A tailored protocol combines pigment removal with clearing, facilitating labeling and whole-body imaging across phylogeny.
Abstract
Tissue clearing combined with deep imaging has emerged as a powerful alternative to classical histological techniques. Whereas current techniques have been optimized for imaging selected nonpigmented organs such as the mammalian brain, natural pigmentation remains challenging for most other biological specimens of larger volume. We have developed a fast DEpigmEntation-Plus-Clearing method (DEEP-Clear) that is easily incorporated in existing workflows and combines whole system labeling with a spectrum of detection techniques, ranging from immunohistochemistry to RNA in situ hybridization, labeling of proliferative cells (EdU labeling) and visualization of transgenic markers. With light-sheet imaging of whole animals and detailed confocal studies on pigmented organs, we provide unprecedented insight into eyes, whole nervous systems, and subcellular structures in animal models ranging from worms and squids to axolotls and zebrafish. DEEP-Clear thus paves the way for the exploration of species-rich clades and developmental stages that are largely inaccessible by regular imaging approaches.
INTRODUCTION
The focus on a handful of well-established “molecular model species” has been instrumental to drive biological discovery and technological development for the past decades. However, it is increasingly recognized that these model species only cover a limited spectrum of ecological diversity, calling for a more systematic effort in establishing novel model systems (1–4). Advances in the establishment of versatile technologies are keys in this effort. This is exemplified by the progress in RNA sequencing technology or the advent of precise genome editing methodology, tools that have begun to pave the way for systematic research into less conventional models (5–12).
In contrast to sequencing or genome editing technologies, other methodologies are still far more restricted in their applicability and use, resulting in substantial limitations concerning the exploration of less established biological models. One critical area is the imaging of cells or molecules in the context of their natural tissue environment and the imaging of complete tissues and organisms. This often involves sectioning and reconstruction, resource-intensive processes that are particularly challenging in cases similar to the nervous system, where dissected nerves and individual neuronal projections are notoriously difficult to reconstruct. Whereas advanced microscope setups provide superior resolution in thin samples, deep imaging of nervous systems, other tissues, or whole organisms therefore remains a key challenge outside few well-established model systems. Major reasons for this are the scattering of light caused by the differences in the refractive indices (RIs) of biological molecules (e.g., water versus lipid versus protein) and the absorption of light by natural pigments (13).
Driven by research on the mammalian brain, various methods have been established to resolve the heterogeneity in RIs in unpigmented samples. This includes the removal of lipids from lipid-rich tissues such as the central nervous system and the use of media that are able to match RIs. Strategies to match the RI and/or to remove lipids have been broadly classified in two categories: those relying primarily on organic solvents, such as the benzyl alcohol with benzyl benzoate (BABB) (14), Three-dimensional imaging of solvent-cleared organs (3DISCO) and immunolabeling-enabled three-dimensional imaging of solvent-cleared organs (iDISCO) (15–17), ultimate DISCO (uDISCO) (18), stabilised DISCO (sDISCO) (19), polyethylene glycol (PEG)-associated solvent system (PEGASOS) (20) or 2nd generation ethyl cinnamate-based clearing (2ECi) (21) protocols; and those primarily relying on hydrophilic reagents and detergents, including the ClearT (22), Rapid clearing method based on Triethanolamine and Formamide (RTF) (23), Clear, unobstructed brain imaging cocktails and computational analysis (CUBIC) (24–27), Scale (28, 29) or SeeDB (30, 31) methods, and variants of the clear lipid-exchanged anatomically rigid imaging/immunostaining-compatible tissue hydrogel (CLARITY) (32, 33) approach [reviewed in (13, 34)].
While each of these techniques offers unique advantages, they are not easily applicable to many species of ecological or evolutionary interest that exhibit pigmentation. Although several existing clearing protocols decolorize heme, the blood pigment, they have not been optimized for removal of other body pigments. Animals produce a variety of additional pigments through conserved biochemical pathways. Several of these pigments are characterized by poor solubility: Melanin, a tyrosine-derived ultraviolet protectant, is very poorly soluble in both lipid- and water-based solvents [reviewed in (35)] yet serves as a prominent pigment of the vertebrate retinal pigment epithelium (RPE), a cell layer acting to diminish photo-oxidative stress and absorb scattered light. Likewise, ommochromes are a class of pigments derived from tryptophan catabolism that are particularly abundant in invertebrate species. They are tricyclic compounds consisting of phenoxazone or phenothiazine subunits and are for instance responsible for yellow, red, brownish, or black pigmentation of insect eyes, cephalopod body pigmentation, or integuments (36). Another class of abundant and poorly soluble pigments are pterins, cyclic compounds biochemically derived from guanosine triphosphate; pterins are cofactors of the enzymatic biosynthesis of ommochromes and thus often found in combination with those pigments, e.g., in invertebrate eyes (35). Whereas natural albinism, the application of pathway inhibitors, or the selection for single or combined mutants in the respective pigment pathways provides advantages for clearing and deep imaging, these are not broadly applicable approaches for many species of interest. Despite some recent efforts, such as the combination of decalcification and bleaching to adapt a CUBIC protocol for use in pigmented crustaceans (37) or the mouse eye–specific EyeCi protocol (38), the variety of body and eye pigments found in animals therefore remains a central challenge for the broad use of clearing and deep imaging. Further, it also remains unclear if any attempt to combine depigmentation and clearing would preserve other biomolecules for single or multiple imaging approaches.
Here, we provide a novel, fast, and simple method that combines depigmentation with tissue clearing in samples from five different species representing four distinct animal clades (annelids, molluscs, bony fishes, and tetrapods) and sizes up to several centimeters in length. This method, which we term DEpigmEntation-Plus-Clearing (DEEP-Clear), combines advantages of methods previously categorized in either organic solvent– or hydrophilic reagent–based clearing approaches, allowing for complete whole-body clearing and RI matching of fixed specimens within less than a day. DEEP-Clear efficiently removes naturally occurring pigments including pterins, ommochromes, heme, carotenoids, and melanin. DEEP-Clear is compatible with immunohistochemical (IHC) analysis in specimens fixed with either paraformaldehyde (PFA) or Bouin’s fixative, thus allowing the visualization of a broad range of epitopes. Moreover, it allows the visualization of 5-ethynyl-2′-deoxyuridine (EdU)–labeled chromatin of proliferative cells and is compatible with transgenically expressed fluorophores. Last, DEEP-Clear retains compatibility with whole-mount fluorescence RNA in situ hybridization (RNA-FISH), a technique that has been left out of the focus of most existing clearing methods (34). DEEP-Clear permits deep imaging of labeled structures across scales, ranging from whole-body imaging using advanced light-sheet microscopy to detailed high-resolution investigation using confocal microscopy. Thus, DEEP-Clear is a highly versatile tool that will make depigmentation, tissue clearing, labeling, and imaging applicable to a broad spectrum of animal model systems and experimental approaches.
RESULTS
DEEP-Clear enables combined depigmentation and clearing across four animal clades
Pende and coworkers (39) have recently developed a CUBIC-based approach (FlyClear) that not only renders different tissues transparent but also causes depigmentation of eyes in Drosophila melanogaster while preserving endogenous transgenic green fluorescent protein (GFP) and mCherry signal. Given that D. melanogaster eyes contain both ommochromes and pterins (35), we hypothesized that this protocol could serve as a suitable basis for developing a depigmentation and tissue clearing approach of broader applicability to other animal model organisms and possibly also be compatible with other detection methods. We decided to focus our work on representatives of four distinct noninsect animal clades:
1) The marine bristle worm Platynereis dumerilii as a representative of annelids (adult samples, around 15 mm in length).
2) The Hawaiian bobtail squid Euprymna scolopes and the longfin inshore squid Doryteuthis pealeii, two cephalopod molluscs (hatchling stage, 3 mm in length).
3) The zebrafish Danio rerio as a representative for bony fishes (from larva to juvenile stages of around 12 mm in length).
4) The axolotl Ambystoma mexicanum as a reference species for tetrapods (juvenile samples, up to 35 mm in length).
Both annelids and molluscs are key groups in the large lophotrochozoan superphylum, while bony fishes and tetrapods are the most species-rich groups of deuterostomes. Our choice of models therefore covers a substantial spectrum of ecologically relevant noninsect animal diversity. Moreover, the selected species and developmental stages also provide exemplary access to interesting neurobiological aspects (such as central nervous system regeneration, cranial nerve complexity, or different types of visual organs) that strongly benefit from a method providing depigmentation, clearing, active labeling, and whole-body imaging.
