Abstract
Background/Aim: Rectal cancer accounts for approximately one-third of all colorectal cancers. Currently, the standard treatment for locally advanced rectal cancer (LARC) is neoadjuvant chemoradiotherapy (CRT) with capecitabine or 5-fluorouracil followed by curative surgery. Unfortunately, only 20% of patients with LARC present complete pathological response after CRT, whereas in 20-40% cases the response is poor or absent. The aim of our study was to evaluate whether microRNAs (miRNAs) in tumor biopsy specimen have the potential to predict therapeutic response in LARC patients. Patients and Methods: In total 87 LARC patients treated by CRT were enrolled in our prospective study. To identify predictive miRNAs, we used small RNA sequencing in 40 tumor biopsy samples of LARC patients (20 responders, 20 non-responders) and qPCR validation of selected miRNA candidates. Results: In the discovery phase of the study, we identified 69 miRNAs to have significantly different expression between the group of responders (TRG 1,2) and a group of non-responders (TRG 4,5) to neoadjuvant CRT. Among these miRNAs, 48 showed a lower expression and 21 showed higher expression in tumor tissues from poorly responding LARC patients. Five miRNAs were selected for validation, but only miR-487a-3p was confirmed to have a significantly higher expression in the tumor biopsy specimens of non-responders to neoadjuvant CRT (p<0.0006, AUC=0.766). Gene Ontology (GO) clustering and pathway enrichment analysis of the miR-487a-3p mRNA targets, revealed potential mechanisms behind miR-487a-3p roles in chemoradioresistance (e.g. TGF-beta signaling pathway, protein kinase activity, double-stranded DNA binding, or microRNAs in cancer). Conclusion: By combination of miRNA expression profiling and integrative computational biology we identified miR-487a-3p as a potential predictive biomarker of CRT response in LARC patients.
Keywords: Rectal cancer, chemoradiotherapy, microRNA, prediction
Colorectal cancer (CRC) is the third most common cancer worldwide and accounts for approximately 10% of all solid tumors. CRC is the second leading cause of cancer-related deaths, and its incidence is on the rise. Tumors of the rectum are generally classified as CRC and arise from colon epithelia located up to 15 cm from the anal verge, and they comprise approximately 40% of all CRC cases (1). Rectal cancer (RC) incidence is dramatically increasing and its incidence in patients under the fourth decade of life quadrupled since 1980 (2). Unfortunately, about half of all RC patients are in the stage of locally advanced rectal carcinoma (LARC), which requires radical treatment. Due to high invasiveness and the risk of local recurrence and metastasis, RC requires different medical management than does colon cancer. The gold-standard treatment of patients with LARC is based on neoadjuvant chemoradiotherapy (CRT) with 5-fluoropyrimidines (5-FU) followed by surgical treatment and, eventually, adjuvant chemotherapy. CRT aims to improve local control of the disease, minimize impact of the surgery on the surrounding tissues and possible avoidance of permanent colostomy.
For a description of the response to CRT, tumor regression grade (TRG) is used to evaluate histological tumor regression after CRT. Nowadays, several TRG classification systems (e.g., Dworak, Mandard, Ryan, AJCC, Modified Dworak) are used, such diversity additionally complicating the precise estimation of tumor regression (3). Tumor regression grading systems classification systems are based on findings from the macroscopic and microscopic examination of the resection by the pathologist. In this study, Mandard [1- Complete regression (fibrosis without detectable tissue of tumor), 2- Fibrosis with scattered tumor cells, 3- Fibrosis and tumor cells with preponderance of fibrosis, 4- Fibrosis and tumor cells with preponderance of tumor cells, 5- Tissue of tumor without changes of regression] TRG systems were used (4). Unfortunately, response to treatment varies dramatically between individuals, ranging from complete pathological response (pCR) to incomplete pathologic response (pIR) to resistance to therapy. Since neoadjuvant CRT is often associated with significant adverse symptoms, toxicity and high medical costs, these negative effects could be avoided or reduced with use of biomarkers predicting response to CRT in LARC patients (5). To date, there is no reliable biomarker that could predict response to CRT in RC patients.
Experience with other types of cancers suggests that microRNAs (miRNAs) might be such biomarkers (6). MicroRNA (miRNA) are highly conserved, small, non-coding RNAs, 18-25 nucleotides in length. MiRNAs are involved in the post-transcriptional regulation of gene expression, and miRNA expression levels were found to be deregulated in gastrointestinal cancers among others (7). MiRNAs were reported to have deregulated expression in tumor tissue in comparison to healthy mucosa in RC patients (8-10). Two studies identified different miRNA expression profiles in RC and CRC and confirmed that RC and CRC differ both anatomically and genetically (11,12). Several studies identified miRNAs (e.g. miR-21, let-7e, miR-31, miR-590-5p, miR-233, miR-19a, miR-630, miR-194) deregulated in tumor tissue of RC patients responding to CRT in comparison to non-responders (13-22). However, there seems to be no overlap in results of independent studies, which could be due to differences in the pre-analytical factors and the number of patients used in exploratory phase which could be crucial for the discovery of novel miRNA biomarker of CRT response.
