RNA binding protein PCBP1 guards effector and antitumor T cell functions.
Abstract
Distinct lineages of T cells can act in response to various environmental cues to either drive or restrict immune-mediated pathology. Here, we identify the RNA binding protein, poly(C)-binding protein 1 (PCBP1) as an intracellular immune checkpoint that is up-regulated in activated T cells to prevent conversion of effector T (Teff) cells into regulatory T (Treg) cells, by restricting the expression of Teff cell–intrinsic Treg commitment programs. This was critical for stabilizing Teff cell functions and subverting immune-suppressive signals. T cell–specific deletion of Pcbp1 favored Treg cell differentiation, enlisted multiple inhibitory immune checkpoint molecules including PD-1, TIGIT, and VISTA on tumor-infiltrating lymphocytes, and blunted antitumor immunity. Our results demonstrate a critical role for PCBP1 as an intracellular immune checkpoint for maintaining Teff cell functions in cancer immunity.
INTRODUCTION
Immune checkpoint blockade (ICB) with antibodies targeting coinhibitory molecules programmed cell death-1 (PD-1) and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) have demonstrated clinical benefits in malignances such as melanoma, non–small cell lung, and cancer head and neck cancer, ultimately changing the practice of medical oncology (1–4). ICB has particularly been successful in melanoma, where up to 40% of patients show evidence of continued durable disease control. Recent evidence, which suggests that blocking one inhibitory receptor allows another to dominate and subvert antitumor immunity, provides further impetus for combination therapy or dual blockade of immune checkpoint to improve response and survival (1, 5). However, there are considerable toxicities associated with this approach, and therefore, optimizing treatments to define responders versus nonresponders is pertinent. Moreover, identifying additional molecular predictors of response to ICB would support more robust predictions and improve treatment outcomes in patients (6, 7).
RNA binding proteins (RBPs) are important posttranscriptional regulators that control RNA biology (splicing, stability, etc.) and translation. RBPs also enable T cells to maintain a poised transcriptome that can be rapidly translated in response to T cell receptor (TCR) stimulation. Thus, RBPs are quickly emerging as critical regulators of gene expression in response to T cell activation and differentiation (8–10). One such RBP is the poly(C)-binding proteins (PCBPs), which are multifunctional adaptor proteins implicated in the regulation of numerous biological processes. Most cell types express PCBP1, a member of a family of four homologous proteins that includes PCBP2, PCBP3, and PCBP4 (11). Besides RNA binding, PCBP1 also functions as an iron-binding protein that delivers iron to ferritin and other iron-dependent proteins (12, 13). Moreover, PCBP1 exhibits DNA and RNA binding properties, as well as controlling protein output by altering the fate of their binding targets (14, 15). Notably, the RNA binding function of PCBP1 is regulated by the phosphorylation state of the protein (16, 17). That is, unphosphorylated PCBP1 is able to bind to selective mRNAs and restrict their translation, thereby repressing its targets. However, in a chronic immune-suppressive setting, such as cancer, PCBP1 is phosphorylated at Ser43 by transforming growth factor–β (TGF-β) via the protein kinase, Bβ/AKT2, causing release of PCBP1 from the target transcript and subsequent mRNA translation, thus derepression of targets (18–20). Notably, the liberated mRNAs subsequently regulate downstream transcription. One such target, moesin, is repressed by PCBP1 in activated T cells but is posttranscriptionally induced in regulatory T cells (Tregs) via TGF-β–mediated release from PCBP1 repression (21). In turn, moesin promotes optimal TGF-β signaling by stabilizing the TGF-β receptors, which consequently augments Treg differentiation (21). Although PCBP1 was recently reported to promote granulocyte-macrophage colony-stimulating factor (GM-CSF) production by T cells (22), the physiological roles of PCBP1 in the development and function of CD4+ and CD8+ T cells, as well as Treg differentiation, are unknown. Because of its sensitivity to TGF-β signaling, inhibition of PCBP1 function may be a critical strategy for tumors to subvert antitumor immunity.
Here, we found that PCBP1 is up-regulated in activated T cells. PCBP1 was critical for stabilizing effector T cell functions by preventing their shift into Tregs and subverting immune-suppressive signals. Specifically, PCBP1 constrained expression of effector T cell–intrinsic Treg commitment programs to safeguard the immune response and antitumor functions. Consequently, T cell–specific deletion of PCBP1 resulted in a significantly greater proportion of forkhead box protein P3–positive (FoxP3+) Tregs that alleviated acute graft-versus-host disease (aGVHD) but blunted antitumor responses in mice. Whole-transcriptome analysis revealed that PCBP1 has a global function in restricting the expression of genes that promote immune tolerance.
RESULTS
PCBP1 is up-regulated in activated T cells
To gain insight into the role of PCBP1 in T cells, we first analyzed the expression of mRNA encoding members of the PCBP family in human CD3+ T cells before and after activation (23). We found that PCBP1 mRNA was abundant in both resting T cells and in activated T cells (Fig. 1A). We next assessed PCBP1 protein expression in naïve/resting (T0 and Tc-0) or anti-CD3/anti-ICOS paramagnetic activated (Th0 and Tc-1) human T cells from healthy individuals. We detected lower PCBP1 expression in naïve CD4+ (Fig. 1B) and CD8+ (Fig. 1C) T cells that was robustly up-regulated upon bead activation. Moesin, which is repressed by PCBP1 (21), was also elevated in resting T cells but was rapidly down-regulated in activated human T cells (Fig. 1, B and C).
Fig. 1. Activated T cells up-regulate PCBP1.
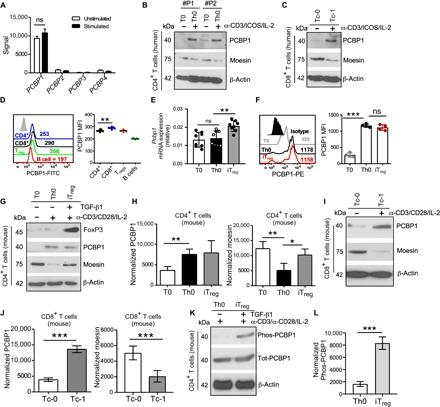
(A) Expression of mRNA encoding PCBP1-4 in human CD3+ T cells before (Unstimulated) and after (Stimulated) stimulation for 24 hours with anti-CD3 and anti-CD28 (obtained from Gene Expression Omnibus accession code GSE13887). n = 4 biologically independent samples. (B and C) Immunoblotting for total PCBP1 and moesin expression in human CD4+ (B) and CD8+ (C) T cells left unstimulated (T0 and Tc-0) or stimulated (Th0 and Tc-1) with antibodies against CD3 and ICOS with IL-2 for 4 days. β-Actin was used as loading control. (D) Flow cytometry (left) and quantification (right) of PCBP1 expression in subsets of splenic lymphocytes from mice. FITC, fluorescein isothiocyanate. (E and F) Relative Pcbp1 mRNA expression (E) and fluorescence-activated cell sorting (FACS) analysis and PCBP1 mean fluorescence intensity (MFI) in subsets of in vitro polarized T cells. (E) n = 8; (F) n = 4. PE, phycoerythrin. (G and H) Immunoblotting of moesin, PCBP1, FoxP3, and β-actin (G) and quantification for PCBP1 and moesin (H) using splenic mouse CD4+ T cells activated with anti-CD3 and anti-CD28 for 3 days in the absence (Th0) or presence (iTreg) of TGF-β in vitro. n = 5. (I and J) Mouse splenic CD8+ T cells from the same experiments as (G and H). n = 5. (K and L) Immunoblotting for phosphorylated and total PCBP1 (K) in Th0 and iTregs and quantification (L). (D) Error bars represent means ± SE and (E, F, H, J, and L) SD; *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test); ns, not significant.
PCBP1 levels in ex vivo mouse splenic lymphocytes were grossly comparable (Fig. 1D). Similar to human T cells, Pcbp1 mRNA was comparable between resting and activated mouse T cells but increased in iTregs (Fig. 1E). PCBP1 protein was markedly elevated in CD4+ and CD8+ T cells after TCR stimulation (Fig. 1, F to J). In addition, PCBP1 was distinctly expressed in CD69low and CD69high CD8+ T cells cultured in vitro with increasing doses of interleukin-2 (IL-2) (fig. S1, A to E). Consistent with PCBP1 repression of moesin translation (21), PCBP1 was reduced in unstimulated T cells with a concomitant increase of moesin in CD4+ (Fig. 1, G and H) and CD8+ T cells (Fig. 1, I and J) isolated from the spleen of wild-type (WT) mice. Activation of T cells up-regulated PCBP1, which then repressed moesin expression (Fig. 1, G to J) (21). iTreg differentiation rescued moesin expression (Fig. 1, G and H) because of TGF-β–mediated phosphorylation of PCBP1, which abrogates its repressive function (Fig. 1, K and L, and fig. S1, F and G) (18, 21). These results indicated that PCBP1 is rapidly up-regulated in response to T cell activation to restrict translation of moesin. Because of the rapid induction of PCBP1 in activated T cells and elevated mRNA in early thymocytes (fig. S1H), we wondered whether PCBP1 has a cell-autonomous role in T cells.