In a systematic set of experiments, we gradually modified the FlyClear protocol and chemistry to achieve decolorization of different kinds of pigments and tissue clearing in all of these species, resulting in a revised DEEP-Clear protocol adaptable for each of the investigated samples (Fig. 1A). Key steps in this process were (i) the combination of FlyClear’s Solution-1, a hyperhydration-based solution containing an aminoalcohol N,N,N′,N′-Tetrakis(2-hydroxyethyl)ethylenediamine (THEED), detergent (Triton X-100), and urea with acetone (an organic solvent) resulting in increased depigmentation speed, thereby reducing possible damage by extended exposure; (ii) the inclusion of a peroxide-bleaching step for melanin-containing samples; and (iii) the variation of urea content and pH in Solution-1 to reduce swelling effects or tissue damage encountered in the squid and zebrafish models. Together, all postfixation clearing steps can be performed within 24 hours or less for each of the samples (Fig. 1A), making DEEP-Clear easy to incorporate into existing workflows. As illustrated in Fig. 1 (B and C), the application of the organic solvent step substantially reduced depigmentation time in the two tested invertebrate models and had synergistic effects on pigment removal (Fig. 1D), allowing us to generate depigmented, transparent samples for each of the species (Fig. 1E).
Fig. 1. A rapid method combining depigmentation with tissue clearing in representatives of four distinct and species-rich animal clades.
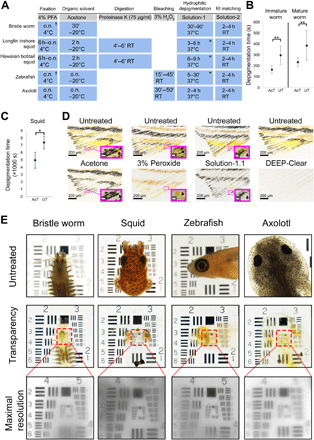
(A) Main steps of the DEEP-Clear protocol, including incubation times for the five main model systems presented in this study. (B) Systematic advancement of eye depigmentation speed by acetone pretreatment in immature and mature worms. Quantified comparisons between acetone-treated (AcT) and untreated (UT) head halves incubated with Solution-1. All values are mean ± SD; statistical significance was determined by a Wilcoxon test, yielding P values of P = 0.00166 (immature worms) and P = 0.00192 (mature worms). (C) Systematic advancement of eye depigmentation speed by acetone pretreatment in squid. Quantification of depigmentation time in acetone-treated and untreated squid halves upon incubation with Solution- 1.1. Values are mean ± SD; statistical significance was determined by a Wilcoxon test (P = 0.01285). (D) Differential and synergistic impact of acetone, peroxide, and Solution-1.1 on zebrafish fin pigments. Panels show fins of untreated (top) and treated (bottom) zebrafish fins. Insets: Magnification of dashed area and impact of different treatments on respective pigments (black arrows). Xanthophore containing pteridine and carotenoid pigments (yellow and orange) and melanophore containing melanin pigment (black). Rightmost panels show the overall impact of the full DEEP-Clear protocol. (E) Wide-field images of specimens placed on top of a USAF 1951 chart. Uncleared samples in PBS (top panels), same samples after depigmentation and refractive index (RI) matching in Solution-2 (middle panels), and higher magnification of red rectangular areas indicating the highest level of transparency reached after RI matching (bottom panels). Scale bars in the insets of (D), 20 μm. In (A), dagger indicates the possibility of fixation with Bouin’s solution; asterisks indicate the use of Solution-1.1 incubation instead of Solution-1. o.n., overnight; RT, room temperature; h, hour; ’, minutes. In (B) and (C), *P < 0.05 and **P < 0.01. Photo credit: Marko Pende, Medical University of Vienna.
With respect to the different pigment types, DEEP-Clear treatment in annelids depigmented the adult eyes that have previously been characterized to contain pterins (fig. S1A) (40), along with the additional red pigments in the body erythrophores (fig. S1, A and B) and the heme-based blood pigments (fig. S1B) (41). We noted that the treatment also destains the chlorophyll of the consumed spinach in the worm gut (fig. S1C). Furthermore, DEEP-Clear removes ommochrome-based pigments in squid (fig. S1D). As exemplified for the zebrafish fin (Fig. 1D), treatment solely with acetone at room temperature causes depigmentation of xanthophores that carry pteridines and carotenoid pigments but does not remove the characteristic black melanin pigments present in melanophores (Fig. 1D). Moreover, melanin in the analyzed vertebrate eyes and body tissues could not be fully solubilized by individual treatment with Solution-1, even with increased pH (13) and extended incubation times. In axolotl eye and body tissues, melanin was merely reduced (fig. S1E), whereas in zebrafish, this prolonged Solution-1 treatment led to severe tissue damage, requiring a change to the urea-reduced Solution-1.1 for subsequent experiments in zebrafish. The difficulty of solubilizing melanin is a known shortcoming of current clearing approaches (13, 27) and prompted us to include the pretreatment step with 3% hydrogen peroxide to achieve full depigmentation of melanin-containing samples (Fig. 1A). Pretreatment solely with hydrogen peroxide efficiently bleached melanin, but not the xanthophore pigments of pteridine and carotenoid nature in zebrafish fins (Fig. 1D), and also diminished pigmentation in the axolotl eye (fig. S1F). By contrast, combining this peroxide treatment with the established acetone and Solution-1(.1) treatment, as well as appropriate RI matching, preserved sample morphology and yielded the highest levels of tissue transparency as assessed on a 1951 United States Air Force resolution test chart (USAF chart) (Fig. 1E and fig. S1G). As the use of peroxide damages signal from transgenic fluorophores, strategies to relabel these fluorophores are presented in a later section. For RI matching, we used Solution-2 containing meglumine diatrizoate; for larger specimens including lipid-rich tissues, this solution was further modified by supplementing with N′-methylnicotinamide and antipyrine (referred to as Solution-2.2), in line with published strategies to raise RI without causing tissue deformation (27).
Together, these results establish that DEEP-Clear is able to efficiently reduce poorly soluble pigments even in large specimens that would otherwise be impossible to use for deep imaging. A qualitative survey of five published methods, applied to specimens of all four clades (fig. S2A), and a systematic quantification of light transmittance through the eyes of cleared and noncleared samples (fig. S2B) provide experimental support for the notion that DEEP-Clear can tackle a spectrum of pigments for which existing methods are not optimized.
Preservation of fine structures across scales and visualization of transgenic signals
We next investigated (i) whether endogenous structures and molecules were preserved in acetone pretreated samples, (ii) whether they could be relabeled in cases where a peroxide bleaching step was included, and (iii) whether fine tissue structures remained intact.
While extended acetone treatment is considered to denature fluorescent proteins, we noted that acetone-treated transgenic zebrafish and mice (fig. S3, A to C) retained their respective fluorescent protein fluorescence: We detect similar or slightly reduced fluorescent signal intensity yields by comparing untreated samples with samples exposed to acetone overnight at −20°C for brn3c::mGFP and HuC::Gal4; UAS::syp-GFP zebrafish and Thy1-YFP-H mice (fig. S3, A to C). In DEEP-Clear–treated pMos{rops::egfp}vbci2 adult worms (Fig. 2A), we could visualize the projection path of enhanced GFP–positive (EGFP+) eye photoreceptor cells (Fig. 2B). Similarly, light-sheet microscopy on complete worms was able to resolve both cell bodies and individual projections of the peripheral EGFP+ cells from individual parapodia onto and along the fibers of the ventral nerve cord of the trunk (Fig. 2, C and D) (42). As signal stability is a relevant aspect both for imaging and sample archiving, we also used DEEP-Clear–treated worm specimens to assess whether EGFP signal could be effectively preserved on a long-term basis. These analyses revealed that, even after several weeks, EGFP+ structures could clearly be distinguished from autofluorescence, exhibiting only limited decay when compared to the first day of recording (fig. S4, A to D). Although pigmentation is no relevant challenge in analyzing mammalian brain samples, we used hemispheres of a Thy1-YFP-H transgenic mouse brain to assess imaging of a large specimen. In this experiment, DEEP-Clear treatment allowed for imaging fluorescent cell bodies across close to 6000 μm of depth, comparable to the established CUBIC approach (fig. S5, A to E) (26). To investigate whether the use of peroxide in the context of melanized sample processing was compatible with signal relabeling, we performed anti-GFP stainings in respective DEEP-Clear–processed brn3c::mGFP zebrafish samples of different developmental stages [6, 10, 17, and 23 days post fertilization (dpf); fig. S6, A to D]. Anti-GFP immunohistochemistry allowed us to visualize the characteristic projections of retinal ganglion cells to the optic tectum (fig. S6, B to D) (43). To address whether finer, subcellular structures would be preserved if a peroxide treatment step was included in the DEEP-Clear procedure, we used HuC::Gal4; UAS::syp-GFP specimens that express a GFP fusion protein of the presynaptic vesicle marker synaptophysin (44). Consistent with previous work on the presence of this marker in arborizations of neurons during tectal development (44, 45), our analyses revealed a dense mesh of punctate structures in the tectum of 3 dpf larvae (fig. S6E). Similar puncta were also found in the mid-hindbrain region and the adjacent cerebellum (fig. S6F).
