Ex Taq™ DNA Polymerase Hot Start Version (Takara Bio Europe, Saint-Germain-en-Laye, France), 1 μl cDNA template, and 0.4 μM of each of the forward and reverse primers. PCR amplifications were run on a C-1000 Thermal cycler (Bio-Rad) and the cycling was at 94˚C for 30 s, followed by 35 cycles of 7 s at 98˚C, 30 s at 60˚C, 30 s at 72˚C, and a final extension for 5 min at 72˚C. Three μl of the PCR products were stained with GelRed (Biotium, Fremont, California, USA), analyzed by electrophoresis through 1% agarose gel, and photographed. The remaining PCR products were purified using the MinElute PCR Purification Kit (Qiagen) and direct sequenced using the dideoxy procedure with the BigDye terminator v1.1 cycle sequencing kit (ThermoFisher Scientific, Waltham, Massachusetts, USA) on the Applied Biosystems SeqStudio Genetic Analyzer system. The basic local alignment search tool (BLAST) software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for computer analysis of sequence data (31).
Results
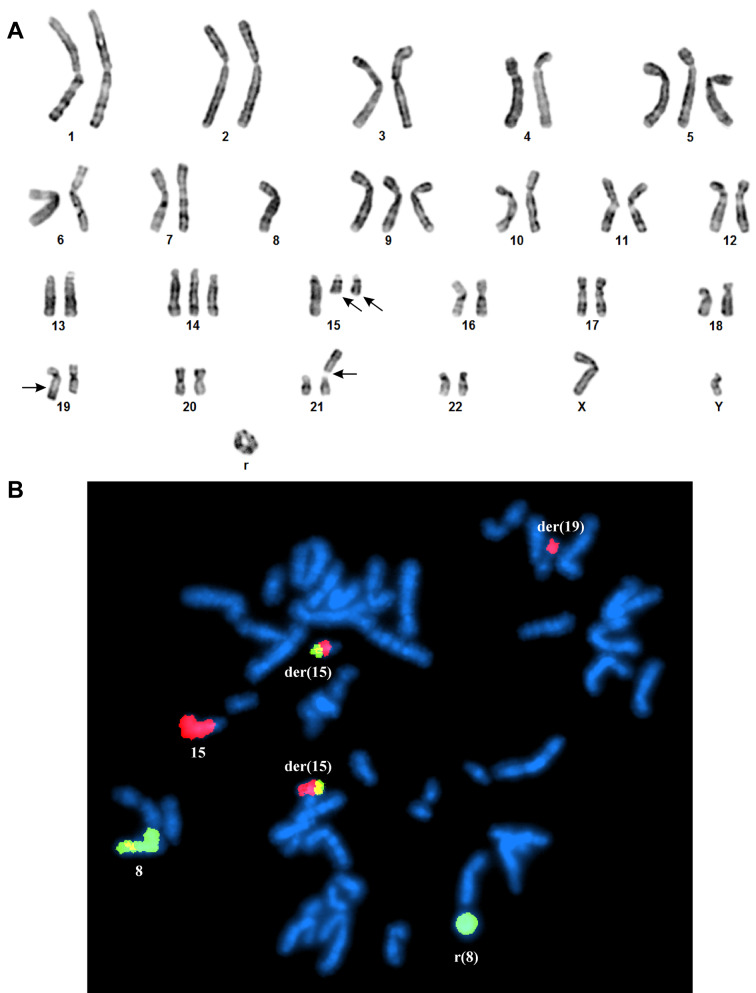
Case 1. The G-banding analysis supported by whole chromosome paint FISH results yielded the karyotype 50,XY,+5,r(8),+9,+14,+15,der(15)t(8;15)(?;?q15)x2, der(19)t(15;19)(q22;q13), add(21)(p13) (Figure 2A and B).
Figure 2. G-banding and FISH analyses of the case 1 of chondroid syringoma. (A) Representative karyogram demonstrating the chromosome aberrations (breakpoints are shown by arrows). (B) FISH using whole chromosome painting probes for chromosome 8 (green signal) and chromosome 15 (red signal) shows the normal chromosomes 8 and 15, the ring chromosome consisting of chromosome 8 material, the two der(15)t(8;15), and the der(19)t(15;19).
Examination of the RNA sequencing data with four different programs did not identify any fusion genes related to chromosome 8 (data not shown). Using the “grep” command and a search term corresponding to the first 20 nt in the exon 3 of PLAG1 on the raw sequencing data, which were in the text-based fastq format, only 5 unique sequences were extracted (Table III). Using the BLAST algorithm on the NCBI National Canter for Biotechnology Information database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) we aligned each of the above-mentioned sequences with the human genomic plus transcript database. The alignment showed that 3 sequences were from the PLAG1 gene whereas 2 sequences were hybrids containing sequences from exon 3 of PLAG1 (sequence with accession number NM_002655.2) and exon 1 of NDRG1 (sequence with accession number NM_006096.4) (Table III).
Table III. The retrieved sequences from the fastq file of the RNA sequencing using the “grep” command and the search term “ATTGGCCAAAATGGGAAGGA” which is the first 20 nt in the exon 3 of PLAG1 to nt 286-305 in the PLAG1 reference sequence with the accession number NM_002655.2.