PCBP1 inhibits optimal differentiation of Tregs
We next sought to examine the physiological roles of PCBP1 in T cells; we mated mice with loxP-flanked alleles (Pcbp1f/f) (12) to mice with transgenic expression of Cre recombinase from the T cell–specific Cd4 promoter (Cd4-Cre) to generate Pcbp1f/fCd4-Cre progeny. Notably, Pcbp1-null embryos are rendered nonviable in the peri-implantation stage (24). We confirmed efficient deletion of Pcbp1 in single-positive (SP) CD4 and CD8, but not in double-negative (DN) thymocytes, by flow cytometry (fig. S2A). We found that Pcbp1fl/flCd4-Cre mice exhibited a lower frequency of SP4 and SP8 cells that resulted in greater than twofold fewer matured thymocytes, although the total number of thymocytes and double-positive cells was unaltered relative to control mice (fig. S2B). As expected, the frequency of DN thymocytes was unaltered in Pcbp1f/fCd4-Cre mice compared to WT littermates (fig. S2C); however, the proportion of SP4 FoxP3+CD25+ mature thymic Tregs (tTregs), as well as Treg precursor cells (pre-Tregs; SP4 CD25+GITR+FoxP3−) (25), was increased in Pcbp1fl/flCd4-Cre mice (fig. S2, D and E). A similar reduction of matured T cells was observed in the spleen and lymph nodes of Pcbp1fl/flCd4-Cre mice (fig. S2, F to H). Notably, Pcbp1fl/flCd4-Cre mice appeared normal and do not exhibit any lymphoproliferative or myeloproliferative disorders. We next examined FoxP3+ Treg subsets in the thymus and peripheral lymphoid organs of Pcbp1fl/flCd4-Cre and WT littermates. As observed in vitro, Pcbp1fl/flCd4-Cre mice contained significantly greater frequencies and absolute number of FoxP3+ Tregs in the thymus and peripheral organs (Fig. 2, A and B). Moreover, Tregs in Pcbp1fl/flCd4-Cre mice expressed elevated CD25, GITR, and NRP1 (fig. S3A); however, CTLA-4 and KLRG-1 were reduced, and Helios expression was unchanged (fig. S3B).
Fig. 2. Loss of PCBP1 causes optimal Treg cell development but is not required for the maintenance of Tregs.
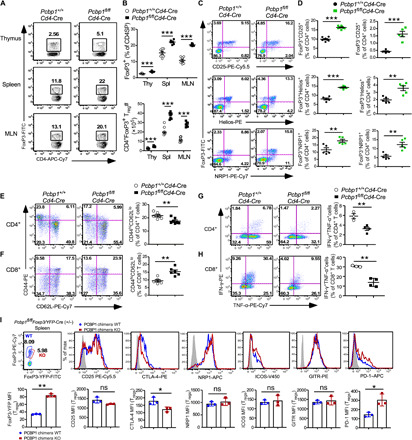
(A and B) Intracellular FoxP3 in CD4+CD8− thymocytes (Thy) and peripheral T cells from the spleen (Spl) and mesenteric lymph nodes (MLN) (A) and quantification (B). n = 7. (C and D) Frequencies of CD25-, Helios-, and NRP1-expressing T cells among CD4+FoxP3+ Tregs (C) and CD4+FoxP3− T cells (D) from the spleen of Pcbp1+/+Cd4-Cre and Pcbp1fl/flCd4-Cre mice. n = 5. (E and F) Representative flow cytometry plots of CD44 and CD62L (left) and quantification (right) in splenic CD4+ (E) and CD8+ (F) T cells from WT and Pcbp1fl/flCd4-Cre mice (n = 6). (G and H) Percentage IFN-γ+TNF-α+–producing T cells (left) and quantification (right) of CD4+ (G) and CD8+ (H) T cells stimulated with phorbol 12-myristate-13-acetate (PMA)/ionomycin in the presence of brefeldin A for 2 hours (WT, n = 3; KO, n = 4). (I) Histograms of CD25, CTLA-4, NRP1, ICOS, GITR, and PD-1 MFI (top) and quantification (bottom) in splenic Tregs from Pcbp1fl/flFoxp3/YFP-Cre+/− chimera female mice. n = 3. (B to I) Error bars represent means ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
We sought to determine why FoxP3+ Tregs are increased in Pcbp1fl/flCd4-Cre mice. We reasoned that induction of PCBP1 by T cell activation is critical to restrict the expression of Treg commitment molecules, thus preventing activated T cells from differentiating toward immunosuppressive Tregs while stabilizing the functions of effector T cells. If so, then deleting PCBP1 from T cells would shift the balance toward increased development of FoxP3+ Tregs (Fig. 2, A and B, and fig. S2D). We examined the proportion of FoxP3+ Tregs that express the tTreg commitment molecules Helios, NRP1, and CD25 (26–28). As expected, the frequencies of FoxP3+CD25+ matured Tregs, as well as Helios (Foxp3+Helioshigh)– and NRP1-expressing (FoxP3+NRP1high) cells, were increased in the spleen of Pcbp1fl/flCd4-Cre mice compared to littermate controls (Fig. 2C). The proportion of FoxP3−CD25+, FoxP3−Helioshigh, and FoxP3−NRP1high cells within the CD4+FoxP3− T cell population was notably increased in Pcbp1fl/flCd4-Cre mice (Fig. 2D), suggesting a poised state in knockout (KO) T cells that potentially supports rapid shift to the Treg lineage in response to TCR signaling. Further analysis revealed a lower frequency of CD4+CD44hiCD62Llo (Fig. 2E) but unexpectedly greater fraction of CD8+CD44hiCD62Llo (Fig. 2F) effector memory T cells in the periphery of Pcbp1fl/flCd4-Cre mice relative to WT. Nonetheless, PCBP1-deficient mice had fewer interferon-γ (IFN-γ) and tumor necrosis factor–α (TNF-α)–producing splenic T cells (Fig. 2, G and H). These data suggested that PCBP1 is crucial for maintaining peripheral T cell homeostasis and CD4+ T cells in Pcbp1fl/flCd4-Cre mice are less activated with reduced functionality.
Despite the decreased T cell functionality and increased Tregs, there was no difference in the frequency of CD4+FoxP3− T cells expressing the transcription factors Tbet, GATA3, and RORγt, which are master transcription factors expressed in T helper 1 (TH1), TH2, and TH17, respectively (fig. S3, C and D). Together, these findings suggest an essential role for PCBP1 in stabilizing the function of effector T cells, as well as maintaining peripheral T cell homeostasis by limiting conversion to Tregs in response to TCR signaling.
PCBP1 expression is not essential for the maintenance of Tregs
Next, we sought to address the role of PCBP1 specifically in Tregs. We deleted Pcbp1 in differentiated Tregs from the FoxP3+ tTreg cell stage onwards by generating Pcbp1fl/flFoxp3-Cre+ mice. These mice did not show any major changes in Treg signature molecules, which indicates that PCBP1 is not required for the maintenance of Tregs (fig. S4A). We also generated chimeric female mice that were homozygous for the floxed Pcbp1 allele and heterozygous for Foxp3YFP-Cre [PCBP1 chimeric KO mice; yellow fluorescent protein (YFP) marks cells with active Cre recombinase]. Following random inactivation of the X chromosome in these mice, the Treg cell compartment contains a mix of PCBP1-sufficient Foxp3-YFP− and PCBP1-deficient Foxp3-YFP+ cells. PCBP1 mixed chimeric WT mice are heterozygous for Foxp3YFP-Cre and contain WT Pcbp1 gene. PCBP1 chimeric KO Tregs showed no major changes in multiple Treg signature molecules, except for CTLA-4 and PD-1, which were decreased and increased, respectively, in the chimeric KO Tregs (Fig. 2I and fig. S4B).
These results indicate that PCBP1 has a fundamental role in subverting Treg phenotype in undifferentiated T cells. That is, deletion of PCBP1 in early thymocytes using the Cd4-Cre system shifts conventional T cells (Tconvs) to the Treg lineage and confer increased Treg hallmark molecules (figs. S2, D and E, and S3A). However, loss of PCBP1 after FoxP3 expression in matured Tregs using the Foxp3-YFP-Cre system does not cause major changes (Fig. 2I and fig. S4, A and B). This dichotomy underscores the importance of phosphorylated PCBP1, which contributes to the RNA binding activity and function of the protein. WT quiescent Tconvs have reduced TGF-β signaling (29), so PCBP1 is unphosphorylated and retains its RNA binding activity, thus preventing the expression of Treg commitment genes by effector T cells upon activation. Deletion of PCBP1 using the Cd4-Cre system abolishes this repressive function, enables the expression of Treg commitment mRNAs, promotes TGF-β signaling via increased moesin expression (21), and shifts Tconvs toward a stable Treg cell lineage (Fig. 2, A and B, and figs. S1G and S4C). In contrast, PCBP1 WT Tregs is phosphorylated because of higher TGF-β signaling (18, 30, 31), and its target mRNAs are derepressed to facilitate efficient Treg cell differentiation (21). Thus, PCBP1 deletion in Tregs caused no major changes since Treg commitment mRNAs were already liberated from PCBP1 control (figs. S1, F and G, and S4D). However, other mechanisms of PCBP1 function may also be involved.
PCBP1 endows effector T cell functions
We examined the possibility that PCBP1-deficient T cells are more poised to convert to Tregs in the right milieu. To examine this possibility, we used the aGVHD model to assess the effects of PCBP1 deletion on the generation of peripheral Tregs, as well as examine the ability of the developed Tregs to attenuate effector T cell responses. We hypothesized that PCBP1-WT naïve T cells transferred into mismatched recipient mice would be activated into effector T cells and drive the pathogenesis of aGVHD (32). In contrast, transfer of T cells lacking PCBP1, which appear poised to convert to Tregs, would shift into this suppressive lineage and confer resistance against aGVHD (33, 34). To this end, T cell–depleted (TCD) bone marrow (BM) cells isolated from B6.CD45.1 mice were cotransferred intravenously with Treg-depleted naïve CD4+ and CD8+ T cells (B6.CD45.2) and isolated from the spleen of WT and Pcbp1fl/flCd4-Cre mice into irradiated BALB/c mice (Fig. 3A). Changes in body weight, clinical disease manifestation, and survival were monitored for up to 80 days after BM transplant (BMT), as reported previously (35). Some BALB/c recipient mice were transplanted with BM lacking only T cells. All recipient mice dropped body weight 2 days after BMT because of the irradiation but quickly recovered. As expected, mice that received only BM cells did not lose body weight nor did they display any clinical signs of aGVHD and remained healthy throughout the experiment (Fig. 3, B to D).
