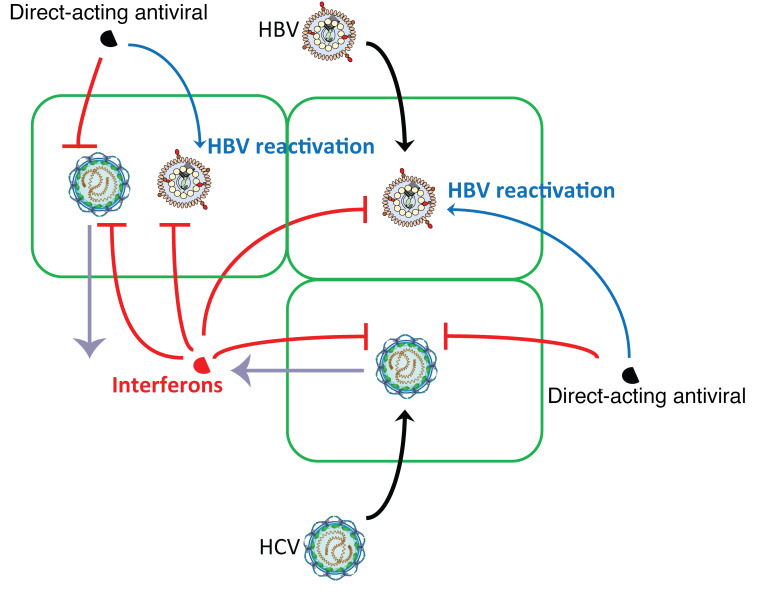
Abstract
In patients with HBV and HCV coinfection, HBV reactivation leading to severe hepatitis has been reported with the use of direct-acting antivirals (DAAs) to treat HCV infection. Here we studied the molecular mechanisms behind this viral interaction. In coinfected cell culture and humanized mice, HBV replication was suppressed by HCV coinfection. In vitro, HBV suppression was attenuated when interferon (IFN) signaling was blocked. In vivo, HBV viremia, after initial suppression by HCV superinfection, rebounded following HCV clearance by DAA treatment that was accompanied by a reduced hepatic IFN response. Using blood samples of coinfected patients, IFN-stimulated gene products including C-X-C motif chemokine 10 (CXCL10), C-C motif chemokine ligand 5 (CCL5), and alanine aminotransferase (ALT) were identified to have predictive value for HBV reactivation after HCV clearance. Taken together, our data suggest that HBV reactivation is a result of diminished hepatic IFN response following HCV clearance and identify serologic markers that can predict HBV reactivation in DAA-treated HBV-HCV–coinfected persons.
Keywords: Hepatology, Virology
Keywords: Hepatitis, Innate immunity
Introduction
Hepatitis B virus (HBV) and hepatitis C virus (HCV) together infect more than 300 million people worldwide. Due to the shared modes of transmission and epidemiological features, they frequently coexist in patients in highly endemic areas or among subjects at high risk of infection, such as persons who inject drug and men having sex with men. The worldwide prevalence of HBV-HCV dual infection is estimated to be 1% to 15%, which varies among geographical regions and study populations (1, 2). Patients coinfected with HBV and HCV have a high risk of more severe liver disease compared with monoinfected individuals (3, 4). Recently, with the advance of direct-acting antivirals (DAAs) in treating HCV, HBV reactivation has been reported in HBV-HCV–coinfected patients (5, 6). In some cases, fulminant hepatic failure occurred and required liver transplantation. Based on these events, the United States Food and Drug Administration (FDA) has issued warnings regarding the risk of HBV reactivation in coinfected patients receiving DAA (7). However, the mechanism of this reactivation remains largely unknown.
Although both HBV and HCV primarily infect hepatocytes, they belong to different virus families. HBV has a partially double-stranded DNA genome, which is transported into the nucleus to form covalently closed circular DNA (cccDNA). Viral replication occurs in the cytoplasm via reverse transcription that is mediated by virus-encoded polymerase (8). During its propagation, the innate immunity of hepatocytes does not efficiently detect the virus (9), which leads to a significantly muted interferon (IFN) response and IFN-stimulated gene (ISG) expression in the infected liver despite high levels of viral replication (10). IFN-α and its pegylated form are approved treatments for chronic HBV infection that function through both direct antiviral and immunomodulatory effects (11, 12). Studies on its direct anti-HBV effects have identified multiple ISGs targeting different steps of the HBV life cycle. For example, APOBEC3G (A3G) from the APOBEC3 family was initially shown to inhibit HBV replication through interfering with viral pregenomic RNA packaging or hypermutation of viral RNA (13, 14). Recently, A3A of the same family was shown to interact with core protein and target cccDNA for degradation (15). IFN-stimulated exoribonuclease gene of 20 kDa (ISG20) has been shown to inhibit HBV replication through degradation of HBV RNA both in vitro and in vivo (16, 17). In addition, a recent study demonstrated that a secreted ISG expressed by hepatocytes can block HBV entry to hepatocytes (18).
In contrast, HCV is an RNA virus that replicates exclusively in the cytoplasm. Unlike the stealth nature of HBV, HCV is recognized by host pattern-recognition receptors (PRRs) upon infection, resulting in a brisk IFN response marked by upregulation of hundreds of ISGs (19).
Many studies have attempted to address the potential interplay of HBV and HCV in coinfection. Available data from cell cultures are inconsistent; several reports proposed that HCV could directly impact HBV infection, with contradictory findings of either enhanced or suppressed HBV replication (20–23), while many other studies claimed HBV and HCV were able to replicate within the same hepatocyte without interference (24, 25). In clinical studies, reciprocal inhibition of HBV and HCV replication in the liver has been suggested (26).
To elucidate the mechanism of viral interference and treatment responses in HBV-HCV coinfection, here we studied various model systems ranging from cell culture to animal models to human studies, and demonstrate a complex interplay between HBV and HCV with the innate immune response at the nexus of virus-virus interactions.
Results
Suppression of HBV replication by HCV in coinfected hepatocytes.
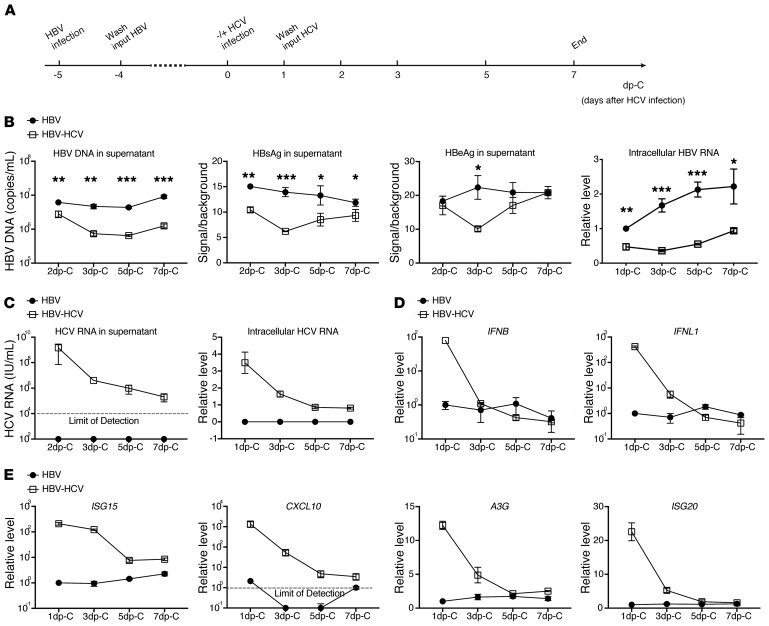
To closely mimic the natural host cell, we used primary human hepatocytes (PHHs) rather than hepatoma cell lines or other hepatocyte-derived systems as our in vitro model for HBV-HCV coinfection (27, 28). PHHs were first infected with HBV for 5 days and then either superinfected with HCV or allowed to persist as HBV monoinfection. Culture media were changed daily and kinetics of intracellular and extracellular viral markers were analyzed at indicated times (Figure 1A). Compared with HBV-monoinfected cells, HBV-HCV–coinfected cells produced fewer HBV virions, less HBV surface antigen (HBsAg) and e antigen (HBeAg) in the supernatant, and had reduced intracellular HBV RNA (Figure 1B). The differential HBV infection was observed as early as the second day after HCV infection and peaked on day 3 after HCV infection and gradually diminished, with HBeAg eventually rebounding to the same level as in HBV monoinfection, suggesting a transient negative effect of HCV on HBV replication. Consistent with this observation, HCV replication in PHHs lasted only for a short period of time, as demonstrated by declining intracellular and extracellular HCV RNA levels over time (Figure 1C). In PHH culture, HCV replication occurs transiently but is rapidly inhibited by the activated IFN response (29).
Figure 1. Kinetics of viral replication and host IFN responses in mono- and coinfected PHHs.
(A) Schematic representation of the experimental setting. PHHs (lot Hu1832) were either monoinfected with HBV for 12 days or superinfected with HCV for 7 days starting on day 5 after HBV infection. (B) HBV replication was analyzed by qPCR for extracellular HBV-DNA, ELISA for HBsAg and HBeAg, and RT-qPCR for intracellular HBV-RNA. (C) HCV replication was measured by RT-qPCR for intracellular and extracellular HCV RNA and concurrent host cell IFN response was measured by RT-qPCR for (D) expression of IFNB and IFNL1, and (E) expression of ISG15, CXCL10, A3G, and ISG20. Time of sample collection is indicated on the x axis. Antigen measurements by ELISA were compared to background level from noninfected cell culture medium. Normalized gene expression relative to that of the HBV-monoinfected cells on 1 day post HCV infection (dp-C) (set as 1) are shown as relative levels for IFNB, IFNL1, ISG15, A3G, and ISG20. For CXCL10, the detection limit was set as 1. Details of RT-qPCR can be found in Supplemental Methods. Means ± SD are shown. Unpaired t test was followed by Hochberg’s procedure to correct for multiple comparisons (B). *P < 0.05; **P < 0.01; ***P < 0.001. The results are representative of 3 separate experiments. Triplicate wells were used for each group.