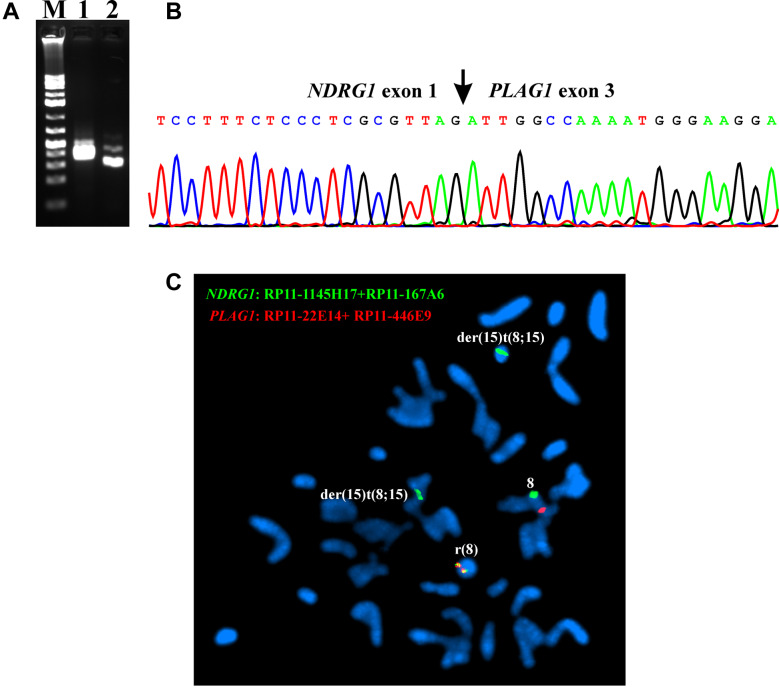
RT-PCR with the primer combinations NDRG1-1F1/PLAG1-498R1 and NDRG1-23F1/PLAG1-458R1 amplified a 352 bp and a 286 bp cDNA fragment, respectively (Figure 3A). Direct sequencing of them showed that they were NDRG1-PLAG1 chimeric cDNA fragments (Figure 3B). The fusion point was identical to that found by the analysis of the RNA sequencing data. Thus, the non-coding sequence exon 1 of NDRG1 (nt 119 in sequence with accession number NM_006096.4) was fused to exon 3 of PLAG1 (nt 286 in NM_002655.2) (Figure 3B).
Figure 3. RT-PCR, Sanger sequencing, and FISH analyses of the case 1 of chondroid syringoma. (A) Gel electrophoresis showing the amplified NDRG1-PLAG1 cDNA fragments using the primer combinations NDRG1-1F1/PLAG1-498R1 (lane 1) and NDRG1-23F1/PLAG1-458R1 (lane 2). M, GeneRuler 1 Kb Plus DNA ladder (ThermoFisher Scientific). (B) Partial sequence chromatograms of the cDNA amplified fragment showing the junction position of exon 1 of NDRG1 with exon 3 of PLAG1. (C) FISH analysis on metaphase spreads with PLAG1 probe (red signal) and NDRG1 probe (green signal) showing that the NDRG1-PLAG1 fusion gene was on the ring chromosome 8 (yellow signal). One copy of PLAG1 (red signal) is on chromosome 8. NDRG1 probe (green signal) hybridized also to chromosome 8 and the two der(15)t(8;15) chromosomes.
FISH analysis on metaphase spreads showed that the NDRG1-PLAG1 fusion gene was on the ring chromosome (Figure 3C).
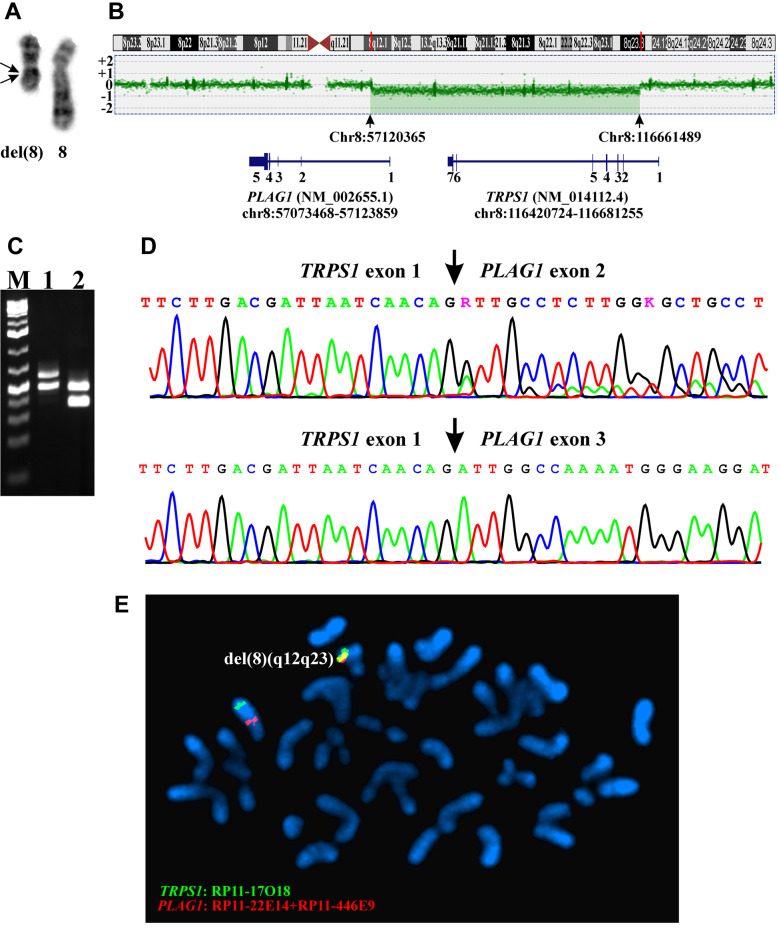
Case 2. The G-banding analysis yielded the karyotype 46,XY,del(8)(q12q23)[10] (Figure 4A). aCGH also detected a large deletion in the q arm of chromosome 8 (Figure 4B). Based on the hg19 assembly, the deletion started at position Chr8:57120365 in intron1 of PLAG1 and ended at Chr8:116661489 in exon 1 of TRPS1 (Figure 4B). Thus, aCGH data were in agreement with the results of G-banding analysis and suggested that a TRPS1-PLAG1 fusion gene had been formed as a result of the deletion.
Figure 4. Genetic analyses of the case 2 of chondroid syringoma. (A) Partial karyotype showing the del(8)(q12q23) and the normal chromosome 8 (breakpoints are shown by arrows). (B) aCGH showing the deletion in the q arm of chromosome 8. Based on the hg19 assembly, the deletion started at position Chr8:57120365 in intron1 of PLAG1 and ended at Chr8:116661489 in exon 1 of TRPS1. (C) Gel electrophoresis showing the amplified TRPS1-PLAG1 fragments using the primer combinations TRPS1-200F1/PLAG1-498R1 (lane 1) and TRPS1-318F1/PLAG1-458R1 (lane 2). (D) Partial sequence chromatograms of the cDNA amplified fragment showing the junction positions of exon 1 of TRPS1 with exon 2 of PLAG1 and exon 1 of TRPS1 with exon 3 of PLAG1. E) FISH analysis on metaphase spreads with PLAG1 probe (red signal) and TRPS1 probe (green signal) showing that the TRPS1-PLAG1 fusion gene was on the del(8)(q12q23) (yellow signal). A copy of PLAG1 (red signal) and TRPS1 (green signal) is on chromosome 8.
