Generation of sensory ganglion organoids and retinal ganglion cells from fibroblasts for research and therapeutic applications.
Abstract
Neural organoids provide a powerful tool for investigating neural development, modeling neural diseases, screening drugs, and developing cell-based therapies. Somatic cells have previously been reprogrammed by transcription factors (TFs) into sensory ganglion (SG) neurons but not SG organoids. We identify a combination of triple TFs Ascl1, Brn3b/3a, and Isl1 (ABI) as an efficient means to reprogram mouse and human fibroblasts into self-organized and networked induced SG (iSG) organoids. The iSG neurons exhibit molecular features, subtype diversity, electrophysiological and calcium response properties, and innervation patterns characteristic of peripheral sensory neurons. Moreover, we have defined retinal ganglion cell (RGC)–specific identifiers to demonstrate the ability for ABI to reprogram induced RGCs (iRGCs) from fibroblasts. Unlike iSG neurons, iRGCs maintain a scattering distribution pattern characteristic of endogenous RGCs. iSG organoids may serve as a model to decipher the pathogenesis of sensorineural diseases and screen effective drugs and a source for cell replacement therapy.
INTRODUCTION
A ganglion is a cluster or group of nerve cells found in the peripheral nervous system (PNS) or central nervous system (CNS). They often interconnect with each other and with other structures in the PNS and CNS to form a complex nervous network. There are three groups of ganglia in the PNS, which are the dorsal root ganglia (DRG), cranial nerve ganglia, and autonomic ganglia, and two types of ganglia in the CNS, which are the basal ganglia in the brain and retinal ganglion in the retina. Unlike other ganglia, which are essentially cell clusters, retinal ganglia consist of a layer/sheet of dispersive retinal ganglion cells (RGCs). Diverse types of neurons in the somatosensory ganglia such as DRG are specialized for different sensory modalities such as proprioception, mechanoreception, nociception (i.e., pain perception), thermoception, and pruriception (i.e., itch perception) (1, 2). Similarly, there are numerous subtypes of RGCs that are specialized for transmitting from the retina different visual information (e.g., color, contrast, and motion direction) to the central visual system in the brain (3). In the human, a variety of pain, itch, neurological, and degenerative disorders affect sensory ganglia (SGs) and RGCs. Mutations in the FXN (frataxin) and IKBKAP genes, for example, result in debilitating Friedreich’s ataxia and familial dysautonomia, respectively (4, 5). Dominant gain-of-function mutations in the sodium channel Nav1.7 gene SCN9A, which is expressed in sensory neurons, are linked to two severe pain syndromes—inherited erythromelalgia and paroxysmal extreme pain disorder, while its recessive loss-of-function mutations cause dangerous congenital insensitivity to pain (6). Recently, peripheral SG dysfunction has also been linked to tactile sensitivity and other behavioral deficits associated with the autism spectrum disorders (7). Both genetic and environmental risk factors contribute to glaucoma, which is a leading cause of blindness worldwide and characterized by progressive degeneration of RGCs and the optic nerve (8).
Despite the difference in morphology and embryonic origin, somatosensory and retinal ganglia share extensive overlap of gene expression and we proposed more than two decades back that both might also share genetic regulatory hierarchies (9, 10). This assumption has largely turned out to be the case. During embryogenesis, somatosensory ganglion neurons arise from the multipotent neural crest (NC) cells through a process of cell migration and coalescence (1). RGCs are also derived from multipotent retinal progenitor cells and destined to the ganglion cell layer by migration. It has been shown that the neurogenic bHLH transcription factors (TFs) Ngn1 and/or Ngn2 are involved in the determination of peripheral sensory neurons (11), and that the homeodomain TFs Isl1 and Brn3a or Brn3b are required for the specification and differentiation of different subtypes of neurons in the somatosensory and retinal ganglia (12–17). Moreover, there is substantial functional redundancy between Ngn1 and Ngn2 as well as between Brn3a and Brn3b in the development of sensory neurons and RGCs (11, 18, 19).
Somatic cell reprogramming by defined TFs into sensory neurons provides a powerful strategy for studying mechanisms of SG development and sensory disease pathogenesis and for generating cells for patient-specific cell replacement therapy, drug screening, and in vitro disease modeling. It has been shown recently that nociceptor and other subtypes of sensory neurons can be directly induced from murine and human fibroblasts by Brn3a and Ngn1 or Ngn2 or by a combination of five TFs including Ascl1, Ngn1, Isl2, Myt1l, and Klf7 (20, 21). The induced sensory neurons express characteristic marker proteins and are electrically active and selectively responsive to various agonists known to activate pain- and itch-sensing neurons (20, 21). However, networked SG did not appear to be consistently generated in these cases, and it is unclear whether RGCs were induced by these combinations of TFs.
Given the advantages of organoids in studying developmental mechanisms and modeling and treating relevant diseases, we sought to generate ganglion organoids and RGCs from mouse and human fibroblasts using TFs controlling in vivo development of sensory and retinal ganglia. The extensive molecular homology between SG neurons and RGCs creates a dilemma as to how to distinguish these two types of neurons. In the past, several RGC markers including Brn3a, Brn3b, Isl1, Thy1.2, Sncg, Math5, Rbpms, and RPF-1 were used to identify RGCs induced from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and somatic cells (22–25). However, this is a questionable practice because although these markers are sufficient to identify RGCs within the retina, they are inadequate as specific markers for identifying induced RGCs (iRGCs), given their expression in SG and other CNS regions as well (10, 15). We thus carefully screened for RGC-specific markers by comparing expression patterns of numerous known markers in the retina and DRG. This analysis revealed Pax6 expression in RGCs but not in DRG and that RGCs can be identified as Pax6+Brn3a+ or Pax6+Brn3b+ double-positive cells. Equipped with this knowledge, we set to generate induced SG (iSG) organoids and iRGCs from fibroblasts by testing the combination of Ascl1, the pioneer neurogenic TF for somatic cell reprogramming of neurons (26), with a variety of SG and retinal TFs. This screen identified a triple-factor combination ABI (Ascl1-Brn3a/3b-Isl1) as the most efficient way to induce self-organized and networked iSG and iRGCs from fibroblasts.
RESULTS
A screen for TFs that reprogram fibroblasts into iSG organoids
Previous studies by our group and others have demonstrated that SGs and RGCs share similar transcriptional regulatory mechanism for their development, for instance, both Brn3 TFs (Brn3a and Brn3b) and Isl1 are involved in the specification and differentiation of DRG neurons and RGCs (12–14, 16). More recently, Ascl1 has been shown to play a pioneering role in induced neuron (iN) reprogramming from somatic cells (26). As a first step to generate iSG and iRGCs directly from somatic cells, we sought to induce SG neurons and RGCs from mouse embryonic fibroblasts (MEFs) by testing the combination of Ascl1 with each of 22 SGs and retinal TFs (Brn3b, Isl1, Math5, Ebf1, Pax6, Tfap2a, Nr4a2, Nrl, Crx, Ptf1a, Neurod1, Lhx2, Ngn1, Ngn2, Chx10, Sox2, Rx, Meis1, Foxn4, Otx2, Sox9, and Six3). When MEFs were infected with doxycycline (Dox)–inducible Ascl1 and Brn3b (AB) or Isl1 (AI) lentiviruses and cultured in the neural differentiation medium containing Dox, they started to change morphology by day 7 and form visible neuronal clusters by day 14 (Fig. 1, C and D). This phenomenon did not occur when Ascl1 acted alone or was combined with each of the rest of 20 TFs (Fig. 1B). Neither did this happen when MEFs were infected with both Brn3b and Isl1 viruses or with only control green fluorescent protein (GFP) viruses (Fig. 1, A and E). When we combined Ascl1 with both Brn3b and Isl1 (ABI), they again induced morphological changes of MEFs but more importantly induced conspicuously more neuronal clusters than either the AB or AI double-factor combinations (Fig. 1, C, D, F, and N, and fig. S1, A and B), suggesting a synergistic effect between Brn3b and Isl1 in reprogramming MEFs into neuronal clusters.
Fig. 1. Combinations of Ascl1, Brn3a/3b, and Isl1 directly convert MEFs into iSG.
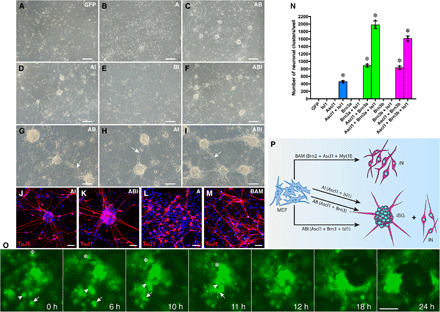
(A to I) Morphological changes of MEFs infected with the indicated lentiviruses (A, Ascl1; B, Brn3b; I, Isl1) and cultured for 14 days. Networked iSGs were induced by combinations of Ascl1 with Brn3b (AB), Isl1 (AI), or both Brn3b and Isl1 (ABI), with the ABI triple-factor combination as the most efficient. Arrows point to the thick fasciculated nerve fibers interconnecting iSG. Scale bars, 160 μm (A to F) and 80 μm (G to I). (J to M) Scattered iNs and clustered iSG induced by AI, ABI, A, or BAM (Brn2 + Ascl1 + Myt1l) were immunolabeled for Tuj1 and counterstained with nuclear 4′,6-diamidino-2-phenylindole (DAPI). Note the morphological differences of Tuj1-immunoreactive neurons between conditions. Scale bars, 40 μm. (N) Quantification of iSG induced by single and combinations of TFs. MEFs (6 × 104) were seeded into each well of 12-well plates and infected with lentiviruses expressing the indicated TFs or GFP, and iSGs in each well were then counted at day 14 following virus infection. Data are means ± SD (n = 3). Asterisks indicate significance in one-way analysis of variance test: *P < 0.0001. (O) Snapshots of a time-lapse video showing how individual neurons induced by ABI self-organized into an iSG. The arrow, arrowhead, and asterisk indicate the positions of three individual iNs at different time points. Scale bar, 62.5 μm. (P) Schematic indicating the outcome (iNs or iSG) of MEFs induced by BAM, AI, AB, or ABI.
The neuronal clusters induced by either double- or triple-factor combinations (AB, AI, and ABI) appeared to be interconnected by thick fasciculated nerve fibers and resemble SG plexus in morphology (Fig. 1, G to I) and thus were designated as iSG organoids. The iSG neurons and associated nerve fibers were highly immunoreactive for the neuronal marker Tuj1 (Fig. 1, J and K, and fig. S2, D to I). Tuj1 immunolabeling also showed that AI- and ABI-induced neurons mostly formed iSG, and only a small number of them were scattered outside the iSG (Fig. 1, J, K, and P). By contrast, Tuj1 immunoreactivity showed that Ascl1 alone induced neurons mostly with an immature morphology and that the BAM (Brn2, Ascl1, and Mytl1) combination induced mature neurons that were scattered instead of clustered (Fig. 1, L, M, and P, and fig. S2, A to C), consistent with previous reports (27). Therefore, we identified the AB, AI, and ABI combinations of TFs capable of inducing MEFs into iSG, with the ABI triple-factor combination as the most efficient.
