Abstract
Single-nucleotide polymorphisms and locus amplification link the NF-κB transcription factor c-Rel to human autoimmune diseases and B cell lymphomas, respectively. However, the functional consequences of enhanced c-Rel levels remain enigmatic. Here, we overexpressed c-Rel specifically in mouse B cells from BAC-transgenic gene loci and demonstrate that c-Rel protein levels linearly dictated expansion of germinal center B (GCB) cells and isotype-switched plasma cells. c-Rel expression in B cells of otherwise c-Rel–deficient mice fully rescued terminal B cell differentiation, underscoring its critical B cell–intrinsic roles. Unexpectedly, in GCB cells transcription-independent regulation produced the highest c-Rel protein levels among B cell subsets. In c-Rel–overexpressing GCB cells this caused enhanced nuclear translocation, a profoundly altered transcriptional program, and increased proliferation. Finally, we provide a link between c-Rel gain and autoimmunity by showing that c-Rel overexpression in B cells caused autoantibody production and renal immune complex deposition.
Keywords: Autoimmunity, Immunology
Keywords: Autoimmune diseases, B cells, NF-kappaB
Introduction
c-Rel is a transcription factor of the NF-κB family strongly linked to human immune pathology. Genome-wide association studies (GWAS) demonstrate that single-nucleotide polymorphisms (SNPs) within the REL locus are associated with human autoimmune diseases including rheumatoid arthritis (1, 2), psoriasis (3), ulcerative colitis (4), celiac disease (5, 6), and psoriatic arthritis (7). B cells are central contributors to autoimmunity through production of autoantibodies and cytokines as well as through antigen presentation. Strikingly, a correlation of autoimmunity-associated SNPs to cell type–specific cis-regulatory elements implicates B cells in all autoimmune conditions linked to REL (8). The first Rel family member, reticuloendotheliosis virus (v-Rel), was discovered because of its ability to malignantly transform lymphoid chicken cells in culture (9). Subsequent studies revealed frequent gains and amplification of the REL gene locus in human B cell lymphomas (10).
Together, these studies clearly implicate c-Rel in human pathology. c-Rel–knockout mice revealed important functions of c-Rel particularly in B and T lymphocytes (11), in line with its expression mostly in hematopoietic cells under normal physiological conditions (12). In B cells, cardinal triggers of c-Rel activation include B cell receptor (BCR) signaling and CD40 ligation as well as engagement of Toll-like receptors (TLRs) (10, 13). These trigger the canonical NF-κB pathway, culminating in the nuclear translocation of NF-κB transcription factors including c-Rel, to drive target gene transcription (14).
Reported c-Rel–dependent genes in B cells include inflammatory mediators, prosurvival proteins, and factors mediating proliferation as well as genes involved in cellular metabolism (10, 15). Accordingly, c-Rel–deficient B cells present strong proliferative defects and reduced survival in response to mitogenic stimulation in vitro (16, 17). Particularly, hallmarks of G1-S transition are dependent on c-Rel (15, 18–21).
In vivo, antigen recognition by B cells in the context of appropriate T cell help results in the formation of germinal centers (GCs), where B cells proliferate and undergo somatic hypermutation as well as class-switch recombination (CSR). GC B (GCB) cells exit the GC to terminally differentiate into antibody-producing plasma cells or memory B cells (22). Corresponding to the in vitro defects of c-Rel–deficient B cells, c-Rel–knockout mice essentially fail to develop GCs in response to immunization (18, 21) and display a severe reduction in antibody titers, especially of the IgG1 and IgG2a isotypes (16). c-Rel affects CSR by regulating B cell proliferation as well as immunoglobulin germline transcription (16, 23). GCB cell–specific gene targeting revealed that GCB cells essentially collapse upon loss of c-Rel as a result of impaired growth and metabolic fitness (15).
Although these studies uncovered important roles of c-Rel in the immune system, direct evidence of a pathophysiological role for gain of c-Rel function is missing to date, mostly owing to the lack of suitable animal models (11). To directly address this fundamental issue, we generated mouse models allowing cell type–specific overexpression of gene loci encoding c-Rel or a GFP–c-Rel fusion protein. These mouse models allowed exploration of the in vivo consequences of c-Rel overexpression and investigation of whether c-Rel gain in B cells constitutes a direct functional link to autoimmunity.
Results
Enhanced c-Rel expression in B cells causes spontaneous expansion of GCB cells.
To generate conditional transgenic (Tg) mouse models for c-Rel overexpression, we modified the mouse Rel gene locus on a bacterial artificial chromosome (BAC). To allow Cre-dependent expression of c-Rel and GFP–c-Rel loci, we introduced a strong CAG promoter followed by a loxP site–flanked STOP cassette upstream of the first translated Rel exon (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI124382DS1). Modified BACs were electroporated into embryonic stem (ES) cells, and clones carrying a single BAC integrant were identified by Southern blot (Supplemental Figure 1).
