Abstract
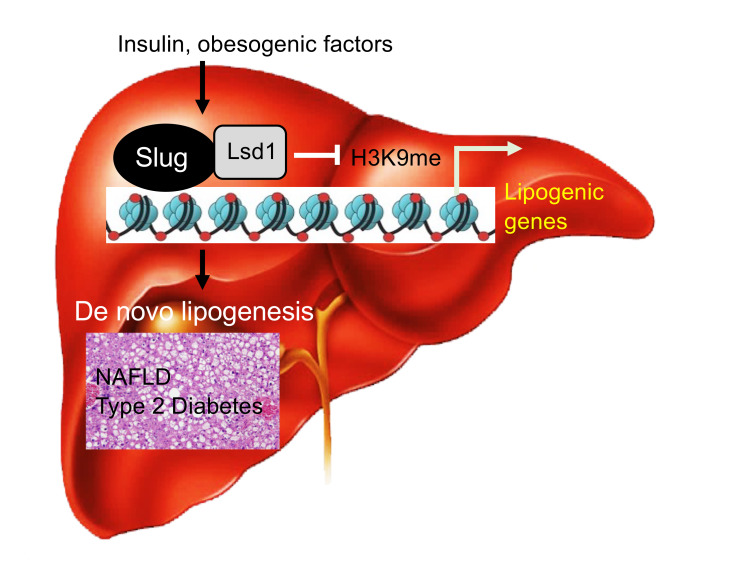
De novo lipogenesis is tightly regulated by insulin and nutritional signals to maintain metabolic homeostasis. Excessive lipogenesis induces lipotoxicity, leading to nonalcoholic fatty liver disease (NAFLD) and type 2 diabetes. Genetic lipogenic programs have been extensively investigated, but epigenetic regulation of lipogenesis is poorly understood. Here, we identified Slug as an important epigenetic regulator of lipogenesis. Hepatic Slug levels were markedly upregulated in mice by either feeding or insulin treatment. In primary hepatocytes, insulin stimulation increased Slug expression, stability, and interactions with epigenetic enzyme lysine-specific demethylase-1 (Lsd1). Slug bound to the fatty acid synthase (Fasn) promoter where Slug-associated Lsd1 catalyzed H3K9 demethylation, thereby stimulating Fasn expression and lipogenesis. Ablation of Slug blunted insulin-stimulated lipogenesis. Conversely, overexpression of Slug, but not a Lsd1 binding-defective Slug mutant, stimulated Fasn expression and lipogenesis. Lsd1 inhibitor treatment also blocked Slug-stimulated lipogenesis. Remarkably, hepatocyte-specific deletion of Slug inhibited the hepatic lipogenic program and protected against obesity-associated NAFLD, insulin resistance, and glucose intolerance in mice. Conversely, liver-restricted overexpression of Slug, but not the Lsd1 binding-defective Slug mutant, had the opposite effects. These results unveil an insulin/Slug/Lsd1/H3K9 demethylation lipogenic pathway that promotes NAFLD and type 2 diabetes.
Keywords: Hepatology, Metabolism
Keywords: Diabetes, Insulin, Insulin signaling
Introduction
Fatty acids serve both as a crucial metabolic fuel and as a core structure component of cell membranes to support life. Fatty acid synthase (Fasn) deficiency results in embryonic lethality (1), attesting to the essential role of de novo lipogenesis. However, excessive lipogenesis leads to lipotoxicity and causes (or worsens) human diseases, including fatty liver disease (2, 3). Liver steatosis is a driving force for nonalcoholic fatty liver disease (NAFLD), alcoholic liver disease, insulin resistance, and type 2 diabetes (2–5). De novo lipogenesis is tightly regulated by metabolic hormone insulin, nutrients, and other metabolic signals. Insulin activates several lipogenic transcription factors (e.g., Srebp1c, Lxrα, USF-1, and E2F1) that stimulate expression of lipogenic enzymes Fasn, acetyl-CoA carboxylase 1 (Acc1), and ATP-citrate lyase (Acl) (6–11). Recently, epigenetic modifications emerge as an important mechanism involved in reprogramming of metabolic pathways. For instance, lysine-specific demethylase-1 (Lsd1) demethylates histone H3 lysine-4 (H3K4) on the Cyp7a1 promoter, thereby suppressing Cyp7a1 expression and bile acid synthesis (12). However, lipogenesis-regulating epigenetic factors remain poorly understood.
Slug (also called Snai2 or Snail2) is a transcriptional regulator that contains an N-terminal SNAG domain and a C-terminal DNA-binding domain. It binds via its DNA binding domain to E2 boxes (CAGGTG or CACCTG) in promoters and enhancers. Slug also binds via its SNAG domain to Lsd1, histone deacetylase 1 (HDAC1), and/or HDAC2, and recruits them to target promoters where these enzymes catalyze epigenetic modifications (13–15). HDAC1/2-mediated histone deacetylation is known to repress gene expression. Lsd1-mediated H3K9 demethylation activates target promoters, and Lsd1-catalyzed H3K4 demethylation has the opposite effects (14–17). Slug and its family member Snail1 are known to promote epithelial-mesenchymal transition (EMT) and stem cell survival and/or proliferation (18–24). Notably, Snail1 emerges as a transcriptional regulator of nutrient metabolism. Adipocyte Snail1 suppresses expression of adipose triacylglycerol lipase (Atgl), thereby inhibiting lipolysis and lipid trafficking (25). Hepatocyte Snail1 suppresses hepatic Fasn expression and lipogenesis (26). In cancer cells, Snail1 suppresses expression of fructose 1,6-bisphosphatase and mitochondrial proteins (27, 28). Unlike Snail1, the metabolic function of Slug is not defined. Interestingly, global Slug knockout attenuates high-fat diet–induced (HFD-induced) obesity and insulin resistance (29); however, Slug target cells and metabolic pathways remain unknown.
In this work, we generated and characterized hepatocyte-specific Slug knockout (SlugΔhep) mice and mice with liver-restricted overexpression of Slug. We identified Slug as a new lipogenic transcription factor that promotes de novo lipogenesis by an epigenetic mechanism. We demonstrated that Slug-associated Lsd1 mediates lipogenesis by demethylating H3K9 on the Fasn promoter. Our results unveil an insulin/Slug/Lsd1/H3K9 demethylation lipogenic pathway that promotes NAFLD and type 2 diabetes.
Results
Hepatic Slug is elevated in NAFLD.