Herein, we report the results of small RNA sequencing of biopsy specimen of tumor and validation of results in a larger cohort of LARC patients. MiR-487a-3p was identified and validated to have deregulated expression in tumor tissue of LARC patients who responded to 5-FU based CRT in comparison to non-responders and could potentially serve as a biomarker of CRT response in LARC patients.
Patients and Methods
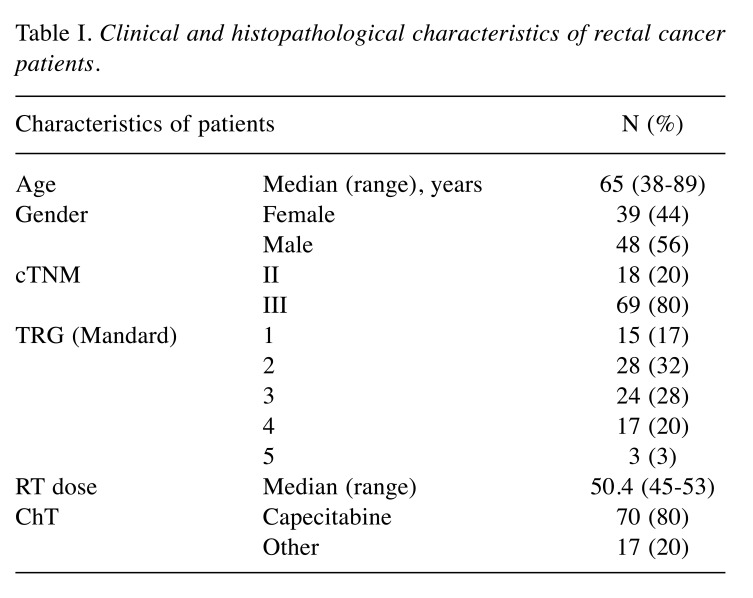
Patients. Eighty-seven patients diagnosed with LARC and treated with neoadjuvant 5-FU based CRT between 2016 and 2019 at University Hospital Brno (Brno, Czech Republic), Masaryk Memorial Cancer Institute (Brno, Czech Republic) and Comprehensive Cancer Centre and AGEL Research and Training Institute Novy Jicin (Novy Jicin, Czech Republic) were enrolled in the study. Pre-treatment and pre-operative staging were based on transrectal ultrasound (TRUS) and magnetic resonance imaging (MRI). According to TRG, patients were divided into two groups of responders to CRT (Mandard TRG 1+2) and non-responders (TRG 3+4+5). Patients characteristics are summarized in Table I. All patients signed informed consent before entering the study. The study was approved by the ethical committee University Hospital Brno (Brno, Czech Republic).
Table I. Clinical and histopathological characteristics of rectal cancer patients.
Sample collection and RNA isolation. Samples of tumor tissues were collected at the time of pre-treatment staging along with TRUS. Samples were stored in RNAlater solution Ambion (Thermo Fisher Scientific, Waltham, MA, USA) at –80˚C until RNA isolation. Total RNA enriched with a fraction of small RNAs was isolated using MirVana MiRNA Isolation Kit Ambion (Thermo Fisher Scientific). Quantity of RNA was measured fluorometrically using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and Qubit RNA BR Assay Kit (Thermo Fisher Scientific) and concentrations were ranging between 93-2445 ng/μl with median 675 ng/μl and average 865 ng/μl. Quality of RNA was measured using NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Isolated RNA was stored at –80˚C.
Small RNA sequencing. In total, 40 samples of RNA isolated from tumor tissue, of which twenty responders to CRT and twenty non-responders were used for cDNA library preparation. RNA was diluted to 50 ng/μl and 100 ng/2 μl was used for cDNA library preparation using CleanTag Small RNA Library Prep Kit (Trilink Biotechnologies, San Diego, CA, USA). Quality control of cDNA libraries was done using the Agilent High Sensitivity D1000 ScreenTape System (Agilent Technologies, Santa Clara, CA, USA) and Tapestation 2200 System (Agilent Technologies). Length of cDNA libraries ranged between 145-163 bp with an average length of 152 bp and median length 152 bp. Concentration was measured using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Concentrations were ranged between 19-371 nM with an average 149 nM and median 150 nM. Libraries were diluted to 4 nM and pooled into final 4 nM pool of cDNA libraries used for sequencing. Sequencing was done using the NextSeq 500/550 High Output Kit v2 (Illumina, San Diego, CA, USA) and NextSeq 500/550 instrument (Illumina) and sequencing setup was single read, 80 cycles.