Fig. 2. Global and focused insight into the adult annelid nervous system.
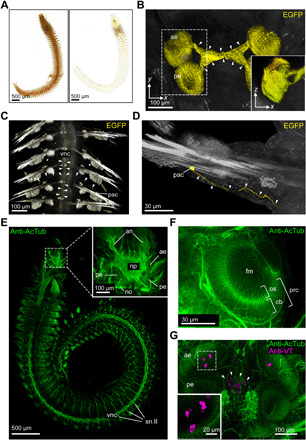
(A) Comparison of uncleared (left) and DEEP-Clear processed (right) bristle worm specimens. (B) Imaging of EGFP signal in photoreceptors of DEEP-Clear–processed pMos{rops::egfp}vbci2 animals by light-sheet microscopy. Arrowheads indicate projections from the eyes into the central-brain neuropil. Inset shows the XZ projection of the boxed area. (C and D) EGFP+ parapodial cell bodies and their projections (arrowheads) into and along the ventral nerve cord as visualized by light-sheet microscopy. (E to G) Anti-acetylated alpha-tubulin (anti-AcTub) immunolabeling revealing the annelid nervous system, visualized either by light-sheet microscopy [(E) whole animal, including the stereotypical trunk nervous system; inset showing major structures of the brain] or confocal microscopy [(F) anterior eye, with photoreceptor cells exhibiting segmentation into outer segments protruding into the central filling mass and basal cell bodies; (G) brain, with the inset showing a magnification of the boxed region of the anterior eye]. The specimen in (G) shows a colabeling by anti-Vasotocin (anti-VT) immunohistochemistry, and arrowheads indicate VT+ puncta (putative dense core vesicles) in deep neurite projections. Corresponding VT+ cell bodies can be seen in close proximity to the anterior eyes. an, antennal nerve; cb, cell body; fm, filling mass; no, nuchal organ; np, neuropil; os, outer segment; pac, parapodial receptor cells; ae, anterior eye; pe, posterior eye; prc, eye photoreceptor cell; sn.II, segmental nerve II; vnc, ventral nerve cord.
Systemic and focal analyses of postlarval invertebrate nervous systems
As endogenously expressed fluorophores are restricted to more established laboratory models, we performed a series of additional experiments to assess whether DEEP-Clear was also compatible with various immunohistochemically labeled epitopes. We focused most of our analyses on the nervous system of the respective species because the complexity of the nervous system presents a particular challenge to classical, section-based labeling efforts, as neural projections extending over different sections are very difficult to reconstruct. Moreover, the nervous system is of special relevance for evolutionary and comparative analyses.
For exploring the compatibility of DEEP-Clear with IHC analyses, we took advantage of antibodies that recognize conserved or specific epitopes. Across all species, we used a monoclonal antibody directed against acetylated alpha-tubulin that has been widely used to visualize animal nervous systems (46–51). In the axolotl, we also used the TuJ1 antibody that is directed against a neuron-specific beta III tubulin isoform (TUBB3) (52–54). We complemented these overall neuroanatomical analyses by performing immunohistochemistry against more specifically expressed epitopes. When required, we used a modified solution for RI matching (Solution-2.1) in which we included four parts of Solution-2 and one part of VECTASHIELD or embedded the samples in pure VECTASHIELD for preventing fluorophore bleaching. For analyzing global neuroanatomy, we used light-sheet microscopy, complementing these analyses with confocal analyses to study cellular or subcellular details.
In Solution-1–treated adult P. dumerilii specimens immunostained with anti-acetylated tubulin antibody, light-sheet microscopy revealed uniform labeling of both cephalic and noncephalic parts of the central nervous system, as well as the diagnostic segmental nerves of the annelid peripheral nervous system (Fig. 2E and movies S1 and S2) (55). Closer inspection of the cephalic region yielded detailed insight into brain structures and a high-resolution view of the adult eye cups (Fig. 2F). The fine structure of the eye that we obtained by confocal microscopy of complete heads matches well with the models previously obtained by laborious analyses of tissue sections by light and electron microscopy (56, 57). We further performed immunostaining with an antibody directed against a P. dumerilii neuropeptide, Vasotocin (VT). In mature adult worms, detection of this antibody consistently labeled punctate structures in a meshwork of neurites reaching deep into the median brain, which prior work has established as the release site of the animal infracerebral gland, where neurohormones are likely emitted into the circulating system (58). We also detected respective VT+ cell bodies in direct proximity to the ventral face of the anterior eye cups, an area that is difficult to image without DEEP-Clear procedure due to massive amount of shielding eye pigments (Fig. 2G).
We next investigated squid hatchlings that had been depigmented and cleared using Solution 1.1 (Fig. 3A), immunolabeled, and lastly matched to an RI of 1.45 with antifade-mounting medium (VECTASHIELD). Despite the size of the specimens (around 3 mm × 1 mm × 1 mm), light-sheet analysis revealed a deep antibody staining of the mitotic cell marker phosphorylated histone H3 (PH3) (Fig. 3B) (59). These analyses revealed, among other domains, mitotic cell clusters usually masked by pigments (Fig. 3B, insets 1 and 2, and movie S3). Furthermore, we could visualize and compare, in undissected specimens, characteristic regions of the complete longfin inshore and the Hawaiian bobtail squid nervous system (Fig. 3C; fig. S7, A to C; and movies S4 and S5) (55). As for octopods, the sepiolid optic lobes are known to have an elaborate structure, with the deep medulla exhibiting large aggregates of perikarya interrupted by thicker fiber tracts. Owing to the strong pigmentation of the mantle, this elaborate, tree-like tissue structure has previously mainly been analyzed by histological analysis of serial sections (55, 60, 61). First accounts of its formation during E. scolopes or Sepia pharaonis development, respectively, have thus been provided using x-ray micro–computed tomography (micro-CT) or magnetic resonance imaging (MRI) (62, 63). Our squid specimens not only revealed individual projections of sensory cells in the retina (fig. S7, B and C) but also allowed us to record high-resolution image stacks reaching deep into the optic lobe. These recordings provided direct insight into the architecture of the optic lobe, including the meshwork of fiber tracts in the deep medulla (Fig. 3, D and E, and movie S6). Moreover, they revealed individual, fine fibers branching between these tracts (arrowheads in Fig. 3E) that are likely part of the tangential arborization of higher-order processing neurons identified in cephalopod squid and octopus lobe sections (60, 61). Together, these results corroborate the versatility of the DEEP-Clear approach and its suitability to provide global and local insight into invertebrate nervous systems using light microscopy at unprecedented level.
Fig. 3. Insight into the nervous system structures in whole-mount specimens of squid hatchlings.
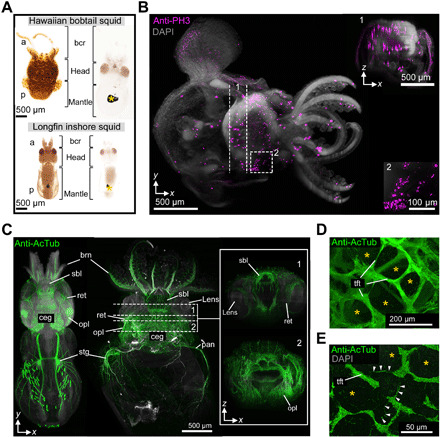
(A) Comparison of uncleared (left) and DEEP-Clear–processed (right) squid hatchlings (anterior up), documenting the removal of eye and body pigments (asterisk indicates remaining ink sac melanin). (B) Light-sheet acquisition of anti–phosphohistone H3 immunolabeling (bobtail squid, dorsal view, anterior to the right; dashed lines and box demarcate the area of the XZ slice shown in inset 1 and the enlargement in inset 2). DAPI, 4′,6-diamidino-2-phenylindole. (C to E) Visualization of the nervous system using anti-AcTub labeling. (C) Light-sheet acquisition (dorsal views, anterior up) in longfin inshore squid (left) and the Hawaiian bobtail squid (right). Inset shows two XZ projections of the respective areas (1 and 2) dashed in the XY view. (D and E) Confocal views of the deep medulla of the bobtail squid optic lobe, revealing the diagnostic structure of thick fiber tracts embracing perikarya (asterisks) and fine transversal fibers (arrowheads) of the Hawaiian bobtail squid. a, anterior; bcr, brachial crown; brn, brachial nerves; ceg, cerebral ganglion; opl, optic lobe; p, posterior; pan, pallial nerve; ret., retina; sbl, superior buccal lobe; stg, stellate ganglion; tft, thick fiber tract.