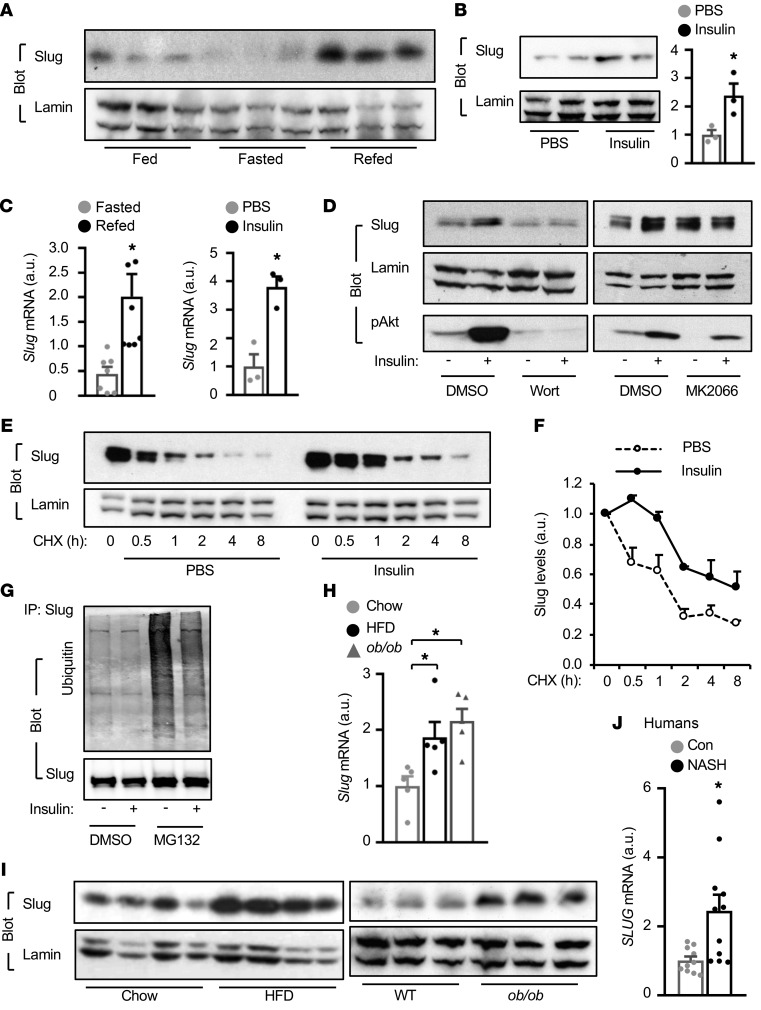
To test if Slug is involved in metabolic regulation, we assessed liver Slug expression in responses to fasting and feeding. Liver Slug levels were lower in overnight-fasted relative to nonfasted states, and were markedly increased by refeeding (Figure 1A). Feeding increased insulin secretion, prompting us to test if insulin is responsible for Slug upregulation. Insulin injection substantially increased liver Slug protein levels in fasted mice (Figure 1B). Liver Slug mRNA levels were also increased by either refeeding or insulin injection (Figure 1C). To gain insight into insulin pathways involved in Slug expression, we inhibited PI3-kinase and Akt in primary hepatocytes using Wortmannin and MK2066, respectively. Inhibition of PI3-kinase or Akt abrogated the ability of insulin to upregulate Slug (Figure 1D), indicating that insulin stimulates Slug expression in a PI3-kinase/Akt-dependent manner. We assessed Slug half-life using protein synthesis inhibitor cycloheximide (CHX). Insulin considerably increased Slug protein stability in human HepG2 hepatocytes (Figure 1, E and F). Baseline Slug ubiquitination was undetectable and dramatically increased in hepatocytes by proteasome inhibitor MG132 treatment (Figure 1G), indicating that Slug is rapidly ubiquitinated and degraded. In accordance with increasing Slug stability, insulin markedly decreased Slug ubiquitination (Figure 1G). Given that obesity is associated with hyperinsulinemia, we assessed hepatic Slug levels in mice with obesity and humans with nonalcoholic steatohepatitis (NASH). Liver mRNA and protein levels of Slug were significantly higher in mice with either HFD-induced (relative to chow-fed) or genetic obesity (ob/ob relative to wild type) (Figure 1, H and I). Importantly, hepatic SLUG expression was also significantly higher in NASH patients (Figure 1J). In publically available human liver data sets, liver expression of both SLUG and FASN is upregulated in subjects with NASH (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI128073DS1). Collectively, these results demonstrate that hepatic Slug is rapidly upregulated by insulin and possibly other metabolic signals.
Figure 1. Hepatic Slug is upregulated by insulin and is elevated in NAFLD.
(A) C57BL/6J males were overnight-fasted and then fed again for 3 hours. Liver nuclear extracts were immunoblotted with the indicated antibodies. (B) C57BL/6J males were fasted overnight and treated with insulin (1 U/kg body weight for 4 hours). Liver nuclear extracts were immunoblotted with indicated antibodies. Slug levels were normalized to lamin A/C levels (n = 3 per group). (C) Liver Slug mRNA abundance (normalized to 36B4 levels; n = 3 per group). (D) Primary hepatocytes were pretreated with wortmannin (100 nM) or MK2066 (100 nM) for 0.5 hours before insulin stimulation (100 nM for 2 hours). Nuclear extracts and cell extracts were immunoblotted with the indicated antibodies. (E and F) Primary hepatocytes were transduced with Slug adenoviral vectors, treated with insulin in the presence or absence of cycloheximide. Nuclear Slug levels were normalized to lamin A/C (n = 3 per group). (G) Primary hepatocytes were transduced with Slug adenoviral vectors for 12 hours, and then stimulated with insulin (100 nM for 1 hour) in the presence or absence of MG132 (5 μM). Cell extracts were immunoprecipitated with antibody against Slug and immunoblotted with the indicated antibodies. (H) Liver Slug mRNA levels (normalized to 36B4 levels). Chow: HFD (n = 5, for 10 weeks); ob/ob (n = 5, 14 weeks of age). (I) Liver nuclear extracts were prepared from WT and ob/ob mice at 14 weeks of age or from WT mice fed a chow diet or HFD for 10 weeks, and immunoblotted with antibodies against Slug and lamin A/C. (J) Liver SLUG mRNA levels in NASH patients (n = 11) and normal subjects (Con) (n = 10) (normalized to GAPDH). Proteins were resolved in parallel gels. Data are presented as mean ± SEM. *P < 0.05, 2-tailed Student’s t test (B, C, and J) or 1-way ANOVA/Sidak posttest (H).
Hepatocyte-specific deletion of Slug protects against liver steatosis.
To study hepatic Slug in vivo, we generated hepatocyte-specific Slug knockout (SlugΔhep) mice using the Cre/loxp system. Loxp sites were inserted into the Slug allele flanking exons 1 to 2 (Slugfl/fl) (Supplemental Figure 1B). Slugfl/fl mice were crossed with albumin-Cre drivers to produce SlugΔhep mice. We confirmed that Slug was ablated specifically in the liver but not other tissues (Supplemental Figure 1C). SlugΔhep mice were grossly normal on standard chow diet. We placed SlugΔhep and Slugfl/fl littermates on HFD. Body weight was slightly lower in male but not female SlugΔhep mice relative to sex-matched Slugfl/fl mice (Figure 2A). Remarkably, the liver was significantly smaller in both male and female SlugΔhep mice relative to sex-matched Slugfl/fl littermates (Figure 2B). Hepatocyte lipid droplets, as assessed by staining liver sections with neutral lipid dye Nile red, were substantially smaller and less abundant in SlugΔhep mice (Figure 2C). Both liver and plasma triacylglycerol (TAG) levels were significantly lower in SlugΔhep relative to sex-matched Slugfl/fl littermates (Figure 2, D and E). Of note, liver TAG content was comparable between SlugΔhep and Slugfl/fl mice on standard chow diet (Supplemental Figure 1D). To further confirm these findings in mice with genetic obesity, we generated Slugfl/fl ob/ob mice by crossing Slugfl/fl with ob+/– mice. Slugfl/fl ob/ob mice were transduced with Cre adenoviral vectors to ablate liver Slug (Figure 2F). Green fluorescent protein (GFP) adenoviral vectors were used as control. Body weight was comparable between the Cre and the GFP groups (Figure 2G). Hepatocyte lipid droplets were smaller and less abundant in Cre relative to GFP expressing mice (Figure 2H). Liver TAG levels were significantly lower in Cre-expressing relative to GFP-expressing Slugfl/fl ob/ob mice (Figure 2I). Therefore, hepatic Slug (elevated in obesity) appears to be critical for liver steatosis development in obesity.
Figure 2. Hepatocyte-specific deletion of Slug protects against liver steatosis in obesity.
(A–E) SlugΔhep and Slugfl/fl mice were fed a HFD for 11 weeks. (A) Growth curves (n = 11 per group). (B) Liver size and weight. Male: n = 9 per group; female: n = 10 per group. (C) Reprehensive liver sections (n = 3–9 mice per group). Scale bar: 100 μm. (D) Liver TAG levels (normalized to liver weight). Male: n = 9 per group; female: n = 6 per group. (E) Overnight fasting plasma TAG levels in male (n = 9 per group). (F–J) Slugfl/fl ob/ob males were transduced with GFP or Cre adenoviral vectors for 3 weeks. (F) Liver extracts were immunoblotted with antibodies against Slug and α-tubulin. (G) Growth curves (n = 6 per group). (H) Representative liver sections (3 pairs). Scale bar: 100 μm. (I) Liver TAG levels (normalized to liver weight). GFP: n = 6; Cre: n = 5. Data are presented as mean ± SEM. *P < 0.05, 2-tailed Student’s t test.
Ablation of hepatic Slug attenuates HFD-induced insulin resistance and glucose intolerance.