Reverse transcription and quantitative real-time PCR. The validation cohort was extended to 87 patients of which forty-three were responders to CRT and 44 non-responders. Samples were diluted to concentration 4 ng/μl and 10 ng/ 2.5 μl was input concentration to reverse transcription. RNA was reverse transcribed using miRCURY LNA RT Kit (Qiagen, Hilden, Germany). Quantitative Real-Time PCR (qRT-PCR) was performed using miRCURY LNA SYBR Green PCR Kit (Qiagen) and miRCURY miRNA PCR Assay (Qiagen) hsa-miR-487a-3p (MIMAT0002178: 5'AAUCAUACAGGGACAUCCAGUU, product no. 339306, cat. no. YP00204381) and SNORD48 (product no. 339306, cat. no. YP00203903). Measurements were performed using Quantstudio 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
MiRNA target prediction, gene ontology and pathway enrichment analysis. We performed in silico analyses on miR-487a-3p and to identify its gene targets we used mirDIP 4.1 (http://ophid.utoronto.ca/mirDIP). For consequent enrichment and network analysis we selected ‘top third’ and ‘top 1%’ predicted targets of miR-487a-3p. Then we performed a functional annotation and comprehensive pathway enrichment analysis of all the proteins of the network using ShinyGO v0.61: Gene Ontology Enrichment Analysis (http://bioinformatics.sdstate.edu/go/). We considered significantly enriched only pathways with q-value [false discovery rate (FDR):Benjamini-Hochberg (BH) method] lower than 0.05. The Gene Ontology Enrichment Analysis was used to classify ‘top third’ and ‘top 1%’ predicted targets of miR-487a-3p accordingly to their molecular function.
Statistical analysis. Sequencing of data was processed using the Chimira online tool, and statistical analysis was done using Bioconductor tool and packages EdgeR and Deseq2. Expression levels of individual miRNAs measured by qRT-PCR were normalized using RNU48 as endogenous control and formula Ct(miR-487a-3p normalized expression)=2–(CtΔ 487a-3p duplicate - Ct Δ RNU48 duplicate). Statistical analysis of expression data was performed using GraphPad Prism 6. Results with a p-value lower than 0.05 were considered statistically significant.
Results
From the group of 87 patients with LARC enrolled in the study, n=43 (49%) were classified as responders (TRG 1,2) and n=44 (51%) were non-responders (TRG 3,4,5). In the validation phase, we analyzed separately the non-responding group with the exclusion of intermediate responders (TRG 3). In this group, based only on TRG 4 and TRG 5 patients remained 20 (23%) patients. There were no significant differences between the two cohorts regarding patient age, gender, treatment and tumor characteristics. Following the TRG classification, n=15 (17%) of them showed a pathological complete response (TRG 1).
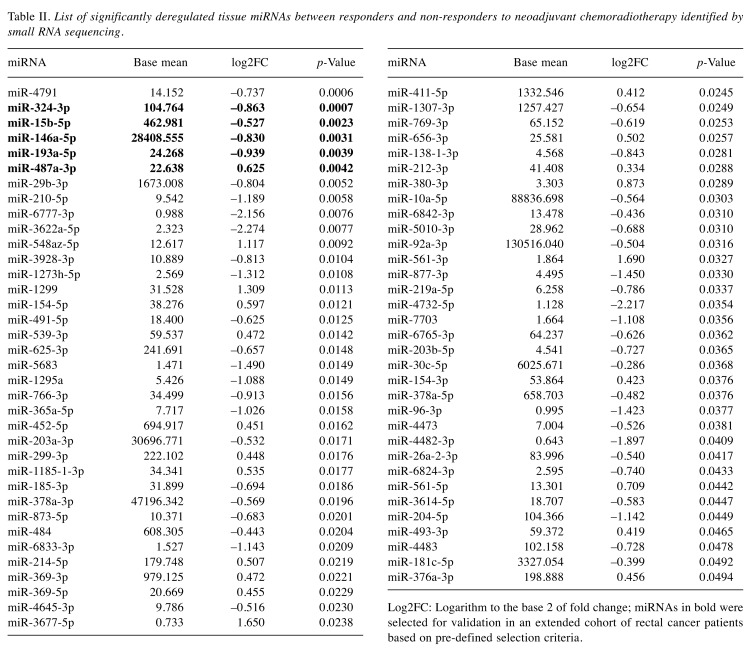
In this discovery phase of the study, we performed small RNA sequencing of the tumor tissue biopsies and identified 69 miRNAs to have a significantly different expression in the group of 20 responders (TRG 1,2) and the group of 20 non-responders (TRG 4,5) to neoadjuvant CRT. Among these miRNAs, 48 showed a lower expression and 21 showed higher expression in tumor tissues from LARC patients with poor response to CRT (Table II). Out of these 69 miRNAs identified in discovery phase, 5 miRNAs (miR-324-3p, miR-15b-5p, miR-146a-5p, miR-193a-5p and miR-487a-3p) were selected for validation phase of the study to evaluate their ability to identify LARC patients with poor response to neoadjuvant CRT. These miRNAs were selected based on the p-value (p<0.005), log2(fold-change)>0.5 and the average number of reads across all sequenced samples (at least 20 reads).
Table II. List of significantly deregulated tissue miRNAs between responders and non-responders to neoadjuvant chemoradiotherapy identified by small RNA sequencing.