The activated IFN response in our coinfected culture was evident, as IFNB and IFNL1 as well as several representative ISGs including C-X-C motif chemokine 10 (CXCL10) that is also known as IFN-γ–inducible protein 10 (IP10), IFN-stimulated gene 15 (ISG15), A3G, and ISG20 were substantially upregulated (Figure 1, D and E). With decreasing HCV replication, the IFN response also diminished. In contrast to the aforementioned differential IFN gene expression, transcription of genes indicative of hepatocyte functions like hepatocyte nuclear factor 4 (HNF4) and transferrin (TF) remained stable during the experiment, eliminating the concern that the PHHs became dedifferentiated or nonviable (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI135616DS1). We previously reported that HBV infection of PHHs does not activate the IFN response (9). To ascertain that activation of the IFN pathway indeed results from authentic HCV replication rather than contaminant in the inoculum, PHHs were infected with HCV in the same way overnight with the addition of sofosbuvir (SOF), a potent DAA against HCV. As expected, blockade of HCV replication greatly reduced IFN activation (Supplemental Figure 1B). Hence, we conclude that de novo HCV infection impairs preexisting HBV replication.
To perform the experiment in a reverse format, we infected PHHs first with HCV for 2 days before subsequent HBV infection (Supplemental Figure 2A). At the time of HBV inoculation, transcriptional analysis showed that IFNL1 and downstream ISG (ISG15, CXCL10, A3G, and ISG20) expression was readily detectable in HCV-infected but not mock-infected cells. Simultaneous SOF treatment effectively abolished induction of IFN and ISGs (Supplemental Figure 2B). Four days after HBV infection, assessment of HBV replication showed that HBV markers were substantially reduced in HBV-HCV–coinfected cells as compared with the HBV-monoinfected cells. In SOF-treated cells in which HCV infection and ISG induction were abolished, HBV replication attained a similar level as the HBV-monoinfected cells (Supplemental Figure 2C).
Thus, in coinfected PHH culture, HCV replication strongly suppresses concurrent HBV infection irrespective of the infection order. This suppression disappeared following either the natural decrease in HCV replication or DAA-induced HCV clearance.
Restoration of HBV replication by inhibiting IFN signaling in HBV-HCV–coinfected hepatocytes.
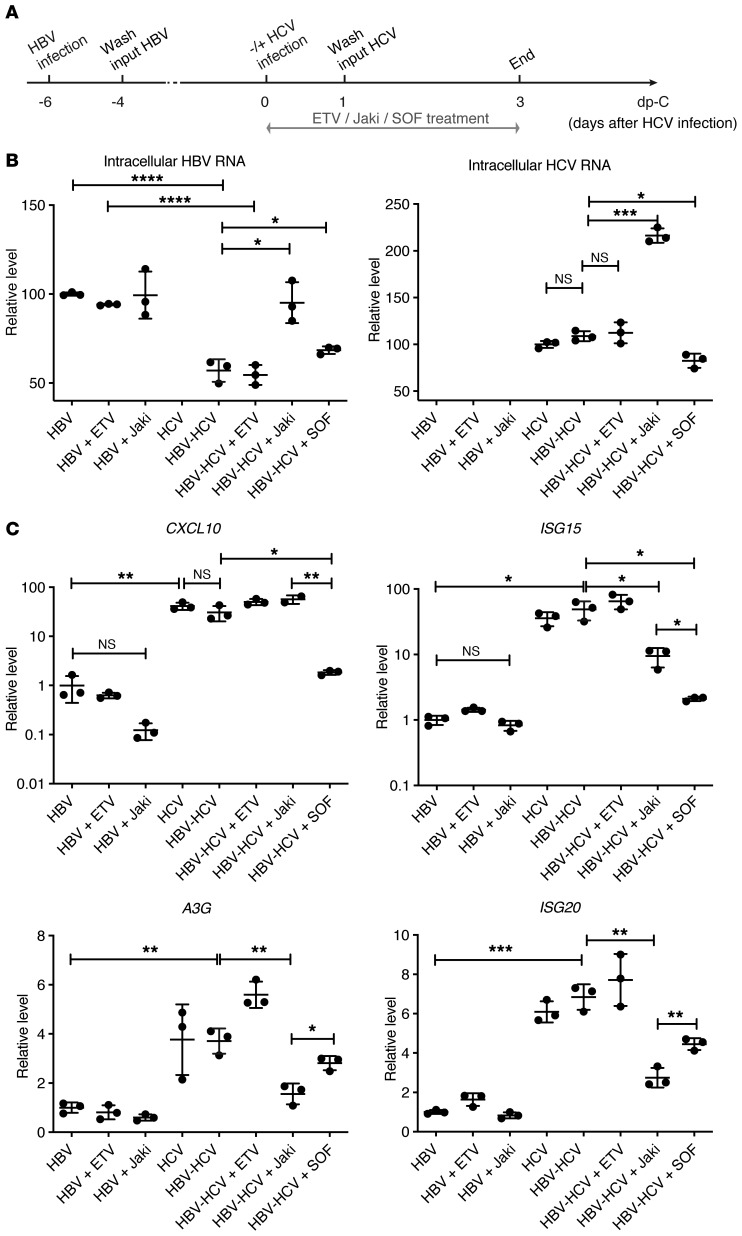
Based on the above findings, we hypothesized that HCV-induced IFN signaling may play a role in inhibiting HBV replication during coinfection. To test this hypothesis, HBV-infected PHHs were superinfected with HCV. Concurrently infected cells were treated with a broad-spectrum Janus kinase inhibitor (Jaki) that dampens ISG expression by inhibiting the Jak family (30), entecavir (ETV) that targets HBV reverse transcription, or SOF as indicated (Figure 2A). As expected, ETV and Jaki had no impact on HBV intracellular RNA levels, as seen in HBV-monoinfected culture. Intracellular HBV RNA in coinfected cells was suppressed to less than 60% of the monoinfected cell level (Figure 2B). When Jaki was added to the HBV-HCV–coinfected cells, HBV replication was fully restored to the same level as in untreated HBV monoinfection, while HCV infection in the same cell culture was also markedly enhanced. This finding strongly supports the idea that HBV suppression by HCV coinfection does not result from direct virus-virus interference. When SOF was added to the coinfection, HCV was significantly suppressed and HBV RNA levels increased. Finally, HBV replication did not have much impact on HCV replication. HCV RNA levels were comparable between HCV-monoinfected and HBV-HCV–coinfected cells with or without ETV treatment (Figure 2B).
Figure 2. Jaki or SOF treatment of mono- and coinfected PHHs.
(A) Schematic representation of the experimental setting. PHHs (lot Hu1574) were infected with HBV or mock infected for 6 days and followed by HCV superinfection at MOI = 1 TCID50/cell for 3 days. One micromolar ETV, 5 μM Jaki, or 10 μM SOF was added to mono- or superinfected cells for 3 days before harvesting the cells for analysis. (B) Relative HBV and HCV levels and (C) ISG expression were measured by RT-qPCR. Relative viral levels were determined by normalizing to monoinfected nontreated samples set as 100 and are shown in linear scale. Relative ISG levels were determined by normalizing to HBV-monoinfected nontreated samples (set as 1) and are shown in log10 scale (CXCL10 and ISG15) or linear scale (A3G and ISG20). Details of RT-qPCR can be found in Supplemental Methods. Means ± SD are shown. Unpaired t test was followed by Hochberg’s procedure to correct for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. NS, not significant. The results are representative of 3 separate experiments. Triplicate wells were used for each group.
To assess the status of the IFN response, transcription of CXCL10, ISG15, A3G, and ISG20 was examined (Figure 2C). In HBV monoinfection, ETV had no effect on ISGs but the CXCL10 level was slightly decreased by Jaki (not significant). The basal expression of certain ISGs may be dependent on a low-level signaling of the Jak/Stat pathway that can be suppressed by Jaki. In uninfected cells, ISG-like 2′-5′-oligoadenylate synthetase 3 (OAS3) but not CXCL10 (lower but not significant) was suppressed by Jaki treatment (Supplemental Figure 3). In HCV-infected cells, all measured ISGs were upregulated regardless of HBV infection status, and downregulated when SOF was applied. With Jaki treatment, ISG15, A3G, and ISG20 expression levels were all reduced compared with coinfected cells without treatment, while CXCL10 expression was not. Interestingly, the magnitudes of reduction for A3G and ISG20, both of which have well-known anti-HBV functions (14, 16), were greater in Jaki-treated coinfected cells compared with SOF-treated cells, potentially explaining the full rescue of HBV replication by Jaki treatment but only a partial restoration in SOF-treated coinfected cells. These findings further support the predominant role of the activated-IFN response to HCV in mediating HBV suppression.
Visualizing HBV-HCV coinfection at the single-cell level.