Whole-mount deep-tissue labeling and imaging in postlarval vertebrates
For our further experiments in zebrafish, we deliberately used larger specimens of around 12 mm × 1 mm × 1 mm (3 to 5 weeks of age), which are characterized by strong body pigmentation, and exhibit a strongly melanized RPE. Specimens were depigmented and cleared as established above (Figs. 1, A and D and 4A). Even in anti-acetylated alpha-tubulin–stained specimens of around 6 and 12 mm in length, light-sheet analysis revealed labeling of both the central and peripheral nervous systems (Fig. 4, B and C, and movie S7). While the neuroanatomy of the adult zebrafish brain has already been characterized in detail using techniques such as histology (64) and super-resolution track density imaging (65), the characterization of eyes and cranial nerves has mainly been restricted to larval and early postlarval stages (51, 66–68). As illustrated by a lateral view of the labeled juvenile head (Fig. 4B, inset, and movie S8), both eyes and cranial nerve branches, including the anterior lateral line system, can be well resolved in the DEEP-Clear–treated juveniles (Fig. 4B, inset), extending the stages accessible to these investigations. Likewise, we resolved the star-like arrangement of individual bundles of retinal ganglion cells whose projections exit the optic disk to form the optic nerve (Fig. 4B, inset). DEEP-Clear thus provides in-depth insight into retinal structures at stages that are traditionally hard to access by light microscopy due to the shielding effect of the RPE. As for bristle worms and squid, we also used DEEP-Clear in combination with additional antisera. Using an anti-serotonin antibody, we could visualize serotonergic cells in the retina, spinal cord, and raphe areas (Fig. 4C and movie S9). Likewise, anti–phosphohistone H3 immunolabeling revealed a number of putative mitotic cells across the juvenile head (Fig. 4D and movie S10).
Fig. 4. DEEP-Clear–enabled analysis of the eyes and cranial nervous system of postlarval zebrafish.
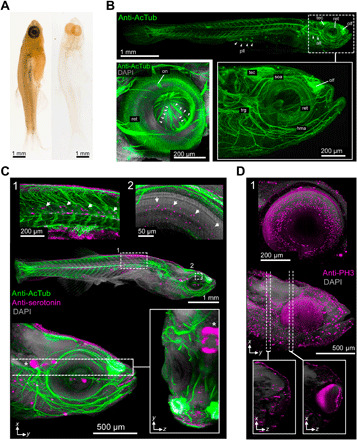
(A) Comparison of uncleared (left) and DEEP-Clear–processed (right) zebrafish juveniles. (B and C) Visualization of the nervous system using anti-AcTub immunohistochemistry (lateral views, anterior to right). (B) Light-sheet acquisition of central and peripheral nervous system (arrowheads indicating lateral line), with the right inset showing a close-up of the head. Left inset: confocal view of retina and bundles of retinal ganglion cells (arrowheads) exiting through the optic disk to form the optic nerve. (C) Light-sheet acquisition of a specimen colabeled by anti-serotonin immunohistochemistry. Magnifications in the top insets show serotonergic cells in (1) the spinal cord and (2) in the eye. The lower image shows a sagittal view of the head and a corresponding YZ slice (inset), with an asterisk close to serotonergic cells of the raphe area. (D) Sagittal view of a light-sheet–acquired juvenile head showing PH3+ cells. Lower insets show XZ projections of the indicated areas, revealing PH3-labeled cells deep in the brain, inset (1) provides an enlarged view of the right eye. All, anterior lateral line; hma, hyomandibular arch; soa, supraorbital arch; tec, tectum; trg, trigeminal ganglion; olf, olfactory epithelium; on, optic nerve; pll, posterior lateral line. Photo credit: Marko Pende, Medical University of Vienna.
Similar to zebrafish, analyses of the neuroanatomy of the axolotl have mainly been restricted to tissue sections (52) (https://msu.edu/course/zol/402/atlas/). After subjecting cleared juvenile specimens (Fig. 5A) to anti-TUBB3 immunohistochemistry, even large axolotl specimens (25 mm in length) allowed light-sheet visualization of deep tissue–labeled neurons (Fig. 5B and movie S11). This allowed us to acquire complete views of the axolotl nervous system (Fig. 5B), distinguish major regions of the axolotl brain and cranial nerves (Fig. 5C), and visualize the exiting of neurons through the optic disk, as in zebrafish (Fig. 5D), or the olfactory system (Fig. 5E and movie S12).
Fig. 5. Whole-body immunolabeling of the juvenile axolotl nervous system.
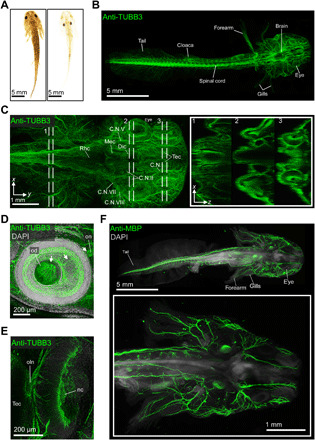
(A) Comparison of uncleared (left) and DEEP-Clear–processed (right) axolotl juveniles (around 3 months of age). (B to E) DEEP-Clear–processed juveniles stained by anti–beta III tubulin (anti-TUBB3) to mark the nervous system. (B) Overview of the juvenile anatomy and nervous system in a DEEP-Clear–processed specimen imaged with light-sheet microscopy. (C) Close-up of the head region, revealing the major brain neuroanatomy and branches of the cranial nerves. The inset shows three XZ projections taken at the indicated positions. (D) Focus on the eye region of a specimen revealing the projection of retinal ganglion cells (arrows) through the optic disk to form the optic nerve. (E) Axolotl nose showing innervation of the olfactory nerve. (F) Labeling of central and peripheral nervous system using anti-MBP antibody and 4′,6-diamidino-2-phenylindole. Inset shows higher magnification of peripheral nerves. nc, nasal cavity; oln, olfactory nerve; od, optic disk; Rhc, rhombencephalon; Mec, mesencephalon; Dic, diencephalon; C.N., cranial nerve. Photo credit: Marko Pende, Medical University of Vienna
To further assess immunolabeling efficiency of large heterogeneous tissue, as well as preservation and accessibility of different cellular compartments, we performed analogous immunostainings with antibodies against the (i) intracellular membrane-associated myelin basic protein (MBP; Fig. 5F), (ii) acetylated tubulin (fig. S8A), (iii) myosin heavy chain (fig. S8B and movies S13 and S14), (iv) phosphorylated histone H3 (PH3; fig. S8C), and (v) the transcription factors sex-determining region Y-related high-mobility group box 9 (SOX9; fig. S8D) and paired-related homeobox protein 1 (PRRX1) (fig. S8, E and F). In all cases, our results were in line with the expected patterns. Anti-MBP immunohistochemistry highlighted presumptive oligodendrocytes and Schwann cells throughout the specimen and the spinal cord (Fig. 5F) (51). Similar to anti-TUBB3 (Fig. 5, B to E), anti-acetylated tubulin immunohistochemistry highlighted the overall neuroanatomy, along with additional small structures in the gills (fig. S8A) that likely reflect ciliated cells found to cover the external gills of amphibians (69). The transcription factors PRRX1 and SOX9 that we investigated in the context of the growing limb showed nuclear expression in the connective tissue and chondrocytes, respectively, in agreement with previous data (70–72), while anti-myosin heavy chain immunohistochemistry highlighted skeletal musculature across the specimen. Together, these results provided direct evidence that DEEP-Clear processing is compatible with the labeling and imaging of epitopes across tissues and subcellular compartments, providing a versatile tool for future studies.