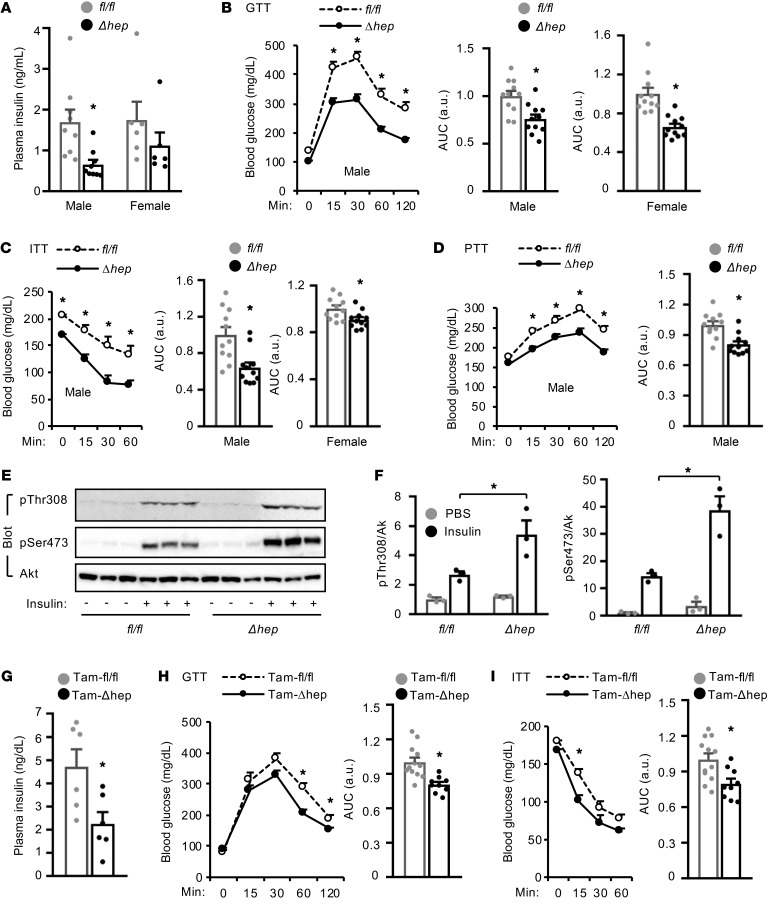
Liver steatosis is known to worsen insulin resistance, prompting us to assess insulin sensitivity in SlugΔhep mice. Mice were fed a HFD for 11 weeks to induce obesity. Overnight-fasted insulin levels were significantly lower in SlugΔhep than in Slugfl/fl littermates (Figure 3A). In glucose (GTT), insulin (ITT), and pyruvate (PTT) tolerance tests, blood glucose levels and AUC were significantly lower in SlugΔhep relative to sex-matched Slugfl/fl mice (Figure 3, B–D). To corroborate these studies, we examined insulin signaling. Insulin-stimulated phosphorylation of hepatic Akt (pThr308 and pSer473) was significantly higher in SlugΔhep than in Slugfl/fl littermates (Figure 3, E and F), indicating that hepatic Slug deficiency improves insulin resistance. To further confirm these findings, we examined mice with adult-onset ablation of hepatic Slug, using Slugfl/fl CreERT2+/– mice generated by crossing Slugfl/fl with albumin-CreERT2 drivers. Adult Slugfl/fl CreERT2+/– mice were injected with tamoxifen to specifically ablate hepatic Slug (TamΔhep). Slugfl/fl mice were similarly treated with tamoxifen (Tamfl/fl) as control. Body weight and fat content were comparable between TamΔhep and Tamfl/fl mice (Supplemental Figure 2, A and B). Liver weight and TAG levels were significantly lower in TamΔhep than in Tamfl/fl mice, and lipid droplets were smaller and less abundant in TamΔhep mice (Supplemental Figure 2, B–D). Overnight-fasted insulin levels were significantly lower in TamΔhep relative to Tamfl/fl mice (Figure 3G). Both glucose and insulin tolerances were also improved in TamΔhep mice (Figure 3, H and I). Collectively, these data suggest that obesity-induced upregulation of hepatic Slug promotes liver steatosis and insulin resistance.
Figure 3. Ablation of hepatic Slug ameliorates diet-induced insulin resistance and glucose intolerance.
(A–E) SlugΔhep and Slugfl/fl mice were fed a HFD for 8 to 11 weeks. (A) Overnight-fasted plasma insulin levels (male: n = 8 per group; female: n = 6 per group). (B–D) GTT, ITT and PTT. Male: n = 11 per group; female: n = 11 per group. AUC: area under curves. (E and F) Mice were fasted overnight and stimulated with insulin (1 U/kg body weight for 5 minutes). Liver extracts were immunoblotted with antibodies against phospho-Akt or Akt. Phospho-Akt levels were normalized to total Akt levels (n = 3). (G–I) Tamfl/fl and TamΔhep males were fed a HFD for 10 weeks. (G) Overnight-fasted plasma insulin levels (n = 6 per group). (H and I) GTT and ITT. Tamfl/fl: n = 12; TamΔhep: n = 9. Data are presented as mean ± SEM. *P < 0.05, 2-tailed Student’s t test (A–D, F–J: AUC) and 2-way ANOVA/Bonferroni’s posttest (B–D, H, I: curves).
Ablation of hepatic Slug suppresses the liver lipogenic program.
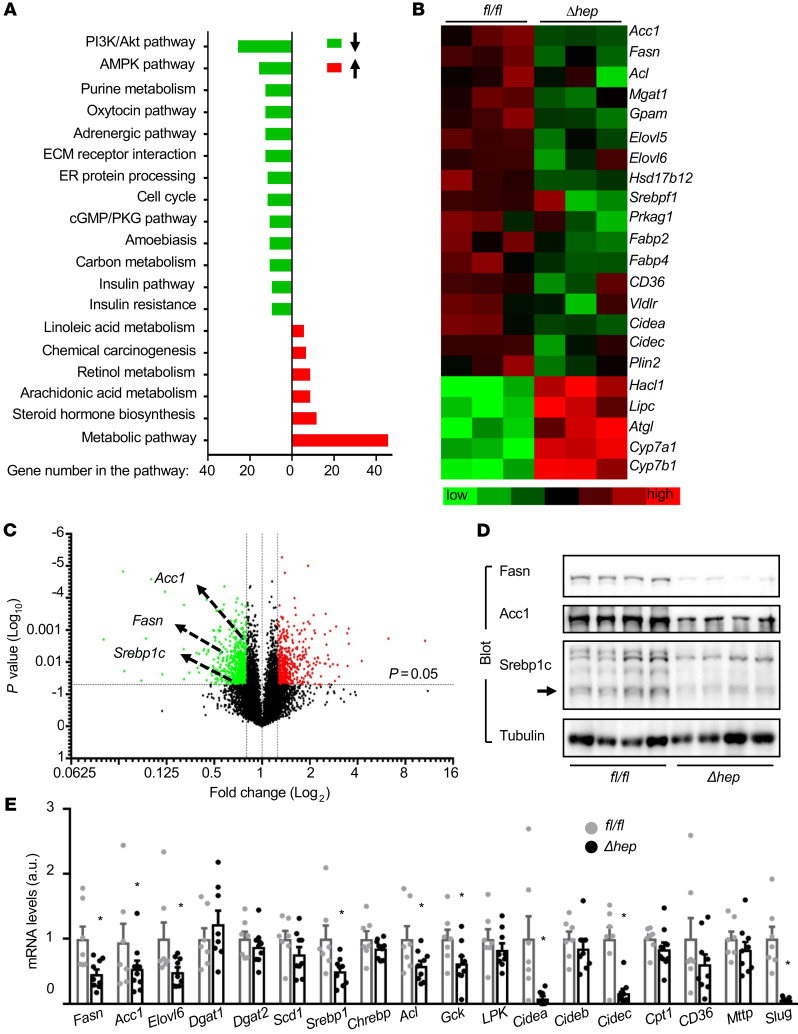
We next sought to identify Slug target genes, using unbiased GeneChIP techniques. SlugΔhep male mice were fed a HFD for 11 weeks, and livers were harvested for Affymetrix analysis. We identified 563 upregulated genes and 710 downregulated genes (>1.25 fold). These genes are involved in many signaling and metabolic pathways (Figure 4A). Notably, expression of lipogenic genes (e.g., Fasn, Acc1, and Srebp1c) was substantially downregulated in SlugΔhep mice (Figure 4, B and C). We confirmed these results by immunoblotting liver extracts. Hepatic Fasn, Acc1, and Srebp1c levels were markedly lower both in SlugΔhep relative to Slugfl/fl mice (Figure 4D) and in TamΔhep relative to Tamfl/fl mice (Supplemental Figure 2E). The mRNA levels of Fasn, Acc1, Srebp1c, Acl, and Elvol6 were lower in SlugΔhep mice (Figure 4E). Expression of lipid droplet proteins (e.g., Cidea, Cidec) was also significantly lower in SlugΔhep mice (Figure 4E). These results suggest that Slug stimulates lipogenic gene expression in the liver. In contrast, expression of the genes that regulate fatty acid uptake (CD36), fatty acid β oxidation (Cpt1α), and very low density lipoprotein secretion (Mttp) was comparable between SlugΔhep and Slugfl/fl mice (Figure 4, B and E). Expression of hepatic Lxrα, Chrebp, and Pparγ was also similar between SlugΔhep and Slugfl/fl mice (Supplemental Figure 2F). Thus, hepatic Slug induces liver steatosis in obesity, presumably by stimulating de novo lipogenesis.