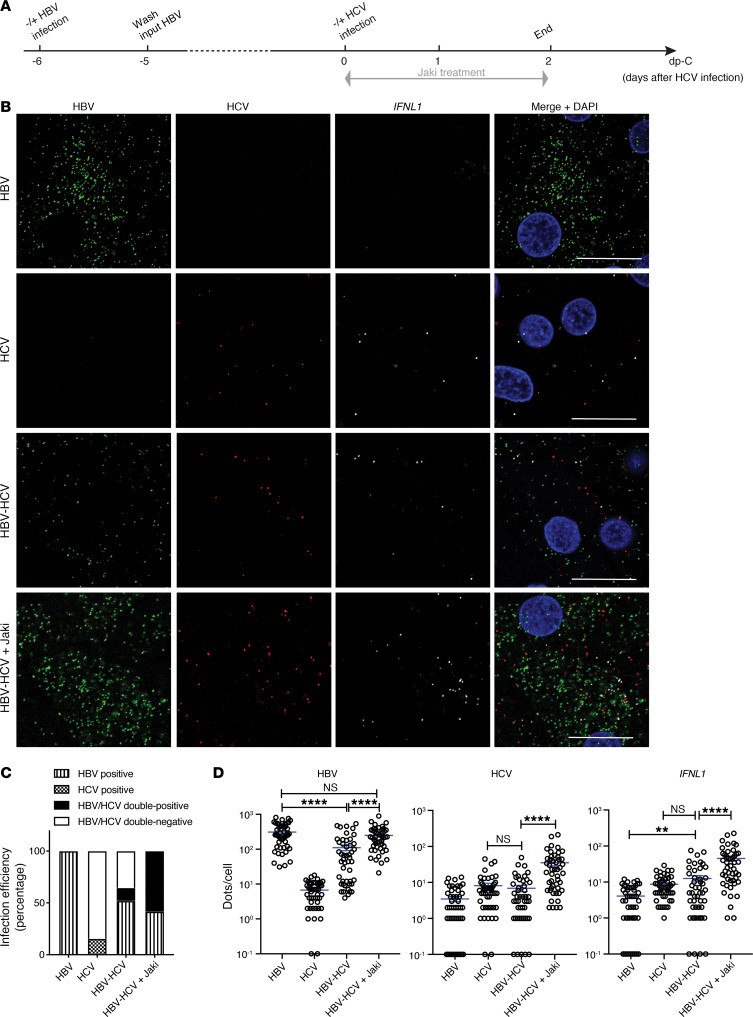
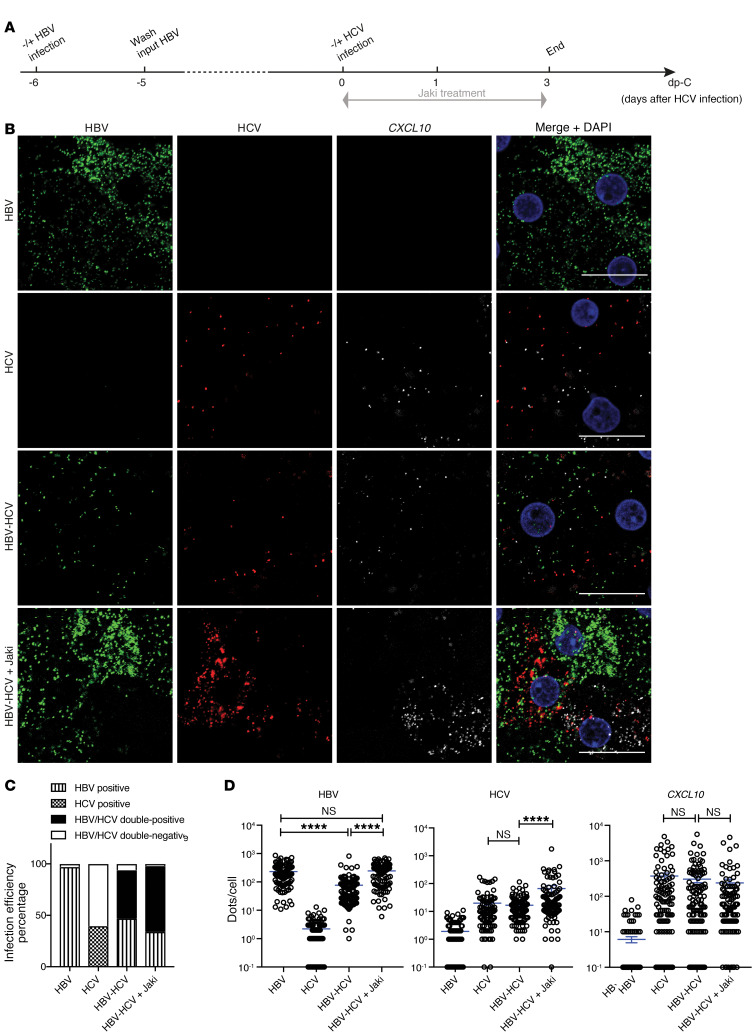
To directly visualize molecular events at the single-cell level, fluorescent multiplex in situ hybridization (RNAScope) was used to stain transcripts of target genes. We first validated the specificity of the assay using HBV- or HCV-monoinfected PHHs (Supplemental Figures 4 and 5). Hybridization to HBV or HCV probe sets resulted in specific signals mainly distributed in the cytoplasm of infected cells. As expected, HBV infection was not accompanied by positive CXCL10 signals, which were evident in HCV infection. Exogenous IFN-α treatment reduced signals of both HBV- and HCV-monoinfected cells and increased CXCL10 staining regardless of the infection status of the cell. Consistent with the preceding qPCR data (Figure 1), Jaki treatment did not change the HBV signal levels but greatly enhanced HCV signal levels in monoinfected cells. CXCL10 levels in the same HCV-infected culture were not visibly altered by Jaki treatment. Of note, HCV staining was much weaker than HBV staining, with only a small fraction of HCV-positive cells. Image quantification supported a low HCV infection efficiency of 20%, and this number increased to 46% with Jaki treatment.
Having confirmed the accuracy and reliability of RNAScope, we proceeded to study HBV-HCV coinfection. PHHs infected with HBV for 6 days were superinfected with HCV (Figure 3A). IFNL1 was stained to directly visualize the endogenous IFN response. As shown, nearly all the cells became positive for HBV signals after 8 days of monoinfection. In line with our previously published data, HBV-monoinfected cells did not induce IFNL1 (9). In HCV-infected cultures, IFNL1 expression was easily visible with its mRNA signals scattered around a cluster of HCV-infected cells (Figure 3B). HCV superinfection caused a general reduction of HBV signals in all the cells irrespective of whether the cells were positive for HCV or not, implying a paracrine signaling–mediated inhibition instead of a direct virus-virus inhibition. Importantly, when Jaki was applied to coinfected culture, not only were HBV signals restored but also HCV and IFNL1 signals were upregulated, making HBV and HCV double-positive cells readily visible. Quantification of infection efficiency by enumerating cells with virus signal dot numbers above those of controls revealed that HBV monoinfection was highly efficient and reached almost 100% positivity. This number decreased to 70% in HBV-HCV–coinfected culture, with 12% being HBV-HCV double positive. In coinfected culture, we did not detect HCV-positive-only cells. In HCV monoinfection, approximately 15% of cells were counted as HCV positive. This number increased to 58% when Jaki was added (Figure 3C). By quantifying dots per cells, we showed that the HBV infection level in individual cells was reduced by HCV superinfection. In the presence of Jaki in coinfected cells, the HBV level was restored to the HBV-monoinfection level, which was also accompanied by enhanced HCV and IFNL1 signals (Figure 3D).
Figure 3. In situ RNA detection of viral replication and IFNL1 expression in mono- and coinfected PHHs.
(A) Schematic representation of the experimental setting. PHHs (lot Hu1794) were first infected with HBV for 6 days, followed by HCV superinfection for 2 days in the presence of 5 μM Jaki as indicated. (B) Probe sets targeting HBV nucleic acids, HCV RNA, and IFNL1 mRNA were used to stain all the cells. Signals are shown as green for HBV, red for HCV, white for IFNL1, and blue for cell nuclei (counterstained by DAPI). Scale bars: 20 μm. (C) Virus-infected cells were defined as having signal dots higher in number than noninfected control cells. Infection efficiency for each virus was calculated as percentage of total number of cells counted in each condition. (D) Numbers of target dots in individual cells were plotted. Cells with 0 signal dots were input as 0.1. Four random views were analyzed. Details of image quantification can be found in Supplemental Methods. Unpaired t test was used for comparisons. Means ± SEM are shown. NS, not significant. **P < 0.01; ****P < 0.0001. The results are representative of 3 separate experiments.
To consolidate our data, we also stained for CXCL10 (Figure 4). After HCV superinfection, enhanced CXCL10 signals were found in both HCV-positive and -negative cells, similar to what was shown above for HCV monoinfection (Supplemental Figure 5). Blockade of ISG expression by Jaki clearly enhanced HCV signals. The number of CXCL10 dots did not change appreciably after Jaki treatment (Figure 4B), which is consistent with the qPCR data described above. Image quantification confirmed our visual inspection and further revealed that with this lot of PHHs, HCV-positive cells reached around 40%. In coinfected cells, 93% of cells were HBV positive, similar to the 97% HBV-positive cells in HBV-monoinfected cultures (Figure 4C). In this experiment, suppression of HBV infection was mainly reflected by a reduced number of signal dots in individual cells (Figure 4D).
Figure 4. In situ RNA detection of viral replication and CXCL10 expression in mono- and coinfected PHHs.
(A) Schematic representation of the experimental setting. PHHs (lot Hu1832) were first infected with HBV for 6 days, followed by HCV superinfection for 3 days in the presence of 5 μM Jaki as indicated. (B) Probe sets targeting HBV nucleic acids, HCV RNA, and CXCL10 mRNA were used to stain all the cells. Signals are shown as green for HBV, red for HCV, white for CXCL10, and blue for cell nuclei (counterstained by DAPI). Scale bars: 20 μm. (C) Infection efficiency for each virus was calculated as percentage of total number of cells counted in each condition. (D) Numbers of target dots in individual cells were plotted. Cells with 0 signal dots were input as 0.1. Six random views were analyzed. Details of image quantification can be found in Supplemental Methods. Unpaired t test was used for comparisons. Means ± SEM are shown. NS, not significant. ****P < 0.0001. The results are representative of 3 separate experiments.
Using this RNAScope technique, we were able to study cells that were positive for both HBV and HCV. The suppression of HBV replication by HCV coinfection was not limited to coinfected cells, but also involved cells negative for HCV, which is consistent with a paracrine suppression via induced IFN. When HCV-induced ISGs were blocked by Jaki, high levels of HBV and HCV genomes could be detected in the same hepatocytes, underscoring the absence of a direct interplay between HBV and HCV.
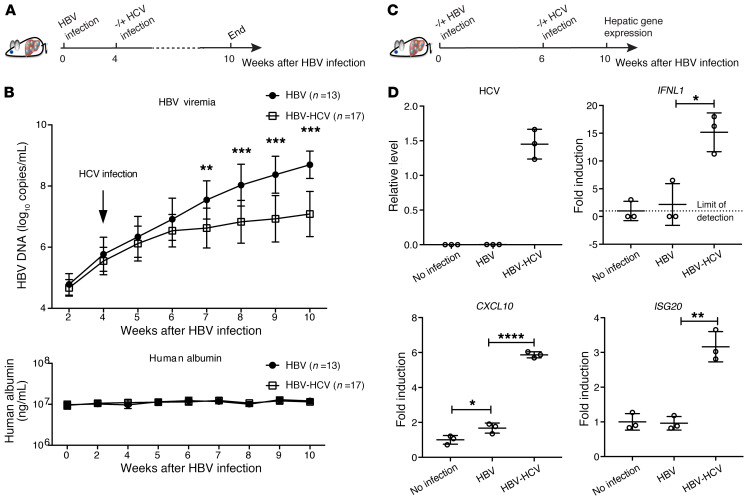
HCV suppresses HBV infection in coinfected humanized cDNA-uPA/SCID mice.