Log2FC: Logarithm to the base 2 of fold change; miRNAs in bold were selected for validation in an extended cohort of rectal cancer patients based on pre-defined selection criteria.
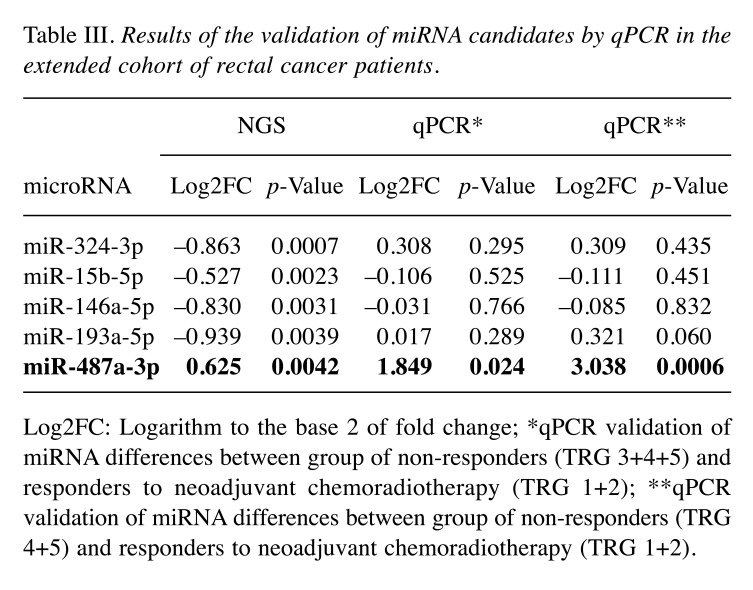
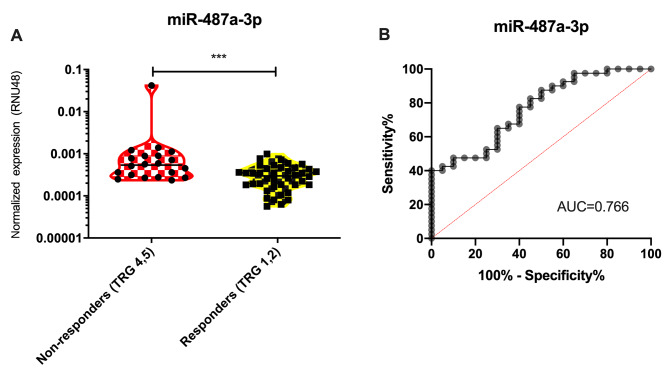
The expression of miRNA candidates was determined by use of miRCURY miRNA PCR Assay (Qiagen) and statistically evaluated between the extended groups of responders (n=43) and non-responders (n=44). By use of non-parametric Mann-Whitney U-test only miR-487a-3p was confirmed to have significantly higher expression in tumor tissue of non-responders (Table III, qPCR*, p<0.024). When non-responders were narrowed only to TRG4 and TRG5 cases (n=20), the significance became much higher (p<0.0006, Figure 1A). Subsequent ROC analysis showed that expression of miR-487a-3p in tumor tissue is indicative of poor response to neoadjuvant CRT in LARC patients and enable to identify these patients (TRG 4,5) with 78% sensitivity and 60% specificity (AUC=0.766; Figure 1B).
Table III. Results of the validation of miRNA candidates by qPCR in the extended cohort of rectal cancer patients.
Log2FC: Logarithm to the base 2 of fold change; *qPCR validation of miRNA differences between group of non-responders (TRG 3+4+5) and responders to neoadjuvant chemoradiotherapy (TRG 1+2); **qPCR validation of miRNA differences between group of non-responders (TRG 4+5) and responders to neoadjuvant chemoradiotherapy (TRG 1+2).
Figure 1. MiR-487a-3p is significantly up-regulated in tumor tissue of rectal cancer patients with poor response to neoadjuvant chemoradiotherapy (A) and based on the ROC analysis miR-487a-3p can distinguish responders (TRG 1,2) and non-responders (TRG 4,5) to this therapy (B). ***pvalue<0.001; AUC, Area under curve.