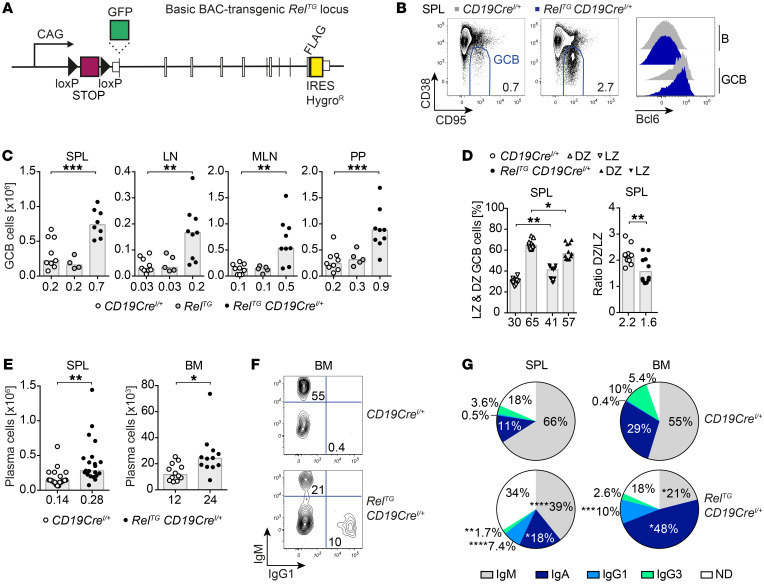
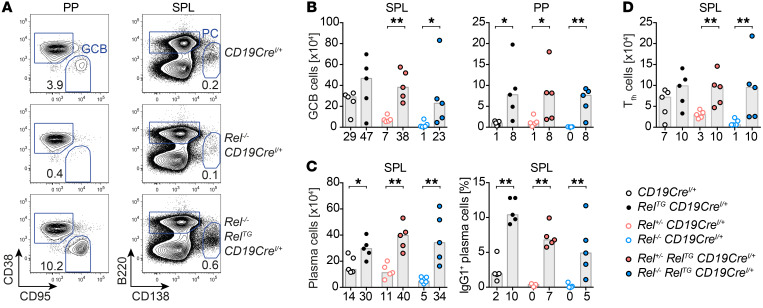
Figure 1. B cell–specific c-Rel overexpression causes spontaneous GCB cell expansion and leads to an accumulation of class-switched plasma cells.
(A) Scheme of the RelTG and GFP-RelTG BAC-transgenic loci. A CAG promoter followed by a loxP-flanked STOP cassette, an N-terminal HA tag or GFP fusion, and a carboxy-terminal FLAG tag were inserted. (B) Representative flow cytometry plots of GCB cells (B220+/CD19+ CD95hiCD38lo). Displayed numbers are median percentages of GCB cells within all B cells (n ≥ 8). Representative flow cytometry histograms of intracellular Bcl6 protein expression. (C) Cell numbers of CD95hiCD38lo GCB cells. Individual data points obtained in 3 or more independent experiments are plotted. Bars and numbers below graphs are median values. **P ≤ 0.01, ***P ≤ 0.001, 1-way ANOVA. (D) Frequencies of light zone (LZ; CXCR4loCD86hi) and dark zone (DZ; CXCR4hiCD86lo) GCB cells and DZ/LZ ratio. Individual data points are plotted. Numbers below graphs and bars are median values for frequencies and geometric means for ratios. *P ≤ 0.05, **P ≤ 0.01, unpaired t test. (E) Cell numbers of CD138+B220lo plasma cells. Individual data points obtained in 6 or more independent experiments are plotted. Bars and numbers below graphs are median values. *P ≤ 0.05, **P ≤ 0.01, unpaired t test. (F and G) Representative flow cytometry plots (F) and pie charts (G) of intracellular Ig isotype staining in plasma cells. Displayed median percentages include data from 8–14 mice analyzed in 5 or more independent experiments. Significant differences with respect to the CD19CreI/+ control genotype are indicated by asterisks adjacent to the respective percentages in the RelTG CD19CreI/+ charts. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, unpaired t test. BAC, bacterial artificial chromosome; HygroR, hygromycin B resistance; SPL, spleen; LN, lymph nodes; MLN, mesenteric lymph nodes; PP, Peyer’s patches; BM, bone marrow; ND, not determined. See Supplemental Figures 1–4.
For B cell–specific c-Rel overexpression, we crossed resulting conditional c-Rel–Tg mice to CD19Cre mice (24). Although the B cell populations in bone marrow, spleen, and peritoneal cavity were largely unchanged in RelTG CD19CreI/+ mice (Supplemental Figure 2, A–C), a higher cellularity of B cells in peripheral organs including lymph nodes (LNs), mesenteric lymph nodes (MLNs), and Peyer’s patches (PPs) was found (Supplemental Figure 2E). The cell-surface levels of B cell activation markers were largely unchanged, except that CD23 was slightly elevated in peripheral B cell populations (Supplemental Figure 2D and data not shown), although not to the same extent as caused by constitutively active canonical NF-κB signaling (25).
Conversely, in striking contrast to the effects of constitutive canonical NF-κB activation, c-Rel overexpression caused spontaneous GCs in spleen and LNs and enhanced GCs in gut-associated lymphoid tissues (GALTs), including MLNs and PPs (Figure 1, B and C, and Supplemental Figure 3, A and B). These expanded GCB cells expressed normal levels of the GC transcriptional regulator Bcl6 and other GC markers including GL7 and PNA (Figure 1B and Supplemental Figure 3, C–E) and showed a trend for a higher proportion of GCB cells with a CXCR4loCD86hi light zone (LZ) phenotype (ref. 26 and Figure 1D).
Enhanced GCs were accompanied by significantly augmented T cell populations in LNs (Supplemental Figure 4, A and B), including specialized CD4+PD-1hiCXCR5hi T follicular helper (Tfh) cells that provide essential molecular cues for effective GC reactions. Tfh cells were expanded in spleen and LNs as well as in the GALT (Supplemental Figure 4, C and D).
This first characterization establishes that gain of c-Rel in B cells causes spontaneous GC reactions.
c-Rel gain induces immunoglobulin class-switched plasma cells.