Although PHH culture is the most physiologically relevant cell culture model to study native hepatocytes, PHHs lose their differentiation status with time in vitro and thus cannot model long-term infection. To validate the observed interplay between HBV and HCV in PHH culture in vivo, we established HBV-HCV coinfection using PHH-transplanted cDNA-uPA/SCID mice, as mouse hepatocytes are not susceptible to HCV infection. Thirty humanized chimeric mice were inoculated intravenously with HBV patient serum (genotype C). Four weeks later, 17 of them were inoculated with HCV patient serum (genotype 1b) (Figure 5A). All mice were monitored continuously until week 10. Mice inoculated with HCV all became HCV viremic at week 3 after HCV infection (Supplemental Figure 6). Notably, at the same time, significantly lower levels of HBV DNA were observed in coinfected mice compared with HBV-monoinfected mice (Figure 5B). HCV replication in those mice reached a plateau around week 6 after HCV inoculation, with a mean titer of 8.4 × 106 copies/mL (range: 4.4 × 105 to 1.3 × 108 copies/mL), which is comparable to the HCV levels in HCV-monoinfected mice (31, 32). Similar levels of human albumin ensured no apparent difference in hepatocyte functionality between HBV-monoinfected mice and superinfected mice (Figure 5B).
Figure 5. Analysis of HBV-HCV coinfection in cDNA-uPA/SCID mice.
(A) Thirty mice were infected with HBV for 4 weeks before 17 mice were randomly selected for HCV inoculation. All mice were maintained until week 10 after HBV inoculation, as shown by the schematic representation. (B) Serum HBV genome (upper) and human albumin (lower) concentrations were determined at the indicated time. (C) In a separate experiment, 9 mice were engrafted with PHHs from the same donor. Three of them were not infected and served as no-infection controls. Six were infected with HBV and on week 6 after HBV infection, 3 mice were coinfected with HCV until week 10. (D) Normalized hepatic HCV levels are presented as relative levels. Negative HCV RNA was input as 0. Normalized human IFNL1, CXCL10, and ISG20 levels relative to those of no-infection control (set as 1) are shown as fold induction. Details of RT-qPCR can be found in Supplemental Methods. Unpaired t test was used alone (D) or followed by Hochberg’s procedure to correct for multiple comparisons (B). Means ± SD are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Having observed HCV suppression of HBV in coinfected mice, we reasoned that human IFN signaling was activated in the liver by HCV to suppress HBV. To test this hypothesis, 9 mice were transplanted with PHHs and 6 of them were inoculated with HBV. Following the establishment of HBV viremia (Supplemental Figure 7A), 3 HBV-infected mice were superinfected with HCV at week 6 after HBV infection (Figure 5C). All 3 mice became HCV positive at week 2 after inoculation (Supplemental Figure 7B). No obvious differences in serum human albumin levels were found among the different groups (Supplemental Figure 7C). All mice were sacrificed at week 4 after superinfection to examine intrahepatic gene expression. In support of our findings in vitro, IFNL1 and ISG (CXCL10 and ISG20) levels were markedly elevated in HBV-HCV–coinfected mouse livers as compared with the HBV-monoinfected livers (Figure 5D). Due to a small group size (n = 3) and variations in HBV viremia in this experiment, a statistically significant difference was not reached when circulating HBV DNA levels were compared between coinfected and HBV-monoinfected mice at the end of the experiment. Alternative analysis based on the aggregate change (area under the curve) of HBV viremia during the coinfection period (4 weeks) was then conducted (Supplemental Figure 7D, left). Such an analysis showed that the aggregate HBV production was significantly reduced by HCV superinfection (Supplemental Figure 7D, right), consistent with the finding from the larger group of mice described above.
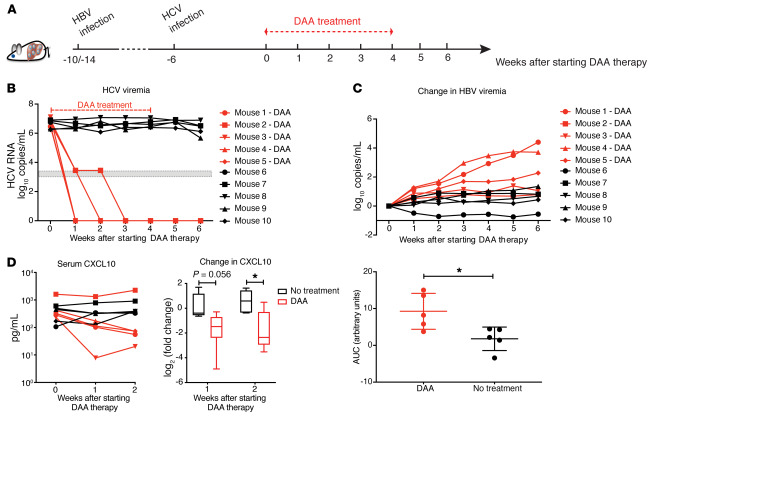
DAA treatment of HCV leads to decreased serum CXCL10 and increased HBV in coinfected cDNA-uPA/SCID mice.
Based on previous findings that suppressed HBV replication was accompanied by an active IFN response in coinfected mice, we reasoned that increased HBV replication should occur after eradication of HCV due to a diminished IFN response in the liver. To address this question, 10 HBV-infected mice were superinfected with HCV (6 mice at week 4 after HBV infection, 4 mice at week 8 after HBV infection). Six weeks after HCV infection, 3 of the 6 mice in the 4-week-post-HBV group, and 2 of the 4 mice in the 8-week-post-HBV group were randomly selected to receive a cocktail of daclatasvir (HCV NS5A inhibitor) and asunaprevir (protease inhibitor) for 4 weeks (Figure 6A). As expected, this DAA regimen efficiently reduced circulating HCV RNA to levels below the detection limit (3 log10 copies/mL) for all 5 mice within 2 weeks with no relapse (Figure 6B). To overcome the heterogeneity of HBV viremia due to differences in the time of HCV inoculation, we again analyzed the change in HBV DNA during the entire treatment period. Other than mouse 2 with DAA treatment showing a marginal increase in HBV viremia, the overall HBV production in DAA-treated mice was significantly higher than that of the untreated group (Figure 6C). We also measured the intrahepatic ISG mRNA levels of the mouse livers, and observed the expected downregulation of IFNL1 and ISGs in DAA-treated coinfected mice compared with the coinfected mice without treatment (Supplemental Figure 8A).
Figure 6. DAA treatment of HBV-HCV–coinfected cDNA-uPA/SCID mice.
(A) Ten mice infected with HBV for 4 (n = 6) or 8 (n = 4) weeks were inoculated with HCV. Six weeks after HCV infection, 5 mice (n = 3, 10 weeks of HBV infection; n = 2, 14 weeks of HBV infection) were treated with daclatasvir (10 mg/kg daily) and asunaprevir (20 mg/kg twice daily) via oral gavage for 4 weeks. Mice were continuously observed for 2 weeks after stopping treatment. (B) Serum HCV RNAs were measured with a lower limit of quantification (3.45 log10 copies/mL) and a lower limit of detection (3 log10 copies/mL) shown as a gray zone on the graph. (C) Changes in HBV viremia (upper ) were calculated by subtracting the baseline levels before DAA initiation from the viral levels at indicated time points. Area under the curve (AUC, lower) for the total change in HBV viremia during the 6 weeks of follow-up was generated by Prism software for individual mice. Means ± SEM are shown. (D) Left: Serum human CXCL10 concentration from mice was measured by ELISA. Right: Fold-change of CXCL10 for each mouse was calculated by dividing its on-treatment levels (week 1 or 2) by the baseline level. Results were grouped based on treatment and are shown in log2 scale. Medians ± 95% confidential intervals are shown. Unpaired t test was used. *P < 0.05.
We further examined changes in human CXCL10 levels in the serum of coinfected mice from week 0 until week 2 on DAA treatment when all treated mice had cleared HCV (Figure 6D). All the nontreated mice maintained a stable high level of CXCL10 over time. In contrast, the DAA-treated mice had a clear CXCL10 reduction by week 2 on treatment compared with baseline, with the exception of mouse 2. We suspected additional factors might account for a higher CXCL10 level in mouse 2, as we noted that starting from the baseline, mouse 2 had 5-fold higher CXCL10 levels compared with the mean values of the other animals. Interestingly, its HBV level also showed the least change after HCV clearance, as mentioned above. To further demonstrate the different dynamics of CXCL10 between groups, the fold-change (FC) of CXCL10 was calculated by comparing the level during treatment to baseline for each mouse (Figure 6D). The DAA-treated mice collectively showed a significantly greater reduction in CXCL10 (borderline in week 1 and significant in week 2). Similar human albumin levels in these mice ruled out the possibility that DAAs might cause significant human hepatocyte death (Supplemental Figure 8B). This observation further supports the idea that the IFN response is the key mediator of HBV suppression in the context of coinfection. Serum CXCL10 level thus can be a surrogate marker for the intrahepatic IFN response. Because of the SCID background of the mouse model, it is reasonable to conclude that host adaptive immunity does not mediate this HBV-HCV interplay, nor is it necessarily involved in the HBV increase after HCV clearance.
Higher pretreatment plasma CXCL10 levels are associated with HBV reactivation in coinfected patients.
Recently, Liu et al. reported a study on 111 HBV-HCV–coinfected patients receiving combined sofosbuvir/ledipasvir treatment (33). HBV reactivation, which is defined as an increase in HBV DNA from a baseline below the lower limit of quantification (LLOQ, 20 IU/mL) to LLOQ or greater, or an increase of greater than 1 log10 IU/mL from a baseline greater than LLOQ following DAA treatment, was identified in a substantial portion of patients (33).
To gain insight into the dynamic changes of immune responses and ISG expression in HBV-HCV–coinfected humans, we first measured various plasma immune mediators in a subgroup of 35 patients randomly selected from the 111 patients reported by Liu et al. (33). Among the 16 cytokines measured by multiplex cytokine ELISA, only CXCL10, CCL5, CCL13, CCL2, and CXCL6 were above the LLOQ for most of the patients and thus used for analyses.