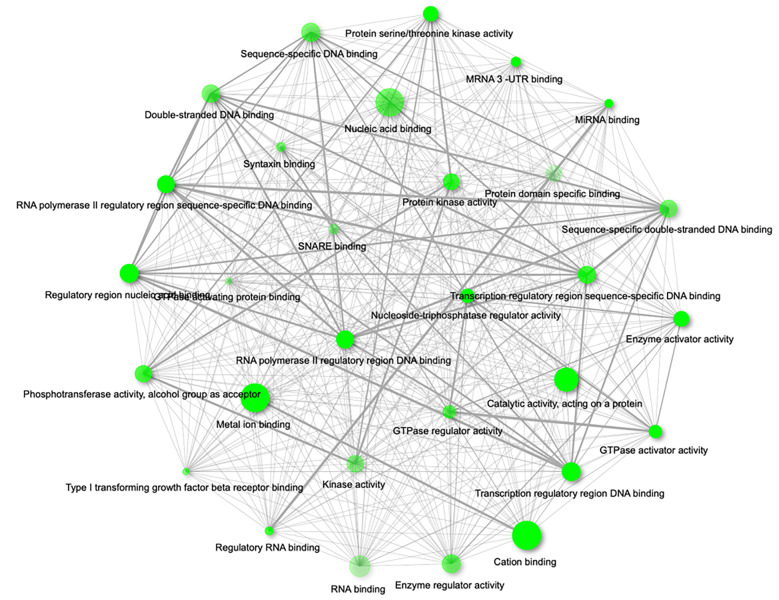
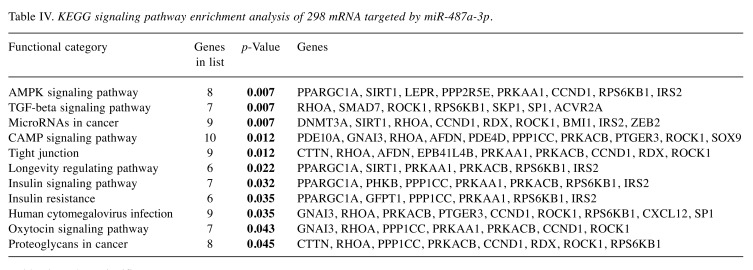
In order to understand the molecular mechanism behind the observed clinical phenotype, we identified 298 predicted mRNAs targeted by miR-487a-3p that passed pre-defined selective filter (data not shown). Further, we performed a Gene Ontology (GO) clustering of the identified targets, considering molecular function (Figure 2) and KEGG signaling pathway enrichment. We have identified 11 significant pathways (p<0.05) including TGF-beta signaling pathway, AMPK signaling pathway, insulin signaling pathway or microRNAs in cancer (summarized in Table IV). The most represented GO molecular functions included
Figure 2. Gene ontology enrichment analysis of miR-487a-3p targets according to their molecular function (network of the 30 most significant terms are shown; p-Value cut-off (FDR)=0.05).
Table IV. KEGG signaling pathway enrichment analysis of 298 mRNA targeted by miR-487a-3p.
Bold values show significance.
RNA polymerase II regulatory region DNA binding (p=0.003), miRNA binding (p=0.003), protein kinase activity (p=0.005), double-stranded DNA binding (p=0.006), type I transforming growth factor beta receptor binding (p=0.006), enzyme regulator activity (p=0.006), GTPase regulator activity (p=0.006) and others (summarized in Figure 2).
Discussion
Administration of neoadjuvant CRT revolutionized the treatment of LARC patients. Unfortunately, a significant part of patients does not benefit from the treatment and might be profiting more from different treatment strategies. Furthermore, patients who respond to CRT and reach pCR might be treated with alternative approaches like the watch-and-wait strategy which might result in preservation of anal sphincter and significant improvement of quality of life of patients. However, the decision whether a patient is eligible for alternative treatment strategies or would benefit from conventional CRT treatment followed by surgery would have to be based on predictive biomarkers that are not available yet. MiRNAs might have the potential to distinguish between responders and non-responders to CRT and serve as predictive biomarkers in the future.
We attempted to find miRNA predictive biomarker of response to CRT with the use of next-generation sequencing. Comparison of global miRNA expression profiles of responders and non-responders to CRT followed by validation of selected miRNAs on a larger independent cohort of patients identified miR-487a-3p as a miRNA with the highest predictive potential. We found miR-487a-3p expression level to be significantly up-regulated (NGS: p=0.0042; qPCR: p=0.0006) in non-responders to CRT in comparison to responders.
Despite many studies evaluating tissue miRNAs with potential to predict response to CRT, results are highly inconsistent. The majority of studies selected miRNAs for validation based on literature search or results from CRC based studies, which might prevent discovery of new predictive biomarkers. Another factor causing inconsistency in the results might be the lack of sufficient amount of patients in the exploratory phases and proper validations on an independent cohort of patients.
We are the first to describe the connection of miR-487a-3p with response to CRT in LARC patients. MiR-487a-3p maps on 14q32 and yet little is known about its function. Nonetheless, the findings of several studies support the relevance of dysregulated expression levels of miR-487a-3p to neoplasia and poor prognosis of patients (23,24). MiR-487a-3p was also reported to be up-regulated in peripheral blood mononuclear cells of type 1 diabetes patients (25). Several studies observed up-regulated expression level of miR-487a to be connected with enhanced invasion, migratory capacity and ability of cancer cell lines to metastasize. Yang et al. described the effects of miR-487a on the progression of gastric cancer (GC) cell lines in vitro and in vivo via inhibition of tumor suppressor T-cell intracellular antigen-1 (TIA1). MiR-487a overexpression was shown to promote progression in AGS cells transfected with miR-487a mimics. Cells showed an increased proliferation rate and decreased cell apoptosis. Moreover, a GC xenograft mouse model confirmed that overexpression of miR-487a enhances tumor growth in vivo (26).