Compatibility with multiple active labeling approaches (IHC, RNA-FISH, and EdU)
In a last set of experiments, we assessed the compatibility of DEEP-Clear with two additional active labeling techniques: picolyl azide-based click chemistry and RNA in situ hybridization. For these experiments, we focused on the adult eyes of P. dumerilii, each of which has an everted, cup-shaped retina. A dense layer of pigments separates the outer segments of the photoreceptors from their basal parts. Like the retina of lower vertebrates (fishes and amphibians), the eye of P. dumerilii is known to grow continuously during the animal’s lifetime, but the details of this growth have not been experimentally assessed yet. We therefore incubated adult animals for 24 hours in seawater supplemented with EdU, a thymidine analog labeling S phase nuclei, and then subjected them to DEEP-Clear. We combined the detection of EdU with in situ hybridization for r-opsin1 expression, detected using a fluorescent tyramide–based amplification strategy (73). A part of these analyses was performed in the background of the aforementioned pMos{rops::egfp}vbci2 strain, allowing us to use EGFP as an independent marker. Cell proliferation was not only consistently detected in the medial brain (asterisks in Fig. 6, A to C) but also revealed within the eye cup (arrows in Fig. 6, D and E). In all tested immature animals and a subset of metamorphosing specimens, EdU-labeled cells were found in the area closest to the pupil, which is normally inaccessible to light microscopy. Our finding is consistent with the hypothesis that the worm retina extends by apposition of new photoreceptor cells along its marginal zone (57), similar to the ciliary marginal zone in teleosts (74).
Fig. 6. Retinal growth patterns and molecular signatures of annelid eye photoreceptors.
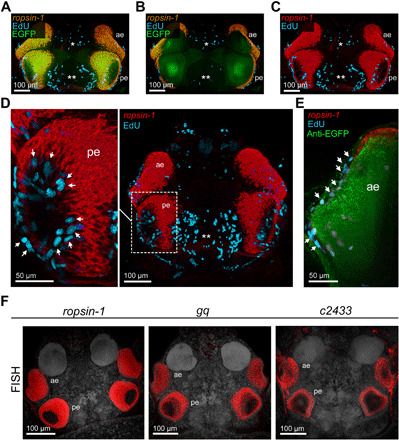
(A to C) Codetection of incorporated EdU (magenta), riboprobes against ropsin-1 (overlay with EGFP is yellow, and pure signal is red), and EGFP (green) in premature pMos{rops::egfp}vbci2 bristle worms. Single and double asterisks indicate regions of cell proliferation in the anterior and posterior ganglionic region, respectively. (D and E) Similar codetection of EdU (magenta), ropsin-1 (red), and EGFP epitopes (green), including close-ups of the posterior (D, left) and anterior (E) eye region. Arrows point to the proliferative cells in either eye. (F) Fluorescent detection of riboprobes against ropsin-1 (left), gq (middle), and tmdc/c2433 (right) in comparative RNA whole-mount hybridizations, revealing expression of all three genes in eye photoreceptors of the worm head. All dorsal views, anterior to the top. ae, anterior eye; pe, posterior eye.
The detection of r-opsin1 transcripts in photoreceptors suggested that DEEP-Clear–processed specimens could be used to explore the expression of additional genes within eye photoreceptor cells, while that was previously impeded by the strong pigmentation of the eyes. We therefore performed analogous RNA in situ hybridizations with riboprobes against gq, a gene encoding a Gq alpha subunit that we previously found to be correlated with r-opsin1+ cells in the trunk (42), and tmdc/c2433, a gene encoding an uncharacterized putative transmembrane protein that we recently identified as a candidate for the EGFP cell-specific transcriptome. Fluorescent detection of riboprobes, followed by DEEP-Clear processing, revealed that gq, as well as the previously unknown transcript tmdc/c2433, was clearly localized to the four adult eyes of the worm, similar to r-opsin1 transcripts (Fig. 6F). This result confirms gq and tmdc/c2433 as parts of the eye photoreceptor signature and provides proof of concept for the compatibility of DEEP-Clear with systematic in situ hybridization assays. We independently confirmed this compatibility also in the zebrafish using a riboprobe directed against a melanopsin gene (opn4.1) expressed in the 6 dpf larval eye. Our results in the fish reproduce the previously reported expression of this gene in horizontal and some photoreceptor cells (75, 76) using the tyramide signal amplification (TSA)/Cy3 detection method bypassing the need to perform stainings on tissue sections (fig. S9). Together, these results support that DEEP-Clear can be readily combined with RNA in situ hybridization and thus a technique that is broadly used not only in standard developmental model species but also typically by researchers exploring new and unconventional models.
DISCUSSION
DEEP-Clear provides a novel, tailored clearing method that harnesses different strategies to remove distinct classes of poorly soluble pigments—including pterins, heme, ommochromes, carotenoids, and melanin—that are abundant in many animal tissues and across phyla. It effectively combines elements and reagents from protocols that have previously been categorized as either organic solvent– or hydrophilic reagent–based approaches (13, 33). This combination has synergistic effects, both by advancing depigmentation speed and removing pigments of different nature, thus shortening the overall time required for all clearing steps of DEEP-Clear in the targeted species to few hours, even for specimens that measure several centimeters in length. This synergy likely reduces the risks posed by extensive detergent, urea (32), and peroxide exposure. A possible limitation of the applicability of DEEP-Clear is thick bone structures, for which dehydration-based whole mouse clearing approaches offer solutions (18, 20). However, these techniques tend to compromise tissue morphology through unavoidable anisotropic shrinkage.
A second aspect that makes DEEP-Clear attractive for a broad use is its applicability to reference species of four distinct, species-rich animal clades. The removal of poorly soluble pigments eliminates a long-standing obstacle for the use of clearing, labeling, and imaging approaches outside the context of lowly pigmented structures like mouse brains, where clearing approaches have been most regularly used. The removal of melanin required peroxide treatment, generally known to cause tissue damage and loss of endogenous fluorophore signal. However, the short peroxide incubation times, which are facilitated by the aforementioned synergistic effects of the other clearing chemicals, as well as the possibility of relabeling bleached fluorescent proteins by IHC, represent suitable solutions. Therefore, DEEP-Clear provides imaging access to diverse tissues and organs that are shielded by natural pigmentation. Our work exemplifies this most directly for the analysis of visual systems across all investigated clades. These usually require researchers to label and image individual tissue sections, followed by reconstruction of their three-dimensional (3D) structures. DEEP-Clear bypasses the need for this time-consuming process by providing direct whole-mount access to these structures that favors easier analysis and higher throughput. Whereas we focused our analysis on nervous system aspects, DEEP-Clear also paves the way for other levels of analysis in the presented clades, such as bacterial symbiosis and bioluminescence, a fundamental postlarval event in squids (77, 78).
A third feature that makes DEEP-Clear attractive as a versatile toolbox is its compatibility with a panel of active molecular labeling techniques. Whereas fluorescent reporters (detected either directly or using IHC relabeling in peroxide-treated samples) require the ability of prior genetic manipulation, the compatibility of DEEP-Clear with immunohistochemistry provides direct benefits for the exploration of a broad panel of target organisms. Provided that suitable immunolabeling conditions are established, DEEP-Clear processing, combined with immunohistochemistry, can reveal structural features ranging from the tissue level (e.g., the architecture of the cephalopod optic lobe) down to subcellular structures such as presynaptic vesicles. Although tissue clearing relies on delipidation, we find membrane-associated epitopes such as MBP or synaptophysin EGFP to remain detectable, likely due to initial cross-linking. The possibility to label and image specific molecular features throughout a specimen distinguishes DEEP-Clear from label-free techniques such as x-ray micro-CT and MRI that have been used for imaging the nervous system of different cephalopod species (62, 63) and the adult zebrafish brain (65).
Compatibility with the detection of RNAs in intact tissues has been rather neglected in the development of current clearing protocols (34), with the exception of the 2ECi protocol (21), and the CLARITY (32) and SWITCH (79) methods. Unlike immunohistochemistry, which requires the availability of high-affinity antibodies for any given target, suitable riboprobes for in situ hybridization can be generated at low costs and for any gene identified in the respective species. DEEP-Clear will thus be applicable to explore gene expression patterns in new model systems and validate cellular fingerprints obtained by single-cell transcriptomics. The additional compatibility of DEEP-Clear with the detection of proliferative cells (EdU and picolyl azide–based click chemistry) opens possibilities for studying regeneration and development, an area that is directly relevant for three of the investigated model species (21, 71, 80, 81).