RT-PCR with the primer combinations TRPS1-200F1/PLAG1-498R1 amplified two, a 570 bp and a 465 bp, cDNA fragments (Figure 4C). Direct sequencing of these fragments showed that both were TRPS1-PLAG1 chimeric cDNA fragments. In the 570 bp long fragment, exon 1 of TRPS1 was fused to exon 2 of PLAG1 whereas in the 465 bp fragment, exon 1 of TRPS1 was fused to exon 3 of PLAG1 (Figure 4D).
RT-PCR with the primer combinations TRPS1-318F1/PLAG1-458R1 also amplified two, a 414 bp and a 309 bp, cDNA fragments (Figure 4C). The sequence of these fragments confirmed the presence of the above-mentioned TRPS1-PLAG1 fusion transcripts (Figure 4D).
FISH analysis on metaphase spreads showed that the TRPS1-PLAG1 fusion gene was on the del(8)(q12q23) chromosome (Figure 4E).
Discussion
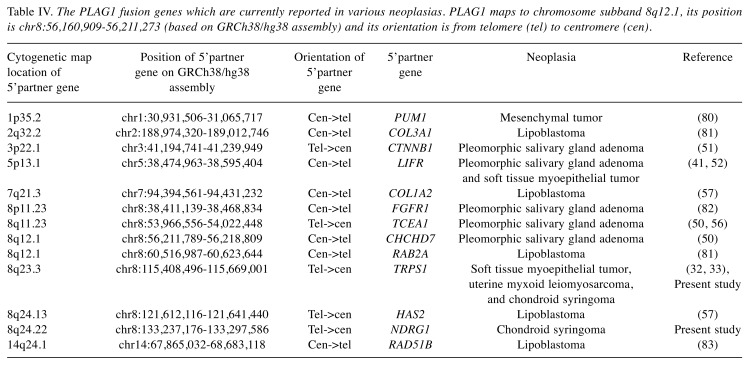
We report the identification of two fusion genes, NDRG1-PLAG1 and TRPS1-PLAG1, in two chondroid syringomas. To the best of our knowledge, the NDRG1-PLAG1 fusion gene is reported here for the first time. However, the TRPS1-PLAG1 gene fusion was recently reported in a myoepithelial tumor of soft tissue and was also found to be recurrent in a subset of uterine myxoid leiomyosarcomas with PLAG1 rearrangements (Table IV) (32,33).
Table IV. The PLAG1 fusion genes which are currently reported in various neoplasias. PLAG1 maps to chromosome subband 8q12.1, its position is chr8:56,160,909-56,211,273 (based on GRCh38/hg38 assembly) and its orientation is from telomere (tel) to centromere (cen).
Chondroid syringoma and its more aggressive counterpart, malignant chondroid syringoma, are included among cutaneous myoepithelial neoplasms (34-38). Recently, we reported a malignant chondroid syringoma which had a t(X;6)(p11;p21) as the sole karyotypic aberration and demonstrated that the molecular consequence of that translocation was fusion of the PHD finger protein 1 (PHF1) gene from 6p21 with the transcription factor binding to IGHM enhancer 3 gene (TFE3) from Xp11 (39). In the present study, we found NDRG1-PLAG1 and TRPS1-PLAG1 in chondroid syringomas. Thus, although the data are extremely limited, malignant chondroid syringoma and benign chondroid syringoma seem to be developed through different pathogenetic mechanisms.
Genetic studies of both cutaneous and soft tissue myoepithelial neoplasms have demonstrated considerable genetic heterogeneity (40-46). In some tumors, rearrangements of the Ewing sarcoma breakpoint region 1 gene (EWSR1) leading to the fusion genes EWSR1-ZNF444, EWSR1-PBX1, EWSR1-PBX3 or EWSR1-POU5F1 were found (40,43-45,47). In benign myoepithelial tumors, rearrangements of PLAG1 have been found (41,42,48,49). Matsuyuama et al. (48) studied 16 cutaneous mixed tumors and found that all of them expressed PLAG1, predominantely in cells with myopithelial or chondroid differentiation. Because they did not detect the fusion genes CTNNB1-PLAG1, LIFR-PLAG1, CHCHD7-PLAG1 which have been reported in pleomorphic adenomas of the salivary gland, they concluded that the mechanism of PLAG1 overexpression in cutaneous mixed tumors may be different from that in pleomorphic adenomas (48,50-52). Bahrami and co-workers (42) found PLAG1 rearrangements in 8 out of 11 (73%) benign myoepitheliomas/mixed tumors of the skin and soft tissue. Antonescu and co-workers (41) detected PLAG1 rearrangements in 13 out of 35 (37%) myoepithelial tumors lacking EWSR1 and FUS rearrangements and identified a LIFR-PLAG1 fusion in one case (Table IV). Recently, Russell-Goldman and co-workers (49) showed that PLAG1 is expressed in skin mixed tumors referred to as being of the apocrine type but not in eccrine-type tumors. The conclusion from the above-mentioned studies was that a subset of benign myoepitheliomas/mixed tumors of the skin and soft tissue exists that are genetically related to their salivary gland counterparts (41,42,46,48,49). PLAG1 together with PLAGL1 (at chromosome subband 6q24.2) and PLAGL2 (at 20q11.21) constitute the PLAG gene family coding for zing finger proteins (53-55). All three members of the family are implicated in neoplasia (54,55). PLAG1 was shown to be rearranged in pleomorphic adenomas of the salivary glands, lipoblastomas, as well as other tumors via chromosomal translocations targeting 8q12 (50-52,56,57) (Table IV). PLAGL1 functions as a suppressor of cell growth and is often deleted or methylated and silenced in cancer cells (58-62). A MYB-PLAGL1 fusion was reported in three acute lymphoblastic leukemias (63-65). PLAGL2 is a proto-oncogene more structurally and functionally similar to PLAG1 than is PLAGL1 (53-55,66-68). Overexpression of PLAGL2 was seen in acute myeloid leukemia, bladder urothelial carcinoma, and colorectal cancer (67-71).