Figure 4. Ablation of hepatic Slug suppresses the hepatic lipogenic program.
SlugΔhep and Slugfl/fl males were fed a HFD for 11 weeks. (A–C) Affymetrix analysis (n = 6 per group). (A) Gene ontology analysis. (B) Volcano plots. (C) Heatmaps. (D) Liver extracts were immunoblotted with the indicated antibodies. Arrow indicates the mature form of Srepb1c. (E) Liver mRNA abundance (normalized to 36B4 levels). Slugfl/fl: n = 7; SlugΔhep: n = 9. Data are presented as mean ± SEM. *P < 0.05, 2-tailed Student’s t test.
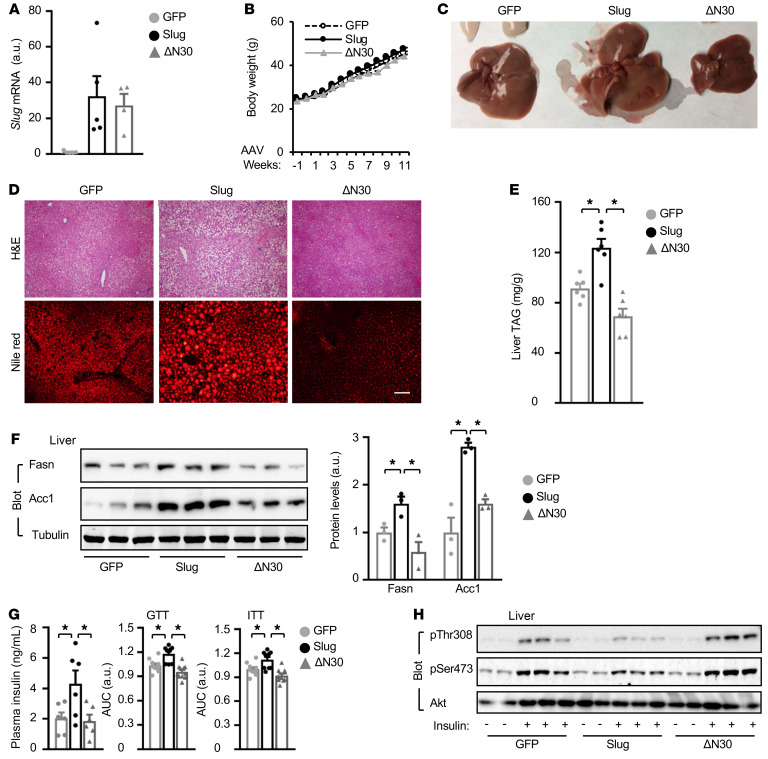
Liver-specific overexpression of Slug but not ΔN30 promotes NAFLD and insulin resistance.
To complement the loss-of-function approach, we tested if liver-restricted overexpression of Slug is sufficient to induce liver steatosis. Considering that the SNAG domain of Slug binds to various epigenetic enzymes, we speculated that this domain might be required for Slug stimulation of lipogenesis. We generated epigenetically defective ΔN30 lacking amino acids 1 to 30. C57BL/6J mice were transduced with adeno-associated viral (AAV) vectors expressing Slug, ΔN30, or GFP (control), and fed a HFD. Liver expression of Slug and ΔN30 was comparable (Figure 5A and Supplemental Figure 3A). Body weight was indistinguishable between AAV-GFP, AAV-Slug, and AAV-ΔN30 transduced mice (Figure 5B). Strikingly, overexpression of Slug but not ΔN30 induced hepatomegaly and severe liver steatosis (Figure 5, C and D). Liver TAG levels were significantly higher in the AAV-Slug but not AAV-ΔN30 groups relative to the AAV-GFP group (Figure 5E). Consistently, both protein and mRNA levels of hepatic Fasn and Acc1 were considerably higher in Slug-overexpressing but not ΔN30-overexpressing mice relative to GFP-expressing mice (Figure 5F and Supplemental Figure 3B). Plasma insulin levels were higher in AAV-Slug–transduced but not AAV-ΔN30–transduced mice relative to AAV-GFP–treated mice (Figure 5G). In GTT and ITT, AUCs were significantly higher in the AAV-Slug but not the AAV-ΔN30 groups relative to the GFP group (Figure 5G). Insulin-stimulated phosphorylation of Akt (pThr308, pSer473) was lower in the AAV-Slug group, but higher in the AAV-ΔN30 group, relative to the AAV-GFP group (Figure 5H and Supplemental Figure 3C). These results suggest that SNAG-associated epigenetic activities are indispensable for hepatic Slug to promote lipogenesis and insulin resistance.
Figure 5. Liver-specific overexpression of Slug but not ΔN30 promotes liver steatosis and insulin resistance.
C57BL/6J males were transduced with AAV-CAG-GFP, AAV-CAG-Slug, or AAV-CAG-ΔN30 vectors, and fed a HFD for 11 weeks. (A) Liver Slug mRNA levels (normalized to 36B4 levels, n = 4–5 per group). (B) Growth curves (n = 10 per group). (C and D) Representative livers and liver sections (n = 3 mice per group). Scale bar: 100 μm. (E) Liver TAG levels (normalized to liver weight); n = 6 per group. (F) Liver extracts were immunoblotted with the indicated antibodies. Fasn and Acc1 levels were normalized to α-tubulin levels. (G) Overnight-fasted plasma insulin levels (n = 6 per group), GTT, and ITT (n = 10, per group) 8 to 9 weeks after AAV transduction. (H) Mice were fasted overnight and stimulated with insulin (1 U/kg body weight for 5 minutes). Liver extracts were immunoblotted with antibodies against phospho-Akt (pThr308, pSer473) and Akt. Data are presented as mean ± SEM. *P < 0.05, 1-way ANOVA/Sidak posttest.
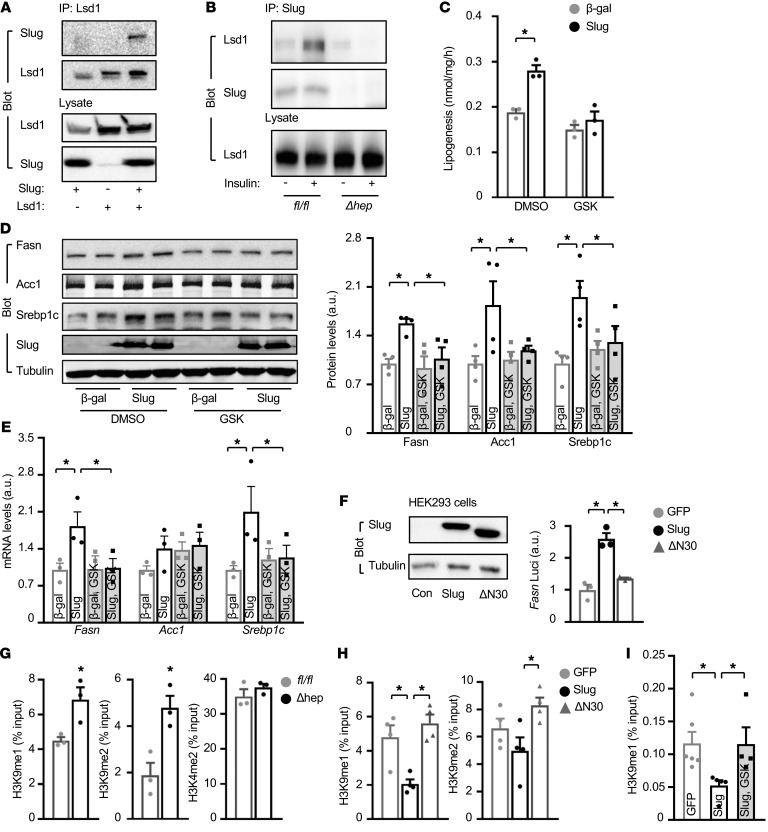
Slug/Lsd1 pathway epigenetically stimulates Fasn expression and lipogenesis.