In line with expanded GCB cells, c-Rel overexpression in B cells also caused a spontaneous significant expansion of CD138+B220lo antibody-producing plasma cells expressing normal levels of the pivotal transcriptional regulators IRF4 and Blimp1 in both spleen and bone marrow (Figure 1E and Supplemental Figure 3, F–H). Although most plasma cells in control mice were of the unswitched IgM subtype, the majority of plasma cells in RelTG CD19CreI/+ mice had switched to other subclasses, including IgG1 and IgA. Particularly, the proportion of IgG1-expressing plasma cells was expanded 14- to 20-fold through c-Rel overexpression (Figure 1, F and G, and Supplemental Figure 3I). Accordingly, RelTG CD19CreI/+ mice presented with higher serum titers of the switched isotypes IgG1, IgG2c, and IgG2b, while IgM serum titers were significantly decreased compared with controls (Supplemental Figure 3J).
In summary, enhanced c-Rel expression specifically in B cells triggers an accumulation of class-switched plasma cells that translates into augmented antibody serum titers of switched isotypes.
c-Rel gain enhances GC reactions in response to immunization.
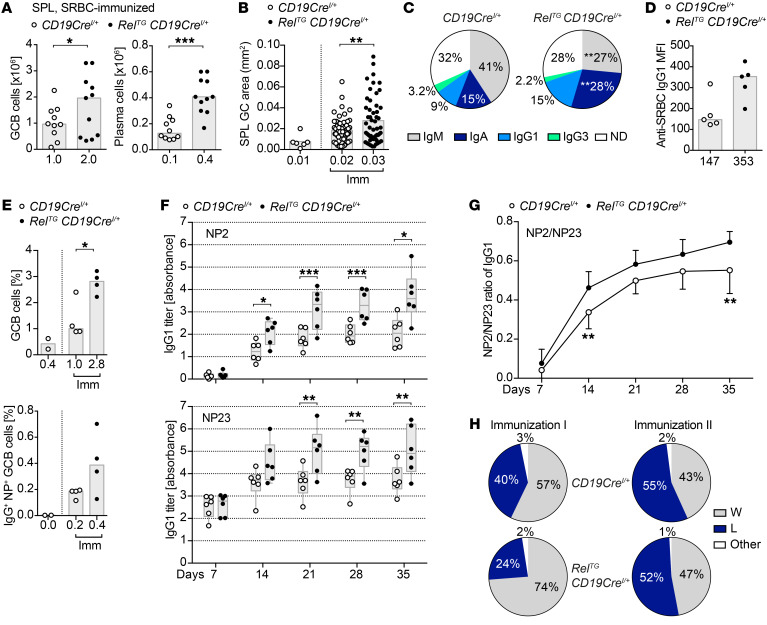
We next investigated consequences of c-Rel overexpression during induced immune reactions. For this purpose, we immunized control and RelTG CD19CreI/+ mice with the T cell–dependent antigen sheep red blood cells (SRBCs). Although splenic total B cell numbers were unaffected (Supplemental Figure 5A), GCB cell, Tfh cell, and plasma cell populations were significantly expanded in RelTG CD19CreI/+ mice following immunization (Figure 2A, Supplemental Figure 4E, and Supplemental Figure 5A). Immunofluorescence microscopy revealed that SRBC-immunized RelTG CD19CreI/+ mice contained enlarged GCs (Figure 2B) accompanied by a trend for a higher number of total GCs (Supplemental Figure 5B). Furthermore, plasma cells of RelTG CD19CreI/+ mice were characterized by a higher fraction of class-switched plasma cells compared with controls (Figure 2C), which was also reflected by enhanced SRBC-specific IgG1 production (Figure 2D), while SRBC-specific IgM levels remained unchanged (Supplemental Figure 5C).
Figure 2. c-Rel gain enhances immune responses upon immunization.
(A) CD95hiCD38lo GCB and CD138+B220lo plasma cell numbers 10–12 days after SRBC immunizations obtained from 4 independent experiments. Bars and numbers below graphs are medians. *P ≤ 0.05, ***P ≤ 0.001, paired t test. (B) Areas of splenic GCs quantified based on Bcl6 immunofluorescence in 5 CD19CreI/+ and 5 RelTG CD19CreI/+ mice 10 days after SRBC immunizations (CD19CreI/+, n = 61; RelTG CD19CreI/+, n = 53). Bars and numbers below graphs are means. **P ≤ 0.01, unpaired t test. (C) Percentages of Ig subtypes in splenic plasma cells obtained by intracellular flow cytometry 10–12 days after SRBC immunization. Displayed medians include data from 6 mice from 2 independent experiments. **P ≤ 0.01, paired t test. (D) Individual anti-SRBC IgG1 titers analyzed by flow cytometry 10 days after immunization of 2 independent experiments. Bars and numbers below graphs are medians. (E) NP-CG–immunized mice analyzed after 14 days: Individual percentages of CD95hiCD38lo GCB and IgG+NP+ GCB cells within all splenic B cells are plotted. Bars and numbers below graphs are medians. *P < 0.05, unpaired t test. (F and G) Specific IgG1 serum titers measured by ELISA and calculated by absorbance summation (61) following NP-CG immunizations. Titers for NP2 (high affinity) and NP23 (high and low affinity) (F) and the NP2/NP23 ratio (G) are displayed from 2 independent immunizations. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, multiple t tests. (H) Affinity maturation 14 days after NP-CG immunizations analyzed by sequencing of IgG1 VH186.2 rearrangements in GCB cells for 2 independently immunized cohorts with 2 mice per genotype: 4 CD19CreI/+ mice: immunization I, n = 126 sequences; immunization II, n = 148 sequences; 4 RelTG CD19CreI/+ mice: immunization I, n = 122 sequences; immunization II, n = 117 sequences. The percentage of sequences with the VH186.2 W33L mutation is shown. SPL, spleen; Imm, immunized; ND, not determined. See Supplemental Figures 5 and 6.