Fig. 3. PCBP1 is essential to drive optimal effector T cell functions but limits Treg cell differentiation.
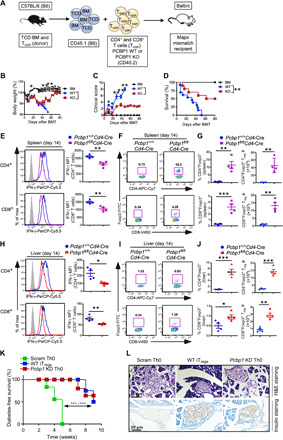
(A to D) Purified CD25-depleted PCBP1 WT or PCBP1 KO donor T cells (7 × 105; H-2b) were transferred into lethally irradiated (700 centigray) BALB/c (H-2d) mice transplanted with 5 × 106 TCD BM cells per mouse. Schematic for aGVHD using PCBP1 WT and KO T cells (A), average body weight of the GVHD mice (B), clinical scores (C), and survival (D) of recipient mice monitored as indicated. (E) FACS analysis and percentage of IFN-γ MFI in CD4+ and CD8+ T cells from spleen cells stimulated with PMA/ionomycin for 4 hours from GVHD mice on day 14 after allo-BMT. (F and G) Frequency of FoxP3+ Tregs (F) and quantification (G) on day 14 after allo-BMT in the spleen of WT and PCBP1 KO T cell recipients. (H) Expression of IFN-γ (left) and quantified plots (right) in the liver day 14 after allo-BMT. (I and J) FoxP3+ Tregs (I) and quantification (J) in the liver of recipient mice on day 14. (K and L) Diabetes-free survival and histology in diabetes model following transfer of scrambled or Pcbp1 knockdown (KD) or WT iTregs. Diabetes-free survival curve (I), and hematoxylin and eosin (H&E; top)– and insulin (bottom)–stained pancreatic sections from recipient mice (J). (B, C, E, G, H, and J) Error bars represent means ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test); (D and K) by log-rank (Mantel-Cox) test.
In contrast, recipients of PCBP1-sufficient T cells showed severe symptoms of aGVHD including rapid body weight loss and hair loss and consequently succumbed to the disease within 50 days after BMT (Fig. 3, B to D). However, the mice infused with PCBP1-deficient T cells displayed less body weight loss with minor clinical signs of aGVHD. Greater than 50% of the mice that received PCBP1-deficient T cells did not develop severe aGVHD and survived beyond 80 days after BMT (Fig. 3, B to D). These data suggest a critical requirement for PCBP1 in driving effector T cell function and pathogenicity.
To understand the relevant mechanisms for pathogenicity, we examined T cell functions based on the production of IFN-γ and TNF-α as the key mediators of aGVHD (36, 37) in the spleen (lymphoid organ) and liver (GVHD target organ) on day 14 after BMT. There was significant reduction of IFN-γ and TNF-α by splenic PCBP1-deficient T cells compared to WT T cells but no difference in Tbet expression (Fig. 3E and fig. S5A). Notably, the proportion and number of FoxP3+ Tregs among splenic CD4+ and CD8+ T cells in KO recipients was markedly increased (Fig. 3, F and G). Similar reduction in IFN-γ and TNF-α and conversion to Tregs by PCBP1-deficient CD4+ and CD8+ T cells were also observed in the liver (Fig. 3, H to J, and fig. S5B).
In another experiment, we determined the function of lentiviral short hairpin RNA (shRNA) Pcbp1 knockdown (KD) CD4+CD25− T cells in a model of type 1 diabetes. Nonobese diabetic (NOD) Rag1−/− recipient mice were infused with WT iTregs or with activated T cells (Th0) transduced with scrambled virus or shRNA targeting Pcbp1. Whereas mice infused with scrambled Th0 cells succumbed to hyperglycemia by week 5, Pcbp1 KD Th0 cells significantly delayed the onset of type 1 diabetes in recipient mice, similar to mice infused with WT iTregs (Fig. 3, K and L). These findings are consistent with our observations that T cells lacking PCBP1 shift toward FoxP3+ Tregs in response to TCR signaling. The generated Tregs from T cells lacking PCBP1 maintain immunosuppressive functions by conferring protection against aGVHD and type 1 diabetes. Together, our data indicate that PCBP1 distinctively regulates effector versus Treg cell differentiation to shape tolerance and immunity.
PCBP1 prevents expression of Treg signatures in effector T cells
To investigate the mechanism for enhanced generation of Tregs in the PCBP1-deficient mice, we performed RNA sequencing (RNA-seq). Total RNA was extracted from splenic CD4+ T cells of 6-to 8-week-old Pcbp1fl/flCd4-Cre and WT littermates. Initial analyses confirmed the quality of the data output (fig. S6, A and B). We next predicted the biological functions of the differentially expressed genes (DEGs) using the ToppGene Suite. We observed extensive alterations in gene expression due to loss of PCBP1 in T cells, and the most significant enrichment of the DEGs was related to Treg cell pathways (Fig. 4, A and B). We found that the enriched Gene Ontology (GO) terms for DEGs were related to the cell cycle and immune system including T cell activation and differentiation, as well as antigen processing and presentation (Fig. 4C). Consistent with the impaired proliferation of T cells and decreased cell numbers observed in PCBP1-deficient mice (fig. S2), we found dysregulation of numerous cell cycle– and apoptosis-related pathways and genes (Fig. 4C and fig. S6C).
Fig. 4. Effector T cell–intrinsic Treg commitment programs are constrained by PCBP1.
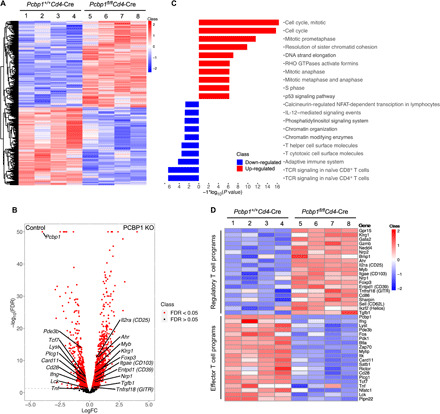
(A) Heat map of DEGs (FDR < 0.1 and absolute log2 fold change > 0.2) in CD4+ cells isolated from the spleen of 6-week-old Pcbp1+/+Cd4-Cre (n = 4) and Pcbp1fl/flCd4-Cre (n = 4) mice. (B) Volcano plot showing differential gene expression in mice. (C) Enrichment of DEGs (FDR < 0.05) in mice for biological process GO terms. GTPases, guanosine triphosphatases. (D) Heat map of up-regulated Treg cell signature and down-regulated effector T cell signature genes (FDR < 0.05) in Pcbp1+/+Cd4-Cre (n = 4) and Pcbp1fl/flCd4-Cre (n = 4) mice.
In addition to the cell cycle–related genes, there was enrichment of several DEGs [false discovery rate (FDR) < 0.05] that play important roles in Treg cell development, stability, and suppressive function. Examples include Foxp3, Klrg1, Nrp1, Nrp2, Ahr, Myb, Il2ra (CD25), Entpd1 (which encodes CD39), Gzmb, Tgfb1, and Bmp1 in CD4+ T cells lacking PCBP1 (Fig. 4, B to D, and fig. S6C) (38, 39). In contrast, but consistent with our observations, genes essential for effector T cell activation and function such as Lck, Cd28, Zap70, and Tcf7 were significantly decreased (FDR < 0.05) in CD4+ T cells lacking PCBP1 expression (Fig. 4D and fig. S6C). In agreement with data from our GVHD experiments, we also found cytokine genes such as Ifng, Tnf, Il21, and Csf2 (which encodes GM-CSF) to be significantly less in CD4+ T cells lacking PCBP1 compared to controls (Fig. 4, C and D). Thus, in the absence of PCBP1, CD4+ T cells expressed less of cytokine genes important for effector T cell functions but enhanced expression of Treg signature genes such as Foxp3, Il2ra, Klrg1, and Nrp1. These results indicate that PCBP1 plays a pivotal role in controlling the expression of effector T cell–intrinsic Treg transcriptional signatures to shape effector versus Tregs. An intriguing concept is that PCBP1 functions as a global intracellular immune checkpoint against Treg cell programs; thus, targeting it may modulate antitumor responses. It also suggests that PCBP1 expression may be necessary to stabilize and sustain the function of effector T cells in cancer.
PCBP1 constrains optimal TGF-β signaling and iTreg differentiation in vitro
To further ascertain that PCBP1 inhibits the conversion of activated T cells to Tregs and, thus, T cells lacking PCBP1 are poised to differentiate to Tregs, we first knocked down Pcbp1 in splenic CD4+CD25− T cells using a lentiviral shRNA system and cultured under iTreg differentiating conditions with anti-CD3/anti-CD28 antibodies and IL-2, in the presence or absence of TGF-β (5 ng/ml) for 3 to 5 days. KD efficiency and liberation of the PCBP1 targets moesin and eiF5A2 (20, 40) were confirmed by immunoblotting (Fig. 5A and fig. S7A). We next assessed the frequency of FoxP3+ iTregs and found that more Pcbp1 KD T cells were converted to iTregs compared to scrambled, even without the addition of exogenous TGF-β (Fig. 5, B and C).