The characteristics of the PLAG1-reported fusion genes found in pleomorphic adenomas of the salivary glands, lipoblastomas, and other tumors are that the chromosome rearrangements result in fusion of the 5’-non-coding region of PLAG1 with the partner gene’s 5’-non-coding region exchanging the two genes’ regulatory elements. Promoter swapping between PLAG1 and the fusion partner thus takes place and the expression of PLAG1 comes under the control of the other gene’s promoter leading to overexpression or ectopic activation of PLAG1 (51,52,54,56,57). Overexpression or ectopic activation of PLAG1 in its turn leads to deregulation of PLAG1-target genes and tumor formation (72-75).
In vitro studies showed that ectopic expression of PLAG1 in NIH3T3 cells results in loss of cell–cell contact inhibition and anchorage-independent growth. PLAG1-expressing NIH3T3 cells induce tumors in nude mice (66). In transgenic mice, targeted PLAG1 overexpression in salivary or mammary glands resulted in tumor development (76,77). PLAG1 was found to regulate the expression of the IGF2 gene coding for insulin-like growth factor 2, to upregulate genes which are associated with IGF and WNT signaling, and to deregulate a plethora of long non-coding RNAs (54,66,72-75,78,79).
NDRG1-PLAG1 and TRPS1-PLAG1 share characteristics of other PLAG1 fusion genes. In the first tumor we analyzed, the untranslated exon 3 of PLAG1 fused with exon 1 of NDRG1. Thus, PLAG1 expression came under the control of NDRG1 promoter. NDRG1 is ubiquitously expressed and codes for a cytoplasmic protein involved in stress responses, hormone responses, cell growth, and differentiation (https://www.ncbi.nlm.nih.gov/gene/10397#gene-expression). In the second tumor, exon 2 or exon 3 of PLAG1 fused with exon 1 of TRPS1 and the expression of PLAG1 came under control of the TRPS1 promoter. TRPS1 is also ubiquitously expressed and codes for a transcription factor that represses GATA-regulated genes and binds to a dynein light-chain protein (https://www.ncbi.nlm.nih.gov/gene/7227#gene-expression).
The number of PLAG1 fusion genes in various neoplasias until now, including the two present ones, is thirteen (Table IV). Worthy of mention is that in seven of them, the 5’ fusion partner maps to chromosome 8: one to the p arm and six to the q arm. Evidently, recombinations involving chromosome 8, particularly the long arm, are for some reason particularly common events in PLAG1 activating fusions.
Conflicts of Interests
No potential conflicts of interest exist.
Authors’ Contributions
IP designed and supervised the research, performed molecular genetic experiments, bioinformatics analysis, and wrote the article. LG performed cytogenetic analysis and evaluated the FISH data. KA performed molecular genetic experiments, FISH analysis, and evaluated the data. ML-I performed the pathological examination. IL performed the pathological examination. FM supervised the research. SH assisted with experimental design and writing of the article. All Authors read and approved the final article.
Acknowledgements
This work was supported by grants from Radiumhospitalets Legater.
References
- 1.Headington JT. Mixed tumors of skin: eccrine and apocrine types. Arch Dermatol. 1961;84(6):989–996. doi: 10.1001/archderm.1961.01580180105016. PMID: 13905736. DOI: 10.1001/archderm.1961.01580180105016. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez FJ. Mixed tumors of the skin of the salivary gland type: a light and electron microscopic study. J Invest Dermatol. 1976;66(1):49–52. doi: 10.1111/1523-1747.ep12478101. DOI: 10.1111/1523-1747.ep12478101. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch P, Helwig EB. Chondroid syringoma. Mixed tumor of skin, salivary gland type. Arch Dermatol. 1961;84(5):835–847. doi: 10.1001/archderm.1961.01580170129018. PMID: 13907712. DOI: 10.1001/archderm.1961.01580170129018. [DOI] [PubMed] [Google Scholar]
- 4.Mills SE. Mixed tumor of the skin: a model of divergent differentiation. J Cutan Pathol. 1984;11(5):382–386. doi: 10.1111/j.1600-0560.1984.tb00395.x. PMID: 6096423. DOI: 10.1111/j.1600-0560.1984.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 5.Stout AP, Gorman JG. Mixed tumors of the skin of the salivary gland type. Cancer. 1959;12(3):537–543. doi: 10.1002/1097-0142(195905/06)12:3<537::aid-cncr2820120313>3.0.co;2-n. PMID: 13652101. DOI: 10.1002/1097-0142(195905/06)12:3<537::aid-cncr2820120313>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 6.Satter EK, Graham BS. Chondroid syringoma. Cutis. 2003;71(1):49–52, 55. PMID: 12553630. [PubMed] [Google Scholar]
- 7.Bekerecioglu M, Tercan M, Karakok M, Atik B. Benign chondroid syringoma: a confusing clinical diagnosis. Eur J Plast Surg. 2002;25(2):316–318. DOI: 10.1007/s00238-002-0385-5. [Google Scholar]
- 8.Chen AH, Moreano EH, Houston B, Funk GF. Chondroid syringoma of the head and neck: clinical management and literature review. Ear Nose Throat J. 1996;75(2):104–108. PMID: 8714424. [PubMed] [Google Scholar]