To further characterize antigen-specific immune responses, we immunized mice with 4-hydroxy-3-nitrophenyl-acetyl chicken γ-globulin (NP-CG). This resulted in enhanced numbers of antigen-specific IgG+NP+ GCB cells and Igλ1+NP+ GCB cells (Figure 2E and Supplemental Figure 6A), correlating with overall elevated GCB cell numbers (Supplemental Figure 6B). Fittingly, the total number of cells producing NP-specific IgG1 (NP23) as well as the number of cells producing high-affinity NP-specific IgG1 (NP2) was also increased in RelTG CD19CreI/+ mice (Supplemental Figure 6C), corresponding to significantly higher serum titers of overall and high-affinity NP–specific IgG1 antibodies starting at 14 days after immunization (Figure 2F). Evaluating NP2/NP23 ratios, we observed a trend toward enhanced affinity maturation in RelTG CD19CreI/+ mice (Figure 2G).
Next, we performed VDJ sequencing on BCRs of the VH186.2 (J558) V gene family in GCB cells purified from mice immunized with NP-CG (Figure 2H and Supplemental Figure 6, D and E). No significant differences in the acquisition of somatic mutations or in the selection of the W33L mutation that increases the affinity of J558-bearing BCRs for the immunizing antigen NP 10-fold were observed.
We conclude that elevated c-Rel significantly enhances GC reactions and ensuing antibody production in response to specific antigens. Enhanced c-Rel levels do not affect somatic hypermutation and have only a minor, if any, impact on affinity maturation.
Enhanced c-Rel expression from the GCB cell stage onward is sufficient for GCB cell and plasma cell expansion.
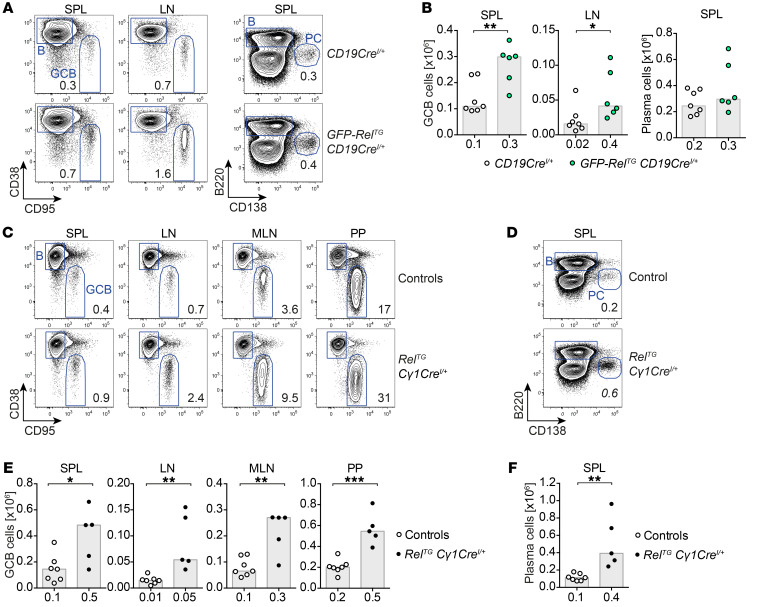
To verify our findings in an independent model, we analyzed single-copy BAC-transgenic mice conditionally expressing a GFP–c-Rel fusion protein. GFP-RelTG CD19CreI/+ mice essentially recapitulated the phenotypes observed in RelTG CD19CreI/+ mice (Figure 3, A and B), demonstrating not only the functionality of the GFP–c-Rel protein but also validating the robust in vivo consequences of c-Rel overexpression.
Figure 3. GFP–c-Rel transgene and c-Rel transgene overexpression limited to the GC stage and beyond phenocopy B cell–specific c-Rel overexpression.
(A) Representative flow cytometry plots of CD95hiCD38lo GCB cells and CD138+B220lo plasma cells in GFP-RelTG CD19CreI/+ and control mice. Displayed numbers are median percentages (n ≥ 6). (B) CD95hiCD38lo GCB cell numbers and CD138+B220lo plasma cell numbers. Individual data points obtained in 2 independent experiments are plotted. Bars and numbers below graphs are median values. *P ≤ 0.05, **P ≤ 0.01, unpaired t test. (C–F) Representative flow cytometry plots and cell numbers of CD95hiCD38lo GCB cells (C and E) and CD138+B220lo plasma cells (D and F) in RelTG Cγ1CreI/+ and control mice. Displayed numbers in plots are median percentages (n ≥ 5). Individual data points obtained in 2 independent experiments are plotted in graphs. Bars and numbers below graphs are median values. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, unpaired t test. SPL, spleen; LN, lymph nodes; MLN, mesenteric lymph nodes; PP, Peyer’s patches. See Supplemental Figure 4F.
We then asked whether the developmental timing of c-Rel overexpression is critical for the observed expansion of terminally differentiated B cells. To address this question, we used RelTG Cγ1CreI/+ mice, in which c-Rel overexpression occurs in GCB cells and derivative cells (27). Interestingly, RelTG Cγ1CreI/+ mice phenocopied RelTG CD19CreI/+ mice with respect to GCB cell and plasma cell expansion (Figure 3, C–F, and Supplemental Figure 4F). These experiments demonstrate that c-Rel overexpression initiated at the GC stage suffices to cause spontaneous terminal B cell differentiation.