Fig. 5. Increased frequency of FoxP3+ Tregs and enhanced TGF-β signaling in T cells expressing lower PCBP1.
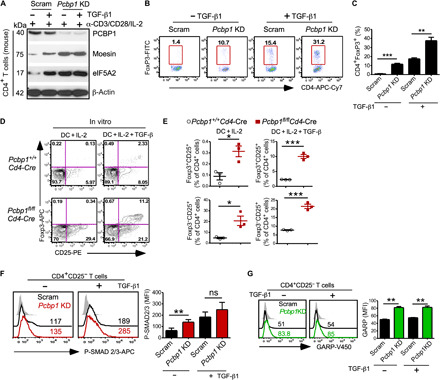
Splenic CD4+CD25− T cells were transduced with scrambled (Scram) or lentiviral shRNA targeting Pcbp1 for 48 hours and cultured in the absence or presence of exogenous TGF-β under iTreg conditions. (A) Immunoblotting of PCBP1 and targets moesin and eIF5A2. β-Actin is shown as loading control. (B and C) FACS analysis (B) and quantification (C) of FoxP3+ T cells 5 days after lentiviral transduction. (D and E) Flow cytometry analyzing CD25-expressing cells among FoxP3+ and FoxP3− T cells (D) and quantification (E) after in vitro culture of splenic CD4+CD25− T cells from Pcbp1f/f and Pcbp1f/fCd4-Cre littermates with allo-DCs in the presence of IL-2 and with or without exogenous TGF-β1 (5 ng/ml). n = 3. (F) Flow cytometry of phospho-Smad2/3 and quantification. (G) Analysis of GARP expression and quantification in scrambled versus Pcbp1 KD T cells. (C and E to G) Error bars represent means ± SD; *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Next, we cocultured naïve CD4+CD25− T cells isolated from the spleen of Pcbp1fl/flCd4-Cre mice or littermate controls in vitro with IL-2 and allogenic dendritic cells (DCs) isolated from BALB/c mice in the presence or absence of TGF-β1. We found that PCBP1-deficient CD4+CD25− T cells differentiated more robustly to mature FoxP3+CD25+ Tregs after 5 days compared to PCBP1-sufficient T cells (Fig. 5, D and E). PCBP1-deficient CD4+CD25− T cells cocultured with DCs in the absence of TGF-β1 also appeared more poised to differentiate into Tregs than WT CD4+CD25− T cells on the basis of the greater frequency of FoxP3−CD25+ pre-Tregs (Fig. 5E). These results further suggested that PCBP1 inhibits differentiation to FoxP3+ Tregs; thus, its deletion confers increased conversion of activated T cells to the Treg cell lineage. Further examination also showed increased phosphorylated Smad2/3 levels in Pcbp1 KD T cells (Fig. 5F and fig. S7B), which was corroborated by increased expression of the TGF-β–docking receptor GARP (Glycoprotein A repetitions predominant) (Fig. 5G) (41). Together, these data suggested that PCBP1 plays important roles in preventing activated T cells from differentiating into Tregs via inhibition of pertinent Treg signaling pathways such as TGF-β signaling.
Loss of PCBP1 in T cells blunts antitumor responses
To test the role of PCBP1 during antitumor responses, we compared the growth of MC38 colon cancer cells in WT mice versus mice lacking PCBP1 in T cells. We observed exponential MC38 tumor growth in both WT and Pcbp1fl/flCd4-Cre mice (Fig. 6A); however, tumor size and weight were significantly greater in Pcbp1fl/flCd4-Cre mice (Fig. 6, A and B). In further agreement with the increased TGF-β signaling under reduced PCBP1 conditions (Fig. 5, F and G) and previous reports that demonstrated that PCBP1 loss promotes tumor metastasis (18), we also observed an enlarged spleen in tumor-bearing PCBP1 KO mice compared to WT littermates (Fig. 6C). Flow cytometry analysis showed increased percentage of CD45− tumor cells in the spleen of Pcbp1fl/flCd4-Cre mice (Fig. 6D). Consistently, WT CD8+ and CD4+ tumor-infiltrating lymphocytes (TILs) expressed higher PCBP1 compared to splenic T cells (Fig. 6E). Absolute numbers of CD4+ and CD8+ T cell populations were decreased in the tumor-draining lymph nodes (TDLNs) between WT and Pcbp1fl/flCd4-Cre mice. Moreover, the frequency and numbers of CD4+ and CD8+ T cells were significantly less in the tumors of Pcbp1fl/flCd4-Cre mice compared to WT (Fig. 6, F and G), suggesting that PCBP1 is crucial for antitumor T cells to proliferate effectively and infiltrate tumors.
Fig. 6. Absence of PCBP1 in T cells compromises antitumor immunity.
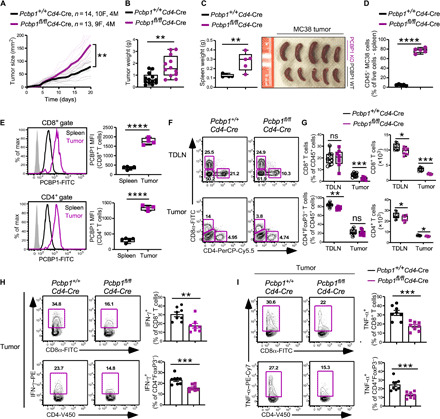
(A) Tumor growth over time following subcutaneous injection of MC38 cells (1 × 106) into Pcbp1+/+Cd4-Cre (n = 14, 10 females and 4 males) and Pcbp1fl/flCd4-Cre (n = 13, 9 females and 4 males) mice. (B) Tumor weight of Pcbp1fl/flCd4-Cre (n = 12) and WT littermate (n = 13) mice as in (A). (C) Enlarged spleens measured (left) and representative examples (right). n = 5. Photo credit: E. A. Ansa-Addo, The Ohio State University. (D) Increased percentage of CD45− cells in the spleen of tumor-bearing Pcbp1f/fCd4-Cre compared with Pcbp1f/f littermates. n = 5. (E) Flow cytometry analysis of PCBP1 expression (left) and quantification (right) in CD8+ (top) and CD4+ (bottom) T cells from spleen and tumor of WT tumor-bearing mice. Spleen, n = 5; tumor, n = 4. (F and G) FACS analysis (F) and quantifications (G) of CD8+ and CD4+FoxP3− T cells among CD45+ T cells from TDLN and tumor of WT and Pcbp1fl/flCd4-Cre mice. TDLN, n = 9; tumor, n = 8. (H) Frequency of IFN-γ–producing cells among CD8+ and CD4+FoxP3− T cells in tumors from Pcbp1fl/flCd4-Cre and WT littermates. n = 8. (I) Frequency of TNF-α–producing cells (left) and quantification (right) among CD8+ and CD4+FoxP3− T cells in tumors from Pcbp1fl/flCd4-Cre and WT littermates determined by flow cytometry. n = 8. (B to I) Error bars represent means ± SE. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (Student’s t test); (A) two-way ANOVA.
We then analyzed FoxP3 expression in CD4+ T cells and found that it increased in the spleen of Pcbp1fl/flCd4-Cre mice but was similar in the tumors and unchanged in CD8+ T cells (fig. S8, A and B). Furthermore, IFN-γ– and TNF-α–producing T cells were significantly impaired in Pcbp1fl/flCd4-Cre mice (Fig. 6, H and I). These data suggested that PCBP1 is up-regulated in tumor-infiltrating T cells to support ongoing antitumor immune responses. However, T cell–specific loss of PCBP1 in the tumor microenvironment due, in part, to increased TGF-β signaling blunts antitumor immunity and promotes tumor progression and metastasis.
Loss of PCBP1 promotes immune suppression and is a potential predictor of response to ICB
The breakdown in the function of PCBP1-deficient T cells, compared to WT, prompted us to investigate whether PCBP1 loss creates an immune-suppressive tumor microenvironment that supports tumor progression. ICB has revolutionized cancer treatment, but further improvements are still required to better predict patients more likely to benefit from this treatment. We analyzed tumor CD8+ and CD4+ non-Tregs from WT and Pcbp1fl/flCd4-Cre mice for expression of inhibitory immune checkpoint receptors and the Treg suppressive molecule CD73. We found a clear increase in PD-1, TIGIT (T Cell Immunoreceptor with Ig and ITIM Domains), VISTA (V-domain Ig suppressor of T cell activation), and CD73, but not CTLA-4, on CD8+ T cells from Pcbp1fl/flCd4-Cre mice compared to WT littermates (Fig. 7A). In addition, TIGIT and VISTA on CD8+ T cells in the TDLN were unchanged or decreased, respectively, in Pcbp1fl/flCd4-Cre mice. Unlike tumor-infiltrating CD8+ T cells, CD8+ T cells from the TDLN of Pcbp1fl/flCd4-Cre mice displayed higher CD73 expression compared to WT cells (fig. S8C). PD-1 and CTLA-4 were higher on CD4+FoxP3+ Tregs from the spleen and tumor of PCBP1 KO mice (fig. S8, D and E).
Fig. 7. Deletion of PCBP1 in T cells increases expression of checkpoint molecules and promotes immunosuppression.