Similarly, Chang et al. revealed that miR-487a is highly expressed in hepatocellular carcinoma (HCC) and correlates with poor postoperative prognosis of HCC patients. Furthermore, miR-487a was found to promote proliferation and metastasis of HCC by suppressing phosphoinositide-3-kinase regulatory subunit 1 and sprouty-related EVH1 domain containing 2 (27). Ma et al. described the role of miR-487a in epithelial–mesenchymal transition (EMT) in breast cancer (BC). TGF-beta-1 treatment, used to induce EMT in cancer cells, significantly increased the expression level of miR-487a in breast cancer (BC) cell lines MDA-MB-231. On other hand, transfection of miR-487a inhibitor significantly decreased the expression of vimentin and increased the expression of E-cadherin in BC cells to repress EMT (28).
In order to understand the molecular mechanism behind the observed clinical phenotype, we performed a Gene Ontology (GO) clustering of the miR-487a-3p targets, considering molecular function (Figure 2) and KEGG signaling pathway enrichment. Among others we identified TGF-beta signaling pathway to be significantly enriched and type I TGF beta receptor binding to be one of the most represented Gene Ontology molecular functions in miR-487a-3p targets. TGF-beta signaling is associated with EMT and therefore could present a potential link between miR-487a-3p and EMT. The process of EMT is closely associated with therapy resistance in various cancers, including CRC (29,30).
Altogether, our bioinformatics predictions and the in vitro observations in other cancer types indicate the involvement of miR-487a-3p in the process of EMT or at least its ability to revert EMT to MET. As there is a clear link between EMT and drug resistance, we speculate, that increased levels of miR-487a-3p could be, in part, functionally linked to poor response to CRT in rectal cancer patients through regulation of the TFG-beta signaling, EMT and consequently also the drug resistance.
In general, miR-487a-3p is described as an oncogene in several cancers, and our data suggest that it may play a similar role in the pathogenesis of RC. Especially, the connection of miR-487a-3p with EMT might be crucial. However, these associations are only hypothetical, and mechanism of miR-487a-3p regulation of treatment resistance remains unclear.
Conclusion
In conclusion, we identified miR-487a-3p as a predictive biomarker of response to neoadjuvant CRT in LARC patients. Validation of the results on larger independent cohort of patients is needed to select the most accurate cut-off value of miR-487a-3p to distinguish between responders and non-responders. Functional studies need to be performed to describe precise role of miR-487a-3p in the regulation of response to CRT in LARC patients.
Conflicts of Interest
The Authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Authors’ Contributions
Conceptualization: OS, ZK; methodology: OS, TM, KT; experimental work: TM, KT; validation: TM, VP, TG, MF, LF, RS, MS, MD, IBN; formal analysis: KT, MS, OS; resources and data collection: IK, MS, OS, ZK; data curation: TM, VP, TG, MF, LF, RS, MS, MD, IBN; writing-original draft preparation: TM, OS; writing-review and editing: TM, OS, MS, IK, ZK; supervision: OS, ZK; funding acquisition: OS, ZK.
Acknowledgements
This study was supported by the Ministry of Health of the Czech Republic by the Czech Health Research Council project No. 16-31765A.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. PMID: 30207593. DOI: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Iskander O, Courtot L, Tabchouri N, Artus A, Michot N, Muller O, Pabst-Giger U, Bourlier P, Kraemer-Bucur A, Lecomte T, Guyetant S, Chapet S, Calais G, Salamé E, Ouaïssi M. Complete pathological response following radiochemotherapy for locally advanced rectal cancer: Short and long-term outcome. Anticancer Res. 2019;39:5105–5113. doi: 10.21873/anticanres.13705. PMID: 31519622. DOI: 10.21873/anticanres.13705. [DOI] [PubMed] [Google Scholar]