To investigate how ABI-reprogrammed neurons are organized into iSG, we used long-term time-lapse microscopy to track them over time in culture. For this purpose, MEFs were prepared from the CAG-GFP transgenic mouse embryos (28) and induced by ABI for 10 days before time-lapse recording. Compared to MEFs, reprogrammed individual neurons appeared to be rounder and neurite-bearing and displayed much higher contrast and brighter GFP fluorescence (Fig. 1O and movies S1 and S2). Over a period of tens of hours, they first formed smaller cellular clusters via migration, which then coalesced into bigger and bigger clusters that resembled SG. We did not observe this self-organization phenomenon for neurons induced by Ascl1 (movies S3 and S4).
The induction of iSG by TFs from MEFs could be through direct cell conversion or might be mediated through an intermediate proliferative progenitor. To distinguish these possibilities, we pulse-labeled cells with 5-ethynyl-2′-deoxyuridine (EdU) for 24 hours at day 14 of reprogramming with AI or ABI and found that almost no Tuj1-positive cells were labeled by EdU, whereas approximately 15% of Tuj1-negative cells (e.g., MEFs) were labeled (fig. S2, G to J and N to P). We then reprogrammed MEFs with ABI in the presence of EdU for 13 days starting from day 1 of reprogramming. In this case, only 6.1% of Tuj1-positive cells were labeled by EdU, whereas 73.1% of Tuj1-negative cells were labeled (fig. S2, J to M), suggesting that iSGs are most likely induced by direct cell transdifferentiation without undergoing a proliferative intermediate state. In agreement with these results, as determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays, we detected no increase of expression levels of the neural progenitor marker genes Nestin and Olig2 over the entire time course (from day 1 to day 12) of ABI reprogramming (fig. S2Q). Similarly, the expression of pluripotent factor genes Oct4, Klf4, and Nanog was not induced during the time course of ABI reprogramming (fig. S2R). Furthermore, immunostaining showed that from day 1 to day 12 of ABI reprogramming, no protein expression was seen for the neural progenitor marker Nestin, pluripotent progenitor markers Nanog and Oct4, or Sox2, a marker for both neural and pluripotent progenitor cells (fig. S2, S and T). Thus, iSGs are most likely induced by direct cell transdifferentiation without undergoing an intermediate state of neural or pluripotent progenitors.
Given the demonstrated functional redundancy and similar DNA binding and transcriptional properties between Brn3a and Brn3b (10, 18, 19), we investigated whether these two factors are interchangeable in somatic cell reprogramming. We tested whether Brn3a was able to replace Brn3b in reprogramming MEFs into iSG and found that this indeed was the case (Fig. 1N and fig. S3, A to I).
iSG neurons exhibit molecular characteristics of peripheral sensory neurons
By immunofluorescent staining and qRT-PCR assays, we examined a variety of molecular neuronal markers, both general and cell type specific, to characterize the iSG reprogrammed from MEFs by ABI (Ascl1 + Brn3b + Isl1 or Ascl1 + Brn3a + Isl1). We found that they were highly immunoreactive for Tuj1 and Map2 (Fig. 2, A and O), two general neuronal hallmarks. They also expressed synapsin and Vamp (synaptobrevin) (Fig. 2, B and C), suggesting that the networked iSG neurons were capable of forming synapses and releasing synaptic vesicles. In the normal SG, the heavy neurofilament NF200 and intermediate neurofilament peripherin are expressed in the A-fiber and C-fiber neurons, respectively, and both were seen to be expressed in the iSG (Fig. 2, D, E, and P). Many neurons in the iSG were also immunoreactive for the vesicular glutamate transporters 1 and 2 (vGLUT1 and vGLUT2) (Fig. 2, F and G), consistent with the fact that peripheral sensory neurons are mostly excitatory glutamatergic neurons. As determined by qRT-PCR, these immunolabeling results were confirmed by the marked up-regulation of expression of Tuj1, Map2, NF200, vGlut1, vGlut2, and vGlut3 genes in the ABI-induced iSG compared to MEFs infected by GFP lentiviruses (Fig. 2W).
Fig. 2. iSGs induced by ABI contain mostly peripheral sensory neurons.
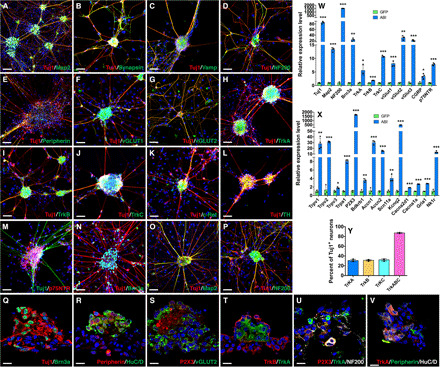
(A to P) iSGs induced by Ascl1, Brn3b, and Isl1 (A to N) or Ascl1, Brn3a, and Isl1 (O and P) were double-immunostained with the indicated antibodies and counterstained with nuclear DAPI. They were immunoreactive for Tuj1, Map2, synapsin, Vamp, NF200, peripherin, vGLUT1, vGLUT2, TrkA, TrkB, TrkC, c-Ret, TH, p75NTR, and Brn3a. Scale bars, 80 μm (A) and 40 μm (B to P). (Q to V) Sections from iSG induced by Ascl1, Brn3b, and Isl1 were immunostained with the indicated antibodies and counterstained with nuclear DAPI. Scale bars, 12.7 μm. (W) qRT-PCR analysis showing that in MEFs infected with ABI (Ascl1 + Brn3b + Isl1) viruses, compared to those infected with GFP viruses, there was a significant increase in expression of the indicated genes, which represent general and subtype-specific sensory neuron markers. Data are means ± SD (n = 3 or 4). Asterisks indicate significance in unpaired two-tailed Student′s t test: *P < 0.05, **P < 0.001, ***P < 0.0001. (X) qRT-PCR analysis showing that in MEFs infected with ABI viruses, compared to those infected with GFP viruses, there was a significant increase in expression of the indicated genes, which represent nociception pathway genes of sensory neurons. Data are means ± SD (n = 3 or 4). Asterisks indicate significance in unpaired two-tailed Student′s t test: *P < 0.05, **P < 0.005, ***P < 0.0005. (Y) Quantification of Tuj1-positive neurons that express each of the three Trk receptors (TrkA, TrkB, or TrkC) individually or combined (TrkABC) in MEFs infected with the ABI viruses. Data are means ± SD (n = 3).
In the DRG, neurotrophin receptor expression marks subtypes of sensory neurons. For instance, TrkA is expressed by cutaneous nociceptive and thermoceptive neurons, TrkB by a subset of cutaneous mechanoreceptive neurons, and TrkC by proprioceptive neurons (1). In the iSG reprogrammed by ABI, qRT-PCR assays revealed that there was a significant up-regulation of TrkA, TrkB, and TrkC gene expression (Fig. 2W). Moreover, immunolabeling confirmed the presence of TrkA, TrkB, and TrkC proteins in both somas and nerve fibers of the induced ganglion neurons (Fig. 2, H to J). Each of the Trk receptors was found in approximately 30% of the iNs, and 87% of the iNs were labeled by costaining for all three Trk receptors (Fig. 2Y), suggesting that each Trk receptor was expressed in a distinct subpopulation of induced ganglion neurons. c-Ret and TH are expressed in subpopulations of nonpeptidergic nociceptors and C-low threshold mechanoreceptors, respectively (1, 2). Correspondingly, we observed expression of both proteins in the iSG and associated nerve fibers (Fig. 2, K and L). In addition, pan-sensory neuron markers Brn3a (for iSG induced by Ascl1 + Brn3b + Isl1) and the nerve growth factor (NGF) receptor p75NTR were also found in iSG neurons (Fig. 2, M and N). qRT-PCR validated the up-regulation of Brn3a and p75NTR expression in the iSG and additionally revealed up-regulation of CGRP, a marker for a subpopulation of peptidergic nociceptive neurons (Fig. 2W).
The TrkA-positive nociceptive neurons in the iSG were further characterized by qRT-PCR analysis. In the iSG, we found significantly up-regulated expression of receptor ion channel genes Trpv1/2/3 and Trpa1, which detect heat and cold, respectively (Fig. 2X) (29). There was also expression of P2X3, Bdkrb1, and Accn1/2, which are receptor genes responsible for damage sensing (Fig. 2X) (29). In addition, induced expression was observed for other pain perception pathway genes including sodium channel gene Scn11a, potassium channel gene Kcnq2, calcium channel genes Cacna1a and Cacna2d1, and neurotransmitter receptor genes Gria1 and Nk1r (Fig. 2X) (29). To further investigate whether distinct types of sensory neurons were aggregated together within the same iSG, we carried out immunostaining analyses of cryosections of ABI-induced iSG. Besides colabeling between Tuj1 and Brn3a in the same iSG, we found that peripherin+ and HuC/D+ neurons, P2X3+ and vGLUT2+ neurons, and TrkA+ and TrkB+ neurons coexisted in the same iSG (Fig. 2, Q to T). Moreover, we detected coexpression of three markers, such as TrkA, P2X3 and NF200, and TrkA, peripherin, and HuC/D, in the same iSG (Fig. 2, U and V), suggesting that individual iSGs are likely aggregated from distinct sensory neuron types.
Over the time course of ABI reprogramming, qRT-PCR assays showed that the expression of general neuronal marker genes Tuj1 and Map2 was progressively induced starting from day 3, whereas other sensory neuronal marker genes including Trpv2, TrkC, and Brn3a were not induced until day 6 or 9 (fig. S1, C to E). Consistent with this, Brn3a-immunoreactive cells did not emerge until day 6 with a mostly scattered pattern, but by day 9 or 12, they mostly coalesced into iSG (fig. S1G). Therefore, as expected, sensory neuronal markers were induced slightly later than general neuronal markers during ABI reprogramming. Concomitant with neuronal induction, the fibroblast marker genes Col1a1 and Twist2 were gradually down-regulated starting from day 3 (fig. S1F).
Immunostaining of iSG induced by AI or AB suggested that they also contained neurons that expressed typical sensory neuronal markers Tuj1, Map2, Dcx (doublecortin), synapsin, NF200, peripherin, vGLUT1, TrkA, TH, HuC/D, and Brn3a (fig. S3, J to S). Together, these data indicate that certain combinations of TFs (ABI, AI, and AB) are capable of reprogramming MEFs into iSG that contain proprioceptive, mechanoreceptive, nociceptive, and thermoceptive sensory neurons.
iSG neurons exhibit physiological characteristics of mature sensory neurons
To assess the electrophysiological properties of neurons within and outside the iSG reprogrammed from MEFs by ABI or AI, we performed whole-cell patch-clamp recordings of cells with neuronal morphology (Fig. 3A). Following 9 days of induction, the recorded neurons (two of two) generated potassium currents and small sodium currents but no action potentials, suggesting that they were functionally immature. At 2 weeks, the great majority of neurons (34 of 37) had typical sodium and potassium currents and exhibited action potential responses (Fig. 3, B to F). Among them, most (70.3%) are multispiking neurons, and the rest (21.6%) are single-spiking (Fig. 3, B, C, E, and K), similar to those reprogrammed from human fibroblasts by Brn3a and Ngn1 or Ngn2 (21). The inward sodium current could be specifically blocked by tetrodotoxin (TTX) and recovered by its removal (Fig. 3, H to J). Moreover, consistent with the synapsin immunoreactivity (Fig. 2B and fig. S3L), some neurons (2 of 37) exhibited spontaneous postsynaptic currents (Fig. 3G), suggesting the formation of functional synapses between iNs. Therefore, the iSG neurons induced by ABI or AI display membrane and physiological properties of mature neurons.