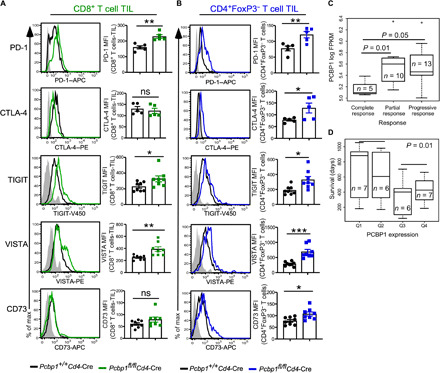
(A and B) Flow cytometric analysis (left) and quantification (right) of PD-1, CTLA-4, TIGIT, VISTA, and CD73 expression in tumor CD8+ (A) and CD4+FoxP3− (B) T cells from Pcbp1+/+Cd4-Cre and Pcbp1fl/flCd4-Cre. (C) Analysis of the clinical relevance of PCBP1 mRNA according to response rate (separating complete responders, partial responders, and progressive responders). FPKM, fragments per kilobase of exon model per million reads mapped. (D) Significance of PCBP1 mRNA expression (Q1, lowest; Q4, highest) and relationship to overall survival. (A and B) Error bars represent means ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test); (C and D) Mann-Whitney test.
In another experiment, WT and Pcbp1fl/flCd4-Cre mice were challenged with MB49 bladder cancer cells. Again, tumor growth was more rapid in PCBP1-deficient mice compared to WT animals (fig. S8F). Consistently, the spleen of KO mice was enlarged with a greater frequency of CD45− cells compared to WT mice (fig. S8, G to I). We detected increased proportions of FoxP3+CD25− cells among CD4+ T cells in the tumors of PCBP1 KO mice (fig. S8, J and K), suggesting that loss of PCBP1 helps expand or maintain this immature Treg subset (42). Similar to the MC38 model and in line with PCBP1 loss promoting immunosuppression, we detected increased PD-1 expression on TILs from Pcbp1fl/flCd4-Cre mice relative to WT littermates (fig. S8, L and M). Together with our RNA-seq data, the increase of inhibitory checkpoint receptors PD-1, TIGIT, and VISTA, as well as the reduced functionality (Figs. 6, H and I, and 7, A and B) in PCBP1-deficient T cells, is indicative of immune exhaustion under reduced PCBP1 conditions. We hypothesized that in such an immune-suppressive setting, ICB would be more effective; thus, PCBP1 could potentially serve as a predictor of response to immunotherapy.
We took advantage of the published RNA-seq database currently available from studies on patients with melanoma before and after anti–PD-1 antibody treatment and examined PCBP1 gene expression. Data from Hugo et al. (43) provide a comprehensive analysis of combined RNA-seq results obtained from melanoma patients. We found that complete responders to PD-1 blockade also had the lowest PCBP1 expression compared to partial or progressive responders (Fig. 7C). Patients with the lowest PCBP1 expression before treatment (Q1 and Q2) responded better to ICB and survived longer compared to melanoma patients with higher PCBP1 (Q3 and Q4) (Fig. 7D). These findings suggest a dichotomy between PCBP1 expression and responsiveness to ICB in cancer. In a nontumor setting, higher PCBP1 stabilizes effector T cell functions to generate immune responses that obstruct tumorigenesis. In cancer, higher PCBP1 levels counteract immune exhaustion by preventing the expression of factors such as moesin, PD-1, and TGF-β signaling that promote immunosuppression. ICB would be less effective under such conditions. In contrast, lower PCBP1 or phosphorylation of PCBP1 elicited by TGF-β liberates expression of multiple factors that drive an immune-suppressive tumor microenvironment. This setting is more conducive to ICB. These data further suggest that assessment of PCBP1 expression before checkpoint blockade therapy could be a potential predictor of response to ICB in patients.
DISCUSSION
The balance between Tregs and effector T cells, or tolerance and immune responses, respectively, is paramount to maintaining homeostasis and preventing the development of autoimmune and/or inflammatory diseases. While Tregs restrict immune function (44, 45), effector T cells promote immune responses against infection and cancer (46, 47). The fundamental roles of RBPs in the immune system have gained considerable attention in recent years because of their involvement in multiple immune processes and emerged as potential therapies (48, 49). Of these RBPs, PCBP1 has mainly been studied in transformed epithelial cells for its role in cancer metastasis and progression (50, 51). In addition, PCBP1 also exhibits iron chaperone activity toward ferritin (11). However, knowledge on PCBP1 function in T cell biology, particularly during differentiation of T cell subsets and T cell–mediated immune responses, is limited.
We recently reported that translational repression by PCBP1 is operational in CD4+ T cells, but how this contributes to T cell fate specification remains unclear. PCBP1 repressed the translation of moesin mRNA and is rescued by TGF-β–mediated phosphorylation of PCBP1, which inhibits its RNA binding function (21). We demonstrated that moesin, in turn, promotes optimal TGF-β signaling, which augments Treg cell differentiation. Deletion of moesin potentiated adoptive T cell therapy and conferred protection against B16 melanoma in mice. In the present study, we have made several new discoveries that enhance our understanding about the roles of PCBP1 in T cell biology. We discovered that PCBP1 is up-regulated in both CD4+ and CD8+ T cells in response to activation. PCBP1 then restricts expression of Treg signature molecules such as moesin in activated T cells to stabilize effector T cell functions.
We show that PCBP1 is an immune checkpoint required for effector T cell functions. PCBP1 is a global regulatory node that subverts immunosuppression in cancer. Because of the recent link between ICB and hyperprogression of cancer in some patients, it is paramount to accurately predict responders to immunotherapy (7). PCBP1 also contributes to efficient thymocyte development and peripheral T cell homeostasis. Mice with PCBP1-deficient T cells exhibited decreased matured thymocytes and peripheral T cells but increased pre-Tregs. We also found that the absence of PCBP1 resulted in increased Treg cell conversion in the thymus and peripheral tissues. These data suggested that PCBP1 suppresses the expression of effector T cell–intrinsic Treg-related molecules to maintain the identity of effector T cells. It also suggests that PCBP1 plays an active role as a negative immune checkpoint for excessive differentiation of FoxP3+ T cells. We demonstrated using the aGVHD model that recipient mice transplanted with PCBP1-sufficient T cells developed severe inflammation and succumbed to the disease. However, mice that received PCBP1-deficient T cells were remarkably protected. The GVHD model has been widely used to examine the functions of effector T cells and Tregs in either promoting or alleviating the pathogenesis of aGVHD, respectively (52).
Notably, PCBP1-deficient T cells robustly converted into peripheral Tregs, which produced less proinflammatory cytokines (IFN-γ and TNF-α) by day 14. However, PCBP1-sufficient T cells produced more cytokines and did not convert to peripheral Tregs. In another study, Pcbp1 KD–activated T cells (Th0) delayed disease onset in a type 1 diabetes model. Further analysis confirmed our finding that absence of PCBP1 results in increased FoxP3+ Tregs due, in part, to derepression of genes essential for commitment to the Treg cell lineage. Previous studies have described mechanisms that constrain effector T cell functions to stabilize the development of Tregs. For example, the transcriptional repressors BACH2 and MiR-125a repress effector T cell programs to stabilize Treg-mediated immune homeostasis (53, 54). By contrast, the nuclear factor κB regulator A20 limits tTreg development to promote effector functions (55). Thus, it is paramount to better characterize the molecular mechanisms that distinctively regulate the fate of effector cells versus Tregs in response to immune activation and against cancer.
By RNA-seq, we found that multiple Treg signature or commitment genes were up-regulated in the absence of PCBP1. We also noted significant up-regulation of molecular pathways that impair T cell activation and proliferation such as Tgfb1, Msn (moesin), Nrp1, etc. Thus, mechanistically, our data suggest that PCBP1 is a global negative regulator of genes that promote the generation and stabilization of Tregs, as well as genes that support an immune-suppressive tumor microenvironment. Effector T cells intrinsically contain Treg signature genes that, if expressed, would destabilize the effector lineage and cause them to switch into Tregs. PCBP1-deficient naïve T cells converted more efficiently to iTregs in vitro compared to WT T cells from littermate mice. Our study also suggests that PCBP1 plays a nonredundant role in ensuring that Treg commitment genes remain inhibited during acquisition of the effector T cell lineage. Such a global function in lymphocyte biology has been described for other RBPs (56, 57).
Notably, PCBP1 is regulated by moesin via TGF-β signaling since moesin controls optimal TGF-β responses to maintain naïve T cell quiescence (21, 29). Loss of PCBP1 in T cells exacerbated tumor growth in the MC38 colon cancer and MB49 bladder cancer models. Moreover, TILs lacking PCBP1 displayed signs of immune exhaustion by up-regulating several inhibitory immune checkpoint receptors and reduced antitumor cytokine production. By analyzing an RNA-seq dataset from a study by Hugo et al. (43), we found that patients with melanoma displaying lower PCBP1 responded better to anti–PD-1 therapy. Notably, this data also includes contributions from T cells and non–T cells. This dichotomy between PCBP1 mRNA expression and response to ICB suggests PCBP1 as a potential predictor of response to immunotherapy and supports our conclusion that under reduced PCBP1 levels, factors that promote immune suppression such moesin, TGF-β signaling, and PD-1 are no longer restrained and therefore inhibit antitumor responses. Although there may be additional mechanisms involved, the results of our study implicate PCBP1 as a critical node for safeguarding effector T cell functions. It also suggests that PCBP1 limits excess differentiation of Tregs and constrains expression of inhibitory immune checkpoint molecules to impair immune exhaustion and immunosuppression. The potential role of RBPs as predictors of response to immunotherapy would offer great advancement to cancer treatment.