- 9.Yavuzer R, Basterzi Y, Sari A, Bir F, Sezer C. Chondroid syringoma: a diagnosis more frequent than expected. Dermatol Surg. 2003;29(2):179–181. doi: 10.1046/j.1524-4725.2003.29045.x. PMID: 12562350. DOI: 10.1046/j.1524-4725.2003.29045.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardisson D, Linares MD, Nistal M. Giant chondroid syringoma of the axilla. J Cutan Med Surg. 1998;3(2):115–117. doi: 10.1177/120347549800300214. PMID: 9822789. DOI: 10.1177/120347549800300214. [DOI] [PubMed] [Google Scholar]
- 11.Kakitsubata Y, Theodorou SJ, Theodorou DJ, Nakahara M, Yuki Y, Yokouchi T. Giant chondroid syringoma presenting as a growing subcutaneous mass in the upper arm: MRI findings with pathologic correlation. Joint Bone Spine. 2009;76(6):711–714. doi: 10.1016/j.jbspin.2009.04.006. PMID: 19793672. DOI: 10.1016/j.jbspin.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Madi K, Attanasio A, Cecunjanin F, Garcia RA, Vidershayn A, Lucido JV. Chondroid syringoma of the foot: a rare diagnosis. J Foot Ankle Surg. 2016;55(2):373–378. doi: 10.1053/j.jfas.2014.12.032. PMID: 26015300. DOI: 10.1053/j.jfas.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 13.Mebazaa A, Trabelsi S, Denguezli M, Sriha B, Belajouza C, Nouira R. Chondroid syringoma of the arm: an unusual localization. Dermatol Online J. 2006;12(1):14. PMID: 16638382. [PubMed] [Google Scholar]
- 14.Okuda H, Tei N, Shimizu K, Imazu T, Yoshimura K, Kiyohara H. Chondroid syringoma of the scrotum. Int J Urol. 2008;15(10):944–945. doi: 10.1111/j.1442-2042.2008.02112.x. PMID: 19138288. DOI: 10.1111/j.1442-2042.2008.02112.x. [DOI] [PubMed] [Google Scholar]
- 15.Pal S, Sengupta S, Jana S, Bose K. Fine-needle aspiration cytology of chondroid syringoma of fore arm: Report of a rare case. J Cytol. 2014;31(3):171–173. doi: 10.4103/0970-9371.145659. PMID: 25538390. DOI: 10.4103/0970-9371.145659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poku JW, Sant GR, Ucci AA. Chondroid syringoma of the scrotum. J Int Med Res. 1996;24(6):482–486. doi: 10.1177/030006059602400606. PMID: 8959533. DOI: 10.1177/030006059602400606. [DOI] [PubMed] [Google Scholar]
- 17.Rogers R, Zhou F, Grunes D, Shapiro RL, Bannan M, Simsir A, Leung A. Chondroid syringoma of the axilla: An unusual tumor diagnosed by fine needle aspiration. Diagn Cytopathol. 2016;44(4):342–346. doi: 10.1002/dc.23424. PMID: 26800260. DOI: 10.1002/dc.23424. [DOI] [PubMed] [Google Scholar]
- 18.Sirivella S, Gielchinsky I. Chondroid syringoma: a rare tumor of the chest wall. Ann Thorac Surg. 2010;89(3):983–985. doi: 10.1016/j.athoracsur.2009.07.062. PMID: 20172178. DOI: 10.1016/j.athoracsur.2009.07.062. [DOI] [PubMed] [Google Scholar]
- 19.Wollina U, Sturm U, Schonlebe J. Benign chondroid syringoma (mixed tumor of skin) on the flank with satellites: case report and literature review. Wien Med Wochenschr. 2014;164(17-18):377–380. doi: 10.1007/s10354-014-0302-0. PMID: 25271109. DOI: 10.1007/s10354-014-0302-0. [DOI] [PubMed] [Google Scholar]
- 20.Panagopoulos I, Gorunova L, Lund-Iversen M, Andersen K, Andersen HK, Lobmaier I, Bjerkehagen B, Heim S. Cytogenetics of spindle cell/pleomorphic lipomas: karyotyping and FISH analysis of 31 tumors. Cancer Genomics Proteomics. 2018;15(3):193–200. doi: 10.21873/cgp.20077. PMID: 29695401. DOI: 10.21873/cgp.20077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panagopoulos I, Bjerkehagen B, Gorunova L, Taksdal I, Heim S. Rearrangement of chromosome bands 12q14~15 causing HMGA2-SOX5 gene fusion and HMGA2 expression in extraskeletal osteochondroma. Oncol Rep. 2015;34(2):577–584. doi: 10.3892/or.2015.4035. PMID: 26043835. DOI: 10.3892/or.2015.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. PMID: 15475419. DOI: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 23.Kangaspeska S, Hultsch S, Edgren H, Nicorici D, Murumagi A, Kallioniemi O. Reanalysis of RNA-sequencing data reveals several additional fusion genes with multiple isoforms. PLoS One. 2012;7(10):e48745. doi: 10.1371/journal.pone.0048745. PMID: 23119097. DOI: 10.1371/journal.pone.0048745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. PMID: 23618408. DOI: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. doi: 10.1186/gb-2011-12-8-r72. PMID: 21835007. DOI: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, Pacheco M, Marra MA, Hirst M, Nielsen TO, Sahinalp SC, Huntsman D, Shah SP. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7(5):e1001138. doi: 10.1371/journal.pcbi.1001138. PMID: 21625565. DOI: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicorici D, Satalan H, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kikku O. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014 DOI: 10.1101/011650. [Google Scholar]