Last, beyond providing optical access to specific tissues and organs, DEEP-Clear also generates substantial potential for whole-animal analyses, provided that fixation conditions retain tissue integrity and that deep penetrance of labels is not prevented (e.g., by titration of antibodies by abundant epitopes at the periphery of specimens). In our experiments, we could observe deep immunolabeling even in large specimens for epitopes of different protein classes, for different subcellular localization (nuclear, membrane-associated, and cytoplasmic), for two major fixation techniques (Bouin’s fixative and PFA), and throughout different body structures. On the acquisition side, the removal of diverse pigments and the high level of overall transparency make DEEP-Clear–processed specimens compatible with advanced light-sheet microscopy. This imaging technique allowed us to capture 3D images of specimens with a thickness of close to 6000 μm and a length of several centimeters, pushing the size limits imposed by regular confocal analysis. Likewise, it offers substantially faster acquisition times and lower data amounts, by passing, or at least strongly limiting, the need to stitch many different acquisitions as they would be generated using separate confocal datasets from different parts of large specimens. Taken together, the speed and broad compatibility of DEEP-Clear remove existing restrictions in the use of tissue clearing and imaging approaches beyond more selective contexts and thereby provides a powerful tool for the exploration of novel model systems across scales.
MATERIALS AND METHODS
Animals
Mouse (Mus musculus)
Thy1-YFP-H mice (82) were bred and kept at the animal care center of the Medical University, Vienna. All experiments and animal handling were carried out in compliance with Austrian ethical guidelines.
Bristle worm (P. dumerilii)
Wild-type and pMos{rops::egfp}vbci2 worms were raised at temperatures between 18° and 20°C and kept in a 16:8-hour light-dark (LD) cycle. Animals were anesthetized using 7.5% MgCl2 (50%) (Roth, A537.1) and artificial seawater (50%) before fixation. All experiments and animal handling were carried out in compliance with Austrian ethical guidelines.
Hawaiian bobtail squid (E. scolopes) and longfin inshore squid (Doryteuthis pealeii)
Squid eggs were obtained from cultures from S. Nyholm (Department of Molecular and Cell Biology, University of Connecticut, Storrs, USA) and J. Foster (University of Florida, Space Life Science Lab, Merritt Island, USA). The Marine Biological Laboratory’s Cephalopod Breeding Initiative (Woods Hole, Massachusetts, USA, affiliated to University of Chicago) provided laboratory-reared specimens that were fixed on site. Egg clutches were shipped in well-oxygenated water to Vienna Schönbrunn Zoo. Eggs were kept in 26° to 27°C warm tanks at 12:12-hour LD cycles at the aquarium facility of the zoo. Live animals were maintained in accordance with the European general guidelines for cephalopod care and welfare (83).
Zebrafish (D. rerio)
Wild-type, brn3c:mGFP, and HuC::Gal4; UAS::syp-GFP fish were raised at 28°C and kept in a 16:8- or 14:10-hour LD cycle. All experiments and animal handling were carried out as per the local Austrian and European ethical guidelines (approved animal protocols GZ: 342445/2016/12 and GZ: BMWFW-66.006/0012-WF/II/3b/2014). Animals were anesthetized using 0.2 to 0.3% tricaine (abcr GmbH, AB142950) before fixation.
Axolotl (A. mexicanum)
Axolotls were raised at 20°C and kept in a 12:12-hour LD cycle. Husbandry followed established standards (84). Animals were anesthetized in 0.03% benzocaine (Sigma-Aldrich, E1501) before sacrifice. All experiments and animal handling were carried out as per the local Austrian and European ethical guidelines (approved animal protocol GZ: 9418/2017/12).
Solutions
Marine phosphate-buffered saline (PBS) consists of 50 mM sodium phosphate buffer with 0.5 M NaCl diluted in dH2O (pH 7.4). Solution-1 consists of 8 to 10% (v/v) THEED (Sigma-Aldrich, 87600-100ML), 5% (v/v) Triton X-100 (Roth, 3051.2), and 25% (w/v) urea (Roth, X999.2) in dH2O, e.g., 4 to 5 ml of THEED, 2.5 ml of Triton X-100, and 12.5 g of urea filled up to 50 ml with dH2O. Solution-1.1 consists of 8 to 10% (v/v) THEED (Sigma-Aldrich, 87600-100ML), 5% (v/v) Triton X-100 (Roth, 3051.2), and 5% (w/v) urea (Roth, X999.2) in dH2O. For Solution-2, a 50% (w/w) meglumine diatrizoate (Sigma-Aldrich, M5266) solution in PBS (pH 9 to 9.3; e.g., 10 g of meglumine diatrizoate added to 10 ml of PBS) was adjusted to an RI of 1.45 by further adding of meglumine diatrizoate. Solution-2.1 was prepared by mixing four parts of Solution-2 with one part of VECTASHIELD (Vector laboratories, H-1000). For Solution-2.2, 11 g of meglumine diatrizoate was added to 9 ml of PBS (pH 9 to 9.3) supplemented with 2 g of antipyrine (Sigma-Aldrich, A5882-100G) and 0.8 g of N′-methylnicotinamide (Santa Cruz Biotechnology, sc-295821A), with an RI adjusted to 1.48.
DEEP-Clear protocol and its combination with other techniques
Details on cloning, the detailed DEEP-Clear protocol in different clades, the combination of DEEP-Clear with immunostaining, and the combination of DEEP-Clear with RNA in situ hybridization and EdU labeling are listed in Supplementary Material and Methods.
Assessing the effect of solutions and comparison with established methods
For details on the assessment of the impact of acetone and for comparison with established techniques, see Supplementary Material and Methods.
Imaging and image processing
For details on light-sheet microscopy, fluorescence stereomicroscopy, laser-scanning confocal and two-photon microscopy, and image processing, see Supplementary Material and Methods.
Qualitative and quantitative assessments and statistical analyses
For quantification of the impact of acetone on depigmentation speed, the quantification of fluorescence signal stability, the qualitative and quantitative assessment of transparency, assessment of contrast, and quantification of signal transmission, see Supplementary Material and Methods.