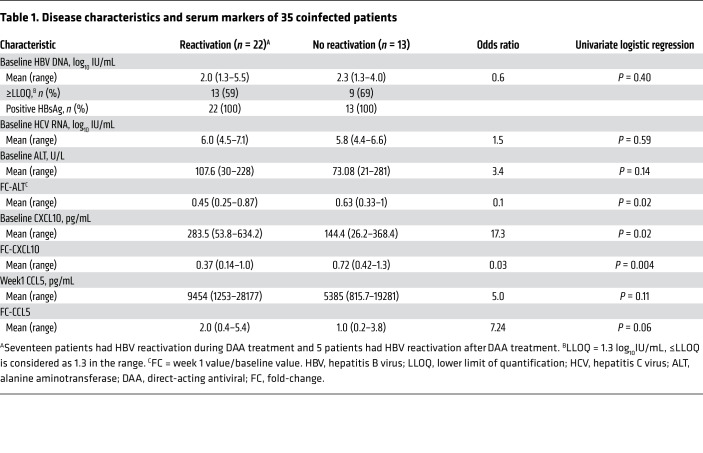
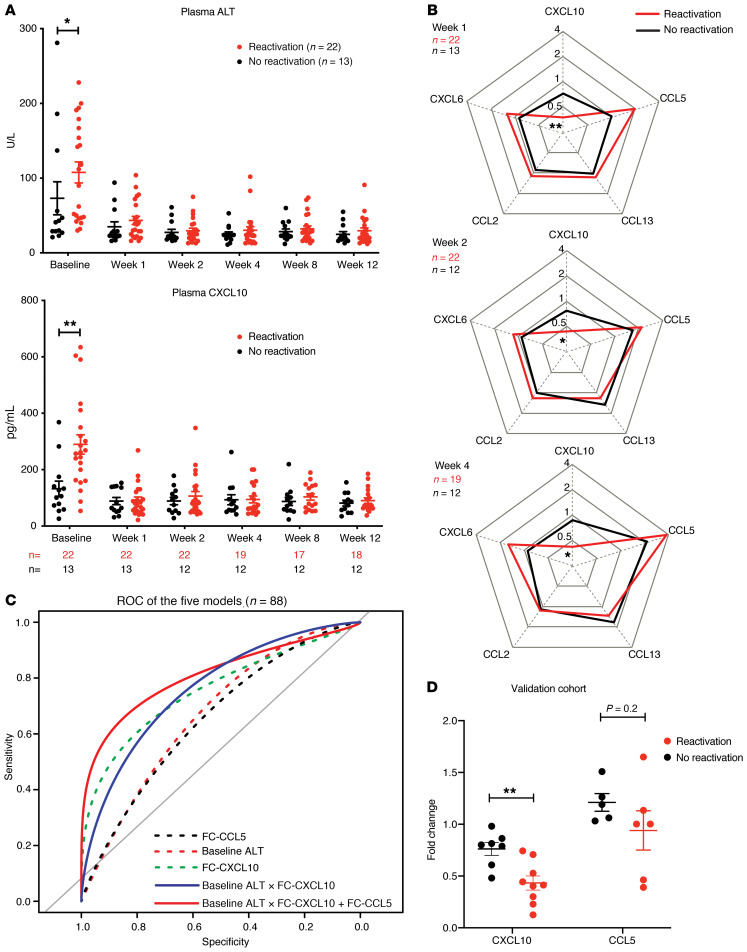
The detailed baseline characteristics of the 35 patients are provided in Table 1. The percentage of patients with unquantifiable HBV DNA was higher (41%) in the group that experienced HBV reactivation than in the group without HBV reactivation (31%) (P = 0.7, χ2 test), and the difference became significant in the larger original cohort of 111 patients (41% versus 13%, P = 0.01, χ2 test). In patients who had quantifiable baseline HBV DNA, their levels were comparable between the HBV-reactivation group and nonreactivation group, and so were the baseline HCV levels. Interestingly, baseline CXCL10 and alanine aminotransaminase (ALT) levels were significantly higher in patients who had HBV reactivation compared with those who did not (Figure 7A and Table 1). Baseline CXCL10 levels correlated with ALT levels (Spearman’s r = 0.6231, P < 0.0001) (Supplemental Figure 9A). Following DAA treatment, CXCL10 and ALT decreased markedly in all patients by week 1, coinciding with a sharp reduction in HCV viremia (Supplemental Figure 9B), which is consistent with published data on HCV monoinfection (34, 35). From week 1 to week 12 on treatment, no significant differences in CXCL10 and ALT levels were observed between the HBV-reactivation and control groups (Figure 7A). After treatment, CXCL10 levels remained low despite ongoing HBV infection and fluctuated when HBV flare occurred in some patients (Supplemental Figure 10). Three patients had hepatitis at various time points after completing the DAA treatment, necessitating anti-HBV nucleoside analog treatment (33). These observations indicated that HCV is the dominant stimulus of hepatic inflammation in coinfection. After HCV eradication by DAA, inflammatory responses including IFN activity marked by circulating CXCL10 as well as ALT released from liver injury diminished quickly, with a much greater net reduction in HBV-reactivation patients because of a higher pretreatment level (Figure 7A).
Table 1. Disease characteristics and serum markers of 35 coinfected patients.
Figure 7. Baseline and longitudinal changes of ALT and immune cytokines in coinfected patients undergoing DAA regimen.
A pilot cohort of 35 coinfected patients were randomly selected for circulating cytokine measurement by multiplex cytokine ELISA. (A) Serum ALT level (upper) and plasma CXCL10 (lower) at indicated times are shown. Sample numbers for CXCL10 measurement are below the x axis. Means ± SEM are shown. Unpaired t test was used. *P < 0.05; **P < 0.01. (B) Dynamic fold-changes of cytokines during the early course of treatment were calculated by comparing to baseline levels. The available sample numbers are given in the upper-left corner of each spider plot. Means are shown. For CXCL10, week 1 P = 0.0007, week 2 P = 0.0036, week 4 P = 0.0012. For CCL5, week 1 P = 0.034. After Bonferroni’s correction for multiple comparisons (5 cytokines), the P values for CXCL10 are still significant (*P < 0.05; **P < 0.01) but not those for CCL5. (C) Performance of ROC curves of CCL5 fold-change (FC-CCL5) (black dotted line; AUC, 0.63; 95% CI, 0.50–0.75), baseline ALT (red dotted line; AUC, 0.68; 95% CI, 0.56–0.80), FC-CXCL10 (green dotted line; AUC = 0.81, 95% CI, 0.71–0.90), baseline ALT × FC-CXCL10 (blue solid line; AUC, 0.81; 95% CI, 0.71–0.90), and combined baseline ALT × FC-CXCL10 with FC-CCL5 (red solid line; AUC, 0.82; 95% CI, 0.73–0.90) for predicting coinfected patients with HBV reactivation. Interactions are denoted by ×. (D) Blood CXCL10 and CCL5 of an independent coinfected-patient cohort were measured. Fold-change of CXCL10 was calculated by comparing the earliest on-treatment level available (10 patients from week 1; 2 patients from week 2; 1 patient from week 3, 4, and 12) to baseline level. Fold-change of CCL5 was calculated by comparing week 1 to baseline level (10 patients have available samples). The final data were grouped based on HBV outcome following the same reactivation criteria. Means ± SEM are shown. Unpaired t test was used. **P < 0.01.
To better illustrate the dynamic changes of the measured cytokines, we determined the FC of the 5 cytokines/chemokines over the baseline plasma levels (Figure 7B). CXCL10 showed the greatest FC (week 1/baseline) of 0.29 for patients with HBV reactivation and 0.63 for patients who did not. In contrast, CCL5, CCL13, CXCL6, and CCL2 did not differ in their baseline levels between HBV-reactivated patients and nonreactivated patients, nor did they show any changes following HCV clearance. Interestingly, CCL5 displayed an early increase in HBV-reactivation patients (week 1 on DAA) (Supplemental Figure 11) and its FC from baseline to week 1 showed a difference, although not significant, between the 2 groups (Table 1). It is not clear why CCL5 levels increased at week 1 after DAA treatment in HBV-reactivation patients or whether such a difference is indeed significant.
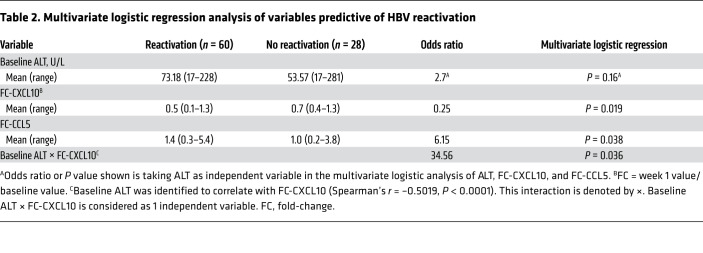
We next evaluated whether CXCL10, ALT, and CCL5 could be used to predict HBV reactivation. Univariate logistic regression analysis suggests CXCL10 FC is of higher predictive value (Table 1). To develop a more accurate model, we examined CXCL10, CCL5, and ALT levels of an additional 53 patients from the same original cohort (Supplemental Table 2). The resulting data confirmed that baseline CXCL10 and ALT are correlated (Spearman’s r = 0.4948, P < 0.001) and higher in HBV-reactivation patients (P = 0.03). When modeled together with the previous 35 patients (Table 2), baseline ALT with interaction with CXCL10 FC can be counted as one independent variable and constitutes the best predictive model of HBV reactivation when added together with CCL5 FC. The modeling was further evaluated by receiver operating characteristic (ROC) curve (Figure 7C), and is detailed further in Supplemental Table 3, which demonstrates that combining baseline ALT interacting with CXCL10 FC and CCL5 FC (AUC, 0.82) has the optimal sensitivity of 0.7 and specificity of 0.9. CXCL10 FC (AUC, 0.81) and baseline ALT interacting with CXCL10 FC are almost as good (AUC, 0.81). The CCL5 FC appears to play a minor role in the prediction model. A scatter plot of baseline ALT and CXCL10 FC was also generated to show the individual data and optimal predictive cutoff values for the entire cohort (Supplemental Figure 12).
Table 2. Multivariate logistic regression analysis of variables predictive of HBV reactivation.