- 28.Vu TN, Deng W, Trac QT, Calza S, Hwang W, Pawitan Y. A fast detection of fusion genes from paired-end RNA-seq data. BMC Genomics. 2018;19(1):786. doi: 10.1186/s12864-018-5156-1. PMID: 30382840. DOI: 10.1186/s12864-018-5156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The “grep” command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13) PLoS One. 2014;9(6):e99439. doi: 10.1371/journal.pone.0099439. PMID: 24950227. DOI: 10.1371/journal.pone.0099439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. Novel KAT6B-KANSL1 fusion gene identified by RNA sequencing in retroperitoneal leiomyoma with t(10;17)(q22;q21) PLoS One. 2015;10(1):e0117010. doi: 10.1371/journal.pone.0117010. PMID: 25621995. DOI: 10.1371/journal.pone.0117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. PMID: 2231712. DOI: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhu G, Benayed R, Ho C, Mullaney K, Sukhadia P, Rios K, Berry R, Rubin BP, Nafa K, Wang L, Klimstra DS, Ladanyi M, Hameed MR. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2019;32(5):609–620. doi: 10.1038/s41379-018-0175-7. PMID: 30459475. DOI: 10.1038/s41379-018-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arias-Stella JA 3rd, Benayed R, Oliva E, Young RH, Hoang LN, Lee CH, Jungbluth AA, Frosina D, Soslow RA, Antonescu CR, Ladanyi M, Chiang S. Novel PLAG1 gene rearrangement distinguishes a subset of uterine myxoid leiomyosarcoma from other uterine myxoid mesenchymal tumors. Am J Surg Pathol. 2019;43(3):382–388. doi: 10.1097/PAS.0000000000001196. PMID: 30489320. DOI: 10.1097/PAS.0000000000001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornick JL, Fletcher CD. Cutaneous myoepithelioma: a clinicopathologic and immunohistochemical study of 14 cases. Hum Pathol. 2004;35(1):14–24. doi: 10.1016/j.humpath.2003.08.016. PMID: 14745720. DOI: 10.1016/j.humpath.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Jo VY, Fletcher CD. Myoepithelial neoplasms of soft tissue: an updated review of the clinicopathologic, immunophenotypic, and genetic features. Head Neck Pathol. 2015;9(1):32–38. doi: 10.1007/s12105-015-0618-0. PMID: 25804378. DOI: 10.1007/s12105-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo VY. Myoepithelial tumors: an update. Surg Pathol Clin. 2015;8(3):445–466. doi: 10.1016/j.path.2015.05.005. PMID: 26297065. DOI: 10.1016/j.path.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Kutzner H, Mentzel T, Kaddu S, Soares LM, Sangueza OP, Requena L. Cutaneous myoepithelioma: an under-recognized cutaneous neoplasm composed of myoepithelial cells. Am J Surg Pathol. 2001;25(3):348–355. doi: 10.1097/00000478-200103000-00009. PMID: 11224605. DOI: 10.1097/00000478-200103000-00009. [DOI] [PubMed] [Google Scholar]
- 38.Mentzel T, Requena L, Kaddu S, Soares de Aleida LM, Sangueza OP, Kutzner H. Cutaneous myoepithelial neoplasms: clinicopathologic and immunohistochemical study of 20 cases suggesting a continuous spectrum ranging from benign mixed tumor of the skin to cutaneous myoepithelioma and myoepithelial carcinoma. J Cutan Pathol. 2003;30(5):294–302. doi: 10.1034/j.1600-0560.2003.00063.x. PMID: 12753168. DOI: 10.1034/j.1600-0560.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- 39.Panagopoulos I, Gorunova L, Lund-Iversen M, Bassarova A, Heim S. Fusion of the genes PHF1 and TFE3 in malignant chondroid syringoma. Cancer Genomics Proteomics. 2019;16(5):345–351. doi: 10.21873/cgp.20139. PMID: 31467228. DOI: 10.21873/cgp.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49(12):1114–1124. doi: 10.1002/gcc.20819. PMID: 20815032. DOI: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonescu CR, Zhang L, Shao SY, Mosquera JM, Weinreb I, Katabi N, Fletcher CD. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52(7):675–682. doi: 10.1002/gcc.22063. PMID: 23630011. DOI: 10.1002/gcc.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51(2):140–148. doi: 10.1002/gcc.20938. PMID: 22038920. DOI: 10.1002/gcc.20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47(7):558–564. doi: 10.1002/gcc.20559. PMID: 18383210. DOI: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- 44.Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48(12):1051–1056. doi: 10.1002/gcc.20706. PMID: 19760602. DOI: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- 45.Flucke U, Palmedo G, Blankenhorn N, Slootweg PJ, Kutzner H, Mentzel T. EWSR1 gene rearrangement occurs in a subset of cutaneous myoepithelial tumors: a study of 18 cases. Mod Pathol. 2011;24(11):1444–1450. doi: 10.1038/modpathol.2011.108. PMID: 21725291. DOI: 10.1038/modpathol.2011.108. [DOI] [PubMed] [Google Scholar]
- 46.Hallor KH, Teixeira MR, Fletcher CD, Bizarro S, Staaf J, Domanski HA, von Steyern FV, Panagopoulos I, Mandahl N, Mertens F. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol. 2008;21(11):1311–1319. doi: 10.1038/modpathol.2008.124. PMID: 18604193. DOI: 10.1038/modpathol.2008.124. [DOI] [PubMed] [Google Scholar]
- 47.Agaram NP, Chen HW, Zhang L, Sung YS, Panicek D, Healey JH, Nielsen GP, Fletcher CD, Antonescu CR. EWSR1-PBX3: a novel gene fusion in myoepithelial tumors. Genes Chromosomes Cancer. 2015;54(2):63–71. doi: 10.1002/gcc.22216. PMID: 25231231. DOI: 10.1002/gcc.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuyama A, Hisaoka M, Hashimoto H. PLAG1 expression in cutaneous mixed tumors: an immunohistochemical and molecular genetic study. Virchows Arch. 2011;459(5):539–545. doi: 10.1007/s00428-011-1149-z. PMID: 21927843. DOI: 10.1007/s00428-011-1149-z. [DOI] [PubMed] [Google Scholar]
- 49.Russell-Goldman E, Dubuc A, Hanna J. Differential expression of PLAG1 in apocrine and eccrine cutaneous mixed tumors: evidence for distinct molecular pathogenesis. Am J Dermatopathol. 2019 doi: 10.1097/DAD.0000000000001393. PMID: 30839344. DOI:10.1097/DAD.0000000000001393. [DOI] [PubMed] [Google Scholar]