Fig. 3. iSG neurons exhibit functional properties of sensory neurons.
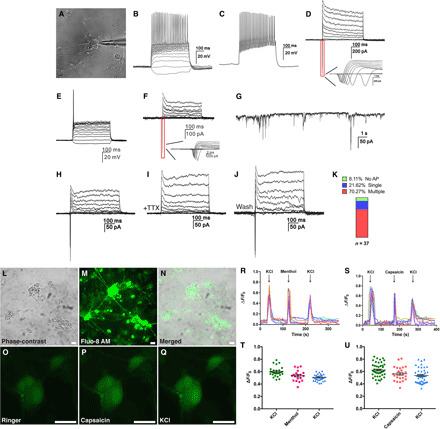
(A) Micrograph showing a typical iSG neuron chosen for patch-clamp recording. (B to D) Current-clamp recordings revealed multiple action potential responses (multiple-spiking) of a differentiated iSG neuron under current injection (B and C). Voltage-clamp recordings of the same neuron indicated fast activated and inactivated inward sodium currents as well as outward potassium currents (D). (E and F) Current injection revealed a single action potential response (single-spiking) of an iSG neuron (E). Voltage-clamp recordings of the same neuron indicated fast activated and inactivated inward sodium currents as well as outward potassium currents (F). (G) Spontaneous postsynaptic currents recorded from a differentiated iSG neuron. (H to J) The sodium currents of an iSG neuron were completely blocked by TTX and were partially restored by its washout. (K) Observed ratios of iSG neurons that are multiple-spiking and single-spiking, or display no action potential (AP). (L to N) iSG induced by ABI and corresponding fluorescent signals after incubation with Fluo-8 AM. Scale bars, 20 μm. (O to Q) Calcium changes indicated by fluorescent intensity in normal Ringer’s solution (O), 10 μM capsaicin (P), and 100 mM KCl (Q). Scale bars, 20 μm. (R) Representative calcium responses to 100 μM menthol and 100 mM KCl. Calcium responses were calculated as the change in fluorescence (∆F) over the initial baseline fluorescence (F0). (S) Representative calcium responses to 10 μM capsaicin and 100 mM KCl. (T and U) Scatter dot plots showing the positive responses of individual cells to menthol, capsaicin, or KCl. Data are means ± SEM (n = 19 to 44).
The nociceptive sensory neurons express ion channels Trpv1, Trpm8, and Trpa1, which respond to heat, cold, and noxious chemicals, respectively (29). By calcium imaging, we used specific agonists capsaicin (10 μM) and menthol (100 μM) for Trpv1 and Trpm8 to confirm the functional expression of these two channels in iSG neurons (20, 21). KCl (100 mM) was transiently perfused to monitor the functional viability of the cells at the beginning and end of recording. Only cells that showed responses to KCl were chosen for analysis. Nearly all the iSG clusters induced by ABI showed green fluorescence following incubation with the calcium indicator Fluo-8 AM (Fig. 3, L to N). We found that among all the recorded cells, 56.8% of them (25 of 44) responded to capsaicin and 70.4% (19 of 27) to menthol (Fig. 3, O to U), suggesting that a large number of iSG neurons express ion channels characteristic of nociceptive sensory neurons.
Integration and innervation properties of iSG neurons
We investigated the ability of iSG neurons to survive and integrate in the DRG by microinjecting dissociated iSG neurons reprogrammed from CAG-GFP mouse embryos (28), into adult rat DRG explants (fig. S4A). Following 2 weeks of culture of the transplanted explants, we found that the GFP+ iSG neurons survived, spread, and integrated in the DRG and were immunoreactive for the pan-sensory neuron marker HuC/D (fig. S4B). Moreover, a large fraction of them were immunoreactive for TrkA, while a small portion expressed TrkB or TrkC (fig. S4, C to E), indicating that iSG neurons maintain subtype specificity in the DRG.
Consistent with their sensory neuron identity, after a week in culture, iSG neurons reprogrammed from Tau-GFP mouse embryos (30) spontaneously aggregated with rhodamine-labeled sensory neurons dissociated from E13.5 mouse DRGs to form DRG-like organoids interconnected by nerve fibers (fig. S4, N to Q). In contrast, when GFP+ iSG neurons were cocultured with P0 mouse skin cells, they did not co-aggregate with skin cells; instead, they projected to and innervated vimentin-immunoreactive epidermal cells with multiple terminal nerve endings (fig. S4, J to M), in agreement with the fact that DRG neurons normally innervate their peripheral targets in the epidermis.
Identification of definitive RGC markers
Previous studies have demonstrated that peripheral SG neurons and RGCs share many common molecular hallmarks, making it difficult to distinguish these two types of sensory neurons in cell culture. During the past decade in stem cell research, a number of supposedly specific molecular markers have been used to identify differentiated or induced SG neurons and RGCs (22–25); unfortunately, however, no efforts have been made to confirm the specificity of these markers, casting doubt on some of the previous conclusions. Because Brn3a, Brn3b, and Isl1 are TFs crucial for retinal cell development, in particular, RGC development (13, 14, 16), there is a possibility that they may also be able to reprogram MEFs into RGCs. We thus set out to identify molecular markers that can definitively distinguish RGCs from peripheral sensory neurons. We postulated that such unique identifiers could be single-molecule markers or a combination of multiple-molecule markers that must be present only in RGCs within the retina but not in peripheral sensory neurons or any other tissues.
In the mammalian retina, our early studies have identified Brn3a and Brn3b as the gold standard markers for RGCs, but meanwhile revealed their expression in peripheral SG and other CNS areas (10, 15). In the mouse, immunolabeling of retinal and DRG sections confirmed the specificity of Brn3a and Brn3b in RGCs within the retina as well as their widespread expression in DRG neurons (fig. S5A), indicating that Brn3a and Brn3b alone cannot distinguish RGCs from DRG neurons outside the retina. Similarly, many other commonly used RGC markers including Thy1.2, RPF-1, Rbpms, HuC/D, Six6, Ebf, Isl1, Zeb2, Lmo4, Ldb1, and Sncg all displayed expression in the DRG (fig. S5A). Expressed in both RGCs and DRGs were also a number of sensory neuron markers including CGRP, peripherin, vGLUT2, vGLUT3, GABA, TrkA, TrkB, TrkC, and P2X3 (fig. S5A). Pax6 appeared to be the only exception among all the tested markers, which is expressed in RGCs and inner nuclear layer within the retina but absent from DRG (fig. S5A). Given the expression of Brn3a, Brn3b, Thy1.2, RPF-1, and Rbpms only in RGCs within the retina, a combination of Pax6 with any of these proteins could serve as a potential unique identifier for RGCs.
The uniqueness of double-positive markers was tested by immunolabeling sections of other CNS areas. Double-immunostaining showed that neuronal cells immunoreactive for both Pax6 and RPF-1, Thy1.2, Rbpms, HuC/D, or Tuj1, albeit absent from the DRG, were present not only in the retina but also in the spinal cord (Fig. 4A), precluding their use as specific RGC markers. The Isl1+Pax6+ double-positive cells were absent from the DRG and spinal cord but present within both the ganglion cell layer and inner nuclear layer in the retina (Fig. 4A), precluding also this combination as a specific RGC identifier. By contrast, Brn3a+Pax6+ and Brn3b+Pax6+ double-positive cells were exclusively RGCs in the retina and were not found in the DRG or spinal cord (Fig. 4A). Given the detection of Brn3a/Brn3b expression in the midbrain and cerebellum (10, 15), we investigated whether there were Brn3a+Pax6+ and Brn3b+Pax6+ double-positive cells in these two brain regions and found none at stages E13.5, P4, and P21 (Fig. 4A). Thus, these results together demonstrate that a combination of Pax6 and Brn3a or Brn3b double markers can serve as specific identifiers for RGCs.
Fig. 4. Identification of iRGCs reprogrammed by ABI.
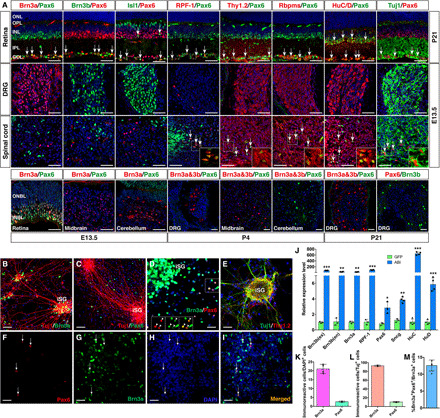
(A) Cryosections from the indicated regions and stages of mice were stained by double immunofluorescence with the indicated antibodies and counterstained with nuclear DAPI. Arrows point to representative double-positive cells. GCL, ganglion cell layer; INBL, inner neuroblastic layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONBL, outer neuroblastic layer; ONL, outer nuclear layer; OPL, outer plexiform layer. Scale bars, 40 μm. (B to E) MEFs were infected with the ABI lentiviruses, cultured for 14 days, and double-immunostained with the indicated antibodies and counterstained with nuclear DAPI. Arrowheads in (D) indicate colocalized cells in the outlined region located outside the iSG. Scale bars, 40 μm (B and C) and 20 μm (D and E). (F to I) MEFs infected with the ABI lentiviruses and cultured for 14 days were dissociated and double-immunostained with anti-Brn3a and anti-Pax6 antibodies and counterstained with nuclear DAPI. Arrows indicate colocalized cells. Scale bars, 20 μm. (J) qRT-PCR analysis of expression levels of the indicated genes (ex, exogenous; en, endogenous) in MEFs infected with ABI or GFP viruses (means ± SD, n = 3 or 4). *P < 0.05, **P < 0.001, ***P < 0.0001. (K and L) Quantification of DAPI- or Tuj1-positive cells that express Brn3a or Pax6 in MEFs infected with the ABI viruses (means ± SD, n = 4). (M) Quantification of Brn3a+Pax6+ iRGCs induced by ABI (means ± SD, n = 4).
We used the single-cell RNA sequencing (scRNA-seq) technology to further confirm the specificity of Brn3a+Pax6+ and Brn3b+Pax6+ double-positive markers. A Brn3b-GFP knockin mouse line was generated, and RGCs were enriched and sequenced by scRNA-seq. In addition, we isolated adult mouse DRG cells, which were then similarly sequenced. Clustering and expression analyses of the sequenced RGCs revealed that most of them expressed Pax6, Brn3a, or Brn3b and both Pax6 and Brn3a or Brn3b; in particular, the great majority of RGCs were positive for both Pax6 and Brn3a (fig. S5B). By contrast, there was a complete absence of DRG cells expressing both Pax6 and Brn3a or Brn3b, although Brn3a and Brn3b were present in most DRG cells (fig. S5B), consistent with the idea that a combination of Pax6 and Brn3a or Brn3b double markers can be used to distinguish RGCs from DRG cells.