Transgenic c-Rel expression in B cells of c-Rel–knockout mice is sufficient to rescue loss of terminal B cell differentiation.
Having discovered that enhanced c-Rel expression in B or GCB cells can drive spontaneous as well as induced GC reactions, we evaluated whether c-Rel expression in B cells alone can rescue the observed loss of GCs in c-Rel–deficient mice. To this end, we crossed our RelTG CD19CreI/+ mice to c-Rel–knockout mice (15). Remarkably, B cell–specific c-Rel expression fully reconstituted the populations of GCB cells, Tfh cells, and plasma cells in heterozygous and homozygous c-Rel–deficient mice (Figure 4, A–D). Interestingly, plasma cells showed a higher frequency of class switching compared with their c-Rel–knockout and CD19CreI/+ counterparts. In particular, switching to the IgG1 isotype, which was completely blocked in heterozygous or homozygous c-Rel–knockout mice, was fully rescued (Figure 4C).
Figure 4. B cell–restricted c-Rel expression in c-Rel–knockout mice fully rescues GC reactions and class switching.
(A) Representative flow cytometry plots of GCB cells (B220+CD95hiCD38lo) and plasma cells (PC, B220loCD138+). Displayed numbers are median percentages (n = 5). (B–D) GCB cell numbers (B); plasma cell numbers and percentages of IgG1-expressing plasma cells (C); and Tfh cell (TCRβ+CD4+CD8–CXCR5hiPD-1hiICOShi) numbers (D). Individual data points obtained in 3 independent experiments are plotted. Bars and numbers below graphs are median values. *P ≤ 0.05, **P ≤ 0.01, Mann-Whitney test. SPL, spleen; PP, Peyer’s patches.
These experiments highlight the in vivo functionality of our transgenically expressed FLAG-tagged c-Rel. Importantly, our data demonstrate that c-Rel expression specifically in B cells is sufficient to drive GC reactions including the generation and expansion of GCB cells and Tfh cells as well as class-switched plasma cells in the absence of c-Rel expression in all other cell types.
GCB cells contain elevated c-Rel, and high c-Rel expression confers a strong competitive advantage in the GC.
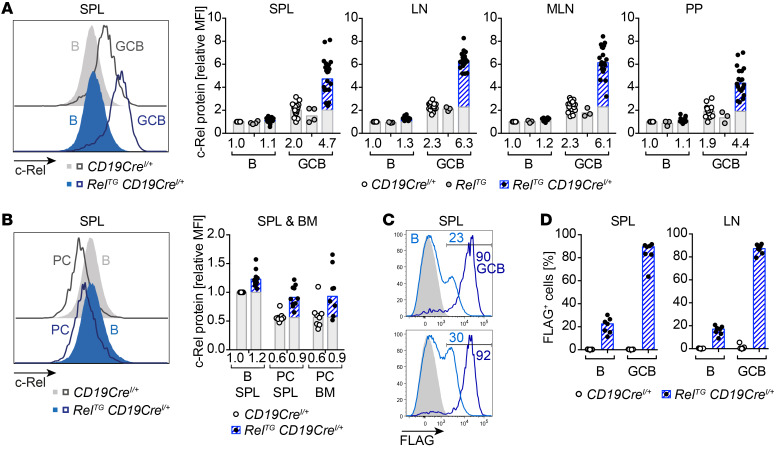
Our data reveal a selected impact of c-Rel overexpression on terminal B cell differentiation. To correlate the observed phenotypes with c-Rel protein abundance, we quantified c-Rel protein levels using intracellular flow cytometry in both c-Rel–Tg and control mice. Intriguingly, we discovered that c-Rel protein levels were upregulated in GCB cells compared with naive B cells (Figure 5A), contrary to what was reported for Rel mRNA levels (28, 29). This finding held true for GCB cells in all analyzed organs, including spleen, LNs, and GALT (Figure 5A). In control mice, c-Rel levels were around 2-fold higher in GCB cells than in naive B cells. In RelTG CD19CreI/+ mice, c-Rel protein levels were 2- to 3-fold higher in GCB cells compared with controls. Within the GC, c-Rel protein levels were moderately higher in LZ compared with dark zone (DZ) GCB cells, irrespective of the genotype (Supplemental Figure 7A). In contrast, in spleen and bone marrow plasma cells, c-Rel protein levels were approximately halved in comparison with naive B cells (Figure 5B). Notably, total c-Rel levels in naive B cells were only marginally higher in RelTG CD19CreI/+ mice compared with CD19CreI/+ controls (Figure 5A). The quantification of c-Rel protein was confirmed using an alternative anti–c-Rel antibody clone (data not shown).
Figure 5. c-Rel protein level dynamics during terminal B cell differentiation.
(A and B) Intracellular flow cytometry data of c-Rel protein abundance. Representative flow cytometry histograms and relative median fluorescence intensities (MFIs) of intracellular c-Rel. Individual data points of 3 or more independent experiments are plotted. Bars and numbers below graphs are geometric means. The c-Rel protein fraction that is additionally present in c-Rel transgenic mice in comparison with respective control populations is highlighted in blue. (A) c-Rel levels in GCB cells (B220+/CD19+ CD95hiCD38lo) normalized to non-GCB (B, B220+/CD19+ CD38+CD95–) of CD19CreI/+ controls. (B) c-Rel expression in plasma cells (PC, B220loCD138+) normalized to B cells (B, B220+CD138–) of CD19CreI/+ controls. (C and D) Intracellular flow cytometry staining of FLAG. (C) Histograms with percentage of FLAG-positive cells of 2 representative RelTG CD19CreI/+ mice. Light blue, B cells (B220+CD38+CD95–); dark blue, GCB cells (B220+CD95hiCD38lo); gray filled, B cells of CD19CreI/+ control. Gate for FLAG-positive B cell population is displayed. (D) Percentage of FLAG-positive subpopulations in B cells and GCB cells. Individual data points (n = 7) and bars representing median values are shown. SPL, spleen; LN, lymph nodes; MLN, mesenteric lymph nodes; PP, Peyer’s patches; BM, bone marrow. See Supplemental Figure 7A.