- 50.Asp J, Persson F, Kost-Alimova M, Stenman G. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer. 2006;45(9):820–828. doi: 10.1002/gcc.20346. PMID: 16736500. DOI: 10.1002/gcc.20346. [DOI] [PubMed] [Google Scholar]
- 51.Kas K, Voz ML, Roijer E, Åström AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15(2):170–174. doi: 10.1038/ng0297-170. PMID: 9020842. DOI: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 52.Voz ML, Åström AK, Kas K, Mark J, Stenman G, Van de Ven WJ. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene. 1998;16(11):1409–1416. doi: 10.1038/sj.onc.1201660. PMID: 9525740. DOI: 10.1038/sj.onc.1201660. [DOI] [PubMed] [Google Scholar]
- 53.Kas K, Voz ML, Hensen K, Meyen E, Van de Ven WJ. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J Biol Chem. 1998;273(36):23026–23032. doi: 10.1074/jbc.273.36.23026. PMID: 9722527. DOI: 10.1074/jbc.273.36.23026. [DOI] [PubMed] [Google Scholar]
- 54.Van Dyck F, Declercq J, Braem CV, Van de Ven WJ. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review) Int J Oncol. 2007;30(4):765–774. PMID: 17332914. [PubMed] [Google Scholar]
- 55.Juma AR, Damdimopoulou PE, Grommen SV, Van de Ven WJ, De Groef B. Emerging role of PLAG1 as a regulator of growth and reproduction. J Endocrinol. 2016;228(2):R45–56. doi: 10.1530/JOE-15-0449. PMID: 26577933. DOI: 10.1530/JOE-15-0449. [DOI] [PubMed] [Google Scholar]
- 56.Åström AK, Voz ML, Kas K, Roijer E, Wedell B, Mandahl N, Van de Ven W, Mark J, Stenman G. Conserved mechanism of PLAG1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: identification of SII as a new fusion partner gene. Cancer Res. 1999;59(4):918–923. PMID: 10029085. [PubMed] [Google Scholar]
- 57.Hibbard MK, Kozakewich HP, Dal Cin P, Sciot R, Tan X, Xiao S, Fletcher JA. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res. 2000;60(17):4869–4872. PMID: 10987300. [PubMed] [Google Scholar]
- 58.Abdollahi A, Pisarcik D, Roberts D, Weinstein J, Cairns P, Hamilton TC. LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24-25, is epigenetically regulated in cancer. J Biol Chem. 2003;278(8):6041–6049. doi: 10.1074/jbc.M210361200. PMID: 12473647. DOI: 10.1074/jbc.M210361200. [DOI] [PubMed] [Google Scholar]
- 59.Valleley EM, Cordery SF, Carr IM, MacLennan KA, Bonthron DT. Loss of expression of ZAC/PLAGL1 in diffuse large B-cell lymphoma is independent of promoter hypermethylation. Genes Chromosomes Cancer. 2010;49(5):480–486. doi: 10.1002/gcc.20758. PMID: 20175198. DOI: 10.1002/gcc.20758. [DOI] [PubMed] [Google Scholar]
- 60.Peille AL, Brouste V, Kauffmann A, Lagarde P, Le Morvan V, Coindre JM, Chibon F, Bresson-Bepoldin L. Prognostic value of PLAGL1-specific CpG site methylation in soft-tissue sarcomas. PLoS One. 2013;8(11):e80741. doi: 10.1371/journal.pone.0080741. PMID: 24260468. DOI: 10.1371/journal.pone.0080741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, Ding Y, Zhu Y, Yin M, Le X, Wang L, Yang Y, Zhang Q. Both gene deletion and promoter hyper-methylation contribute to the down-regulation of ZAC/PLAGL1 gene in gastric adenocarcinomas: a case control study. Clin Res Hepatol Gastroenterol. 2014;38(6):744–750. doi: 10.1016/j.clinre.2013.06.007. PMID: 25091631. DOI: 10.1016/j.clinre.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Godlewski J, Krazinski BE, Kowalczyk AE, Kiewisz J, Kiezun J, Kwiatkowski P, Sliwinska-Jewsiewicka A, Maslowski Z, Kmiec Z. PLAGL1 (ZAC1/LOT1) expression in clear cell renal cell carcinoma: correlations with disease progression and unfavorable prognosis. Anticancer Res. 2016;36(2):617–624. PMID: 26851016. [PubMed] [Google Scholar]
- 63.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, Zhou X, Ma X, Sioson E, Li Y, Rusch M, Gupta P, Pei D, Cheng C, Smith MA, Auvil JG, Gerhard DS, Relling MV, Winick NJ, Carroll AJ, Heerema NA, Raetz E, Devidas M, Willman CL, Harvey RC, Carroll WL, Dunsmore KP, Winter SS, Wood BL, Sorrentino BP, Downing JR, Loh ML, Hunger SP, Zhang J, Mullighan CG. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211–1218. doi: 10.1038/ng.3909. PMID: 28671688. DOI: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian M, Zhang H, Kham SK, Liu S, Jiang C, Zhao X, Lu Y, Goodings C, Lin TN, Zhang R, Moriyama T, Yin Z, Li Z, Quah TC, Ariffin H, Tan AM, Shen S, Bhojwani D, Hu S, Chen S, Zheng H, Pui CH, Yeoh AE, Yang JJ. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27(2):185–195. doi: 10.1101/gr.209163.116. PMID: 27903646. DOI: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, Wang C, Lin L, Kon A, Suzuki H, Shiozawa Y, Kataoka K, Fujii Y, Shiraishi Y, Chiba K, Tanaka H, Shimamura T, Masuda K, Kawamoto H, Ohki K, Kato M, Arakawa Y, Koh K, Hanada R, Moritake H, Akiyama M, Kobayashi R, Deguchi T, Hashii Y, Imamura T, Sato A, Kiyokawa N, Oka A, Hayashi Y, Takagi M, Manabe A, Ohara A, Horibe K, Sanada M, Iwama A, Mano H, Miyano S, Ogawa S, Takita J. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1274–1281. doi: 10.1038/ng.3900. PMID: 28671687. DOI: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 66.Hensen K, Van Valckenborgh IC, Kas K, Van de Ven WJ, Voz ML. The tumorigenic diversity of the three PLAG family members is associated with different DNA binding capacities. Cancer Res. 2002;62(5):1510–1517. PMID: 11888928. [PubMed] [Google Scholar]