Characterization of iRGCs
To assess whether ABI and AI are able to induce iRGCs in addition to iSG, we immunostained ABI-reprogrammed MEF cells with antibodies against Tuj1, Brn3a, and/or Pax6. Double-labeling between Tuj1 and Brn3a or Pax6 showed that Brn3a-expressing cells were concentrated in the iSG, whereas the great majority of Pax6-expressing cells were distributed outside of the iSG and only few of them were seen in the iSG (Fig. 4, B and C). Moreover, all Pax6-positive cells coexpressed Brn3a and most of them displayed relatively weak Brn3a expression (Fig. 4, D and F to I), indicating that iRGCs were reprogrammed from MEFs by ABI. Similar to the distribution of endogenous RGCs that are spread throughout the RGC layer, the Brn3a+Pax6+ iRGCs were scattered and did not organize into clustered mini-ganglia (Fig. 4, C and D), unlike the induced peripheral SG neurons. Quantification of immunoreactive cells indicated that approximately 21.1% of all cells were induced by ABI into Brn3a+ neurons, whereas only about 2.6% of them were reprogrammed into Pax6+ cells (Fig. 4K). Furthermore, there were 93.1% of Tuj1+ cells coexpressing Brn3a, 10.5% of Tuj1+ cells coexpressing Pax6, and 12.6% Brn3a+ cells coexpressing Pax6 (Fig. 4, L and M), suggesting that only a small fraction of the ABI-reprogrammed neurons are Brn3a+Pax6+ double-positive iRGCs and that most of them are Brn3a+ iSG neurons. Similarly, a small number of Brn3a+Pax6+ iRGCs were induced by AI (fig. S3, T and U).
In agreement with the induction of a small proportion of iRGCs by ABI, immunostaining showed that some cells outside the iSG were positive for Thy1.2 (Fig. 4E). qRT-PCR assays revealed a significant up-regulation of several commonly used RGC marker genes including the endogenous Brn3b, Brn3a, RPF-1, Pax6, Sncg, HuC, and HuD in MEFs infected with ABI lentiviruses compared to those infected with GFP viruses (Fig. 4J), consistent with the induction of iRGCs by ABI from MEFs. Moreover, during the time course of ABI reprogramming, we were able to show by qRT-PCR assay that Pax6 expression was progressively induced starting from day 9 (fig. S1E).
Bulk and single-cell transcriptome profiling of iSG neurons and iRGCs
We further characterized the iSG neurons and iRGCs by bulk and single-cell transcriptome profiling. First, we carried out bulk RNA-seq analysis of ABI- and GFP-transduced MEFs after 2 weeks of induction (Fig. 5A). Scatter plot and hierarchical cluster analyses showed that there were numerous genes whose expression was down-regulated or up-regulated in ABI-transduced compared to GFP-transduced MEFs (fig. S6, A to C, and table S1). We performed gene set enrichment analysis (GSEA) of the altered genes followed by network visualization (31), and one major group of clustered networks emerged (fig. S6D). This group encompasses only up-regulated genes that are enriched for GO (gene ontology) terms relevant to neural function and development such as synaptic signaling, synaptic vesicle, synapse organization, neurotransmitter transport, regulation of neurotransmitter levels, exocytosis, calcium ion binding, ligand-gated channel activity, neuron projection, axon, and nervous system development. These results are consistent with the induction of functional SG and retinal ganglion neurons by ABI from MEFs. In agreement with this and qRT-PCR assays (Figs. 2, W and X, and 4J), bulk RNA-seq confirmed up-regulation of many SG and retinal ganglion genes in ABI-transduced MEFs, including NF200, Brn3a, TrkB, vGlut3, Trpv1, P2X3, Gria1, Pax6, Sox11, Sncg, and Thy1 (fig. S6, E and F).
Fig. 5. Single-cell transcriptome profiling of iSG neurons.
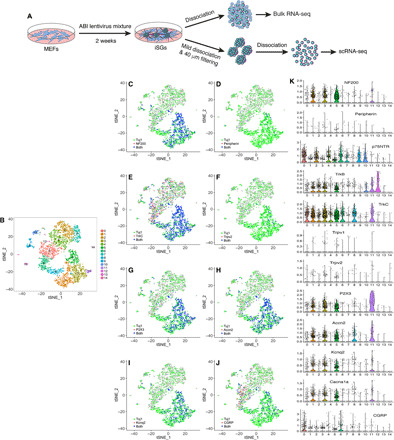
(A) Schematic illustration of the processes for bulk RNA-seq and scRNA-seq analyses. (B) t-distributed Stochastic Neighbor Embedding (t-SNE) plot of the 15 cell clusters generated from the sequenced single iSG neurons. (C to J) t-SNE plots colored by expression of the indicated conventional SG marker genes. (K) Violin plots showing expression patterns of the indicated conventional SG marker genes in single-cell clusters.
We separated ABI-induced iSG from MEFs by mild dissociation and filtering and then carried out scRNA-seq analysis of single iSG cells using the 10× Genomics Chromium platform (Fig. 5A) (32). After processing the sequencing data by the Cell Ranger software pipeline, we clustered the 3231 sequenced single cells into 15 clusters using the Seurat software package (Fig. 5B), which is an R toolkit for single-cell genomics (33). Investigation of gene expression patterns showed high levels of expression of general neuronal marker genes such as Tuj1, Tau, and Map2 in clusters 1, 3, 5, 8, and 11, whereas they are expressed much more weakly in the rest of the clusters (fig. S7, A and C to E). By contrast, many of the previously identified MEF marker genes (34) including Klf4, Mmp2, and Postn have, in general, an opposite expression pattern, displaying little expression in clusters 1, 3, 5, 8, and 11 but obvious expression in the rest of the clusters (fig. S7, A and F to H). Pseudotime trajectory of the sequenced cells constructed using Monocle (35) yielded three presumptive states along which Klf4 expression progressively decreases, while the expression of Tuj1, Tau, and Map2 progressively increases (fig. S7, I and J). Thus, in iSG induced from MEFs by ABI for 2 weeks, there are still some cells that express both neuronal and MEF markers, suggesting that MEFs undergo a transitional intermediate stage that exhibits both MEF and neuronal characteristics before completely reprogrammed into mature iSG neurons (fig. S7B). Consistent with this idea, there were many cells coexpressing both Tuj1 and the fibroblast marker gene vimentin in a number of the clusters (fig. S7K). By days 6 to 12 of ABI reprogramming, we also detected by immunolabeling some cells and nerve bundles that were immunoreactive for both Tuj1 and vimentin proteins (fig. S7L).
Consistent with the induction of iSG neurons, there is expression of NF200, peripherin, p75NTR, TrkB, TrkC, Trpv1, Trpv2, P2X3, Accn2, Kcnq2, Cacna1a, and CGRP in various clusters of sequenced iSG cells (Fig. 5, C to K). In particular, NF200, P2X3, Accn2, Kcnq2, and Cacna1a are primarily expressed in clusters 1, 3, 5, 8, and 11, and peripherin, Trpv1, and Trpv2 are mainly present in a small number of cells in clusters 1, 3, and 5 (Fig. 5, C, D, F to I, and K), indicating their expression in mature iSG neurons and their expression specificity. Many genes that are markers for both SG neurons and RGCs, such as Thy1, Sncg, Rbpms, Gap43, HuC, Sox11, Sox12, Zeb2, Brn3a, Brn3c, and RPF-1, are also expressed in various clusters of sequenced iSG cells (Fig. 6). However, the RGC marker Pax6 is only enriched in small cell clusters 12 and 13 and expressed in few cells in other clusters, consistent with the observation that only a very small number of iSG cells were immunoreactive for Pax6 (Figs. 4C and 6I). The Pax6+ cells in clusters 12 and 13 do not appear to be iRGCs because they lack expression of RGC markers Brn3a, Brn3c, and RPF-1 (Fig. 6I). In agreement with the observation that iRGCs were scattered and rarely present in iSG, there are only a small number of cells coexpressing both Pax6 and Tuj1, Thy1, Gap43, HuC, Sox11, Brn3a, or RPF-1, primarily in clusters 5 and 6 (Fig. 4C and fig. S8, A to H).
Fig. 6. Expression patterns of conventional RGC marker genes in single iSG neurons.
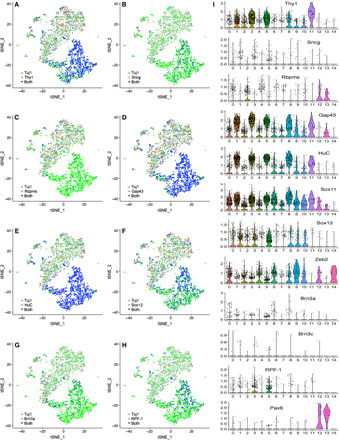
(A to H) t-SNE plots colored by expression of the indicated conventional RGC marker genes. (I) Violin plots showing expression patterns of the indicated conventional RGC marker genes in single-cell clusters.
Reprogramming of HSFs into iSG and iRGCs
We reprogrammed human skin fibroblasts (HSFs) into iSG with a mixture of the three individual ABI lentiviruses only at a low efficiency. To increase the reprogramming efficiency, we created a Dox-inducible lentiviral construct containing Ascl1, Isl1, and Brn3b in a single open reading frame (ORF) tethered by the P2A and T2A self-cleaving peptide sequences (Fig. 7A). HSFs infected by these single ABI-expressing viruses readily formed well-networked iSG in approximately 45 days in the neural differentiation medium (Fig. 7B). Immunostaining of these iSG showed that they contained typical sensory neurons expressing TUJ1, MAP2, NF200, PERIPHERIN, SYNAPSIN, VGLUT1, TRKA, TRKB, TH, and BRN3A (Fig. 7, C to K). Moreover, similar to MEFs, a small number of iRGCs were induced from HSFs by ABI that were immunoreactive for both PAX6 and BRN3A (Fig. 7, L and M). qRT-PCR assays showed that TUJ1 expression was gradually induced by ABI starting from day 10 but the more mature neuron marker gene MAP2 was not induced until day 20 (Fig. 7N). In contrast, the fibroblast marker genes COL1A1 and TWIST2 were progressively down-regulated starting from day 10 (Fig. 7O), concurrent with TUJ1 induction.
Fig. 7. iSG and iRGCs induced by ABI from HSFs.