Our BAC-Tg mouse models were designed with a FLAG tag added to the c-Rel C-terminus, which allows specific detection of transgenic c-Rel expression. Intracellular anti-FLAG staining revealed variegated expression of the transgene in naive B cells, of which 9%–30% displayed detectable FLAG expression. In striking contrast, within the GCB cell population around 90% of cells were positive for FLAG expression, indicating strong positive selection pressure favoring GCB cells containing high c-Rel levels (Figure 5, C and D).
Accordingly, we demonstrate that c-Rel protein abundance is specifically upregulated at the GCB cell stage and that B cells expressing high levels of c-Rel have a very strong competitive advantage in the GC.
GCB cells contain predominantly nuclear c-Rel.
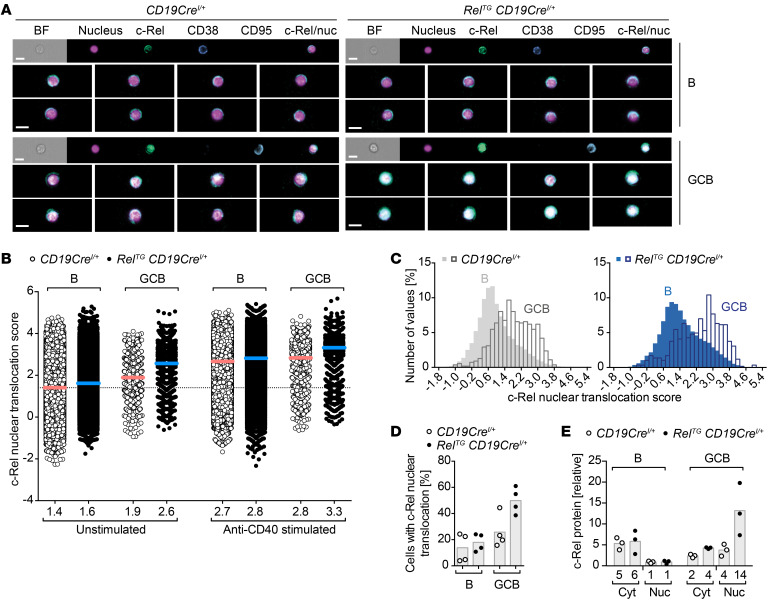
Elevated protein levels of the c-Rel transcription factor per se might not necessarily translate into enhanced function. As a correlate for c-Rel function, we therefore assessed c-Rel nuclear localization in B cells and GCB cells of both control and RelTG CD19CreI/+ mice by imaging flow cytometry. In agreement with published immunofluorescence data (30, 31), we found that in naive resting B cells c-Rel was predominantly cytosolic with only a fraction of cells showing partial nuclear stain, while in unstimulated control GCB cells c-Rel was more strongly localized to the nucleus based on a comparably higher nuclear localization score, defined as described in Methods below (ref. 32, Figure 6, A and B, and Supplemental Figure 7B). In RelTG CD19CreI/+ mice, c-Rel nuclear localization in GCB cells was even more pronounced than in control mice (Figure 6, A–C).
Figure 6. c-Rel protein nuclear translocation during terminal B cell differentiation.
(A–D) Imaging flow cytometry analyses to assess c-Rel nuclear translocation in B cells (B220+CD38+CD95–) and GCB cells (B220+CD95hiCD38lo) in LNs. Cells were stimulated with anti-CD40 for 60 minutes or left unstimulated in the presence of the pan-caspase inhibitor Q-VD-OPh to prolong survival of GCB cells. (A) Representative images of c-Rel nuclear translocation in unstimulated cells. Images were selected based on average nuclear localization score. Scale bars: 10 μm. (B) Quantified c-Rel nuclear translocation score in unstimulated or anti-CD40–stimulated B cells and GCB cells. Individual data points were pooled from 4 mice per genotype obtained in 3 independent experiments (pooled data points for CD19CreI/+: B unstimulated, n = 67,148; B stimulated, n = 75,884; GCB unstimulated, n = 1129; GCB stimulated, n = 1267; pooled data points for RelTG CD19CreI/+: B unstimulated, n = 80,494; B stimulated, n = 89,012; GCB unstimulated, n = 1861; GCB stimulated, n = 1951). Bars and numbers below graphs represent mean values calculated from the mean nuclear translocation scores for each mouse. (C) Histograms to visualize the frequency distribution of nuclear translocation scores for a representative mouse for each genotype. (D) Percentage of cells within the population of unstimulated B and GCB cells with a nuclear translocation score of 2.7 or more. Individual data points for 4 mice per genotype obtained in 3 independent experiments and bars representing mean values are shown. (E) c-Rel protein analyzed by capillary Western blot. Non-GCB (B) and GCB cells were FACS-sorted 7–9 days after SRBC immunization followed by nuclear fractionation. Each data point represents 1 sample of 1 × 106 cells sorted from 2–3 mice. For quantification, c-Rel protein signals were normalized to PTEN (cytosol) or lamin B2 (nucleus). BF, bright field; Nuc, nucleus; Cyt, cytosol. See Supplemental Figure 7.