MATERIALS AND METHODS
Human peripheral blood mononuclear cells
Peripheral blood cells from healthy, deidentified individuals were purchased as a buffy coat (Plasma Consultants) or a leukophoresis (Research Blood Components). Lymphocyte enrichment was performed by centrifugation with Lymphocyte Separation Medium (Mediatech). Untouched CD4+ and CD8+ T cells were isolated by magnetic bead separation (Dynabeads, Thermo Fisher Scientific) and activated at a 1:5 bead–to–T cell ratio using magnetic beads (Dynabeads, Thermo Fisher Scientific) in complete T cell medium supplemented with recombinant human IL-2 (100 IU/ml; National Institutes of Health repository). T cells were harvested after 3 to 4 days, and lysates were prepared for immunoblotting. All procedures were performed under Institutional Review Board–approved protocols from the Medical University of South Carolina.
Mice
Pcbp1f/fCd4-Cre, Pcbp1f/fFoxp3YFP-Cre+, and Pcbp1f/fFoxp3YFP-Cre+/− mice were generated in-house. Pcbp1fl/fl, Foxp3YFP-Cre, Cd4-Cre [Tg (Cd4-cre)1Cwi/BfluJ], and NODRag1−/−, C57BL/6J have been described (12, 21, 58). Mice used for all experiments were on the C57BL/6 background, except for GVHD and diabetes model experiments. GVHD experiments were performed using BALB/c mice as donors for BM and were also used as recipients, and type 1 diabetes studies used NOD Rag1−/−. In all studies comparing WT to PCBP1 KO mice, healthy, sex-matched, and age-matched littermates were used (male and female, 6 to 10 weeks of age unless stated otherwise). Notably, spleen and lymph nodes were kept separately throughout the study. Mice were housed in a specific pathogen–free barrier facility in ventilated cages with at most five animals per cage and provided with ad libitum food and water. With the exception of type 1 diabetes studies, which used NOD Rag1−/− immunodeficient mice, all studies used immune-competent mice that were treatment naïve until the start of study. No differences in phenotype were noted between WT and PCBP1 KO in T cells or Treg-specific cells, and animals were genotyped for floxed alleles and CRE allele. All mouse procedures were performed under Institutional Animal Care and Utilization Committee–approved protocols from The Ohio State University and the Medical University of South Carolina (tumor, type 1 diabetes, and GVHD models) and conformed to all relevant regulatory standards.
Cell lines and in vitro cultures
Primary mouse T cell cultures from the spleen of male and female mice were used to set up T cell in vitro experiments in T cell media at 37°C with 5% CO2 humidified conditions. T cell media contained RPMI 1640 media (catalog no. 10-40-CV) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, Hepes, β-mercaptoethanol, sodium pyruvate, and nonessential amino acids. shRNA lentiviral particles were generated using human embryonic kidney (HEK) 293FT cell lines [American Type Culture Collection (ATCC)] cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, USA). Tumor experiments were performed using MC38 colon cancer (ATCC) and MB49 bladder cancer cell lines (gift from S. Li, Harvard University, USA) all maintained in DMEM, supplemented with 10% FBS heat-inactivated with penicillin-streptomycin.
T cell culture and differentiation in vitro
Briefly, CD4+ T cells were isolated (catalog no. 130-104-454) from WT animal and Pcbp1f/fCd4-Cre mice and activated for 3 to 4 days via anti-CD3/anti-CD28 antibodies (5 μg/ml) plate bound (CD3, catalog no. 16-0031-85; CD28, catalog no. 16-0281-85). For differentiation experiments, naïve CD4+ T cells from WT or KO animals were plated with α-CD3/CD28 antibodies (as above) and IL-2 in the presence of subset-specific cytokines and antibodies (iTreg: TGF-β1, α–IFN-y, and α–IL-4). In other experiments, allo-DCs were used instead of anti-CD28 antibody for polarization to iTregs. Th0 experiments were performed under differentiation conditions (α-CD3/CD28 antibodies and IL-2) without additional cytokines. After 3 days, cells were harvested and analyzed by flow cytometry or Western blotting.
Lymphocyte staining and flow cytometry
Thymi, spleens, and mesenteric lymph nodes were minced into single-cell suspensions in phosphate-buffered saline (PBS), and splenocytes were depleted of red blood cells using ACK lysis buffer (homemade). For cell surface staining, cells were washed with staining buffer (2% FBS heat-inactivated in PBS with 1 mM EDTA and 0.1% sodium azide) and incubated with the indicated antibodies on ice for 30 min. Cells were washed two more times before being analyzed on a flow cytometer. For intracellular cytokine staining, cells were stimulated for 4 hours with PMA (phorbol 12-myristate-13-acetate; 50 ng/ml) and ionomycin (1 μg/ml; Sigma-Aldrich) in the presence of brefeldin A (5 μg/ml; BD Biosciences). Intracellular staining, including FoxP3 (FJK-16) was performed with the eBioscience Foxp3 staining buffer set (00-5523-00, eBioscience). For detection of phosphorylated signaling molecules, cells were serum-starved for 1 hour in serum-free RPMI 1640. Cells were stimulated with TGF-β (5 ng/ml; PeproTech), fixed with Phosflow Lyse/Fix buffer, followed by permeabilization with Phosflow Perm Buffer III (both from BD Biosciences), and stained with antibodies against P-SMAD2/3-APC (1:40 dilution; BD Biosciences). To determine expression of PCBP1, cell surface antigens were first stained before fix/perm with the FoxP3 staining buffer set (eBioscience, Thermo Fisher Scientific) and stained with Pcbp1 primary antibodies [followed by a fluorescein isothiocyanate–conjugated anti-rabbit immunoglobulin G (IgG) (BD Biosciences, USA)]. Expression of surface and intracellular markers was analyzed with a flow cytometer (BD FACSVerse) and FlowJo software (Tree Star Inc.).
Immunoblotting
T cells were stimulated under Th0 or iTreg conditions as described previously (21). Th0 cells are defined as naïve CD4+ T cells activated in the presence of anti-CD3/anti-CD28 antibodies and IL-2, but without TGF-β1, and maintained in complete T cell medium. Cells were harvested on day 3 or 4 and then counted and washed with cold serum-free RPMI 1640 medium. Cells were lysed with radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors and prepared for SDS–polyacrylamide gel electrophoresis. For immunoblotting, polyvinylidene difluoride membranes were blocked at room temperature for 1 hour with 6% nonfat milk blocking buffer and then probed with rabbit anti-PCBP1 and rabbit anti–phospho-PCBP1 (homemade), rabbit anti-moesin (Q480) and rabbit anti-eIF5A2 (D8L8Q) (Cell Signaling Technology), rat anti-FOXP3 (FJK-16s, eBioscience), rabbit anti–phospho-Smad3 (EP823Y) and rabbit anti-Smad3 (EP568Y) (both from Abcam), and mouse anti–β-actin (Sigma-Aldrich). Secondary anti-mouse (A8924), anti-rat (A5795), and anti-rabbit (A0545) IgG–horseradish peroxidase antibodies were obtained from Sigma-Aldrich.
RNA isolation, cDNA synthesis, and quantitative real-time polymerase chain reaction
Total RNA was isolated with RNeasy Plus kit (QIAGEN, CA), and cDNA was synthesized using the QuantiTect Reverse Transcription Kit (QIAGEN) according to the manufacturer’s instructions or SuperScript reverse transcriptase reagent kit (Invitrogen). Quantitative polymerase chain reaction (PCR) was performed using SYBR Premix Ex Taq (Bio-Rad, USA) on a system (Life Technologies, USA). mRNA levels of Pcbp1 and Actb were determined by the primers listed below. Gene expression was normalized to β-actin using the ΔΔCt. method (Pcbp1: forward, 5′-GGAAGAAAGGGGAGTCGGTG-3′ and reverse, 5′-AAGATGGCATTGGTAGGCCC-3′; Actb: forward, 5′-TTGCTGACAGGATGCAGAAG-3′ and reverse, 5′-ACATCTGCTGGAAGGTGGAC-3′).
RNA-seq and analysis
Total naïve CD4+ T cells were isolated from the spleen of WT and Pcbp1f/f Cd4-Cre mice using the naïve T cell isolation kit (Miltenyi, USA). Total RNA was extracted from the cells and sent to Novogene for RNA-seq analysis. Libraries were prepared using 50 ng of total RNA using the NEBNext Ultra RNA Library Kit for Illumina (catalog no. E7530) and sequenced on HiSeq 3000 at 75 base pair paired-end. Each sample was analyzed in quadruplet. Sequencing reads were first evaluated for quality control using the FastQC software and then aligned against the mouse mm10 reference genome using the TopHat2 software. Mapped reads were assigned to gene features and quantified using the HTSeq software and further evaluated for quality control using correlation and multidimensional scaling plots. Normalization and differential expression were performed using the edgeR software. The significantly DEGs (FDR < 0.05) were visualized using heat maps and volcano plot representations and considered for subsequent functional enrichment analysis using ToppGene Suite.
Lentiviral production and transfection
Lentiviral particles were produced in HEK293FT cells according to standard protocols. Spleen-isolated primary CD4+CD25− T cells (2 × 105 cells per well) were transduced with lentiviral supernatants. Murine lentiviral DNA plasmid against Pcbp1 (hnRNP E1) (sequence: CCG GCC ATG ATC CAA CTG TGT AAT TCT CGA GAA TTA CAC AGT TGG ATC ATG GTT TTT G) (Sigma-Aldrich, TRC1-pLKO.1-puromycin) were transduced into CD4+ T cells activated for 48 hours with anti-CD3 and anti-CD28 with polybrene (8 μg/ml; Sigma-Aldrich).