Supplementary Material
Acknowledgments
We would like to acknowledge the following colleagues for providing help, input, or reagents: S. Nyholm (Department of Molecular and Cell Biology, University of Connecticut, Storrs, USA); J. Foster (University of Florida, Space Life Science Lab, Merritt Island, USA); The Marine Biological Laboratory’s Cephalopod Breeding Initiative (Woods Hole, Massachusetts, USA; affiliated to University of Chicago); A. Weissenbacher (Tiergarten Schönbrunn, Vienna); M. Wullimann (LMU, Munich); W. Haubensak, D. Keays, G. Nordmann, S. Nimpf, E. P. Malkemper, L. Landler, T. Hochstoeger, A. Pauli, K. Panser, and members of Elly Tanaka laboratory [Research Institute of Molecular Pathology (IMP), Vienna], T. Lendl and K. Aumayr (IMP/IMBA/GMI Biooptics Facility); J. Knoblich, D. Reumann, S. Bian, F. Bonnay, J. Sidhaye, O. Wueseke, J. Penninger, M. Orthofer, and M. Onji (Institute of Molecular Biotechnology/IMBA, Vienna); M. Borysova and A. Belokurov (Max Perutz Laboratories, Vienna); Max Perutz Laboratories Central Facility Biooptics-Light Microscopy (Vienna); K. Elsayad, L. Zhang, A. Kavirayani, and T. Engelmaier [Vienna BioCenter Core Facilities (VBCF), Vienna]; and Y. Albert Pan (Virginia Tech Carilion, Roanoke). We also acknowledge members of the Raible and Tessmar-Raible laboratories, as well as two anonymous reviewers, for detailed and constructive feedback that helped to improve this study. Funding: The study was funded by the Austrian Science Fund (FWF; projects P30035, P28970, P23102, P30686, I2972, F78, and M1674). The Advanced Microscopy Facility of the Vienna Biocenter Core Facilities (VBCF) acknowledges support from the City of Vienna and the Austrian Ministry of Science (Vision 2020). Author contributions: M.P. conceived parts of the study, developed DEEP-Clear protocols, performed light-sheet imaging and parts of confocal imaging, tested compatibility with endogenous fluorescence, and wrote parts of the manuscript. K.V. conceived parts of the study; tested compatibility of DEEP-Clear with immunolabeling, in situ hybridization, EdU labeling approaches; did parts of confocal imaging; and wrote parts of the manuscript. M.P. and K.V. performed the principal experiments for the study. H.S. contributed to the experiments in the Hawaiian bobtail squid and wrote parts of the manuscript. A.W.S. performed experiments to detect the proliferative compartment in the bristle worm eyes. P.M. characterized antibodies for axolotl tissues, contributed to the interpretation of experiments in axolotl, and wrote parts of the manuscript. S.S. was responsible for ultramicroscope and objective design. M.P.S.D. performed fluorescent in situ hybridization in zebrafish. K.B. developed imaging processing tools and performed statistics. R.R.-i-D. contributed to the identification of the eye photoreceptor signature genes. S.-C.P. assisted with DEEP-Clear experiments. P.P. assisted with confocal imaging and image processing. M.Z. performed and analyzed anti-VT immunostainings. O.S. contributed to the analysis of Hawaiian bobtail squid experiments. E.M.T. contributed to the analysis of axolotl experiments. F.R. conceived parts of the study, supervised the study, analyzed results, and wrote parts of the manuscript. H.-U.D. supervised parts of the study and provided light-sheet infrastructure and DEEP-Clear reagents. All authors read and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw microscopy datasets are available from the corresponding authors upon request. The sequence of P. dumerilii tmdc/c2433 has been submitted to GenBank (MK330892).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaba0365/DC1
REFERENCES AND NOTES
- 1.Bolker J., Model organisms: There's more to life than rats and flies. Nature 491, 31–33 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Brenowitz E. A., Zakon H. H., Emerging from the bottleneck: Benefits of the comparative approach to modern neuroscience. Trends Neurosci. 38, 273–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tessmar-Raible K., Arendt D., New animal models for evolution and development. Genome Biol. 6, 303 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren G., In praise of other model organisms. J. Cell Biol. 208, 387–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achim K., Pettit J.-B., Saraiva L. R., Gavriouchkina D., Larsson T., Arendt D., Marioni J. C., High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat. Biotechnol. 33, 503–509 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Albertin C. B., Simakov O., Mitros T., Wang Z. Y., Pungor J. R., Edsinger-Gonzales E., Brenner S., Ragsdale C. W., Rokhsar D. S., The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook C. E., Chenevert J., Larsson T. A., Arendt D., Houliston E., Lénárt P., Old knowledge and new technologies allow rapid development of model organisms. Mol. Biol. Cell 27, 882–887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liscovitch-Brauer N., Alon S., Porath H. T., Elstein B., Unger R., Ziv T., Admon A., Levanon E. Y., Rosenthal J. J. C., Eisenberg E., Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169, 191–202.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moroz L. L., Kohn A. B., Single-neuron transcriptome and methylome sequencing for epigenomic analysis of aging. Methods Mol. Biol. 1048, 323–352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belhaj K., Chaparro-Garcia A., Kamoun S., Patron N. J., Nekrasov V., Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Gilles A. F., Averof M., Functional genetics for all: Engineered nucleases, CRISPR and the gene editing revolution. EvoDevo 5, 43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander J. D., Joung J. K., CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tainaka K., Kuno A., Kubota S. I., Murakami T., Ueda H. R., Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol. 32, 713–741 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Dodt H.-U., Leischner U., Schierloh A., Jährling N., Mauch C. P., Deininger K., Deussing J. M., Eder M., Zieglgänsberger W., Becker K., Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Becker K., Jährling N., Saghafi S., Weiler R., Dodt H. U., Chemical clearing and dehydration of GFP expressing mouse brains. PLOS ONE 7, e33916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ertürk A., Becker K., Jährling N., Mauch C. P., Hojer C. D., Egen J. G., Hellal F., Bradke F., Sheng M., Dodt H.-U., Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Renier N., Wu Z., Simon D. J., Yang J., Ariel P., Tessier-Lavigne M., iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Pan C., Cai R., Quacquarelli F. P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Ertürk A., Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Hahn C., Becker K., Saghafi S., Pende M., Avdibašić A., Foroughipour M., Heinz D. E., Wotjak C. T., Dodt H.-U., High-resolution imaging of fluorescent whole mouse brains using stabilised organic media (sDISCO). J. Biophotonics 12, e201800368 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Jing D., Zhang S., Luo W., Gao X., Men Y., Ma C., Liu X., Yi Y., Bugde A., Zhou B. O., Zhao Z., Yuan Q., Feng J. Q., Gao L., Ge W.-P., Zhao H., Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masselink W., Reumann D., Murawala P., Pasierbek P., Taniguchi Y., Bonnay F., Meixner K., Knoblich J. A., Tanaka E. M., Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 146, dev166884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuwajima T., Sitko A. A., Bhansali P., Jurgens C., Guido W., Mason C., ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu T., Zhu J., Li Y., Ma Y., Wang J., Cheng X., Jin S., Sun Q., Li X., Gong H., Luo Q., Xu F., Zhao S., Zhu D., RTF: A rapid and versatile tissue optical clearing method. Sci. Rep. 8, 1964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami T. C., Mano T., Saikawa S., Horiguchi S. A., Shigeta D., Baba K., Sekiya H., Shimizu Y., Tanaka K. F., Kiyonari H., Iino M., Mochizuki H., Tainaka K., Ueda H. R., A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Susaki E. A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T. M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S., Abe T., Kiyonari H., Shimizu Y., Miyawaki A., Yokota H., Ueda H. R., Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Susaki E. A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H. R., Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Tainaka K., Murakami T. C., Susaki E. A., Shimizu C., Saito R., Takahashi K., Hayashi-Takagi A., Sekiya H., Arima Y., Nojima S., Ikemura M., Ushiku T., Shimizu Y., Murakami M., Tanaka K. F., Iino M., Kasai H., Sasaoka T., Kobayashi K., Miyazono K., Morii E., Isa T., Fukayama M., Kakita A., Ueda H. R., Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A., Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Hama H., Hioki H., Namiki K., Hoshida T., Kurokawa H., Ishidate F., Kaneko T., Akagi T., Saito T., Saido T., Miyawaki A., ScaleS: An optical clearing palette for biological imaging. Nat. Neurosci. 18, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Ke M.-T., Fujimoto S., Imai T., SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Ke M.-T., Nakai Y., Fujimoto S., Takayama R., Yoshida S., Kitajima T. S., Sato M., Imai T., Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep. 14, 2718–2732 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., Deisseroth K., Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B., Treweek J. B., Kulkarni R. P., Deverman B. E., Chen C.-K., Lubeck E., Shah S., Cai L., Gradinaru V., Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigouroux R. J., Belle M., Chédotal A., Neuroscience in the third dimension: Shedding new light on the brain with tissue clearing. Mol. Brain 10, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shamim G., Ranjan S. K., Pandey D. M., Ramani R., Biochemistry and biosynthesis of insect pigments. Eur. J. Entomol. 111, 149–164 (2014). [Google Scholar]