An essential signal for GCB cells is stimulation of the CD40 receptor that in vivo is provided by CD40L expression on Tfh cells. Upon in vitro anti-CD40 stimulation, c-Rel translocated to the nucleus in B cells and GCB cells of both control and RelTG CD19CreI/+ mice (Figure 6B and Supplemental Figure 7C). Remarkably, the nuclear localization score of 2.6 in unstimulated c-Rel–overexpressing GCB cells reached a range similar to the score of 2.7 obtained in anti-CD40–stimulated control B cells (Figure 6B). Based on the nuclear localization score, we found that elevated expression of c-Rel in RelTG CD19CreI/+ mice increased the proportion of GCB cells with nuclear c-Rel localization to 50% compared with 26% in control mice and to 18% in non-GCB cells compared with 14% in control non-GCB cells (Figure 6D).
In line with the imaging flow cytometry data, the analysis of nuclear and cytosolic fractions of flow cytometry–purified B cell subsets from SRBC-immunized mice by capillary Western blot demonstrated higher c-Rel protein levels in the nucleus of GCB cells compared with non-GCB cells in CD19CreI/+ control mice and the highest nuclear c-Rel levels in GCB cells of RelTG CD19CreI/+ mice (Figure 6E).
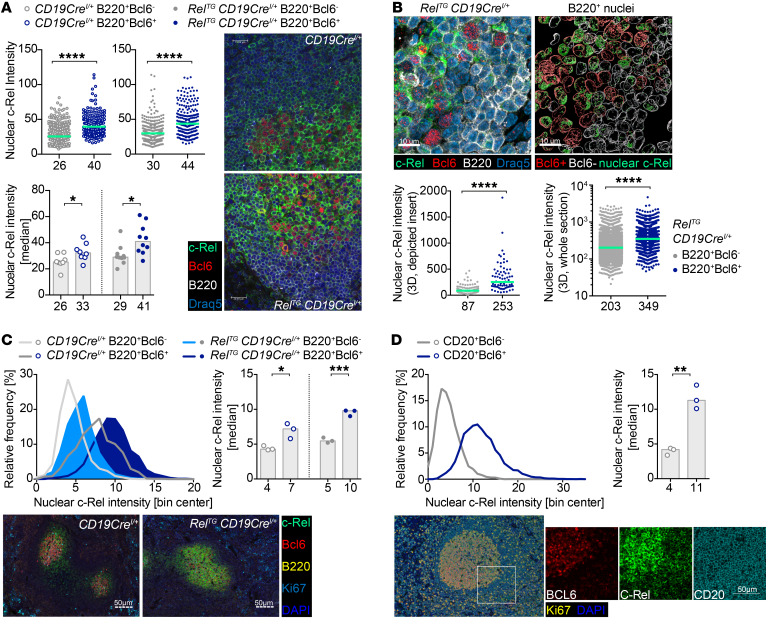
In order to verify these results with a method that does not require organ disruption to generate single-cell suspensions, we performed immunofluorescence microscopy analyses on spleen cryosections of SRBC-immunized RelTG CD19CreI/+ and control mice, including c-Rel–knockout mice. Our analysis revealed higher c-Rel staining in GCB cells compared with mantle zone (MZ) B cells and strong cytoplasmic c-Rel staining in all cases, but also clearly detectable nuclear c-Rel staining. The exact boundaries of the individual tightly packed MZB and GCB cells could not be reliably established, which precluded a systematic analysis of cytoplasmic c-Rel levels. Ilastik-based (33) quantification demonstrated higher c-Rel staining intensities in GCB cell nuclei compared with MZB cell nuclei in CD19CreI/+ control mice and the highest intensities in c-Rel–Tg GCB cell nuclei (Figure 7A and Supplemental Figure 8, A and B). We confirmed the elevated nuclear presence of c-Rel in GCB cells in RelTG CD19CreI/+ spleens by 3D reconstruction analysis of B cell nuclei in thick splenic cryosections using the Imaris surface object creation module (Figure 7B and Supplemental Figure 8C). Independent analyses with a different anti–c-Rel antibody of FFPE-fixed splenic sections using the Vectra Polaris system essentially recapitulated the results (Figure 7C and Supplemental Figure 9A). In agreement with our mouse studies, Vectra Polaris analysis of human tonsillar sections demonstrated much higher c-Rel staining in CD20+BCL6+ GCB cells compared with CD20+BCL6– MZB cells, and quantification showed clearly elevated nuclear c-Rel levels in GCB compared with MZB cells (Figure 7D and Supplemental Figure 9B). All 3 fluorescent imaging approaches yielded a consistent result: Both mouse and human GCB cells are generally characterized by higher nuclear c-Rel staining intensities compared with MZB cells, and elevated expression of c-Rel in RelTG CD19CreI/+ mice is sufficient to cause a further increase in nuclear c-Rel in GCB cells.
Figure 7. Immunofluorescence of c-Rel nuclear presence in B and GCB cells.