Diabetes model
Splenocytes from NOD mice were depleted of CD25+ cells by incubating with anti-CD25 MACS beads (Miltenyi) and passing through the MACS LD column (Miltenyi). CD25-depleted splenocytes were adoptively transferred in recipient mice at 6 × 106 CD25− T cells per mouse, together with 2 × 105 scrambled, Pcbp1 KD CD4+CD25− T cells, or WT in vitro–induced Tregs, intravenously into NOD Rag1−/− mice. Blood glucose concentrations were then monitored kinetically. A glucose value of 200 mg/dl or more was defined as diabetic. Autoimmune destructions of pancreatic islet cells were verified by histology and insulin staining.
Histology
Mouse organs were fixed in 4% formalin, embedded in paraffin, sectioned, and stained following standard hematoxylin and eosin (H&E) staining protocol. For histology and multiplex cytokine assay, organs from KO and heterozygous mice were fixed with 3.7% formalin. After dehydration with 30% sucrose, tissues were then embedded in optimal cutting temperature compound, sectioned, and processed for H&E staining or insulin staining according to standard protocols.
Tumor models
WT or Pcbp1f/fCd4-Cre mice, aged 6 to 10 weeks, littermates were challenged subcutaneously with MC38 cells (1 × 106) or MB49 tumor cells (5 × 105). For all tumor models, tumors were measured regularly with a caliper by personnel blinded to the genotype of the mice. Tumor area was calculated using the equation (L × W), where L denotes length and W denotes width. The maximal tumor burden was 400 mm2, and this limit was not exceeded in any of the experiments. Tumor-bearing mice were euthanized at 20 days after inoculation and harvested for TIL analysis. In all flow cytometry experiments, tumors of similar sizes were used for comparison. All animals were housed under specific pathogen–free conditions in accordance with institutional and federal guidelines.
Study design
The aim of this study was to characterize the roles of PCBP1 in effector T cell functions. The studies were performed using groups of age- and gender-matched mice, 6 to 10 weeks old. We generated a Pcbp1 conditional KO mouse model (Pcbp1fl/flCd4-Cre) and tested their responses against Pcbp1+/+Cd4-Cre littermate controls. The number of replicates for each experiment is indicated in the figure legends. No animals were excluded in the studies. Histological analyses were performed double-blinded where appropriate.
GVHD model in vivo
Major histocompatibility complex–mismatched (B6 → BALB/c) BMT model was used as previously established. Briefly, recipient BALB/c mice (7 to 8 weeks old) were conditioned with total body irradiation at 700 centigray (single dose) using X-RAD 320 irradiators (Precision X-ray Inc., North Branford, CT). Within 24 hours after irradiation, recipients were intravenously injected with 5 × 106 TCD BM alone or with 7 × 105 T cells (CD25+ depleted) from WT or KO, which were purified from the spleen and lymph node by negative depletion using magnetic beads as previously described (35). Recipient mice were monitored for survival, body weight loss, and GVHD clinical score for 80 days. The GVHD clinical scores were tabulated as five parameters: weight loss, posture, activity, fur texture, and skin integrity.
Statistical analyses
Statistical parameters are all reported in the figure legends. Statistical analyses were performed with Prism software version 5.0 or 8.0 (GraphPad Software, La Jolla, CA, USA; https://graphpad.com), and significances were determined using Student’s t test, one-way analysis of variance (ANOVA), log-rank test, and Mann-Whitney nonparametric test where specified. All data are presented as means ± SE or SD unless otherwise stated. All experiments were carried out in triplicate technical and biological replicates unless otherwise stated. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank A. Henderson (Ansa-Addo Lab) and members of the Li, Liu, and Howe laboratories for assistance and insightful discussions on the project. We also thank O. Protchenko in the Philpott laboratory for help in obtaining the Pcbp1-flox mice. We also thank members of the Pelotonia Institute for Immuno-Oncology at Ohio State University (OSU) and the Cancer Immunology Program at the Hollings Cancer Center (MUSC) for helpful discussions. Funding: This research was supported by the NIH R01CA188419, R01AI070603, and P01CA186866 (to Z.L.), the American Cancer Society Intra-institutional Grant 129806-IRG-16-185-17-IRG (to E.A.A.-A.), and the Flow Cytometry & Cell Sorting Shared Resource Unit at the OSU and Hollings Cancer Center. Author contributions: E.A.A.-A. and Z.L. designed and oversaw the study. E.A.A.-A., H.-C.H., B.R., S.I., D.B., and M.H.N. performed experiments and data analysis. J.H.N. and D.C. analyzed RNA-seq data and generated graphs. C.M.P., B.L., X.-Z.Y., C.P., and P.H.H. provided reagents and expertise in experimental design and data interpretation. E.A.A.-A. and Z.L. wrote the manuscript with input from all coauthors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: RNA-seq data have been deposited in the GEO database under accession number GSE131826. The Pcbp1 mutant mouse strain is available through a material transfer agreement with OSU. Requests for mice should be directed to Z.L. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz3865/DC1
REFERENCES AND NOTES
- 1.Sharma P., Allison J. P., Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 161, 205–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., Song W., Rubinstein M., Liu D., Recent updates in cancer immunotherapy: A comprehensive review and perspective of the 2018 China Cancer Immunotherapy Workshop in Beijing. J. Hematol. Oncol. 11, 142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanmamed M. F., Chen L., A paradigm shift in cancer immunotherapy: From enhancement to normalization. Cell 175, 313–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamoto K., Al-Habsi M., Honjo T., Role of PD-1 in immunity and diseases. Curr. Top. Microbiol. Immunol. 410, 75–97 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Gide T. N., Quek C., Menzies A. M., Tasker A. T., Shang P., Holst J., Madore J., Lim S. Y., Velickovic R., Wongchenko M., Yan Y., Lo S., Carlino M. S., Guminski A., Saw R. P. M., Pang A., McGuire H. M., Palendira U., Thompson J. F., Rizos H., Silva I. P. D., Batten M., Scolyer R. A., Long G. V., Wilmott J. S., Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 35, 238–255.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Davar D., Kirkwood J. M., PD-1 immune checkpoint inhibitors and immune-related adverse events: Understanding the upside of the downside of checkpoint blockade. JAMA Oncol. 5, 942–943 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kamada T., Togashi Y., Tay C., Ha D., Sasaki A., Nakamura Y., Sato E., Fukuoka S., Tada Y., Tanaka A., Morikawa H., Kawazoe A., Kinoshita T., Shitara K., Sakaguchi S., Nishikawa H., PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 9999–10008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner M., Díaz-Muñoz M. D., RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 19, 120–129 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Techasintana P., Ellis J. S., Glascock J., Gubin M. M., Ridenhour S. E., Magee J. D., Hart M. L., Yao P., Zhou H., Whitney M. S., Franklin C. L., Martindale J. L., Gorospe M., Davis W. J., Fox P. L., Li X., Atasoy U., The RNA-binding protein HuR posttranscriptionally regulates IL-2 homeostasis and CD4+ Th2 differentiation. Immunohorizons 1, 109–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel K. U., Bell L. S., Galloway A., Ahlfors H., Turner M., The RNA-binding proteins Zfp36l1 and Zfp36l2 enforce the thymic β-selection checkpoint by limiting DNA damage response signaling and cell cycle progression. J. Immunol. 197, 2673–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leidgens S., Bullough K. Z., Shi H., Li F., Shakoury-Elizeh M., Yabe T., Subramanian P., Hsu E., Natarajan N., Nandal A., Stemmler T. L., Philpott C. C., Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J. Biol. Chem. 288, 17791–17802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu M. S., Zhang D., Protchenko O., Shakoury-Elizeh M., Philpott C. C., PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Invest. 127, 1786–1797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philpott C. C., Ryu M. S., Frey A., Patel S., Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J. Biol. Chem. 292, 12764–12771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji X., Humenik J., Yang D., Liebhaber S. A., PolyC-binding proteins enhance expression of the CDK2 cell cycle regulatory protein via alternative splicing. Nucleic Acids Res. 46, 2030–2044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripathi V., Sixt K. M., Gao S., Xu X., Huang J., Weigert R., Zhou M., Zhang Y. E., Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-β. Mol. Cell 64, 1010 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Hwang C. K., Wagley Y., Law P. Y., Wei L. N., Loh H. H., Phosphorylation of poly(rC) binding protein 1 (PCBP1) contributes to stabilization of mu opioid receptor (MOR) mRNA via interaction with AU-rich element RNA-binding protein 1 (AUF1) and poly A binding protein (PABP). Gene 598, 113–130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howley B. V., Howe P. H., TGF-beta signaling in cancer: Post-transcriptional regulation of EMT via hnRNP E1. Cytokine 118, 19–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhury A., Hussey G. S., Ray P. S., Jin G., Fox P. L., Howe P. H., TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussey G. S., Chaudhury A., Dawson A. E., Lindner D. J., Knudsen C. R., Wilce M. C. J., Merrick W. C., Howe P. H., Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41, 419–431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussey G. S., Link L. A., Brown A. S., Howley B. V., Chaudhury A., Howe P. H., Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PLOS ONE 7, e52624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansa-Addo E. A., Zhang Y., Yang Y., Hussey G. S., Howley B. V., Salem M., Riesenberg B., Sun S., Rockey D. C., Karvar S., Howe P. H., Liu B., Li Z., Membrane-organizing protein moesin controls Treg differentiation and antitumor immunity via TGF-β signaling. J. Clin. Invest. 