We next set out to identify SNAG-elicited epigenetic modifications responsible for Slug stimulation of lipogenesis. The SNAG domain is known to bind to Lsd1 (16, 30). We confirmed that Slug but not ΔN30 bound to Lsd1, using coimmunoprecipitation (Figure 6A and Supplemental Figure 3D). Importantly, insulin further increased Slug association with Lsd1 (Figure 6B). Overexpression of Slug markedly increased expression of Fasn, Acc1, and Srebp1c as well as lipogenesis in primary hepatocytes (Figure 6, C–E), further confirming that Slug directly stimulates de novo lipogenesis. To examine the role of Slug-associated Lsd1, we treated primary hepatocytes with Lsd1 inhibitor GSK2879552 (GSK). GSK abrogated the ability of Slug to stimulate lipogenic gene expression and lipogenesis (Figure 6, C–E). To verify that Lsd1 mediates Slug lipogenic action in vivo, we transduced C57BL/6J mice with Slug adenoviral vectors and then treated the mice with GSK. Slug overexpression markedly increased Fasn expression in the liver as expected, and GSK treatment significantly inhibited the ability of Slug to stimulate Fasn expression (Supplemental Figure 4A). The Fasn promoter contains 2 and 3 putative Slug-binding motifs in mice and humans, respectively. Overexpression of Slug but not Lsd1 binding-defective ΔN30 increased Fasn luciferase reporter activities (Figure 6F). Together, these observations suggest that Lsd1 mediates Slug stimulation of lipogenic gene expression.
Figure 6. Slug/Lsd1/H3K9 demethylation pathway stimulates lipogenesis.
(A) Coimmunoprecipitation of Slug with Lsd1 in HEK293 cells. (B) Primary hepatocytes were stimulated with insulin (100 nM for 1 hour). Cell extracts were immunoprecipitated with anti-Slug antibody and immunoblotted with antibodies against Lsd1 and Slug. (C–E) Primary hepatocytes were transduced with Slug or β-gal adenoviral vectors and treated with GSK2879552 (1 μM) or DMSO for 24 hours. (C) Lipogenesis rates (normalized to protein levels) (n = 3 per group). (D) Cell extracts were immunoblotted with the indicated antibodies. Fasn and Acc1 levels were normalized to α-tubulin levels (n = 3). (E) Fasn, Acc1, and Srebp1c mRNA abundance (normalized to 36B4 levels) (n = 3 per group). (F) HEK293 cells were transfected with AAV-CAG-Slug or AAV-CAG-ΔN30 vectors. Cell extracts were immunoblotted with antibodies against Slug and α-tubulin. Fasn luciferase reporter activity (normalized to β-gal internal control) in HepG2 cells (n = 3). (G) SlugΔhep (n = 3) and Slugfl/fl (n = 3) males were fed a HFD for 11 weeks. Fasn promoter H3K9 and H3K4 methylation levels were measured in the liver by ChIP-qPCR. (H) Liver Fasn promoter H3K9 and H3K4 methylation levels (n = 4 per group). C57BL/6J males were transduced with AAV-CAG-GFP, AAV-CAG-Slug, or AAV-CAG-ΔN30 vectors, and fed a HFD for 11 weeks. (I) C57BL/6J mice were transduced with GFP or Slug adenoviral vectors and treated with GSK2879552. Fasn promoter H3K9me1 levels were assessed in the liver using ChIP (exclusion criteria: greater than 3 times SD). Data are presented as mean ± SEM. *P < 0.05, 2-tailed Student’s t test (G) or 1-way ANOVA/Sidak posttest (C–F and I).
We attempted to elucidate Lsd1-catalyzed epigenetic modifications on the Fasn promoter, using chromatin immunoprecipitation–quantitative PCR (ChIP–qPCR). H3K9 monomethylation (H3K9me1) and dimethylation (H3K9me2) levels were significantly higher in SlugΔhep relative to Slugfl/fl mice (Figure 6G). Conversely, liver-specific overexpression of Slug substantially decreased Fasn promoter H3K9me1 and H3K9me2 levels (Figure 6H). Deletion of the SNAG domain completely abolished the ability of ΔN30 to decrease Fasn promoter H3K9 methylation levels in mice (Figure 6H). In contrast, H3K4 dimethylation (H3K4m2) levels were similar between SlugΔhep and Slugfl/fl mice (Figure 6G). To corroborate these studies, we transduced C57BL/6J mice with Slug adenoviral vectors and treated them with Lsd1 inhibitor GSK2879552. Slug overexpression decreased Fasn promoter H3K9me1 levels as expected, and GSK2879552 treatment dramatically suppressed the ability of Slug to decrease Fasn promoter H3K9me1 levels (Figure 6I). Collectively, these results suggest that Slug-bound Lsd1 catalyzes H3K9 demethylation, thereby stimulating Fasn expression.
Slug deficiency decreased Srepb1c expression (Figure 4, D and E), prompting us to test if Srepb1c acts downstream to mediate Slug lipogenic action. We silenced Srepb1c in primary hepatocytes using shRNA adenoviral vectors as described previously (31). Srebp1c expression was dramatically suppressed by Srebp1c shRNA adenoviral vectors compared with scramble adenoviral vectors (Supplemental Figure 4B). Slug overexpression increased de novo lipogenesis in both scramble and shRNA adenoviral vector–transduced hepatocytes, but lipogenesis rates were lower in Srebp1c-silenced hepatocytes (Supplemental Figure 4C). These data suggest that Slug stimulates lipogenesis by both Srebp1c-dependent and Srebp1c-independent mechanisms. Aside from the Fasn promoter, we also observed that Slug occupied Srebp1c and Acc1 promoters (Supplemental Figure 4D), suggesting that Slug likely activates expression of multiple lipogenic genes.
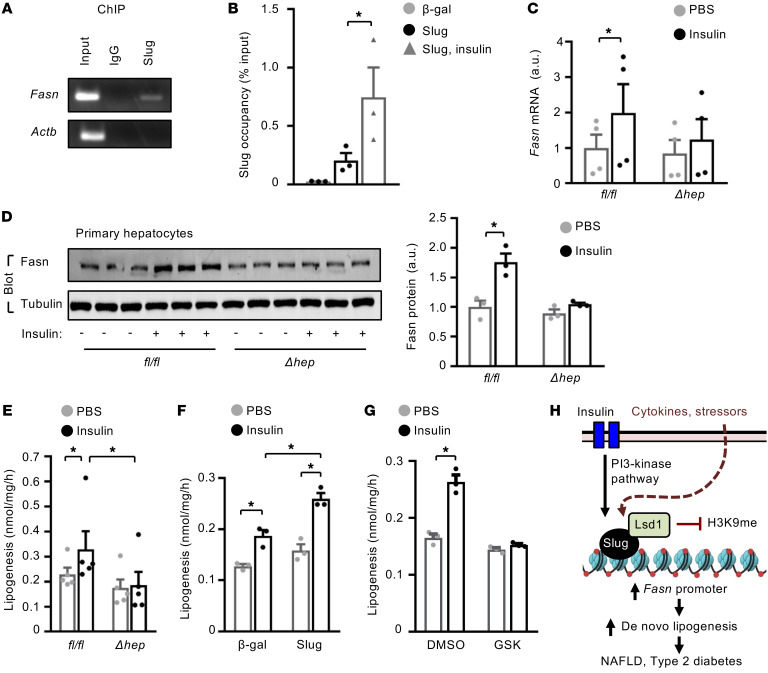
Slug/Lsd1 pathway mediates insulin stimulation of lipogenesis.