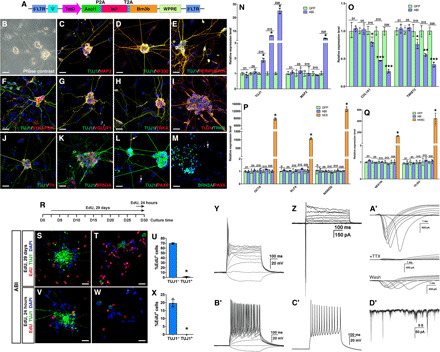
(A) Schematic of the lentiviral construct. (B to M) Networked iSG induced by ABI from HSFs (B) and iSG and iRGCs double-immunostained with the indicated antibodies and counterstained with DAPI (C to M). (N to Q) qRT-PCR assay showing the time course [days 1 (D1) to 20 (D20)] of expression changes of the indicated marker genes in HSFs infected with ABI or GFP viruses (means ± SD, n = 4). *P < 0.0001 for (N), (P), and (Q) and *P < 0.01, **P < 0.001, ***P < 0.0001 for (O). hES, human embryonic stem cell; hiNSC, human neural stem cell. (R) Schematic of EdU labeling schedule. (S to U) ABI-transduced HSFs were labeled by EdU for 29 days and colabeled for both TUJ1 and EdU before (S) and after dissociation (T). (U) Corresponding quantification (means ± SD, n = 4). *P < 0.0001. (V to X) ABI-transduced HSFs were labeled by EdU for 24 hours and colabeled for both TUJ1 and EdU before (V) and after dissociation (W). (X) Corresponding quantification (means ± SD, n = 4). *P < 0.0001. (Y, Z, and A′) Current-clamp recordings revealed single action potential responses (single-spiking) of a differentiated iSG neuron (Y). Voltage-clamp recordings of the same neuron indicated fast activated and inactivated inward sodium currents as well as outward potassium currents (Z and A′). The sodium currents of the iSG neuron were effectively blocked by TTX and were partially restored by its washout (A′). (B′ and C′) Current-clamp recordings revealed an iSG neuron with multiple action potential responses (multiple-spiking). (D′) Spontaneous postsynaptic currents recorded from a differentiated iSG neuron. Scale bars, 80 μm (B) and 20 μm (C to M, S, T, V, and W).
To determine whether iSG induction was mediated by a pluripotent or neural progenitor intermediate, we investigated by qRT-PCR assay expression of pluripotent factor genes and neural progenitor marker genes during HSF reprogramming by ABI. We found no significant change in expression levels of pluripotent factor genes OCT4, KLF4, and NANOG during the reprogramming process (from day 1 to day 20) (Fig. 7P). Similarly, there was no induction of NESTIN and OLIG2 expression in the reprogramming process (Fig. 7Q), suggesting that iSGs were reprogrammed from HSFs by ABI without an intermediate state of pluripotent or neural progenitors. Consistent with this, by day 30 of reprogramming, almost no reprogrammed TUJ1+ neurons were labeled by EdU when EdU was added to the reprogramming cell culture for 29 days or 24 hours (Fig. 7, R to X), confirming that iSG reprogramming occurred in the absence of an intermediate state of proliferative progenitors.
The electrophysiological properties of reprogrammed human iSG neurons were evaluated by whole-cell patch-clamp recording. At day 60, most neurons (15 of 17) exhibited typical sodium and potassium currents and showed action potential responses (Fig. 7, Y, Z, and A′). In addition, the inward sodium current could be specifically and completely blocked by TTX and partially recovered by its removal (Fig. 7A′). Similar to mouse iSG neurons, some (4 of 17) were multi-spiking, while the others (11 of 17) were single-spiking (Fig. 7, Y, B′, and C′), although in human iSG single-spiking neurons appeared to be more abundant than those in mouse iSG (Fig. 3K). Among all neurons recorded from day 25 to day 39, a small fraction (4 of 44) displayed spontaneous postsynaptic activities (Fig. 7D′), indicating the ability for human iSG neurons to form functional synapses, in agreement with their synapsin labeling (Fig. 7F). Thus, the human iSG neurons induced by ABI from HSFs have the physiological properties characteristic of mature neurons.
We further investigated the ability of human iSG neurons to survive and integrate in the DRG by microinjecting GFP-tagged human iSG neurons into adult rat DRG explants (fig. S4A). Two weeks after transplantation, we found that the GFP+ neurons survived and integrated in the DRG, and were all (99 of 99) immunolabeled by an anti-human nuclei antibody (fig. S4F), indicating that material transfer did not occur between the transplanted and host cells. The transplanted GFP+ cells were immunoreactive for pan-sensory neuron markers, and some of them were immunoreactive for TrkA, TrkB, or TrkC (fig. S4, G to I), suggesting that similar to mouse iSG neurons, transplanted human iSG neurons can also survive in the DRG and maintain sensory neuron subtypes.
DISCUSSION
Formation of self-organized iSG organoids by ABI induction of fibroblasts
Although scattered sensory neurons (iSNs) were previously induced from fibroblasts by TFs (20, 21), to our knowledge, this is the first time to demonstrate that self-organized iSG organoids can be consistently induced directly from somatic cells by defined TFs. The bHLH TF Ascl1 has been shown to be a pioneer neurogenic TF in converting fibroblasts into neurons in in vitro somatic cell reprogramming (26). However, the neurons reprogrammed by Ascl1 alone are mostly slow-maturing and excitatory (36). Addition of Brn2 and Myt1l (BAM) improved the reprogramming efficiency, maturing speed, and varieties of the iNs (27, 36, 37). The iNs induced by BAM were rather generic but motor neurons could be specifically induced when BAM were combined with four other TFs (Lhx3, Hb9, Isl1, and Ngn2) (38). Similarly, when trying BAM with other combinations, Wainger et al. (20) found that the combination of five factors (Ascl1, Myt1l, Ngn1, Isl2, and Klf7) could successfully convert fibroblasts into nociceptor neurons. Notably, all of these reprogramming formulas include Ascl1 as a key component. Alternatively, the bHLH TFs Ngn1 and Ngn2 were combined with Brn3a to reprogram fibroblasts into mature iSNs (21).
Our experiments in this study have demonstrated that the ABI TF combination is most effective in inducing MEFs into self-organized mini-SG, while the AI and AB combinations have a weaker activity (fig. S8J). Thus, Brn3a/3b appears to act synergistically with Isl1 to improve the induction efficiency of iSG organoids. As revealed by time-lapse microscopy, the larger iSG organoids are formed by cell migration and coalescing smaller cell aggregates. The mini-SG induced from both murine and human fibroblasts contain mature and functional sensory neurons. They exhibit typical inward sodium currents, which can be blocked by TTX and recover after TTX removal, and are a mixture of neurons displaying multiple-spiking action potentials or single-spiking action potential. They also show calcium responses to potassium chloride, capsaicin, and menthol. All these features closely resemble their endogenous counterparts.
The iSG neurons reprogrammed by ABI display extensive cell diversities in their expression of characteristic receptors, ligands, ion channels, neuropeptides, neurotransmitters, and so on, similar to the endogenous sensory neurons. In agreement with iSNs induced by Ngn1/2 and Brn3a (21), the iSGs contain roughly equivalent percentages (~30%) of TrkA+, TrkB+, and TrkC+ neurons, supporting the notion that Trk receptors may arise in a stochastic manner such that each donor cell has an approximately equivalent chance to express one of the Trk receptor genes. By bulk RNA-seq, scRNA-seq, and/or qRT-PCR analyses, we investigated the characteristic markers involved in sensory signaling pathways including transduction, conduction, and synaptic transmission of sensory signals. At the transduction level, we found up-regulated expression of genes responsible for perceptions to stimuli such as heat (Trpv1, Trpv2, Trpv3), cold (Trpa1), damage (P2X3, Bdkrb1), and touch (Trpc1, Trpc4, Asic2/Accn1, Accn2). Trpv1, also known as capsaicin receptor that is expressed mainly in the nociceptive neurons (29), has been shown to be present and functional in iSG neurons by capsaicin stimulation. The signaling conduction of sensory neurons is primarily mediated by sodium channels, which propagate the signals, and potassium channels, which usually act to reduce excitability. We found that the expression of many Na+ channels (Scn1a, 2a1, 2b, 3a, 3b, 7a, 11a) and K+ channels (Kcnq2, 4; Kcna2, 3, 4, 5, 6; Kcnb2, c1, d2, e4, f1, h2, j2, k3, s3, t1, etc.) were up-regulated in iSG neurons. For synaptic transmission, neurotransmitter receptors and presynaptic voltage-gated Ca2+ channels are two groups of important regulatory molecules. Correspondingly, the expression of a variety of neurotransmitter receptors (Nk1r, Nr3c2; Gria1, 2, 4; Grid1, k1, k2, k4, k5; Grin1, 2a, etc.) and Ca2+ channels (Cacna1a, 1b, 1d, 2d1, 2d2, 2d3; Cacnb1, g4, etc.) were significantly up-regulated in iSG neurons.
Apart from the molecular and electrophysiological properties, ABI-reprogrammed iSG neurons also have salient cellular and innervation characteristics of sensory neurons. For instance, when transplanted, they can survive, integrate, and maintain the nociceptive, mechanoreceptive, and proprioceptive subtypes in the DRG. Moreover, the iSG neurons exhibit strong affinity for endogenous DRG neurons and spontaneously aggregate with them to form interconnected DRG-like organoids in culture. In addition, we have demonstrated by coculture that the iSG neurons have the capacity to innervate the peripheral targets of sensory neurons, i.e., epidermal cells, indicating that the iSGs contain bona fide sensory neurons reprogrammed from fibroblasts by ABI.
Therefore, the combination of ABI TFs is able to reprogram murine and human fibroblasts into self-organized iSG organoids composed of heterogeneous sensory neurons, closely resembling the endogenous SG. Previously, Ascl1 in combination with Brn3a, Brn3b, or Brn3c was shown to induce iNs from MEFs (39). Although the sensory neuron identity of the iNs was not investigated, some of the data suggest the formation of iSG organoids by the Ascl1 and Brn3a combination (39). This is consistent with our work that showed that the AB combination enabled induction of iSG organoids, albeit fewer than those induced by the ABI combination (Fig. 1). Similarly, the data reported in a previous study also suggest the formation of iSG organoids by the nociceptive neurons reprogrammed from MEFs using a 5-TF combination (20). However, unlike the ABI combination, the 5-TF combination did not appear to induce iSG organoids from human fibroblasts (20), suggesting a difference in reprogramming capacity and/or efficiency by different combinations of TFs.
iSG organoids as a tool to study and treat sensorineural diseases
The peripheral ganglia, including cranial ganglia, DRG, trigeminal ganglia, enteric system ganglia, autonomic ganglia, and others, are derived from migrating NC cells. The NC is thought to be a unique cell population found in vertebrates and is initially induced at the neural plate border as a result of neural plate folding and fusion (40). After undergoing an epithelial-to-mesenchymal transition, the NC cells delaminate from the neuroepithelium and become highly migratory. Most NC cells migrate as a chain or group in a so-called collective cell migration, in which cell contact and cooperation allow them to migrate directionally. Guided by local cues and long-range chemoattractants, NC cells reach their destination and differentiate into ganglia and other tissue types.
Mutations in crucial genes controlling the migration and differentiation of NC cells may cause aganglionosis such as Hirschsprung’s disease, which may occur by itself or in association with other genetic disorders such as Down syndrome, Waardenburg-Shah syndrome, Mowat-Wilson syndrome, or Bardet-Biedl syndrome (41). This group of genes includes RET, ZEB2, EDNRB, SOX10, and PHOX2B, and mutations of them or their regulatory sequences may increase the risk of Hirschsprung’s disease more than 1000-fold (41). In our RNA-seq data, the expression of Ret, Zeb2, Ednrb, and Sox10 was significantly up-regulated in the iSG neurons, in agreement with their importance in the differentiation and formation of SG. Other known risk genes—Bbs4, Bbs10, Edn3, Gfra1, and Arvcf (41)—were also significantly elevated in iSG. The hereditary sensory and autonomic neuropathies (HSANs) consist of several clinically heterogeneous disorders characterized by defective development and maintenance, and progressive degeneration of sensory and autonomic nervous systems. Mutations in the SPTLC1, WNK1, IKBKAP, and TRKA genes have been shown to cause HSAN types I to IV, respectively (42). In addition, loss-of-function mutations in SCN9A and PRDM12 result in congenital insensitivity to pain (6, 43). Indifference to pain appears to be desirable but risks the loss of a vital protective mechanism with dangerous consequences such as unknowingly chewing tongues and lips and damaging digits and joints. On the other hand, pain hypersensitivity reduces the quality of life and may increase susceptibility to chronic pain.