127, 1321–1337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Yin W., Zhu L., Li J., Yao Y., Chen F., Sun M., Zhang J., Shen N., Song Y., Chang X., Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity 49, 80–92.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Fernandez D. R., Telarico T., Bonilla E., Li Q., Banerjee S., Middleton F. A., Phillips P. E., Crow M. K., Oess S., Muller-Esterl W., Perl A., Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J. Immunol. 182, 2063–2073 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghanem L. R., Kromer A., Silverman I. M., Chatterji P., Traxler E., Penzo-Mendez A., Weiss M. J., Stanger B. Z., Liebhaber S. A., The Poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol. Cell. Biol. 36, 304–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lio C. W., Hsieh C. S., A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimoto N., Oida T., Hirota K., Nakamura K., Nomura T., Uchiyama T., Sakaguchi S., Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int. Immunol. 18, 1197–1209 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Weiss J. M., Bilate A. M., Gobert M., Ding Y., Curotto de Lafaille M. A., Parkhurst C. N., Xiong H., Dolpady J., Frey A. B., Ruocco M. G., Yang Y., Floess S., Huehn J., Oh S., Li M. O., Niec R. E., Rudensky A. Y., Dustin M. L., Littman D. R., Lafaille J. J., Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209, 1723–1742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa Y., Ohkura N., Kidani Y., Vandenbon A., Hirota K., Kawakami R., Yasuda K., Motooka D., Nakamura S., Kondo M., Taniuchi I., Kohwi-Shigematsu T., Sakaguchi S., Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu E., Chia C. P. Z., Chen W., Zhang D., Park S. A., Jin W., Wang D., Alegre M. L., Zhang Y. E., Sun L., Chen W. J., T cell receptor-regulated TGF-β type I receptor expression determines T cell quiescence and activation. Immunity 48, 745–759.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W., Konkel J. E., Development of thymic Foxp3+ regulatory T cells: TGF-β matters. Eur. J. Immunol. 45, 958–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konkel J. E., Zhang D., Zanvit P., Chia C., Zangarle-Murray T., Jin W., Wang S., Chen W. J., Transforming growth Factor-β signaling in regulatory T cells controls T helper-17 cells and tissue-specific immune responses. Immunity 46, 660–674 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Betts B. C., Bastian D., Iamsawat S., Nguyen H., Heinrichs J. L., Wu Y., Daenthanasanmak A., Veerapathran A., O’Mahony A., Walton K., Reff J., Horna P., Sagatys E. M., Lee M. C., Singer J., Chang Y. J., Liu C., Pidala J., Anasetti C., Yu X. Z., Targeting JAK2 reduces GVHD and xenograft rejection through regulation of T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, 1582–1587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fondi C., Nozzoli C., Benemei S., Baroni G., Saccardi R., Guidi S., Nicoletti P., Bartolozzi B., Pimpinelli N., Santucci M., Bosi A., Massi D., Increase in FOXP3+ regulatory T cells in GVHD skin biopsies is associated with lower disease severity and treatment response. Biol. Blood Marrow Transplant. 15, 938–947 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Daenthanasanmak A., Iamsawat S., Chakraborty P., Nguyen H. D., Bastian D., Liu C., Mehrotra S., Yu X. Z., Targeting Sirt-1 controls GVHD by inhibiting T-cell allo-response and promoting Treg stability in mice. Blood 133, 266–279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iamsawat S., Daenthanasanmak A., Voss J. H., Nguyen H., Bastian D., Liu C., Yu X. Z., Stabilization of Foxp3 by targeting JAK2 enhances efficacy of CD8 induced regulatory T cells in the prevention of Graft-versus-Host disease. J. Immunol. 201, 2812–2823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leclerc M., Naserian S., Pilon C., Thiolat A., Martin G. H., Pouchy C., Dominique C., Belkacemi Y., Charlotte F., Maury S., Salomon B. L., Cohen J. L., Control of GVHD by regulatory T cells depends on TNF produced by T cells and TNFR2 expressed by regulatory T cells. Blood 128, 1651–1659 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Wang H., Yang Y. G., The complex and central role of interferon-γ in graft-versus-host disease and graft-versus-tumor activity. Immunol. Rev. 258, 30–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aida K., Miyakawa R., Suzuki K., Narumi K., Udagawa T., Yamamoto Y., Chikaraishi T., Yoshida T., Aoki K., Suppression of Tregs by anti-glucocorticoid induced TNFreceptor antibody enhances the antitumor immunity of interferon-α gene therapy for pancreatic cancer. Cancer Sci. 105, 159–167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samstein R. M., Arvey A., Josefowicz S. Z., Peng X., Reynolds A., Sandstrom R., Neph S., Sabo P., Kim J. M., Liao W., Li M. O., Leslie C., Stamatoyannopoulos J. A., Rudensky A. Y., Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 151, 153–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussey G. S., Howley B. V., Howe P. H., Post-transcriptional mapping reveals critical regulators of metastasis. Oncoscience 2, 831–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rachidi S., Metelli A., Riesenberg B., Wu B. X., Nelson M. H., Wallace C., Paulos C. M., Rubinstein M. P., Garrett-Mayer E., Hennig M., Bearden D. W., Yang Y., Liu B., Li Z., Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci Immunol. 2, eaai7911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tai X., Erman B., Alag A., Mu J., Kimura M., Katz G., Guinter T., McCaughtry T., Etzensperger R., Feigenbaum L., Singer D. S., Singer A., Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity 38, 1116–1128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hugo W., Zaretsky J. M., Sun L., Song C., Moreno B. H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., Seja E., Lomeli S., Kong X., Kelley M. C., Sosman J. A., Johnson D. B., Ribas A., Lo R. S., Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontenot J. D., Gavin M. A., Rudensky A. Y., Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Hori S., Nomura T., Sakaguchi S., Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Govindaraj C., Scalzo-Inguanti K., Madondo M., Hallo J., Flanagan K., Quinn M., Plebanski M., Impaired Th1 immunity in ovarian cancer patients is mediated by TNFR2+ Tregs within the tumor microenvironment. Clin. Immunol. 149, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Roychoudhuri R., Eil R. L., Clever D., Klebanoff C. A., Sukumar M., Grant F. M., Yu Z., Mehta G., Liu H., Jin P., Ji Y., Palmer D. C., Pan J. H., Chichura A., Crompton J. G., Patel S. J., Stroncek D., Wang E., Marincola F. M., Okkenhaug K., Gattinoni L., Restifo N. P., The transcription factor BACH2 promotes tumor immunosuppression. J. Clin. Invest. 126, 599–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman R., McHugh J., Turner M., RNA binding proteins as regulators of immune cell biology. Clin. Exp. Immunol. 183, 37–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner M., Monzón-Casanova E., RNA-binding proteins mind the GAPs. Nat. Immunol. 18, 146–148 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Guo J., Jia R., Splicing factor poly(rC)-binding protein 1 is a novel and distinctive tumor suppressor. J. Cell. Physiol. 234, 33–41 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zhou M., Tong X., Downregulated Poly-C binding protein-1 is a novel predictor associated with poor prognosis in acute myeloid leukemia. Diagn. Pathol. 10, 147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Fu J., Wang D., Yu Y., Heinrichs J., Wu Y., Schutt S., Kaosaard K., Liu C., Haarberg K., Bastian D., McDonald D. G., Anasetti C., Yu X. Z., T-bet is critical for the development of acute graft-versus-host disease through controlling T cell differentiation and function. J. Immunol. 194, 388–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roychoudhuri R., Hirahara K., Mousavi K., Clever D., Klebanoff C. A., Bonelli M., Sciumè G., Zare H., Vahedi G., Dema B., Yu Z., Liu H., Takahashi H., Rao M., Muranski P., Crompton J. G., Punkosdy G., Bedognetti D., Wang E., Hoffmann V., Rivera J., Marincola F. M., Nakamura A., Sartorelli V., Kanno Y., Gattinoni L., Muto A., Igarashi K., O’Shea J. J., Restifo N. P., BACH2 represses effector programs to stabilize Treg-mediated immune homeostasis. Nature 498, 506–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan W., Zhu S., Dai D., Liu Z., Li D., Li B., Gagliani N., Zheng Y., Tang Y., Weirauch M. T., Chen X., Zhu W., Wang Y., Chen B., Qian Y., Chen Y., Fang J., Herbst R., Richman L., Jallal B., Harley J. B., Flavell R. A., Yao Y., Shen N., MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat. Commun. 6, 7096 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Fischer J. C., Otten V., Kober M., Drees C., Rosenbaum M., Schmickl M., Heidegger S., Beyaert R., van Loo G., Li X. C., Peschel C., Schmidt-Supprian M., Haas T., Spoerl S., Poeck H., A20 restrains thymic regulatory T cell development. J. Immunol. 199, 2356–2365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galloway A., Saveliev A., ukasiak S., Hodson D. J., Bolland D., Balmanno K., Ahlfors H., Monzon-Casanova E., Mannurita S. C., Bell L. S., Andrews S., Diaz-Munoz M. D., Cook S. J., Corcoran A., Turner M., RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 352, 453–459 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Diaz-Muñoz M. D., Bell S. E., Fairfax K., Monzon-Casanova E., Cunningham A. F., Gonzalez-Porta M., Andrews S. R., Bunik V. I., Zarnack K., Curk T., Heggermont W. A., Heymans S., Gibson G. E., Kontoyiannis D. L., Ule J., Turner M., The RNA-binding protein HuR is essential for the B cell antibody response. Nat. Immunol. 16, 415–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Wu B. X., Metelli A., Thaxton J. E., Hong F., Rachidi S., Ansa-Addo E., Sun S., Vasu C., Yang Y., Liu B., Li Z., GP96 is a GARP chaperone and controls regulatory T cell functions. J. Clin. Invest. 125, 859–869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz3865/DC1