Considering that insulin increases Slug expression and Slug binding to Lsd1, we tested if Lsd1-elicited H3K9 demethylation mediates insulin stimulation of lipogenesis. We confirmed that Slug physically bound to the Fasn promoter in primary hepatocytes, using ChIP assays (Figure 7A). Importantly, insulin further increased Slug occupancy on the Fasn promoter (Figure 7B). Strikingly, deletion of Slug markedly attenuated the ability of insulin to stimulate Fasn expression in hepatocytes prepared from SlugΔhep mice (Figure 7, C and D). Accordingly, ablation of Slug decreased both baseline and insulin-stimulated lipogenesis (Figure 7E). Conversely, overexpression of Slug markedly increased de novo lipogenesis under both basal and insulin-stimulated conditions (Figure 7F). To examine the role of Slug-bound Lsd1, we pretreated primary hepatocytes with Lsd1 inhibitor GSK2879552 before insulin stimulation. Remarkably, Lsd1 inhibition, like Slug ablation, abolished the ability of insulin to stimulate de novo lipogenesis (Figure 7G). Based on these findings, we propose an epigenetic lipogenesis model (Figure 7H). Insulin stimulates Slug expression, Slug interaction with Lsd1, and recruitment of Slug/Lsd1 complexes to lipogenic promoters/enhancers. Lsd1 in turn catalyzes H3K9 demethylations, thereby stimulating expression of lipogenic genes and subsequent lipogenesis.
Figure 7. Insulin stimulated lipogenesis via Slug/Lsd1 epigenetic pathway.
(A and B) Primary hepatocytes were transduced with Slug or β-gal adenoviral vectors, and stimulated with insulin (100 nM for 2 hours). Slug occupancy on the Fasn promoter was assessed by ChIP-qPCR and normalized to inputs (n = 3 per group). (C–E) Primary hepatocytes were stimulated with insulin (50 nM) for 3 hours (C) or 12 hours (D and E). (C) Fasn mRNA abundance (normalized to 36B4 levels) (n = 3 per group). (D) Cell extracts were immunoblotted with the indicated antibodies. Fasn levels were normalized to α-tubulin levels. (E) Lipogenesis rates (n = 3 per group). (F) Lipogenesis (normalized to protein levels, n = 3 per group). Primary hepatocytes were transduced with Slug or β-gal adenoviral vectors and stimulated with insulin (50 nM for 12 hours). (G) Lipogenesis (n = 3 per group). Primary hepatocytes were pretreated with GSK2879552 (4 μM) or DMSO and stimulated with insulin (50 nM for 5 hours). (H) Insulin stimulates Slug expression, Slug-Lsd1 interactions, and recruitment of Slug/Lsd1 to the Fasn promoter where Lsd1 demethylates H3K9, thereby activating Fasn expression and de novo lipogenesis. Cytokines and metabolic stressors similarly activate the Slug/Lsd1 lipogenic pathway. Data are presented as mean ± SEM. *P < 0.05, 1-way ANOVA/Sidak posttest.
Discussion
In this study, we have identified Slug as an important lipogenic transcription factor. We found that Slug binds to the promoters of lipogenic genes Fasn, Acc1, and Srebp1c. In primary hepatocytes, overexpression of Slug suppressed Fasn, Acc1, and Srebp1c expression and de novo lipogenesis, and ablation of Slug had the opposite effects. In vivo, hepatocyte-specific ablation of Slug decreased expression of hepatic Fasn, Acc1, and Srebp1c and dramatically attenuated obesity-associated liver steatosis in SlugΔhep mice. SlugΔhep mice were resistant to HFD-induced insulin resistance and glucose intolerance, presumably owing to protection against liver steatosis. Conversely, liver-restricted overexpression of Slug markedly worsened HFD-induced liver steatosis and insulin resistance. Insulin potently upregulated Slug levels by increasing both Slug expression and stability, and also stimulated binding of Slug to the Fasn promoter. Ablation of Slug markedly decreased insulin-stimulated lipogenesis. These results reveal an insulin/Slug lipogenic pathway. Importantly, hepatic Slug levels were aberrantly higher in mice and humans with obesity and NAFLD. Taken together, these findings suggest that the insulin/Slug lipogenic pathway contributes to NAFLD and liver steatosis-related metabolic disease.
We confirmed that Slug binds to Lsd1 via its SNAG domain. Interestingly, insulin stimulation further increased Slug-Lsd1 interactions. Remarkably, in primary hepatocytes, Lsd1 inhibitor treatment abrogated the ability of Slug to stimulate expression of lipogenic genes and de novo lipogenesis. Similarly, Lsd1 inhibitor treatment also decreased the ability of Slug to stimulate liver Fasn expression in mice. In HFD-fed mice, deletion of the SNAG domain (ΔN30) abolished the ability of Slug to stimulate expression of lipogenic genes, liver steatosis, and insulin resistance. Lsd1 is able to activate target promoters through catalyzing H3K9 deacetylation. We found that liver Fasn promoter H3K9me1 and H3K9me2 levels were significantly higher in SlugΔhep mice relative to Slugfl/fl mice, and were significantly lower in Slug-overexpressing mice (restricted to the liver) relative to GFP-overexpressing mice. Strikingly, deletion of Lsd1-binding SNAG domain (ΔN30) abrogated the ability of Slug to decrease liver Fasn promoter H3K9me1 and H3K9me2 levels. Likewise, Lsd1 inhibitor treatment also blocked the ability of Slug to decrease liver Fasn promoter H3K9me1 levels. Based on these findings, we proposed a Slug epigenetic model of de novo lipogenesis (Figure 7H). Slug recruits, via its SNAG domain, Lsd1 to Fasn and other lipogenic gene promoters where Lsd1 catalyzes H3K9 demethylation, thereby epigenetically increasing expression of lipogenic genes and lipogenesis. Insulin potently stimulates the Slug epigenetic pathway which mediates, at least in part, insulin-stimulated lipogenesis. It is worth mentioning that many cytokines and cellular stressors, including TNF-α, IL-6, hypoxia, and oxidative stress, which are elevated in obesity, also stimulate Slug expression (32–36). We are tempted to propose that Slug serves as an epigenetic integrator of these obesogenic factors to orchestrate pathogenic lipogenesis, leading to NAFLD and related metabolic disease.
We previously reported that Snail1 suppresses de novo lipogenesis in hepatocytes (26). Snail1 elicits repressive deacetylation of both H3K9 and H3K27, but not demethylation of H3K9, on the Fasn promoter (26). Clearly, Slug and Snail1 have the opposing actions on de novo lipogenesis. Slug and Snail1 likely recruit distinct epigenetic enzymes to lipogenic promoters, resulting in functionally opposite histone modifications. Therefore, hepatic lipogenesis is likely to be governed by a Slug/Snail1 epigenetic balance. Hepatic Slug/Snail1 imbalance may contribute to aberrant lipogenesis and NAFLD. Of note, multiple transcription factors (e.g., Srebp1c, Lxrα, USF-1, and E2F1) have been identified to be involved in de novo lipogenesis (6–11). We postulate that Slug-triggered epigenetic modifications confer permissive chromatin conformations on which other transcription factors act to stimulate expression of lipogenic genes. This hypothesis warrants additional investigation in the future.
In conclusion, we unravel an insulin/Slug/Lsd1/H3K9 demethylation lipogenic pathway. This epigenetic pathway is aberrantly activated in the liver under obesity condition, and promotes NAFLD and insulin resistance. The Slug/Lsd1 pathway may serve a potential therapeutic target for the treatment of NAFLD and type 2 diabetes.
Methods
Generation of SlugΔhep mice.