- 67.Landrette SF, Kuo YH, Hensen K, Barjesteh van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ, Delwel R, Castilla LH. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 2005;105(7):2900–2907. doi: 10.1182/blood-2004-09-3630. PMID: 15585652. DOI: 10.1182/blood-2004-09-3630. [DOI] [PubMed] [Google Scholar]
- 68.Li N, Li D, Du Y, Su C, Yang C, Lin C, Li X, Hu G. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncol Rep. 2019;41(2):875–884. doi: 10.3892/or.2018.6914. PMID: 30535429. DOI: 10.3892/or.2018.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qu G, Xu Y, Wan SP, Yang G. Expression of PLAGL2 in bladder urothelial carcinoma and its relationship to lymph node metastasis and survival. Sci Rep. 2018;8(1):6044. doi: 10.1038/s41598-018-24526-5. PMID: 29662235. DOI: 10.1038/s41598-018-24526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su C, Li D, Li N, Du Y, Yang C, Bai Y, Lin C, Li X, Zhang Y. Studying the mechanism of PLAGL2 overexpression and its carcinogenic characteristics based on 3’-untranslated region in colorectal cancer. Int J Oncol. 2018 doi: 10.3892/ijo.2018.4305. PMID: 29512763. DOI:10.3892/ijo.2018.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J, Liu H, Zhang L, Liu X, Zhang C, Wang Y, He Q, Zhang Y, Li Y, Chen Q, Zhang L, Wang K, Bu Y, Lei Y. DJ-1 promotes colorectal cancer progression through activating PLAGL2/Wnt/BMP4 axis. Cell Death Dis. 2018;9(9):865. doi: 10.1038/s41419-018-0883-4. PMID: 30158634. DOI: 10.1038/s41419-018-0883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voz ML, Agten NS, Van de Ven WJ, Kas K. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 2000;60(1):106–113. PMID: 10646861. [PubMed] [Google Scholar]
- 73.Voz ML, Mathys J, Hensen K, Pendeville H, Van Valckenborgh I, Van Huffel C, Chavez M, Van Damme B, De Moor B, Moreau Y, Van de Ven WJ. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene. 2004;23(1):179–191. doi: 10.1038/sj.onc.1207013. PMID: 14712223. DOI: 10.1038/sj.onc.1207013. [DOI] [PubMed] [Google Scholar]
- 74.Declercq J, Van Dyck F, Van Damme B, Van de Ven WJ. Upregulation of Igf and Wnt signalling associated genes in pleomorphic adenomas of the salivary glands in PLAG1 transgenic mice. Int J Oncol. 2008;32(5):1041–1047. PMID: 18425330. [PubMed] [Google Scholar]
- 75.Wang Y, Shang W, Lei X, Shen S, Zhang H, Wang Z, Huang L, Yu Z, Ong H, Yin X, Yang W, Zhang C. Opposing functions of PLAG1 in pleomorphic adenoma: a microarray analysis of PLAG1 transgenic mice. Biotechnol Lett. 2013;35(9):1377–1385. doi: 10.1007/s10529-013-1213-7. PMID: 23690029. DOI: 10.1007/s10529-013-1213-7. [DOI] [PubMed] [Google Scholar]
- 76.Declercq J, Skaland I, Van Dyck F, Janssen EA, Baak JP, Drijkoningen M, Van de Ven WJ. Adenomyoepitheliomatous lesions of the mammary glands in transgenic mice with targeted PLAG1 overexpression. Int J Cancer. 2008;123(7):1593–1600. doi: 10.1002/ijc.23586. PMID: 18649356. DOI: 10.1002/ijc.23586. [DOI] [PubMed] [Google Scholar]
- 77.Declercq J, Van Dyck F, Braem CV, Van Valckenborgh IC, Voz M, Wassef M, Schoonjans L, Van Damme B, Fiette L, Van de Ven WJ. Salivary gland tumors in transgenic mice with targeted PLAG1 proto-oncogene overexpression. Cancer Res. 2005;65(11):4544–4553. doi: 10.1158/0008-5472.CAN-04-4041. PMID: 15930271. DOI: 10.1158/0008-5472.CAN-04-4041. [DOI] [PubMed] [Google Scholar]
- 78.Zhao X, Ren W, Yang W, Wang Y, Kong H, Wang L, Yan L, Xu G, Fei J, Fu J, Zhang C, Wang Z. Wnt pathway is involved in pleomorphic adenomas induced by overexpression of PLAG1 in transgenic mice. Int J Cancer. 2006;118(3):643–648. doi: 10.1002/ijc.21400. PMID: 16108035. DOI: 10.1002/ijc.21400. [DOI] [PubMed] [Google Scholar]
- 79.Xu W, Liu L, Lu H, Fu J, Zhang C, Yang W, Shen S. Dysregulated long noncoding RNAs in pleomorphic adenoma tissues of pleomorphic adenoma gene 1 transgenic mice. Mol Med Rep. 2019;19(6):4735–4742. doi: 10.3892/mmr.2019.10149. PMID: 31059011. DOI: 10.3892/mmr.2019.10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, Lang FM, Martinez-Ledesma E, Lee SH, Zheng S, Verhaak RGW. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46(D1):D1144–D1149. doi: 10.1093/nar/gkx1018. PMID: 29099951. DOI: 10.1093/nar/gkx1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshida H, Miyachi M, Ouchi K, Kuwahara Y, Tsuchiya K, Iehara T, Konishi E, Yanagisawa A, Hosoi H. Identification of COL3A1 and RAB2A as novel translocation partner genes of PLAG1 in lipoblastoma. Genes Chromosomes Cancer. 2014;53(7):606–611. doi: 10.1002/gcc.22170. PMID: 24700772. DOI: 10.1002/gcc.22170. [DOI] [PubMed] [Google Scholar]
- 82.Persson F, Winnes M, Andrén Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27(21):3072–3080. doi: 10.1038/sj.onc.1210961. PMID: 18059337. DOI: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 83.Deen M, Ebrahim S, Schloff D, Mohamed AN. A novel PLAG1-RAD51L1 gene fusion resulting from a t(8;14)(q12;q24) in a case of lipoblastoma. Cancer Genet. 2013;206(6):233–237. doi: 10.1016/j.cancergen.2013.05.019. PMID: 23890983. DOI: 10.1016/j.cancergen.2013.05.019. [DOI] [PubMed] [Google Scholar]