- 3.Kim SH, Chang HJ, Kim DY, Park JW, Baek JY, Kim SY, Park SC, Oh JH, Yu A, Nam B-H. What is the ideal tumor regression grading system in rectal cancer patients after preoperative chemoradiotherapy. Cancer Res Treat. 2016;48:998–1009. doi: 10.4143/crt.2015.254. PMID: 26511803. DOI: 10.4143/crt.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680–2686. doi: 10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c. PMID: 8194005. DOI: 10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 5.Du B, Wang X, Wu D, Wang T, Yang X, Wang J, Shi X, Chen L, Zhang W. MicroRNA expression profiles identify biomarkers for predicting the response to chemoradiotherapy in rectal cancer. Mol Med Rep. 2018 doi: 10.3892/mmr.2018.9215. PMID: 29956755. DOI:10.3892/mmr.2018.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slattery ML, Herrick JS, Mullany LE, Samowitz WS, Sevens JR, Sakoda L, Wolff RK. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56:769–787. doi: 10.1002/gcc.22481. PMID: 28675510. DOI: 10.1002/gcc.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Zhang Y, Wong SH, Law PTY, Zhao S, Yu J, Chan MTV, Wu WKK. Common deregulation of seven biological processes by MicroRNAs in gastrointestinal cancers. Sci Rep. 2018;8:3287. doi: 10.1038/s41598-018-21573-w. PMID: 29459716. DOI: 10.1038/s41598-018-21573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W. Identification of aberrantly expressed miRNAs in rectal cancer. Oncol Rep. 2012 doi: 10.3892/or.2012.1769. PMID: 22576798. DOI: 10.3892/or.2012.1769. [DOI] [PubMed] [Google Scholar]
- 9.Gaedcke J, Grade M, Camps J, Sokilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Moller S, Beissbarth T, Ried T, Litman T. The rectal cancer microRNAome - microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res. 2012;18:4919–4930. doi: 10.1158/1078-0432.CCR-12-0016. PMID: 22850566. DOI: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okugawa Y, Shirai Y, Nodono H, Matsutani F, Itoh M, Hishida A, Morimoto Y, Nishikawa R, Yokoe T, Tanaka K, Urata H, Toiyama Y, Araki T, Inoue Y, Tanaka M, Kusunoki M, Miki C. Objective predictive score as a feasible biomarker for short-term survival in terminalIy Ill patients with cancer. Anticancer Res. 2017;37:267–275. doi: 10.21873/anticanres.11317. PMID: 28011502. DOI: 10.21873/anticanres.11317. [DOI] [PubMed] [Google Scholar]
- 11.Orosz E, Kiss I, Gyöngyi Z, Varjas T. Expression of circulating miR-155, miR-21, miR-221, miR-30a, miR-34a and miR-29a: Comparison of colonic and rectal cancer. In Vivo. 2018;32:1333–1337. doi: 10.21873/invivo.11383. PMID: 30348685. DOI: 10.21873/invivo.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullock MD, Pickard KM, Nielsen BS, Sayan AE, Jenei V, Mellone M, Mitter R, Primrose JN, Thomas GJ, Packham GK, Mirnezami AH. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis. 2013;4:e684–e684. doi: 10.1038/cddis.2013.213. PMID: 23788041. DOI: 10.1038/cddis.2013.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotchi M. microRNA expression is able to predict response to chemoradiotherapy in rectal cancer. Mol Clin Oncol. 2012 doi: 10.3892/mco.2012.9. PMID: 24649136. DOI: 10.3892/mco.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svoboda M, Sana J, Fabian P, Kocakova I, Gombosova J, Nekvindova J, Radova L, Vyzula R, Slaby O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol. 2012;7 doi: 10.1186/1748-717X-7-195. PMID: 23167930. DOI: 10.1186/1748-717X-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kheirelseid EAH, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G, Kerin MJ. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int J Colorectal Dis. 2013;28:247–260. doi: 10.1007/s00384-012-1549-9. PMID: 22903298. DOI: 10.1007/s00384-012-1549-9. [DOI] [PubMed] [Google Scholar]
- 16.Lopes-Ramos CM, Habr-Gama A, Quevedo B de S, Felício NM, Bettoni F, Koyama FC, Asprino PF, Galante PA, Gama-Rodrigues J, Camargo AA, Perez RO, Parmigiani RB. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med Genomics. 2014;7 doi: 10.1186/s12920-014-0068-7. PMID: 25496125. DOI: 10.1186/s12920-014-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caramés C, Cristobal I, Moreno V, Marín J, González-Alonso P, Torrejón B, Minguez P, Leon A, Martín J, Hernández R, Pedregal M, Martín M, Cortés D, García-Olmo D, Fernández M, Rojo F, García-Foncillas J. MicroRNA-31 emerges as a predictive biomarker of pathological response and outcome in locally advanced rectal cancer. Int J Mol Sci. 2016;17:878. doi: 10.3390/ijms17060878. PMID: 27271609. DOI: 10.3390/ijms17060878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvi S, Molinari C, Foca F, Teodorani N, Saragoni L, Puccetti M, Passardi A, Tamberi S, Avanzolini A, Lucci E, Calistri D. miR-17-92a-1 cluster host gene (MIR17HG) evaluation and response to neoadjuvant chemoradiotherapy in rectal cancer. OncoTargets Ther. 2016;9:2735–2742. doi: 10.2147/OTT.S105760. PMID: 27226732. DOI: 10.2147/OTT.S105760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millino C, Maretto I, Pacchioni B, Digito M, De Paoli A, Canzonieri V, D’Angelo E, Agostini M, Rizzolio F, Giordano A, Barina A, Rajendran S, Esposito G, Lanfranchi G, Nitti D, Pucciarelli S. Gene and MicroRNA expression are predictive of tumor response in rectal adenocarcinoma patients treated with preoperative chemoradiotherapy: Combined miRNAS and gene expression. J Cell Physiol. 2017;232:426–435. doi: 10.1002/jcp.25441. PMID: 27225591. DOI: 10.1002/jcp.25441. [DOI] [PubMed] [Google Scholar]