Snai2 genomic DNA was prepared from 129X1/SvJ mice using PCR, confirmed by DNA sequencing, and used to prepare target vectors (Supplemental Figure 1B). Slug target vectors were introduced by electroporation into 129/Sv ES cells that were subsequently selected by G418 and FIAU. Slug targeting (Slugfl-Neo) was verified by PCR and DNA sequencing analyses. Slugfl-Neo ES cells were injected into C57BL/6J blastocysts to generate Slugfl-Neo mice. The Neo cassette was removed to generate Slugfl/+ mice by crossing Slugfl-Neo mice with TgACT-FLPe mice (Jackson Laboratory) (37). Slugfl/+ mice were backcrossed with C57BL/6J mice over 6 generations (remove the Flp gene) and crossed with albumin-Cre drivers to produce SlugΔhep (Slugfl/fl Cre+/–) mice. Slugfl/fl mice were crossed with albumin-CreERT2 drivers to generate Slugfl/fl CreER+/– mice. Adult Slugfl/fl CreER+/– mice were intraperitoneally injected with tamoxifen (Cayman Chemical) (0.5 mg/mouse, twice 2 days apart) to ablate Slug specifically in hepatocytes. Slugfl/fl mice were crossed with ob+/– mice to generate Slugfl/fl ob/ob mice. Adult Slugfl/fl ob/ob mice were transduced with Cre or GFP (control) adenoviral vectors (1011 viral particles/mouse) via tail vein injection to ablate hepatocyte Slug. Mice were housed on a 12-hour light-dark cycle in the Unit for Laboratory Animal Medicine at the University of Michigan (ULAM) and fed ad libitum either a chow diet (9% fat in calories; TestDiet) or a HFD (60% fat in calories; Research Diets).
Human samples.
Human liver samples were provided by the Liver Tissue Cell Distribution System at the University of Minnesota (Minneapolis, Minnesota, USA) and were described previously (38, 39). Both males and females were included. Individuals with an alcohol-drinking history (2–3 drinks/day) or liver cancer were excluded. The average ages for healthy individuals and NASH patients were approximately 56 and 53, respectively.
Liver-specific overexpression of Slug and Lsd1 inhibitor treatment.
Murine Slug or ΔN30 was inserted into AAV-CAG vectors. C57BL/6J males (8 weeks) were transduced with AAV-CAG-GFP, AAV-CAG-Slug, or AAV-CAG-ΔN30 vectors (1011 viral particles/mouse) via tail vein injection. After 1 week of recovery, they were placed on a HFD for 8 to 11 weeks. C57BL/6J males (9–10 weeks) were transduced with Slug or GFP adenoviral vectors via tail veins. Five days later, mice were treated with Lsd1 inhibitor GSK2879552 (10 mg/kg body weight, daily) for an additional 5 days. Livers were harvested for ChIP and immunoblotting assays.
Glucose, insulin, and pyruvate tolerance tests.
For glucose tolerance test (GTT), mice were fasted overnight and intraperitoneally injected with glucose (1 g/kg body weight). For insulin tolerance test (ITT), mice were fasted for 4 hours and intraperitoneally injected with human insulin (0.75 U/kg body weight). For pyruvate tolerance test (PTT), mice were fasted for 6 hours and intraperitoneally injected with pyruvate (1 g/kg body weight). Blood samples were collected from tail veins 0, 15, 30, 60, and 120 minutes after injection and used to measure blood glucose levels. Plasma insulin levels were measured using mouse insulin ELISA kits (Crystal Chem).
Nile red staining and TAG levels.
Liver frozen sections were fixed in 4% paraformaldehyde for 20 minutes, stained with Nile red (1 μg/mL in PBS) for approximately 30 minutes, and visualized using fluorescence microscopy (40). Liver samples were homogenized in 1% acetic acid. Lipids were extracted using 80% chloroform/methanol (2:1). Organic fractions were dried in a chemical hood, resuspended in a KOH (3 M)/ethanol solution, incubated at 70°C for 1 hour, and mixed with MgCl2 (0.75 M). Aqueous fractions were used to measure TAG levels using Free Glycerol Reagent (MilliporeSigma) (25).
Fasn luciferase reporter assays.
The rattus Fasn promoter (from –225 to +45) was prepared by PCR (forward primer: 5′-AGTGCCTCTCATGTATGCTTAA-3′ and reverse primer: 5′-TCCCGCAGTCTCGATACCTTGG-3′) and inserted into pGL3 vectors. HepG2 cells were grown in DMEM containing 5 mM glucose and 10% calf serum at 5% CO2 and 37°C, and transiently cotransfected with Fasn luciferase reporter plasmids using polyethylenimine (Sigma-Aldrich) (41). Luciferase activities were measured 72 hours after transfection using a kit (Promega) and normalized to β-gal internal control.
De novo lipogenesis and shRNA knockdown.
Primary hepatocytes were isolated using liver perfusion with type II collagenase (Worthington Biochem) (42), and were grown in William’s E Medium (Sigma-Aldrich) supplemented with 2% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Hepatocytes were transduced with Slug or GFP adenoviral vectors as described previously (43), and treated with GSK2879552 (1 μM) for 24 hours. Do novo lipogenesis was assessed using [3H]-acetate and normalized to total protein levels as described previously (44). Primary hepatocytes were isolated from SlugΔhep and Slugfl/fl mice at 8 to 10 weeks of age, deprived of serum overnight in the presence of 5 mM glucose, and stimulated with 50 nM insulin for 12 hours. Lipogenesis was measured as described above. Primary hepatocytes were isolated from overnight-fasted C57BL/6J males, grown in William’s E Medium pretreated with GSK2879552 (4 μM) for 4 hours, and subsequently stimulated with insulin (50 nM) in the presence of GSK2879552. Lipogenesis was measured 4 hours later. Primary hepatocytes were transduced with Srebp1c shRNA (GTCTTCTATCAATGACAAGA) adenoviral vectors (scramble RNA vectors as control) to silence Srebp1c as described previously (31). Concomitantly, hepatocytes were also transduced with Slug or β-gal adenoviral vectors, and subjected to lipogenesis or immunoblotting assays 2 days later.
Immunoblotting, immunoprecipitation, and protein stability.
Tissues or cells were homogenized in a lysis buffer (50 mM Tris-HCl, pH 7.5, 1.0% NP-40, 150 mM NaCl, 2 mM EGTA, 1 mM Na3VO4, 100 mM NaF, 10 mM Na4P2O7, 1 mM PMSF, 10 mg/mL aprotinin, and 10 mg/mL leupeptin). Tissue or cell extracts were immunoprecipitated and/or immunoblotted with the indicated antibodies. Primary hepatocytes were transduced with Slug adenoviral vectors. Forty hours later, the cells were derived of serum overnight and treated with cycloheximide (100 μg/mL) in the presence of either insulin (100 nM) or PBS (control) for 0 to 8 hours. Cell extracts were immunoblotted with antibodies against Slug or α-tubulin. Slug protein was quantified and normalized to α-tubulin levels. Slug abundance was presented as a ratio to its baseline levels before cycloheximide treatment and plotted against cycloheximide treatment duration. In some figures, proteins were blotted in parallel gels because their molecular weights or their abundance were drastically different. Antibody information is listed in Supplemental Table 1.
Quantitative real-time RT-PCR.
Total RNAs were extracted using TRIzol reagent (Invitrogen Life Technologies). The first-strand cDNAs were synthesized using random primers and M-MLV reverse transcriptase (Promega). Quantitative real-time RT-PCR (qPCR) was performed using Radiant SYBR Green 2X Lo-ROX qPCR Kits (Alkali Scientific) and StepOnePlus RT PCR Systems (Life Technologies Corporation). qPCR primers are listed in Supplemental Table 2.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were described previously (25). Briefly, liver samples were treated with 1% formaldehyde for 10 minutes to cross-link DNA protein complexes. Genomic DNA was extracted and sheared to 200- to 500-bp fragments using a sonicator (Model Q800R, QSONICA). DNA protein complexes were immunoprecipitated with the indicated antibodies. Cross-link was reversed by heating at 65°C for 4 hours. DNA was recovered using commercial kits or chemical purifications and used for PCR or qPCR analysis. Fasn promoter primers flanking the putative Slug-binding motifs are listed in Supplemental Table 2. ChIP antibody information is listed in Supplemental Table 1.
Affymetrix microarray analysis.
SlugΔhep and Slugfl/fl males were fed a HFD for 11 weeks. Variable transcripts were analyzed using The KEGG pathway analysis and DAVID Bioinformatics Resources version 6.8 (http://david.ncifcrf.gov).
Statistics.