To validate our finding in an independent patient cohort, we collected baseline and on-treatment blood samples from DAA-treated coinfected patients in Japan and Germany, and measured the CXCL10 and CCL5 levels. We did not analyze the ALT levels because they were done by various clinical laboratories with nonuniform standards at various times. The detailed patient information is listed in Supplemental Table 4 and HBV reactivation was defined by the same criteria as mentioned above. Although we could not verify the upregulation of CCL5 with the current small number of patients who had available week 1 samples, we could indeed validate the higher reduction of CXCL10 in HBV-reactivation patients as compared with that of no-reactivation patients (Figure 7D). With an FC cutoff of 0.55 for CXCL10 to predict HBV reactivation based on the above model, the calculated sensitivity is 78%, specificity 86%, the positive predictive value 88%, and negative predictive value 75% in this smaller validation cohort.
Discussion
To understand the molecular mechanism of HBV reactivation in DAA-treated patients, we investigated HBV-HCV interactions in established coinfection models in vitro and in vivo. We demonstrated that in both cell culture and mouse models lacking adaptive immunity, HBV was suppressed by HCV infection regardless of infection sequence. Those observations are consistent with data from chimpanzees showing that HBV replication was greatly attenuated by HCV irrespective of the inoculation sequence (36, 37). Previous in vitro studies failed to reveal this interaction mainly because of the usage of hepatoma cell lines (24, 38), which are known to have impaired innate immune function in both viral sensing and IFN signaling (28). Instead, several in vitro studies have claimed direct viral interactions. One study reported that HCV NS5A protein enhanced HBV replication and other studies showed that the HCV core protein directly suppressed HBV enhancer 1 and 2 (20, 22, 23). Many of those studies relied on overexpression of viral proteins in hepatoma cell lines. In contrast, our in vitro study used an infectious system in PHHs, which closely resembles natural infection. We observed that PHHs can support high levels of HBV and HCV in the same cells when ISG expression is blocked, thus supporting the notion that HBV and HCV do not interfere with each other’s replication via direct interaction of their viral components.
In line with our finding that HBV infection did not affect HCV replication in coinfected PHHs, studies in chronically HCV-infected chimpanzees superinfected with HBV indicated that HCV viremia did not change significantly during HBV superinfection (36). Yet in chronically coinfected patients, HCV levels were reported to be lower compared with HCV-monoinfected patients (415,000 IU/mL vs. 750,000 IU/mL, respectively) (39). It is possible that, during chronic infection, viruses may shape the host environment and cellular responses in a way that is suboptimal for either virus. Several studies have shown that HBV or HCV infection modifies hepatic gene expression profiles differently (40, 41). In livers of HBV-infected patients, genes involved in cell cycle arrest, induction of apoptosis, and hepatocyte differentiation are preferentially expressed, whereas cell cycle promotion, lipid metabolism, and epidermal growth factor receptor signaling are upregulated in livers of chronic hepatitis C patients (40). Unveiling such indirect interference experimentally is challenging due to the short infection duration with any experimental model.
Using a Jaki to specifically block ISG expression, we observed that IFNL1 expression was enhanced in HCV-infected cells, which could be explained by enhanced HCV replication or blockage of ISGs involved in negative feedback of the IFN response (42). With blockage of anti-HBV ISG expression, we could show that the HCV-induced IFN response is the main mediator of HBV suppression. Our in vitro data showed that coinfected cells produced fewer HBV transcripts, progeny viruses, and antigens, which are indicative of hampered HBV replication and transcription. This finding is consistent with previous reports that IFN can inhibit different steps of the HBV life cycle including viral entry, viral RNA transcription, nucleocapsid formation, and cccDNA degradation (15–18, 43).
CXCL10 is a highly upregulated ISG in HCV monoinfection (19). In our study, we observed that CXCL10 was not reduced by the addition of Jaki like other ISGs, nor was its level enhanced, like for IFNL1, following an increase in HCV replication, indicating both IFN-dependent and -independent CXCL10 production. The IFN-independent CXCL10 production is mediated by a direct transcriptional activation of the CXCL10 promoter following Toll-like receptor 3 (TLR3) or retinoic acid–inducible protein I (RIG-I) engagement in infected cells (44, 45), which can serve as a serum marker for hepatic IFN responses.
Serum CXCL10 has been shown to be mainly derived from hepatocytes in HCV patients and has been proposed as a marker for activated hepatic inflammation (46). Previous studies showed that CXCL10 decreased rapidly following the sharp decrease in serum HCV RNA by DAA treatment (34, 35, 47). Our study is the first to our knowledge reporting a similarly rapid decrease in CXCL10 levels in the blood of coinfected mice and patients within weeks of DAA initiation. This CXCL10 decrease strongly suggests a reduced intrahepatic IFN activity after HCV clearance in coinfected liver, which likely contribute to HBV reactivation, similar to patients undergoing immunosuppression.
Our study does not completely rule out the role of cell-mediated immunity in HBV reactivation of DAA-treated coinfected patients. In particular, NK cells exhibit increased cytotoxicity driven mainly by IFN (48). Thus, NK cells may also participate in repressing HBV replication through IFN effects in coinfection. HBV-specific T cells are either depleted or in an exhausted state in HBV patients (49). HCV coinfection has not been shown to affect the T cell response of HBV and vice versa (50, 51). In addition, HCV clearance following DAA treatment in HCV patients does not affect other T cell responses (52). Thus, the role of T cells in HBV and HCV interactions may be minor. Our studies in the cDNA-uPA/SCID mice also support the hypothesis that the observed HBV-HCV interaction is mostly a result of an IFN effect because of the absence of cellular immunity in this model.
The lack of involvement of HBV-specific adaptive immunity in the clinical setting probably explains why a large percentage of treated patients had increased HBV viremia, but few had ALT greater than 2 times the upper limit of normal after DAA treatment. However, more severe outcomes of HBV reactivation in patients after HCV clearance have been reported (5). In our patient cohort, serum cytokine/chemokine levels and the timing and severity of the HBV reactivation seem to be quite variable. Similar to spontaneous or immunosuppression-associated HBV reactivation observed in chronic hepatitis B patients, the mechanism whereby HBV replication increases and results in clinically relevant outcome in DAA-induced HBV reactivation is likely more complicated than the observed IFN effect and reflects the multifactorial nature of immunologically mediated viral control and disease. Interestingly, Jakis developed clinically to treat malignancies have been associated with HBV reactivation (53, 54). It is tempting to speculate that endogenous IFN signaling may be an intrinsic component of viral control in HBV infection and blocking this pathway with Jakis can lead to HBV reactivation.
Our comprehensive analyses of a large panel of serum cytokines and chemokines in this study point to a high pretreatment serum CXCL10 level, a marker of the magnitude of endogenous IFN response, or its FC from baseline to week 1 is highly associated with HBV reactivation. This observation together with the additional association of higher baseline ALT levels and HBV reactivation suggests that a highly inflammatory state of the coinfected liver with high endogenous IFN activity may be prone to HBV reactivation after HCV is eliminated. A previous cross-sectional study of HBV-HCV–coinfected patients suggested an association of lower HBsAg levels with higher serum CXCL10 levels (55). HBsAg levels were not available in this cohort so we could not validate this finding, but we noted more patients with baseline HBV DNA below the LLOQ in the HBV reactivation group that also had high pretreatment CXCL10. A high pretreatment serum CXCL10 level has been used as a negative predictor for response to IFN-based therapy in HCV patients (56, 57), for which a high-baseline IFN response may result in a refractory state or nonresponse to exogenous IFN treatment (58). The mechanisms whereby these 2 phenomena can be explained may be similar. During chronic HBV-HCV coinfection, an equilibrium state of host antiviral functions and viral escape mechanisms of persistence exists and the natural course of coinfection depends on the balance of these opposing forces. A higher baseline IFN response may contribute to a stronger anti-HBV state in the coinfected liver. A rapid and marked reduction in the IFN response by DAA treatment in some patients may trigger an imbalance of the anti-HBV state and thus an increase in HBV replication.
Of interest, a high baseline ALT level in HBV-monoinfected patients is a predictor for better response to IFN treatment (59). In those patients, ALT elevation is mainly caused by cytolytic immune responses targeting HBV. In the coinfected patients, the situation is more complicated. As mentioned above, coinfected patients with a higher baseline ALT and/or CXCL10 levels may have a more inflammatory and endogenous IFN response to HCV, which keeps HBV replication under better control, and once the endogenous IFNs diminish with DAA treatment, it is more likely that HBV will rebound. At present, we are not clear about the importance of CCL5 in HBV reactivation. It does not seem to play a prominent role in the predicative model nor has it been validated in an indepedent cohort.
Previous studies have identified single-nucleotide polymorphisms (SNPs) in the IFN-λ genes to be highly associated with IFN-based treatment response and baseline hepatic ISG levels (8, 60). In the clinical study of HBV-HCV coinfection undergoing DAA treatment described here (33), the majority of patients carry the favorable SNP (77% homozygous and 23% heterozygous) and no correlation with HBV reactivation was observed.
Although HCV infection is curable, treatment of HBV-HCV–coinfected patients can be challenging because of potential HBV reactivation. In addition, the fundamental science behind the intricate and complex interactions between 2 common pathogenic human viruses infecting the same cell type is highly intriguing. Our study elucidates a potentially unique aspect of these interactions and provides a molecular mechanism that explains why HCV is the so-called “dominant” virus in coinfection and treatment of HCV by DAAs leads to HBV reactivation. Along the same line with official recommendations by professional medical societies (AASLD HCV Guidance; ref. 61) and the FDA (62), our data support the idea that HBV-HCV–coinfected patients with low or undetectable HBV DNA levels should be either prophylactically treated with anti-HBV nucleoside analogs, or monitored at regular intervals during DAA treatment and treated as soon as HBV reactivation occurs. Based on our multivariate model, serum CXCL10 and baseline ALT may serve as a useful tool to assess the risk of HBV reactivation and provide guidance for managing these patients.
Methods
PHH culture and virus infection.
Cryopreserved PHHs from different donors (Hu1894, Hu1574, Hu1832, Hu1663, and Hu8196) were purchased from Thermo Fisher Scientific. Cell thawing and culture were done as described previously (63). Infection was typically initiated 24 hours after cell plating. HBV infection was performed at a multiplicity of infection (MOI) of 400 genomes/cell. HCV (JFH1) inoculation was performed at an MOI of 0.5 or 1 TCID50/cell, as indicated. Entecavir (1 μM, Sigma-Aldrich), sofosbuvir (10 μM, Advanced ChemBlock Inc.), or Jaki (CAS 457081-03-7, Sigma-Aldrich) was added to the culture media as indicated.