- 20.Eriksen AHM, Sørensen FB, Andersen RF, Jakobsen A, Hansen TF. Association between the expression of microRNAs and the response of patients with locally advanced rectal cancer to preoperative chemoradiotherapy. Oncol Lett. 2017;14:201–209. doi: 10.3892/ol.2017.6141. PMID: 28693154. DOI: 10.3892/ol.2017.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Angelo E, Zanon C, Sensi F, Digito M, Rugge M, Fassan M, Scarpa M, Pucciarelli S, Nitti D, Agostini M. miR-194 as predictive biomarker of responsiveness to neoadjuvant chemoradiotherapy in patients with locally advanced rectal adenocarcinoma. J Clin Pathol. 2018;71:344–350. doi: 10.1136/jclinpath-2017-204690. PMID: 28870889. DOI: 10.1136/jclinpath-2017-204690. [DOI] [PubMed] [Google Scholar]
- 22.Campayo M, Navarro A, Benítez JC, Santasusagna S, Ferrer C, Monzó M, Cirera L. miR-21, miR-99b and miR-375 combination as predictive response signature for preoperative chemoradiotherapy in rectal cancer. PLoS One. 2018;13:e0206542. doi: 10.1371/journal.pone.0206542. PMID: 30388154. DOI: 10.1371/journal.pone.0206542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Yu W, Gao J, Ma W, Frentsch M, Thiel A, Liu M, Rahman N, Qin Z, Li X. MicroRNA-487a-3p functions as a new tumor suppressor in prostate cancer by targeting CCND1. J Cell Physiol. 2020;235:1588–1600. doi: 10.1002/jcp.29078. PMID: 31309555. DOI: 10.1002/jcp.29078. [DOI] [PubMed] [Google Scholar]
- 24.González-Vallinas M, Rodríguez-Paredes M, Albrecht M, Sticht C, Stichel D, Gutekunst J, Pitea A, Sass S, Sánchez-Rivera FJ, Lorenzo-Bermejo J, Schmitt J, De La Torre C, Warth A, Theis FJ, Müller NS, Gretz N, Muley T, Meister M, Tschaharganeh DF, Schirmacher P, Matthäus F, Breuhahn K. Epigenetically regulated chromosome 14q32 miRNA cluster induces metastasis and predicts poor prognosis in lung adenocarcinoma patients. Mol Cancer Res. 2018;16:390–402. doi: 10.1158/1541-7786.MCR-17-0334. PMID: 29330288. DOI: 10.1158/1541-7786.MCR-17-0334. [DOI] [PubMed] [Google Scholar]
- 25.Zurawek M, Dzikiewicz-Krawczyk A, Izykowska K, Ziolkowska-Suchanek I, Skowronska B, Czainska M, Podralska M, Fichna P, Przybylski G, Fichna M, Nowak J. miR-487a-3p upregulated in type 1 diabetes targets CTLA4 and FOXO3. Diabetes Res Clin Pract. 2018;142:146–153. doi: 10.1016/j.diabres.2018.05.044. PMID: 29859273. DOI: 10.1016/j.diabres.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Wang M, Lin B, Yao D, Li J, Tang X, Li S, Liu Y, Xie R, Yu S. miR-487a promotes progression of gastric cancer by targeting TIA1. Biochimie. 2018;154:119–126. doi: 10.1016/j.biochi.2018.08.006. PMID: 30144499. DOI: 10.1016/j.biochi.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Chang R-M, Xiao S, Lei X, Yang H, Fang F, Yang L-Y. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017;23:2593–2604. doi: 10.1158/1078-0432.CCR-16-0851. PMID: 27827315. DOI: 10.1158/1078-0432.CCR-16-0851. [DOI] [PubMed] [Google Scholar]
- 28.Ma M, He M, Jiang Q, Yan Y, Guan S, Zhang J, Yu Z, Chen Q, Sun M, Yao W, Zhao H, Jin F, Wei M. MiR-487a promotes TGF-β1-induced EMT, the migration and invasion of breast cancer cells by directly targeting MAGI2. Int J Biol Sci. 2016;12:397–408. doi: 10.7150/ijbs.13475. PMID: 27019625. DOI: 10.7150/ijbs.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skarkova V, Kralova V, Vitovcova B, Rudolf E. Selected aspects of chemoresistance mechanisms in colorectal carcinoma – A focus on epithelial-to-mesenchymal transition, autophagy, and apoptosis. Cells. 2019;8:234. doi: 10.3390/cells8030234. PMID: 30871055. DOI: 10.3390/cells8030234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ieda T, Tazawa H, Okabayashi H, Yano S, Shigeyasu K, Kuroda S, Ohara T, Noma K, Kishimoto H, Nishizaki M, Kagawa S, Shirakawa Y, Saitou T, Imamura T, Fujiwara T. Visualization of epithelial-mesenchymal transition in an inflammatory microenvironment–colorectal cancer network. Sci Rep. 2019;9:16378. doi: 10.1038/s41598-019-52816-z. PMID: 31705021. DOI: 10.1038/s41598-019-52816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]