The ability to reprogram somatic cells into iSG organoids by ABI presents new possibilities for modeling sensorineural diseases, studying their pathogenesis, screening for counteractive drugs, and developing cell replacement therapies. For example, patient-derived iSG organoids may be used as an in vitro model for pain to screen and evaluate potential drug treatments. In the future, iSG organoids and neurons may also be used in transplantation as a cell replacement therapy for damaged or degenerated SG. In this respect, we found that transplanted iSG neurons were able to integrate and maintain the nociceptive, mechanoreceptive, and proprioceptive subtypes in the DRG. It has long been recognized that genetic factors are a major contributor to personalized pain perception and the efficacy of analgesic drugs (29). Generation of iSG organoids from autologous somatic cells may thus provide an exciting novel approach to model personalized pain and sensory pathology and help to achieve precision medicine for pain.
Definitive molecular markers to identify RGCs, especially iRGCs in vitro
In this study, we made efforts to define specific molecular markers to identify RGCs both in vitro and in vivo. This is important because it is impossible to apply commonly used RGC markers to distinguish RGCs from SG neurons in vitro given the high molecular similarity between these two cell types. Since the 1990s, we have established the Brn3 family of TFs, Brn3a, Brn3b, and Brn3c, as the gold standards to identify RGCs in the retina (10, 15). However, Brn3 proteins are not unique to the retina but expressed in other sensory and CNS tissues as well, e.g., trigeminal ganglia, DRG, spiral ganglia, and midbrain (10, 15, 44). Apart from Brn3 proteins, Thy1.2, Sncg, and Rbpms are also commonly used as specific RGC markers. But here again, we show their abundant expression in DRG neurons. Therefore, although because of the spatial separation of the retina from SG in the organism, these so called RGC-specific markers are able to distinguish RGCs from SG neurons in vivo, they are unable to do so in vitro. Unfortunately, however, a number of previous studies used these supposedly RGC-specific markers to identify RGCs induced from ESCs, iPSCs, and somatic cells in vitro (22–25), casting doubt on some of the arrived conclusions.
To avoid misidentifying iRGCs and iSG neurons in vitro, we screened for molecular markers that can definitively distinguish RGCs from SG neurons. A rigorous criterion was set that these unique identifiers should be single-molecule markers or a combination of multiple-molecule markers that must be present only in RGCs within the retina but not in SGs or any other tissues. Following a careful examination of a large number of known RGC and SG neuron markers, it became apparent that none of them alone were specific to RGCs. Further double-immunolabeling analysis indicated that a combination of Pax6 and Brn3a or Brn3b double markers satisfied the criterion of specific RGC identifiers. Brn3a+Pax6+ and Brn3b+Pax6+ double-positive cells were found exclusively in RGCs of the retina but not in the DRG, spinal cord, midbrain, or cerebellum, where Brn3a, Brn3b, or Pax6 is normally expressed. Moreover, scRNA-seq analysis confirmed Brn3a+Pax6+ and Brn3b+Pax6+ cells as RGCs and their complete absence in the DRG. Thus, we are able to define the combination of Pax6 with either Brn3a or Brn3b double protein markers as specific identifiers for RGCs. Armed with this knowledge, we found that ABI TFs had the capacity to reprogram MEFs into a small number of Brn3a+Pax6+ iRGCs, representing about 13% of all Brn3a+ neurons. Unlike iSG organoids resembling endogenous SG, iRGCs did not coalesce into clusters but remained scattered, similar to the dispersive distribution pattern of endogenous RGCs in the retina (fig. S8, I and J). Therefore, ABI-induced iSG and iRGCs maintain the morphology characteristic of their endogenous equivalents.
In summary, in a screen of multiple SG and RGC TFs, we have identified a triple-factor combination ABI as the most efficient combination to reprogram self-organized and networked iSG organoids from mouse and human fibroblasts. By immunostaining, qRT-PCR, whole-cell patch-clamp recording, calcium imaging, and bulk and scRNA-seq approaches, we are able to demonstrate that the iSG organoids display molecular and cellular features, subtype diversity, electrophysiological properties, and peripheral innervation patterns characteristic of peripheral SGs. Furthermore, using immunolabeling and scRNA-seq analyses, we have identified bona fide RGC-specific molecular markers to demonstrate that the ABI combination has the additional capacity to induce from fibroblasts a small number of iRGCs. Unlike the ABI-reprogrammed iSG organoids characteristic of endogenous SG, iRGCs maintain a dispersive distribution pattern resembling that of endogenous RGCs in the retina. The iSG organoids and iRGCs may be used to model sensorineural/retinal diseases, to screen for effective drugs and potentially, as cell-based replacement therapy.
MATERIALS AND METHODS
Animals
All experiments on rodents were performed according to the IACUC (Institutional Animal Care and Use Committee) standards and approved by Sun Yat-sen University and Zhongshan Ophthalmic Center. The C57BL/6 mice were purchased from the Vital River Laboratories (Beijing, China).
Construction of viral plasmids and preparation of lentiviruses
The full-length ORFs of Brn3a, Brn3b, Isl1, Math5, Ebf1, Pax6, Tfap2a, Nr4a2, Nrl, Crx, Ptf1a, Neurod1, Lhx2, Ngn1, Ngn2, Chx10, Sox2, Rx, Meis1, Foxn4, Otx2, Sox9, or Six3 were subcloned into the Eco RI site of the FUW-TetO vector (45). In addition, by overlapping PCR subcloning, Ascl1, Isl1, and Brn3b were tethered by P2A and T2A self-cleaving peptide sequences into a single ORF, which was inserted into the same FUW-TetO backbone. Lentiviruses were prepared as previously described (34).
Preparation of fibroblasts and epidermal cells
The MEFs were prepared as previously described (34). For isolation of mouse epidermal cells, P0 C57BL/6 mice were anesthetized with ice for 5 min and the brain was removed using a sterilizing razor in a 10-cm culture dish containing Hanks’ balanced salt solution (HBSS) (Gibco). The epidermis was isolated from the remaining tissue using a pair of fine-tip forceps under a dissection microscope, transferred into a fresh 6-cm culture plate containing 1 ml of 0.25% trypsin, thoroughly minced using a pair of surgical scissors and forceps, and incubated for 15 min at 37°C in a CO2 incubator. Six-milliliter MEF medium containing Dulbecco’s modified Eagle’s medium (DMEM)/High Glucose (HyClone) supplemented with 10% fetal bovine serum (Gibco), 1× penicillin/streptomycin (Gibco), 1× MEM nonessential amino acids (NEAA) (Gibco), and 0.008% (v/v) 2-mercaptoethanol (Sigma-Aldrich) was added into the plate to terminate the reaction. After being mixed using a 10-ml pipette, the digested tissue was transferred to a 15-ml fresh tube, centrifuged at 1000 rpm for 5 min, and resuspended in 5-ml fresh MEF medium. The isolated epidermal cells were expanded by culture in the MEF medium at 37°C in a CO2 incubator. The HSFs were purchased from the American Type Culture Collection (CRL1502, 12-week gestation). MEFs, mouse epidermal cells, and HSFs were all maintained and expanded in the MEF medium.
Fibroblast reprogramming
To induce iSG and iRGCs from MEFs, 3 × 104 MEF cells (at passage 3) were cultured in 500-μl MEF medium in a well of a 24-well plate containing a glass coverslip precoated with Matrigel (Corning). They were infected the next day with 500-μl mixture of lentiviruses and fresh MEF medium in the presence of polybrene (10 μg/ml). After 16-hour infection, the virus and medium mixture was removed. The cells were induced for 4 days in the neuron basic medium [(DMEM/F12 (1:1) (Life Technologies) supplemented with 1× B27 (Gibco) and basic fibroblast growth factor (bFGF) (10 ng/ml) (R&D Systems)] in the presence of Dox (2 ng/ml) (Sigma-Aldrich) and then for another 4 days in the neuron maintenance medium containing the neuron basic medium supplemented with insulin-like growth factor 1 (IGF-1) (100 ng/ml), brain-derived neurotrophic factor (BDNF) (10 ng/ml), and glial cell line–derived neurotrophic factor (GDNF) (10 ng/ml) in the presence of Dox (2 μg/ml). The medium was replaced with the neuron maintenance medium without Dox following the 8-day induction period. By 14 days after infection with Ascl1, Brn3b/3a, and Isl1 (ABI), Ascl1 and Brn3b/3a (AB), or Ascl1 and Isl1 (AI) lentiviruses, many visible neuronal clusters were formed.
With modifications, the HSFs were similarly induced. In brief, after virus infection, the human cells were cultured in the neuron basic medium with Dox for 10 days and then in the neuron maintenance medium without Dox for another 10 days. On day 21, the medium was replaced with the neuron mature medium, which is the maintenance medium supplemented with NGF (20 ng/ml), NT-3 (20 ng/ml), and 10 μM forskolin. Thirty days after viral infection, many neuronal clusters were visible, which were usually smaller than those induced from MEFs. To improve the induction efficiency of the HSFs, we created a Dox-inducible lentiviral construct containing Ascl1, Isl1, and Brn3b in a single ORF as described above.
qRT-PCR, immunohistochemistry, and immunocytochemistry
RNA extraction and qRT-PCR analysis were carried out as previously described (34). The qRT-PCR primers used are shown in table S2.