Human chimeric mice, virus infection, and human cytokine ELISA.
Generation of the cDNA-uPA/SCID mice and transplantation of PHHs were performed by PhoenixBio Co., Ltd., as previously described under an approved animal protocol (64, 65). For infection, male mice at 12 weeks after transplantation were injected intravenously with 100 μL of diluted HBV-positive patient serum (genotype C, 1 × 106 genomes) and 100 μL of diluted HCV-positive patient serum (genotype 1b, 1 × 106 genomes) at indicated times. Coinfected mice receiving DAA treatment were administered asunaprevir (40 mg/kg, twice a day, Bristol-Myers Squibb) combining daclatasvir (10 mg/kg, once a day, Bristol-Myers Squibb) for 4 weeks, as previously described (66).
Human CXCL10 and CCL5 in mouse serum were measured using Human Quantikine ELISA kits (DIP100 for CXCL10, DRN00B for CCL5; R&D Systems). Species specificity was validated by the manufacturer.
Patient samples and multiplex cytokine ELISA.
Serial plasma samples of HBV-HCV–coinfected patients from a previously published study had been collected at multiple time points including a pretreatment baseline, on drug at weeks 1, 2, 4, 8, and 12, and posttreatment week 4, 12, and every 12 weeks thereafter up to posttreatment week 108 (33). For the current study, an initial 35 patients who had available plasma samples were randomly selected from the original cohort. Samples from 28 patients were analyzed until posttreatment week 96 when available. After initial analysis, 7 remaining patients were tested only for the baseline and on-treatment samples. Multiplex cytokine ELISAs to quantify CCL5, CXCL6, CXCL11, IL-28A, IL-6, CXCL10, CCL13, CCL4, CXCL1, IFN-γ, IL-28B, CXCL8, CCL2, CXCL9, TNF-α, and IFN-β were performed following the manufacturer’s instructions (LXSAHM, R&D Systems). Briefly, plasma was thawed on ice 1 hour before the experiment and diluted 5-fold for measurement of CCL5 and 2-fold for the rest of the cytokines. Cytokine concentrations were measured using a standard curve of each specific cytokine provided by the kit. The plate was read in a Bio-Plex 200 system (Bio-Rad) using Bio-Plex Manager software 5.0. Cytokine analysis was performed according to the manufacturer’s instructions. A second round of measurement of CXCL10 and CCL5 in baseline and week 1 samples of the additional 53 patients was performed with the same standard Luminex kit containing one analyte.
Statistics.
Continuous variables are reported as means ± SD or means ± SEM as indicated. Differences between groups were compared by Student’s 2-sample t tests if the data were normally distributed; otherwise, Mann-Whitney U tests were used, followed by the application of Hochberg’s multiple-comparisons procedure under circumstances where more than 3 simultaneous inferences (comparisons) were performed (67). Spearman’s coefficient was used to evaluate association. Logistic regression analysis was used to identify variables independently associated with HBV reactivation. ROC analysis was used to determine the prediction strengths of the potential biomarkers in identifying HBV reactivation, and it was internally validated using a 10-fold cross-validation approach. All statistical tests were 2-sided and conducted at a 0.05 nominal level of statistical significance. Analyses were performed with GraphPad Prism (version 7.0), R (version 3.5.0), and SAS (version 9.4 TS1M6).
Study approval.
All experiments with mice were approved by the Ethics Review Committee for Animal Experimentation of the Graduate School of Biomedical Sciences, Hiroshima University (A18-64). All experiments with human samples were approved under NTUH-REC 201812063RINC and OHSRP 18-NIDDK-00585.
Author contributions
XC and TJL designed the project. XC, TU, and YX conducted research. TU, MI, and KC performed the mouse infection experiments. CJL, PJC, AG, VS, MMM, JV, SZ, YT, MO, HA, KT, NM, SH, YK, MI, and KC assisted with clinical sample collection and data acquisition. RU conducted statistical analysis on patient data. XC and TJL wrote the manuscript; all authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the Flow Cytometry Core of the National Heart, Lung, and Blood Institute of the NIH for providing instrument and technical support for multiplex cytokine ELISA. We thank Paul Juneau from the NIH library for statistics consultation and data analysis. XC, YX, TJL are supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH. TU, MI, and KC are supported by the Research Program on Hepatitis from the Japanese Agency for Medical Research and Development (AMED) (grant numbers 19fk0310109h0003 and 19fk0210020h0003). PJC and CJL are partially supported by the Ministry of Science and Technology, Taiwan (MOST 107-3017-F-002-002), and by the Center of Precision Medicine from the Ministry of Education (MOE) in Taiwan (NTU-107L9014-1).
Version 1. 03/12/2020
In-Press Preview
Version 2. 05/18/2020
Electronic publication
Version 3. 06/01/2020
Print issue publication
Footnotes
YX’s present address is: State key Laboratory of Virology, School of Basic Medical Sciences, Wuhan University, Wuhan, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):3205–3220.https://doi.org/10.1172/JCI135616.
See the related Commentary at When viruses collide: hepatitis B virus reactivation after hepatitis C treatment.
Contributor Information
Xiaoming Cheng, Email: xiaoming.cheng@nih.gov.
Takuro Uchida, Email: takuro.uchida@nih.gov.
Yuchen Xia, Email: yuchenxia@whu.edu.cn.
Regina Umarova, Email: regina.umarova@nih.gov.
Chun-Jen Liu, Email: cjliu@ntu.edu.tw.
Pei-Jer Chen, Email: peijerchen@ntu.edu.tw.
Anuj Gaggar, Email: anuj.gaggar@gilead.com.
Vithika Suri, Email: vithika.suri@gilead.com.
Johannes Vermehren, Email: vermehren@medicum-wiesbaden.de.
Stefan Zeuzem, Email: zeuzem@em.uni-frankfurt.de.
Yuji Teraoka, Email: teraoka22@hiroshima-u.ac.jp.
Mitsutaka Osawa, Email: m-osawa-m@hiroshima-u.ac.jp.
Hiroshi Aikata, Email: aikata@hiroshima-u.ac.jp.
Keiji Tsuji, Email: k-tsuji@hiroshima-med.jrc.or.jp.
Nami Mori, Email: nami7373@star.ocn.ne.jp.
Shuhei Hige, Email: shuhei.hige@ja-hokkaidoukouseiren.or.jp.
Yoshiyasu Karino, Email: ykarino@ja-hokkaidoukouseiren.or.jp.
Michio Imamura, Email: mimamura@hiroshima-u.ac.jp.
Kazuaki Chayama, Email: chayama@hiroshima-u.ac.jp.
T. Jake Liang, Email: JakeL@bdg10.niddk.nih.gov.
References
- 1.Konstantinou D, Deutsch M. The spectrum of HBV/HCV coinfection: epidemiology, clinical characteristics, viralinteractions and management. Ann Gastroenterol. 2015;28(2):221–228. [PMC free article] [PubMed] [Google Scholar]
- 2.Mavilia MG, Wu GY. HBV-HCV coinfection: viral interactions, management, and viral reactivation. J Clin Transl Hepatol. 2018;6(3):296–305. doi: 10.14218/JCTH.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pol S, et al. The negative impact of HBV/HCV coinfection on cirrhosis and its consequences. Aliment Pharmacol Ther. 2017;46(11–12):1054–1060. doi: 10.1111/apt.14352. [DOI] [PubMed] [Google Scholar]
- 4.Squadrito G, Cacciola I, Alibrandi A, Pollicino T, Raimondo G. Impact of occult hepatitis B virus infection on the outcome of chronic hepatitis C. J Hepatol. 2013;59(4):696–700. doi: 10.1016/j.jhep.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Bersoff-Matcha SJ, et al. Hepatitis B virus reactivation associated with direct-acting antiviral therapy for chronic hepatitis C virus: a review of cases reported to the U.S. Food and Drug Administration adverse event reporting system. Ann Intern Med. 2017;166(11):792–798. doi: 10.7326/M17-0377. [DOI] [PubMed] [Google Scholar]
- 6.Mücke MM, et al. Hepatitis B virus reactivation during direct-acting antiviral therapy for hepatitis C: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2018;3(3):172–180. doi: 10.1016/S2468-1253(18)30002-5. [DOI] [PubMed] [Google Scholar]
- 7.Pockros PJ. Black box warning for possible HBV reactivation during DAA therapy for chronic HCV infection. Gastroenterol Hepatol (N Y) 2017;13(9):536–540. [PMC free article] [PubMed] [Google Scholar]
- 8.Noureddin M, et al. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: A phenotype-genotype correlation study. Genes Immun. 2015;16(5):321–329. doi: 10.1038/gene.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng X, et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66(6):1779–1793. doi: 10.1002/hep.29348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A. 2004;101(17):6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen F, et al. Hepatitis B virus sensitivity to interferon-α in hepatocytes is more associated with cellular interferon response than with viral genotype. Hepatology. 2018;67(4):1237–1252. doi: 10.1002/hep.29609. [DOI] [PubMed] [Google Scholar]
- 12.Micco L, et al. Differential boosting of innate and adaptive antiviral responses during pegylated-interferon-alpha therapy of chronic hepatitis B. J Hepatol. 2013;58(2):225–233. doi: 10.1016/j.jhep.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Liang G, et al. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2013;110(6):2246–2251. doi: 10.1073/pnas.1221921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303(5665):1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 15.Lucifora J, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343(6176):1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 2017;13(4):e1006296. doi: 10.1371/journal.ppat.1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leong CR, et al. Interferon-stimulated gene of 20 kDa protein (ISG20) degrades RNA of hepatitis B virus to impede the replication of HBV in vitro and in vivo. Oncotarget. 2016;7(42):68179–68193. doi: 10.18632/oncotarget.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Y, Cheng X, Blossey CK, Wisskirchen K, Esser K, Protzer U. Secreted interferon-inducible factors restrict hepatitis B and C virus entry in vitro. J Immunol Res. 2017;2017:4828936. doi: 10.1155/2017/4828936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas E, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142(4):978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan Y, et al. NS5A protein of HCV enhances HBV replication and resistance to interferon response. Biochem Biophys Res Commun. 2007;359(1):70–75. doi: 10.1016/j.bbrc.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Chen SY, et al. Mechanisms for inhibition of hepatitis B virus gene expression and replication by hepatitis C virus core protein. J Biol Chem. 2003;278(1):591–607. doi: 10.1074/jbc.M204241200. [DOI] [PubMed] [Google Scholar]