Differences between 2 groups were analyzed by 2-tailed Student’s t test. Comparisons among more than 3 groups were analyzed by 1-way ANOVA/Sidak posttest (GraphPad Prism 7). Longitudinal data (GTT, ITT, and PTT) were analyzed by 2-way ANOVA/Bonferroni’s posttest (GraphPad Prism 7). A P value less than 0.05 was considered significant. Complete antibody and primer source data are presented in Supplemental Information.
Study approval.
Animal experiments were conducted following the protocols approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC).
Author contributions
YL, HL, LJ, and QS conducted the experiments. YL and LR designed the experiments and wrote the paper. YL, HL, QS, LJ, LY, JDL, WSW, and LR performed data analysis and edited the paper.
Supplementary Material
Acknowledgments
We thank Mark J. Canet, Zhiguo Zhang, Xin Tong, Yatrik M. Shah, and M. Bishr Omary for assistance and discussions. This study was supported by grants R01 DK094014, R01 DK114220, and R01 DK115646 (to LR) and DK102456 (to JDL) from the National Institutes of Health; the American Diabetes Association 1-18-IBS-189 (to LR); and the American Heart Association Postdoctoral Fellowship 14POST20230007 (to YL). This work used the cores supported by the Michigan Diabetes Research and Training Center (NIH DK20572) and the University of Michigan Gut Peptide Research Center (NIH DK34933).
Version 1. 02/25/2020
In-Press Preview
Version 2. 05/04/2020
Electronic publication
Version 3. 06/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):2992–3004.https://doi.org/10.1172/JCI128073.
See the related Commentary at Insulin-stimulated lipogenesis gets an epigenetic makeover.
Contributor Information
Yan Liu, Email: liuyan.umich@gmail.com.
Haiyan Lin, Email: lhaiyan@umich.edu.
Lin Jiang, Email: godspeedj2002@gmail.com.
Qingsen Shang, Email: shangqs@umich.edu.
Lei Yin, Email: leiyin@umich.edu.
Jiandie D. Lin, Email: jdlin@umich.edu.
Wen-Shu Wu, Email: wuwenshu@uic.edu.
Liangyou Rui, Email: ruily@umich.edu.
References
- 1.Chirala SS, et al. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci U S A. 2003;100(11):6358–6363. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert JE, Ramos-Roman MA, Browning JD, Parks EJ. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014;146(3):726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabbrini E, et al. Physiological mechanisms of weight gain-induced steatosis in people with obesity. Gastroenterology. 2016;150(1):79–81.e2. doi: 10.1053/j.gastro.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erion DM, et al. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metab. 2009;10(6):499–506. doi: 10.1016/j.cmet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jornayvaz FR, Shulman GI. Diacylglycerol activation of protein kinase Cε and hepatic insulin resistance. Cell Metab. 2012;15(5):574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15(5):725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobin KA, et al. Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J Biol Chem. 2002;277(12):10691–10697. doi: 10.1074/jbc.M109771200. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A. 2004;101(31):11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136(6):1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denechaud PD, et al. E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. J Clin Invest. 2016;126(1):137–150. doi: 10.1172/JCI81542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, Fang S, Byun S, Seok S, Kemper B, Kemper JK. Farnesoid X receptor-induced lysine-specific histone demethylase reduces hepatic bile acid levels and protects the liver against bile acid toxicity. Hepatology. 2015;62(1):220–231. doi: 10.1002/hep.27677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inukai T, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4(3):343–352. doi: 10.1016/S1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 14.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari-Amorotti G, et al. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 2013;73(1):235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambeat A, et al. LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. 2016;15(11):2536–2549. doi: 10.1016/j.celrep.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Dong C, Zhou BP. Epigenetic regulation of EMT: the Snail story. Curr Pharm Des. 2014;20(11):1698–1705. doi: 10.2174/13816128113199990512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desgrosellier JS, et al. Integrin αvβ3 drives slug activation and stemness in the pregnant and neoplastic mammary gland. Dev Cell. 2014;30(3):295–308. doi: 10.1016/j.devcel.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue A, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell. 2002;2(4):279–288. doi: 10.1016/S1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, Feinberg T, Keller ET, Li XY, Weiss SJ. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat Cell Biol. 2016;18(9):917–929. doi: 10.1038/ncb3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu WS, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123(4):641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 25.Sun C, et al. Adipose Snail1 regulates lipolysis and lipid partitioning by suppressing adipose triacylglycerol lipase expression. Cell Rep. 2016;17(8):2015–2027. doi: 10.1016/j.celrep.2016.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Insulin/Snail1 axis ameliorates fatty liver disease by epigenetically suppressing lipogenesis. Nat Commun. 2018;9(1):2751. doi: 10.1038/s41467-018-05309-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SY, et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72(14):3607–3617. doi: 10.1158/0008-5472.CAN-12-0006. [DOI] [PubMed] [Google Scholar]
- 28.Dong C, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23(3):316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Mancera PA, et al. Adipose tissue mass is modulated by SLUG (SNAI2) Hum Mol Genet. 2007;16(23):2972–2986. doi: 10.1093/hmg/ddm278. [DOI] [PubMed] [Google Scholar]
- 30.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic breast cancer 1, early onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109(41):16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong X, et al. E4BP4 is an insulin-induced stabilizer of nuclear SREBP-1c and promotes SREBP-1c-mediated lipogenesis. J Lipid Res. 2016;57(7):1219–1230. doi: 10.1194/jlr.M067181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storci G, et al. TNFalpha up-regulates SLUG via the NF-κB/HIF1α axis, which imparts breast cancer cells with a stem cell-like phenotype. J Cell Physiol. 2010;225(3):682–691. doi: 10.1002/jcp.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao JW, Liu LJ, Huang J. Interleukin-6-induced epithelial-mesenchymal transition through signal transducer and activator of transcription 3 in human cervical carcinoma. Int J Oncol. 2014;45(1):165–176. doi: 10.3892/ijo.2014.2422. [DOI] [PubMed] [Google Scholar]
- 34.Huang CH, et al. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11(12):1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha HS, Bae EK, Ahn JK, Lee J, Ahn KS, Koh EM. Slug suppression induces apoptosis via Puma transactivation in rheumatoid arthritis fibroblast-like synoviocytes treated with hydrogen peroxide. Exp Mol Med. 2010;42(6):428–436. doi: 10.3858/emm.2010.42.6.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MC, Cui FJ, Kim Y. Hydrogen peroxide promotes epithelial to mesenchymal transition and stemness in human malignant mesothelioma cells. Asian Pac J Cancer Prev. 2013;14(6):3625–3630. doi: 10.7314/APJCP.2013.14.6.3625. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Morris DL, Jiang L, Liu Y, Rui L. SH2B1 in β-cells regulates glucose metabolism by promoting β-cell survival and islet expansion. Diabetes. 2014;63(2):585–595. doi: 10.2337/db13-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, et al. Hepatic neuregulin 4 signaling defines an endocrine checkpoint for steatosis-to-NASH progression. J Clin Invest. 2017;127(12):4449–4461. doi: 10.1172/JCI96324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Sheng L, Xiong Y, Shen H, Liu Y, Rui L. Liver NF-κB-inducing kinase promotes liver steatosis and glucose counterregulation in male mice with obesity. Endocrinology. 2017;158(5):1207–1216. doi: 10.1210/en.2016-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang B, Shen H, Chen Z, Yin L, Zan L, Rui L. Carboxyl terminus of HSC70-interacting protein (CHIP) down-regulates NF-κB-inducing kinase (NIK) and suppresses NIK-induced liver injury. J Biol Chem. 2015;290(18):11704–11714. doi: 10.1074/jbc.M114.635086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheng L, et al. NF-κB–inducing kinase (NIK) promotes hyperglycemia and glucose intolerance in obesity by augmenting glucagon action. Nat Med. 2012;18(6):943–949. doi: 10.1038/nm.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen H, et al. Mouse hepatocyte overexpression of NF-κB-inducing kinase (NIK) triggers fatal macrophage-dependent liver injury and fibrosis. Hepatology. 2014;60(6):2065–2076. doi: 10.1002/hep.27348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng L, Cho KW, Zhou Y, Shen H, Rui L. Lipocalin 13 protein protects against hepatic steatosis by both inhibiting lipogenesis and stimulating fatty acid β-oxidation. J Biol Chem. 2011;286(44):38128–38135. doi: 10.1074/jbc.M111.256677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.