Immunostaining of tissue sections and cells was carried out as previously described (34, 46). The following antibodies (with dilution information) were used: mouse anti-Brn3a (Santa Cruz Biotechnology, sc-390780; 1:1000), mouse anti-Brn3a (Santa Cruz Biotechnology, sc-8429; 1:100), goat anti-Brn3b (Santa Cruz Biotechnology, sc-6026; 1:1000), rat anti-Thy1.2 (BD Biosciences, 550543), goat anti–RPF-1 (Santa Cruz Biotechnology, sc-104627; 1:100), rabbit anti-Rbpms (PhosphoSolutions, 1830-RBPMS; 1:500), mouse anti-HuC&D (Life Technologies, A-21271; 1:500), rabbit anti-Pax6 (BioLegend, 901301; 1:2000), mouse anti-Pax6 (Developmental Studies Hybridoma Bank, Pax6; 1:1000), rabbit anti-Six6 (Sigma-Aldrich, HPA001403; 1:500), rabbit anti-Ebf (Santa Cruz Biotechnology, sc-33552; 1:1000), mouse anti-Isl1 (Abcam, ab20670; 1:2000), rabbit anti-Zeb2 (Santa Cruz Biotechnology, sc-48789; 1:1000), rat anti-Lmo4 (1:1000; (47), rabbit anti-Ldb1 (Abcam, ab96799; 1:1000), rabbit anti-Sncg (GeneTex, GTX110483; 1:200), rabbit anti-CGRP (Neuromics, RA24112; 1:200), rabbit anti-peripherin (Millipore, ab1530; 1:1000), rabbit anti-vGLUT1 (Synaptic System,135303; 1:500), mouse anti-vGLUT2 (Abcam, ab79157; 1:500), mouse anti-vGLUT3 (Sigma-Aldrich, SAB5200312; 1:500), rabbit anti-GABA (Sigma-Aldrich, A-2052; 1:1000), goat anti-TrkA (Abcam, ab76291; 1:500), rabbit anti-TrkA (Abcam, ab76291; 1:500), goat anti-TrkB (R&D Systems, AF1494; 1:500), goat anti-TrkC (R&D Systems, AF1404; 1:500), rabbit anti-P2X3 (Millipore, AB5895; 1:100), mouse anti-Tuj1 (Millipore, MAB5564; 1:500), rabbit anti-Tuj1 (Abcam, ab18207; 1:2000), mouse anti-Map2 (Sigma-Aldrich, M1406; 1:2000), rabbit anti-synapsin (Calbiochem, 574778; 1:500), goat anti-Dcx (Santa Cruz Biotechnology, sc-8066; 1:500), mouse anti-NF200 (Millipore, MAB5266; 1:500), rabbit anti-TH (Protos Biotech, CA-101bTHrab; 1:1000), rabbit anti-Vamp (Synaptic System, 104203; 1:500), rabbit anti-p75NTR (Abcam, ab8874; 1:500), mouse anti-c-Ret (Sigma-Aldrich, o4886; 1:1000), goat anti-GFP (Abcam, ab6673; 1:2000), rabbit anti-GFP (MBL, 598; 1:2000), chicken anti-GFP (Abcam, ab13970; 1:2000), rabbit anti-vimentin (Abcam, ab92547; 1:2000), and mouse anti-human nuclei (Millipore, MAB1281; 1:200). The secondary antibodies used included donkey anti-rabbit, donkey anti-goat, and donkey anti-mouse Alexa 488 immunoglobulin G (IgG), Alexa 594 IgG, Alexa 546 IgG, Alexa 647 IgG, or Alexa 594 IgM (1:1000; Invitrogen). 4′,6-Diamidino-2-phenylindole (DAPI) (Invitrogen) was used for nuclear counterstaining. Images were captured with a laser scanning confocal microscope (Carl Zeiss, LSM700).
EdU labeling
One day following infection with ABI lentiviruses, the MEFs were cultured in the presence of 10 μM EdU (Life Technologies) for 13 days, or 13 days after infection with AI or ABI viruses, the MEFs were cultured for 24 hours in the presence of 10 μM EdU. The cells were then fixed, and EdU staining was carried out according to the manufacturer’s instruction (Life Technologies). For HSF reprogramming by ABI, EdU was added to the reprogramming cell culture for 29 days starting from day 1 of reprogramming or for 24 hours starting at day 29. Images were captured with a confocal microscope.
Time-lapse recording
For time-lapse recording, we used the JuLI Stage (NanoEntek) with a motorized stage, computer-controlled lens change, and a built-in incubator that supplied humidified 5% CO2 at 37°C for live cell recording. MEFs (5 × 104) derived from the CAG-GFP transgenic mice (28) were induced for 10 days by infection with the ABI lentiviruses or Ascl1 lentiviruses in a well of a 12-well plate precoated with Matrigel. The plate was then placed into the incubator of the JuLI Stage for time-lapse recording for 50 hours. A series of pictures were taken from each well of the 12-well plate in a period of 50 hours under the control of the JuLI EDIT software, which can also edit and replay these pictures in a continuous mode like a movie.
Single iSG cell harvesting
To prepare single iSG cells, MEFs were infected with the ABI (Ascl1, Brn3b, and Isl1) lentiviruses and induced for 2 weeks. Following addition of 500-μl Accutase (Millipore) into a well of a 12-well plate, neuronal clusters were suspended by gently pipetting up and down several times using a 1-ml pipette and transferred into a 70-μm cell strainer (Falcon) to collect neuronal clusters. Most neuronal clusters attached to the Nylon membrane of the 70-μm cell strainer, which was cut from the cell strainer using a pair of scissors and placed into a low-adhesion 6-cm plate containing 4-ml neuron basic medium. To separate the neuronal clusters from the Nylon membrane, the plate was shaken left and right 10 times. The neuronal clusters were then transferred into a 15-ml tube, centrifuged at 1000 rpm for 5 min, resuspended with 1-ml Accutase, and incubated for 5 min at 37°C in a CO2 incubator. The neuronal clusters were dissociated into many single cells, which were subsequently used for injection of DRG explants, qRT-PCR, and scRNA-seq analysis.
DRG explant culture and transplantation
After euthanization of the rat by the asphyxiation method (CO2 inhalation), the vertebral columns were isolated from the rest of the tissue using a pair of sharp scissors and washed three times with HBSS in a 10-cm culture dish. Both sides of the vertebral columns were mounted onto a surgical mat using needles, and a double cut was made using a pair of surgical scissors to expose the ventral side of the spinal cord. After removal of the spinal cord, DRGs were exposed in the contralateral dorsal spinal roots and pulled out using a pair of fine tweezers. They were collected into a 6-cm culture dish containing HBSS after removal of the attached excessive fibers and connective tissues under a dissection microscope. Four DRGs were transferred onto a Millipore Millicell-CM Low Height Culture Plate Insert using a 3-ml Pasteur pipette, and the rest of HBSS was removed using a 200-μl pipette. Then, the insert was placed into a well of a six-well plate containing 1-ml DRG culture medium [BEM (Gibco) supplemented with 20 mM glucose, 1× KIT (Gibco), putrescine (16 ng/ml) (Sigma-Aldrich), 10 mM vitamin C (Sigma-Aldrich), NGF (20 ng/ml) (PeproTech), and 10 mM 5-fluoro-2-deoxyuridine (FDU) (Sigma-Aldrich)]. After being cultured for 1 day, each DRG was injected with 4 × 103 GFP-labeled single iSG cells. Two weeks following iSG cell injection, the explants were processed and immunostained as described above.
Electrophysiological analysis
Whole-cell patch-clamp recordings of the iNs were performed with the EPC 10 USB amplifier (HEKA Electronik, Lambrecht, Germany) as previously described (34). Neurons induced from MEFs for 9 or 14 days or from HSFs for 25 to 60 days were used for patch-clamp recordings. In brief, coverslips with adhered cells were transferred into a recording chamber and bathed with Ringer’s containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, and 20 mM glucose, bubbled with 95% O2 and 5% CO2. Cell responses were recorded with 6- to 9-megohm resistance pipettes that were filled with an internal solution containing 105 mM K-gluconate, 5 mM KCl, 5 mM NaOH, 15 mM KOH, 0.5 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 2 mM adenosine 5′-triphosphate, 0.5 mM guanosine 5′-triphosphate, 10 mM Hepes, and 2 mM ascorbate (pH 7.2). The cells and recording pipettes were viewed on a monitor that was coupled to a charge-coupled device camera (Evolve, Photometrics, Tucson, USA) mounted on an upright microscope. Oxygenated external solution was continuously perfused into the recording chamber at a flow rate of 1.5 to 2 ml/min by a peristaltic pump (LEAD-2, Longer Pump, Hebei, China). Capacitive transients were compensated via the Patch Master software (PatchMaster, HEKA), and the series resistance was compensated by ~50%. For current-clamp recording, a small, constant holding current was injected to maintain resting membrane potential (Vrest) at −70 mV and current pulses with a step size of 10 pA were applied to induce action potentials. Voltage-clamp recordings were performed on the same cells directly following current clamp recordings. A simple step protocol from −90 to +30 mV for 200 ms was applied to assess the voltage-gated sodium channels and voltage-gated potassium channels. TTX (Tocris, USA) was added to the bath solution to a final concentration of 0.5 μM and perfused into the recording chamber for 5 min. After recording of the currents again, TTX was washed out, followed by the third time of recording.
Calcium imaging
The fluorescent probe Fluo-8 AM (AAT Bioquest, Sunnyvale, Canada) was used to detect the changes of intracellular calcium. As described above, MEFs were induced for 2 to 3 weeks to form neuronal clusters by infection with the ABI lentiviruses. Under dark environment, glass coverslips with adhered neuronal clusters were loaded with Fluo-8 AM (10 μm) for 25 to 30 min at room temperature. After three rinses with Ringer’s solution, the coverslip was placed into a recording chamber. An upright microscope (Olympus, BX51W1) equipped with a mercury lamp with a 488-nm filter was used to excite Fluo-8. A digital camera (Hamamatsu Photonics, Japan) that was also equipped on the microscope was used to record the fluorescent signal. The software HCImage Live (Hamamatsu Corporation, USA) was used to control the camera and ImageJ for data analysis. Following a 30-s recording of the baseline (F0), 100 mM KCl was puffed to detect the activity of the cells. After a 2-min wash with Ringer’s, fluorescent signals were decreased to the baseline. Then, 100 μM menthol or 10 μM capsaicin was puffed to stimulate the iNs. KCl (100 mM KCl) was applied again after menthol/capsaicin to confirm the viability of the tested cells. Only the cells that responded to KCl two times successively were chosen for analysis.
RNA-seq analysis
Bulk RNA-seq analysis was performed with modification as previously described (48). Two weeks after infection of MEFs with lentiviruses, total RNA was extracted from GFP-transduced and ABI (Ascl1, Brn3b, and Isl1)–transduced MEFs using the TRIzol reagent according to the manufacturer’s instruction. Ribosomal RNA was depleted before preparation of RNA-seq libraries, which were subsequently sequenced using an Illumina HiSeq 4000 sequencer (Biomarker Technologies, China). The obtained sequence reads were trimmed and mapped to the mouse reference genome (mm10) using HISAT2 (https://daehwankimlab.github.io/hisat2/), and gene expression and changes were analyzed using Cufflinks and Cuffdiff. Hierarchical cluster and scatter plot analyses of gene expression levels were performed using the R software (http://cran.r-project.org). GSEA was carried out as described (31), which was followed by network visualization in Cytoscape using the EnrichmentMap plugin (https://enrichmentmap.readthedocs.io/en/latest/).
scRNA-seq analysis
Single iSG cells were prepared as described above. Single adult mouse DRG cells were prepared as described previously (49). In brief, DRGs were collected, transferred into a low-adhesion 6-cm pate with 2 ml of DMEM/F12 medium containing collagenase IV (1.25 mg/ml), and incubated at 37°C in a 5% CO2 incubator for 50 min. Then, the medium was replaced with 2-ml DMEM/F12 medium containing 0.025% trypsin and incubated for 30 min. Following the addition of 2-ml DMEM/F12 medium containing 33% fetal bovine serum, all the medium was removed using a 10-ml pipette. After being washed three times with 2-ml HBSS, the DRGs were transferred into a 1.5-ml tube containing 1.2-ml DMEM/F12 and triturated by pipetting up and down several times using a 1-ml pipette to obtain single DRG cells. A Brn3b-GFP reporter mouse line was created using the CRISPR-Cas9 gene editing system to label adult RGCs by GFP, which were enriched by fluorescence-activated cell sorting. A more detailed description of this mouse line and RGC enrichment procedure will be published elsewhere.