- 22.Schüttler CG, Fiedler N, Schmidt K, Repp R, Gerlich WH, Schaefer S. Suppression of hepatitis B virus enhancer 1 and 2 by hepatitis C virus core protein. J Hepatol. 2002;37(6):855–862. doi: 10.1016/S0168-8278(02)00296-9. [DOI] [PubMed] [Google Scholar]
- 23.Shih CM, Lo SJ, Miyamura T, Chen SY, Lee YH. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Virol. 1993;67(10):5823–5832. doi: 10.1128/JVI.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellecave P, et al. Hepatitis B and C virus coinfection: a novel model system reveals the absence of direct viral interference. Hepatology. 2009;50(1):46–55. doi: 10.1002/hep.22951. [DOI] [PubMed] [Google Scholar]
- 25.Yang D, et al. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci U S A. 2014;111(13):E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raimondo G, et al. Longitudinal evaluation reveals a complex spectrum of virological profiles in hepatitis B virus/hepatitis C virus-coinfected patients. Hepatology. 2006;43(1):100–107. doi: 10.1002/hep.20944. [DOI] [PubMed] [Google Scholar]
- 27.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8(3):443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melén K, Julkunen I. Impaired antiviral response in human hepatoma cells. Virology. 1999;263(2):364–375. doi: 10.1006/viro.1999.9983. [DOI] [PubMed] [Google Scholar]
- 29.Helle F, et al. Permissivity of primary human hepatocytes and different hepatoma cell lines to cell culture adapted hepatitis C virus. PLoS One. 2013;8(8):e70809. doi: 10.1371/journal.pone.0070809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiraga N, et al. Absence of viral interference and different susceptibility to interferon between hepatitis B virus and hepatitis C virus in human hepatocyte chimeric mice. J Hepatol. 2009;51(6):1046–1054. doi: 10.1016/j.jhep.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Hiraga N, et al. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis C virus and its susceptibility to interferon. FEBS Lett. 2007;581(10):1983–1987. doi: 10.1016/j.febslet.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 33.Liu CJ, et al. Efficacy of ledipasvir and sofosbuvir treatment of HCV infection in patients coinfected with HBV. Gastroenterology. 2018;154(4):989–997. doi: 10.1053/j.gastro.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Meissner EG, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest. 2014;124(8):3352–3363. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burchill MA, et al. Rapid reversal of innate immune dysregulation in blood of patients and livers of humanized mice with HCV following DAA therapy. PLoS One. 2017;12(10):e0186213. doi: 10.1371/journal.pone.0186213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland SF, Asabe S, Engle RE, Purcell RH, Chisari FV. Limited hepatitis B virus replication space in the chronically hepatitis C virus-infected liver. J Virol. 2014;88(9):5184–5188. doi: 10.1128/JVI.03553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradley DW, Maynard JE, McCaustland KA, Murphy BL, Cook EH, Ebert JW. Non-A, non-B hepatitis in chimpanzees: interference with acute hepatitis A virus and chronic hepatitis B virus infections. J Med Virol. 1983;11(3):207–213. doi: 10.1002/jmv.1890110304. [DOI] [PubMed] [Google Scholar]
- 38.Eyre NS, et al. Hepatitis B virus and hepatitis C virus interaction in Huh-7 cells. J Hepatol. 2009;51(3):446–457. doi: 10.1016/j.jhep.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Marot A, et al. Characteristics of patients with hepatitis B virus and hepatitis C virus dual infection in a Western European country: Comparison with monoinfected patients. Clin Res Hepatol Gastroenterol. 2017;41(6):656–663. doi: 10.1016/j.clinre.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Kaneko S. Different signaling pathways in the livers of patients with chronic hepatitis B or chronic hepatitis C. Hepatology. 2006;44(5):1122–1138. doi: 10.1002/hep.21383. [DOI] [PubMed] [Google Scholar]
- 41.Branda M, Wands JR. Signal transduction cascades and hepatitis B and C related hepatocellular carcinoma. Hepatology. 2006;43(5):891–902. doi: 10.1002/hep.21196. [DOI] [PubMed] [Google Scholar]
- 42.Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36(3):150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A. 2005;102(28):9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brownell J, et al. Independent, parallel pathways to CXCL10 induction in HCV-infected hepatocytes. J Hepatol. 2013;59(4):701–708. doi: 10.1016/j.jhep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brownell J, et al. Direct, interferon-independent activation of the CXCL10 promoter by NF-κB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol. 2014;88(3):1582–1590. doi: 10.1128/JVI.02007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey CE, et al. Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J Leukoc Biol. 2003;74(3):360–369. doi: 10.1189/jlb.0303093. [DOI] [PubMed] [Google Scholar]
- 47.Alao H, et al. Baseline intrahepatic and peripheral innate immunity are associated with hepatitis C virus clearance during direct-acting antiviral therapy. Hepatology. 2018;68(6):2078–2088. doi: 10.1002/hep.29921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahlenstiel G, et al. Natural killer cells are polarized toward cytotoxicity in chronic hepatitis C in an interferon-alfa-dependent manner. Gastroenterology. 2010;138(1):325–35.e1. doi: 10.1053/j.gastro.2009.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehermann B, Thimme R. Insights from antiviral therapy into immune responses to hepatitis B and C virus infection. Gastroenterology. 2019;156(2):369–383. doi: 10.1053/j.gastro.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbani S, et al. Acute phase HBV-specific T cell responses associated with HBV persistence after HBV/HCV coinfection. Hepatology. 2005;41(4):826–831. doi: 10.1002/hep.20614. [DOI] [PubMed] [Google Scholar]
- 51.Boni C, et al. Antiviral CD8-mediated responses in chronic HCV carriers with HBV superinfection. Hepatology. 2004;40(2):289–299. doi: 10.1002/hep.20332. [DOI] [PubMed] [Google Scholar]
- 52.Martin B, et al. Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol. 2014;61(3):538–543. doi: 10.1016/j.jhep.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 53.Lai GM, Yan SL, Chang CS, Tsai CY. Hepatitis B reactivation in chronic myeloid leukemia patients receiving tyrosine kinase inhibitor. World J Gastroenterol. 2013;19(8):1318–1321. doi: 10.3748/wjg.v19.i8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda K, et al. Fatal hepatitis B virus reactivation in a chronic myeloid leukemia patient during imatinib mesylate treatment. Leuk Lymphoma. 2006;47(1):155–157. doi: 10.1080/14639230500236818. [DOI] [PubMed] [Google Scholar]
- 55.Wiegand SB, et al. Dominance of hepatitis C virus (HCV) is associated with lower quantitative hepatitis B surface antigen and higher serum interferon-γ-induced protein 10 levels in HBV/HCV-coinfected patients. Clin Microbiol Infect. 2015;21(7):710.e1–710.e9. doi: 10.1016/j.cmi.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Diago M, et al. Association of pretreatment serum interferon gamma inducible protein 10 levels with sustained virological response to peginterferon plus ribavirin therapy in genotype 1 infected patients with chronic hepatitis C. Gut. 2006;55(3):374–379. doi: 10.1136/gut.2005.074062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero AI, et al. Interferon (IFN)-gamma-inducible protein-10: association with histological results, viral kinetics, and outcome during treatment with pegylated IFN-alpha 2a and ribavirin for chronic hepatitis C virus infection. J Infect Dis. 2006;194(7):895–903. doi: 10.1086/507307. [DOI] [PubMed] [Google Scholar]
- 58.Feld JJ, Liang TJ. Hepatitis C -- identifying patients with progressive liver injury. Hepatology. 2006;43(2 suppl 1):S194–S206. doi: 10.1002/hep.21065. [DOI] [PubMed] [Google Scholar]
- 59.Viganò M, Grossi G, Loglio A, Lampertico P. Treatment of hepatitis B: Is there still a role for interferon? Liver Int. 2018;38 suppl 1:79–83. doi: 10.1111/liv.13635. [DOI] [PubMed] [Google Scholar]
- 60.Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV--response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9(7):406–417. doi: 10.1038/nrgastro.2012.101. [DOI] [PubMed] [Google Scholar]
- 61. American Association for the Study of Liver Disease, Infectious Diseases Society of America. HCV Guidance: Recommendations for testing, managing, and treating hepatitis C. http://www.hcvguidelines.org Updated November 6, 2019. Accessed March 30, 2020.
- 62. U.S. Food and Drug Administration. FDA Drug Safety Communication: FDA warns about the risk of hepatitis B reactivating in some patients treated with direct-acting antivirals for hepatitis C. http://www.fda.gov/Drugs/DrugSafety/ucm522932.htm Updated October 4, 2016. Accessed March 30, 2020.
- 63.Xia Y, Cheng X, Li Y, Valdez K, Chen W, Liang TJ. Hepatitis B virus deregulates the cell cycle to promote viral replication and a premalignant phenotype. J Virol. 2018;92(19):e00722-18. doi: 10.1128/JVI.00722-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohshita H, Tateno C. Propagation of human hepatocytes in uPA/SCID mice: producing chimeric mice with humanized liver. Methods Mol Biol. 2017;1506:91–100. doi: 10.1007/978-1-4939-6506-9_6. [DOI] [PubMed] [Google Scholar]
- 65.Tateno C, et al. Generation of novel chimeric mice with humanized livers by using hemizygous cDNA-uPA/SCID mice. PLoS One. 2015;10(11):e0142145. doi: 10.1371/journal.pone.0142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kan H, et al. Combination therapies with daclatasvir and asunaprevir on NS3-D168 mutated HCV in human hepatocyte chimeric mice. Antivir Ther (Lond) 2016;21(4):307–315. doi: 10.3851/IMP3009. [DOI] [PubMed] [Google Scholar]
- 67.Wright SP. Adjusted P-values for simultaneous inference. Biometrics. 1992;48(4):1005–1013. doi: 10.2307/2532694. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.