- 36.Figon F., Casas J., Ommochromes in invertebrates: Biochemistry and cell biology. Biol. Rev. Camb. Philos. Soc. 94, 156–183 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Konno A., Okazaki S., Aqueous-based tissue clearing in crustaceans. Zool. Lett. 4, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henning Y., Osadnik C., Malkemper E. P., EyeCi: Optical clearing and imaging of immunolabeled mouse eyes using light-sheet fluorescence microscopy. Exp. Eye Res. 180, 137–145 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Pende M., Becker K., Wanis M., Saghafi S., Kaur R., Hahn C., Pende N., Foroughipour M., Hummel T., Dodt H.-U., High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 9, 4731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viscontini M., Hummel W., Fischer A., Pigmente von Nereiden (Annelida, Polychaeten). 1., Vorläufige Mitteilung. Isolierung von Pterindimeren aus den Augen von Platynereis dumerilii (Audouin & Milne Edwards) 1833. Helvetica Chimica Acta 53, 1207–1209 (1970). [Google Scholar]
- 41.C. Hauenschild, A. Fischer, Großes Zoologisches Praktikum (Gustav Fischer, 1969). [Google Scholar]
- 42.Backfisch B., Veedin Rajan V. B., Fischer R. M., Lohs C., Arboleda E., Tessmar-Raible K., Raible F., Stable transgenesis in the marine annelid Platynereis dumerilii sheds new light on photoreceptor evolution. Proc. Natl. Acad. Sci. U.S.A. 110, 193–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao T., Roeser T., Staub W., Baier H., A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development 132, 2955–2967 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Niell C. M., Meyer M. P., Smith S. J., In vivo imaging of synapse formation on a growing dendritic arbor. Nat. Neurosci. 7, 254–260 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Meyer M. P., Smith S. J., Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J. Neurosci. 26, 3604–3614 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arendt D., Tessmar K., Medeiros de Campos-Baptista M.-I., Dorresteijn A., Wittbrodt J., Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development 129, 1143–1154 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Piperno G., Fuller M. T., Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101, 2085–2094 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shigeno S., Yamamoto M., Organization of the nervous system in the pygmy cuttlefish, Idiosepius paradoxus Ortmann (Idiosepiidae, Cephalopoda). J. Morphol. 254, 65–80 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Williams R. T., Bridges J. W., Fluorescence of solutions: A review. J. Clin. Pathol. 17, 371–394 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L. Bally-Cuif, P. Vernier, in Fish Physiology, S. F. Perry, M. Ekker, A. P. Farrell, C. J. Brauner, Eds. (Academic Press, 2010), vol. 29, pp. 25–80. [Google Scholar]
- 51.Brösamle C., Halpern M. E., Nogo–Nogo receptor signalling in PNS axon outgrowth and pathfinding. Mol. Cell. Neurosci. 40, 401–409 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Amamoto R., Huerta V. G. L., Takahashi E., Dai G., Grant A. K., Fu Z., Arlotta P., Adult axolotls can regenerate original neuronal diversity in response to brain injury. eLife 5, e13998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee M. K., Tuttle J. B., Rebhun L. I., Cleveland D. W., Frankfurter A., The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil. Cytoskeleton 17, 118–132 (1990). [DOI] [PubMed] [Google Scholar]
- 54.McHedlishvili L., Epperlein H. H., Telzerow A., Tanaka E. M., A clonal analysis of neural progenitors during axolotl spinal cord regeneration reveals evidence for both spatially restricted and multipotent progenitors. Development 134, 2083–2093 (2007). [DOI] [PubMed] [Google Scholar]
- 55.T. Bullock, G. A. Horridge, Structure and Function in the Nervous Systems of Invertebrates (San Francisco, Freeman, 1965). [Google Scholar]
- 56.Fischer A., On the structure and light-dark adaptation of eyes of the polychaete Platynereis dumerilii. Z. Zellforsch. Mikrosk. Anat 61, 338–353 (1963). [PubMed] [Google Scholar]
- 57.Fischer A., Brökelmann J., The eye of Platynereis dumerilii (Polychaeta): its fine structure in ontogenetic and adaptive change. Z. Zellforsch. Mikrosk. Anat. 71, 217–244 (1966). [PubMed] [Google Scholar]
- 58.Hofmann D. K., Regeneration and endocrinology in the polychaete Platynereis dumerilii: An experimental and structural study. Wilhelm Roux. Arch. Dev. Biol. 180, 47–71 (1976). [DOI] [PubMed] [Google Scholar]
- 59.Verduzco D., Amatruda J. F., Analysis of cell proliferation, senescence, and cell death in zebrafish embryos. Methods Cell Biol. 101, 19–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young J. Z., Regularities in the retina and optic lobes of octopus in relation to form discrimination. Nature 186, 836–839 (1960). [DOI] [PubMed] [Google Scholar]
- 61.Young J. Z., The central nervous system of Loligo. I. The optic lobe. Philos. Trans. R. Soc. B 267, 263–302 (1974). [DOI] [PubMed] [Google Scholar]
- 62.Kerbl A., Handschuh S., Nödl M.-T., Metscher B., Walzl M., Wanninger A., Micro-CT in cephalopod research: Investigating the internal anatomy of a sepiolid squid using a non-destructive technique with special focus on the ganglionic system. J. Exp. Mar. Biol. Ecol. 447, 140–148 (2013). [Google Scholar]
- 63.Liu Y.-C., Liu T.-H., Su C.-H., Chiao C.-C., Neural organization of the optic lobe changes steadily from late embryonic stage to adulthood in cuttlefish Sepia pharaonis. Front. Physiol. 8, 538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. F. Wullimann, B. Rupp, H. Reichert, Neuroanatomy of the Zebrafish Brain (Birkhäuser, 1996). [DOI] [PubMed] [Google Scholar]
- 65.Ullmann J. F. P., Calamante F., Collin S. P., Reutens D. C., Kurniawan N. D., Enhanced characterization of the zebrafish brain as revealed by super-resolution track-density imaging. Brain Struct. Funct. 220, 457–468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Higashijima S.-i., Hotta Y., Okamoto H., Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 20, 206–218 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Icha J., Kunath C., Rocha-Martins M., Norden C., Independent modes of ganglion cell translocation ensure correct lamination of the zebrafish retina. J. Cell Biol. 215, 259–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wan Y., Almeida A. D., Rulands S., Chalour N., Muresan L., Wu Y., Simons B. D., He J., Harris W. A., The ciliary marginal zone of the zebrafish retina: Clonal and time-lapse analysis of a continuously growing tissue. Development 143, 1099–1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nokhbatolfoghahai M., Downie J. R., The external gills of anuran amphibians: Comparative morphology and ultrastructure. J. Morphol. 269, 1197–1213 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Currie J. D., Kawaguchi A., Traspas R. M., Schuez M., Chara O., Tanaka E. M., Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev. Cell 39, 411–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., Currie J. D., Gray Camp J., Tanaka E. M., Treutlein B., Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nacu E., Gromberg E., Oliveira C. R., Drechsel D., Tanaka E. M., FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407–410 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Tessmar-Raible K., Steinmetz P. R. H., Snyman H., Hassel M., Arendt D., Fluorescent two-color whole mount in situ hybridization in Platynereis dumerilii (Polychaeta, Annelida), an emerging marine molecular model for evolution and development. BioTechniques 39, 460–464 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Johns P. R., Growth of the adult goldfish eye. III. Source of the new retinal cells. J. Comp. Neurol. 176, 343–357 (1977). [DOI] [PubMed] [Google Scholar]
- 75.Davies W. I., Zheng L., Hughes S., Katherine Tamai T., Turton M., Halford S., Foster R. G., Whitmore D., Hankins M. W., Functional diversity of melanopsins and their global expression in the teleost retina. Cell. Mol. Life Sci. 68, 4115–4132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matos-Cruz V., Blasic J., Nickle B., Robinson P. R., Hattar S., Halpern M. E., Unexpected diversity and photoperiod dependence of the zebrafish melanopsin system. PLOS ONE 6, e25111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McFall-Ngai M., Hawaiian bobtail squid. Curr. Biol. 18, R1043–R1044 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Nyholm S. V., McFall-Ngai M. J., The winnowing: Establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Murray E., Cho J. H., Goodwin D., Ku T., Swaney J., Kim S.-Y., Choi H., Park Y.-G., Park J.-Y., Hubbert A., McCue M., Vassallo S., Bakh N., Frosch M. P., Wedeen V. J., Seung H. S., Chung K., Scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 163, 1500–1514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C.-H., Poss K. D., Regeneration genetics. Annu. Rev. Genet. 51, 63–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gazave E., Béhague J., Laplane L., Guillou A., Préau L., Demilly A., Balavoine G., Vervoort M., Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev. Biol. 382, 246–267 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., Nerbonne J. M., Lichtman J. W., Sanes J. R., Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Fiorito G., Affuso A., Basil J., Cole A., de Girolamo P., D’Angelo L., Dickel L., Gestal C., Grasso F., Kuba M., Mark F., Melillo D., Osorio D., Perkins K., Ponte G., Shashar N., Smith D., Smith J., Andrews P. L. R., Guidelines for the care and welfare of cephalopods in research –A consensus based on an initiative by CephRes, FELASA and the Boyd Group, FELASA and the Boyd Group. Lab. Anim. 49, 1–90 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Khattak S., Murawala P., Andreas H., Kappert V., Schuez M., Sandoval-Guzmán T., Crawford K., Tanaka E. M., Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat. Protoc. 9, 529–540 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Becker K., Saghafi S., Pende M., Sabdyusheva-Litschauer I., Hahn C. M., Foroughipour M., Jährling N., Dodt H.-U., Deconvolution of light sheet microscopy recordings. Sci. Rep. 9, 17625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Costantini I., Ghobril J.-P., Di Giovanna A. P., Mascaro A. L. A., Silvestri L., Müllenbroich M. C., Onofri L., Conti V., Vanzi F., Sacconi L., Guerrini R., Markram H., Iannello G., Pavone F. S., A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 5, 9808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaba0365/DC1