The number and viability of prepared single cells were quantified using Countess II (Thermo Fisher Scientific, AMQAX1000). Next, single-cell libraries were generated with the Chromium Single Cell 3’ V2 Chemistry Library Kit, Gel Bead & Multiplex Kit, and Chip Kit from 10x Genomics. In brief, cell suspension at concentration of 1.2 million/ml was loaded in a Single Cell 3′ Chip along with the RT Single Cell 3′ Gel Beads and the Partitioning oil, and Single Cell Gel Bead-In-Emulsions were generated in the Chromium Controller. Reverse transcription reaction was run to obtain complementary DNA (cDNA), which was amplified by PCR. To generate the libraries, Enzymatic Fragmentation, End Repair, and A-tailing Double Sided Size Selection were used to incorporate the barcodes and index read sequences. The libraries were qualified by bioanalyzer (Agilent Technologies) and quantified by a Qubit dsDNA High Sensitivity Assay kit (Invitrogen) and then sequenced on Illumina X Ten platform in 150 paired-end configuration.
Raw reads were processed using the 10x Genomics Cell Ranger pipeline (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest) with the mm10 as the reference. Cell Ranger can cluster the single cells, identify the marker genes of each cluster, and export a matrix with unique molecular identifier (UMI) values of each gene in a single cell. The R software package Seurat (https://satijalab.org/seurat, version 2.2) (33) was used for further analysis. Default parameters were used for most of the Seurat analyses. For the FeaturePlot function, max.cutoff was 0.5. The pseudotime trajectory analysis of iSG cells was performed using Monocle 2 (http://cole-trapnell-lab.github.io/monocle-release/) (35).
Statistics
Statistical analysis was performed using the GraphPad Prism 7 and Microsoft Excel computer programs. The results are expressed as means ± SD for experiments conducted at least in triplicates. Unpaired two-tailed Student’s t test or one-way analysis of variance test were used to assess differences between two groups, and a value of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank E. Shiang for help with the artwork. Funding: This work was supported, in part, by the National Natural Science Foundation of China (81670862, 81721003, 31871497, 81870682, and 31700900), National Key R&D Program of China (2017YFA0104100, 2018YFA0108300, and 2017YFC1001300), National Basic Research Program (973 Program) of China (2015CB964600), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program, Science and Technology Planning Projects of Guangzhou City (201904020036 and 201904010358), China Postdoctoral Science Foundation (2019 M650223), and the Fundamental Research Funds of the State Key Laboratory of Ophthalmology, Sun Yat-sen University. Author contributions: D.X., K.J., Y.S., and M.X. conceived and designed the research. D.X., Q.D., Y.G., X.H., M.Z., J.Z., P.R., Z.X., Y.L., and Y.H. performed the experiments and analyzed the data. D.X., K.J., and M.X. interpreted the data and wrote the manuscript. All authors contributed to critical reading of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The RNA-seq and scRNA-seq data have been deposited in the NCBI Gene Expression Omnibus database under accession codes PRJNA595403 and PRJNA597624, respectively.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz5858/DC1
REFERENCES AND NOTES
- 1.Marmigere F., Ernfors P., Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 8, 114–127 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Lallemend F., Ernfors P., Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 35, 373–381 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Baden T., Berens P., Franke K., Román Rosón M., Bethge M., Euler T., The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaugenhaupt S. A., Blumenfeld A., Gill S. P., Leyne M., Mull J., Cuajungco M. P., Liebert C. B., Chadwick B., Idelson M., Reznik L., Robbins C. M., Makalowska I., Brownstein M. J., Krappmann D., Scheidereit C., Maayan C., Axelrod F. B., Gusella J. F., Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68, 598–605 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J.-L., Cocozza S., Koenig M., Pandolfo M., Friedreich's ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Dib-Hajj S. D., Cummins T. R., Black J. A., Waxman S. G., From genes to pain: Nav 1.7 and human pain disorders. Trends Neurosci. 30, 555–563 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Orefice L. L., Mosko J. R., Morency D. T., Wells M. F., Tasnim A., Mozeika S. M., Ye M., Chirila A. M., Emanuel A. J., Rankin G., Fame R. M., Lehtinen M. K., Feng G., Ginty D. D., Targeting peripheral somatosensory neurons to improve tactile-related phenotypes in ASD models. Cell 178, 867–886.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quigley H. A., Glaucoma. Lancet 377, 1367–1377 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Xiang M., Zhou H., Nathans J., Molecular biology of retinal ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 93, 596–601 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang M., Zhou L., Macke J. P., Yoshioka T., Hendry S. H., Eddy R. L., Shows T. B., Nathans J., The Brn-3 family of POU-domain factors: Primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 15, 4762–4785 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Q., Fode C., Guillemot F., Anderson D. J., Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 13, 1717–1728 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y., Dykes I. M., Liang X., Eng S. R., Evans S. M., Turner E. E., A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 11, 1283–1293 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang M., Gan L., Zhou L., Klein W. H., Nathans J., Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc. Natl. Acad. Sci. U.S.A. 93, 11950–11955 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan L., Xiang M., Zhou L., Wagner D. S., Klein W. H., Nathans J., POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 93, 3920–3925 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang M., Zhou L., Peng Y.-W., Eddy R. L., Shows T. B., Nathans J., Brn-3b: A POU domain gene expressed in a subset of retinal ganglion cells. Neuron 11, 689–701 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Mu X., Fu X., Beremand P. D., Thomas T. L., Klein W. H., Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. U.S.A. 105, 6942–6947 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badea T. C., Cahill H., Ecker J., Hattar S., Nathans J., Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61, 852–864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan L., Yang Z., Feng L., Gan L., Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development 132, 703–712 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Liu W., Khare S. L., Liang X., Peters M. A., Liu X., Cepko C. L., Xiang M., All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development 127, 3237–3247 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Wainger B. J., Buttermore E. D., Oliveira J. T., Mellin C., Lee S., Saber W. A., Wang A. J., Ichida J. K., Chiu I. M., Barrett L., Huebner E. A., Bilgin C., Tsujimoto N., Brenneis C., Kapur K., Rubin L. L., Eggan K., Woolf C. J., Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 18, 17–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard J. W., Eade K. T., Szűcs A., Lo Sardo V., Tsunemoto R. K., Williams D., Sanna P. P., Baldwin K. K., Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 18, 25–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng F., Wang X., Gu P., Wang Z., Guo W., Induction of retinal ganglion-like cells from fibroblasts by adenoviral gene delivery. Neuroscience 250, 381–393 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Langer K. B., Ohlemacher S. K., Phillips M. J., Fligor C. M., Jiang P., Gamm D. M., Meyer J. S., Retinal ganglion cell diversity and subtype specification from human pluripotent stem cells. Stem Cell Rep. 10, 1282–1293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teotia P., Chopra D. A., Dravid S. M., Van Hook M. J., Qiu F., Morrison J., Rizzino A., Ahmad I., Generation of functional human retinal ganglion cells with target specificity from pluripotent stem cells by chemically defined recapitulation of developmental mechanism. Stem Cells 35, 572–585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T., Yokoi T., Tamalu F., Watanabe S.-I., Nishina S., Azuma N., Generation of retinal ganglion cells with functional axons from human induced pluripotent stem cells. Sci. Rep. 5, 8344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F., Drechsel D., Martynoga B., Castro D. S., Webb A. E., Südhof T. C., Brunet A., Guillemot F., Chang H. Y., Wernig M., Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Sudhof T. C., Wernig M., Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y., 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Foulkes T., Wood J. N., Pain genes. PLOS Genet. 4, e1000086 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker K. L., Meyer M., Barde Y.-A., Neurotrophins are required for nerve growth during development. Nat. Neurosci. 4, 29–37 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., McFarland C., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao D., Liu X., Zhang M., Zou M., Deng Q., Sun D., Bian X., Cai Y., Guo Y., Liu S., Li S., Shiang E., Zhong H., Cheng L., Xu H., Jin K., Xiang M., Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 9, 2865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A., Trapnell C., Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chanda S., Ang C. E., Davila J., Pak C., Mall M., Lee Q. Y., Ahlenius H., Jung S. W., Südhof T. C., Wernig M., Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., Wernig M., Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., Eggan K., Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsunemoto R., Lee S., Szűcs A., Chubukov P., Sokolova I., Blanchard J. W., Eade K. T., Bruggemann J., Wu C., Torkamani A., Sanna P. P., Baldwin K. K., Diverse reprogramming codes for neuronal identity. Nature 557, 375–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shellard A., Mayor R., Chemotaxis during neural crest migration. Semin. Cell Dev. Biol. 55, 111–118 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Heuckeroth R. O., Hirschsprung disease—Integrating basic science and clinical medicine to improve outcomes. Nat. Rev. Gastroenterol. Hepatol. 15, 152–167 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Rotthier A., Baets J., Timmerman V., Janssens K., Mechanisms of disease in hereditary sensory and autonomic neuropathies. Nat. Rev. Neurol. 8, 73–85 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Chen Y.-C., Auer-Grumbach M., Matsukawa S., Zitzelsberger M., Themistocleous A. C., Strom T. M., Samara C., Moore A. W., Cho L. T.-Y., Young G. T., Weiss C., Schabhüttl M., Stucka R., Schmid A. B., Parman Y., Graul-Neumann L., Heinritz W., Passarge E., Watson R. M., Hertz J. M., Moog U., Baumgartner M., Valente E. M., Pereira D., Restrepo C. M., Katona I., Dusl M., Stendel C., Wieland T., Stafford F., Reimann F., von Au K., Finke C., Willems P. J., Nahorski M. S., Shaikh S. S., Carvalho O. P., Nicholas A. K., Karbani G., McAleer M. A., Cilio M. R., McHugh J. C., Murphy S. M., Irvine A. D., Jensen U. B., Windhager R., Weis J., Bergmann C., Rautenstrauss B., Baets J., De Jonghe P., Reilly M. M., Kropatsch R., Kurth I., Chrast R., Michiue T., Bennett D. L. H., Woods C. G., Senderek J., Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 47, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang E. J., Liu W., Fritzsch B., Bianchi L. M., Reichardt L. F., Xiang M., Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development 128, 2421–2432 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D., Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Li S., Mo Z., Yang X., Price S. M., Shen M. M., Xiang M., Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43, 795–807 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Sum E. Y., Peng B., Yu X., Chen J., Byrne J., Lindeman G. J., Visvader J. E., The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem. 277, 7849–7856 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Jin K., Jiang H., Xiao D., Zou M., Zhu J., Xiang M., Tfap2a and 2b act downstream of Ptf1a to promote amacrine cell differentiation during retinogenesis. Mol. Brain 8, 28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fornaro M., Sharthiya H., Tiwari V., Adult mouse DRG explant and dissociated cell models to investigate neuroplasticity and responses to environmental insults including viral infection. J. Vis. Exp. 9, 56757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz5858/DC1