(A) Confocal immunofluorescence quantification of nuclear c-Rel intensities of B220+Bcl6– and B220+Bcl6+ cells for exemplary spleen sections from SRBC-immunized RelTG CD19CreI/+ and CD19CreI/+ mice is shown as scatter plots. Histograms depict median nuclear c-Rel intensities for all analyzed mice showing individual data points for each mouse. Green lines, bars, and numbers below graphs are median values (scale bars: 20 μm). *P ≤ 0.05, ****P ≤ 0.0001, unpaired t test. (B) 3D quantification of nuclear c-Rel in splenic B cell nuclei of an exemplary SRBC-immunized RelTG CD19CreI/+ mouse. Insert of a stained section (left; see Supplemental Figure 8C) shows cytoplasmic and nuclear c-Rel staining. Reconstructed surface objects (right) show abundant localization of c-Rel protein (green) in nuclei of Bcl6+ (red, n = 95) versus Bcl6– (white, n = 125) B cells. Graphs below show mean c-Rel 3D intensities in individual nuclei for the depicted insert (left) and for an entire spleen section (right) of B220+Bcl6– mantle zone cells (n = 5980) and B220+Bcl6+ cells (n = 1767); green lines and numbers below graphs depict median values (scale bars: 10 μm). ****P ≤ 0.0001, unpaired t test. (C) Nuclear c-Rel assessed by immunohistochemistry. Graphs display histogram of nuclear c-Rel intensities in B220+Bcl6– and B220+Bcl6+ cells in 3 RelTG CD19CreI/+ and 3 CD19CreI/+ mice (SRBC-immunized) and median nuclear c-Rel intensities for each mouse. Bars and numbers below bars are median values. Exemplary images are shown (scale bars: 50 μm). *P ≤ 0.05, ***P ≤ 0.001, unpaired t test. (D) Frequency distribution of nuclear c-Rel intensities and median nuclear c-Rel intensities in CD20+Bcl6– and CD20+Bcl6+ cells in tonsils of 3 human individuals assessed by immunohistochemistry. Exemplary images are shown (scale bar: 50 μm). **P ≤ 0.01, unpaired t test. See Supplemental Figures 8 and 9.
Together, our data derived from imaging flow cytometry data, Western blot, and immunofluorescence microscopy demonstrate that the extent of nuclear and therefore most likely transcriptionally active c-Rel in GCB cells is higher than that in naive B cells. These experiments emphasize the critical functional role of c-Rel protein abundance in GCB cells.
Gene expression profiling shows enrichment of cell cycle, cell growth, and Myc signatures in c-Rel–overexpressing GCB cells.
In order to assess changes in gene expression that underlie the observed expansion of GCB cells and plasma cells caused by c-Rel overexpression, we performed RNA sequencing analysis. For this purpose, we FACS-purified splenic naive B cells, GCB cells, and plasma cells of 8 SRBC-immunized RelTG CD19CreI/+ and CD19CreI/+ control mice.
Comparing c-Rel–Tg and control naive resting B cells yielded only 9 differentially expressed genes (adjusted P value [padj] < 0.05) (Supplemental Information). In contrast, in GCB cells enhanced c-Rel protein levels significantly affected the expression of 282 genes, of which 137 were upregulated and 145 were downregulated. In plasma cells we found 87 significantly upregulated and 138 significantly downregulated genes (Supplemental Information). The expression of 28 genes was commonly regulated in both GCB and plasma cells (Figure 8A).
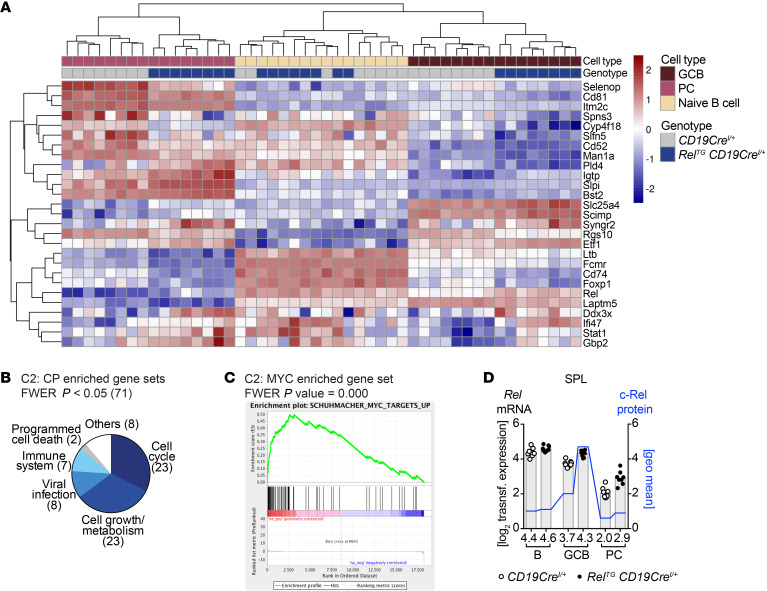
Figure 8. Strong enrichment of cell cycle and cell growth pathways in gene expression of c-Rel–overexpressing GCB cells.
(A) Common significantly differentially expressed genes (padj < 0.05) in GCB cells and plasma cells (PC) of RelTG CD19CreI/+ compared with control mice. Z-transformed expression values for each of 8 individual mice per genotype are shown in a heatmap representation. (B) Categories of significantly enriched gene sets in RelTG CD19CreI/+ GCB cell population evaluated by GSEA of C2: CP data set. (C) Enrichment plot of enriched gene sets in RelTG CD19CreI/+ GCB cell population with the highest normalized enrichment score among C2 gene sets containing the term MYC. (D) On the left x axis, log2 of transformed RNA expression values for Rel are plotted (gray bars). Individual data points are plotted. Bars and numbers below graphs are geometric means (n = 8). On the right x axis, geometric means of c-Rel protein levels are plotted (blue line) based on flow cytometry data presented in Figure 5A. FWER, family-wise error rate. See Supplemental Figure 10 and Supplemental Information.
In GCB cells, gene set enrichment analysis (GSEA) (34, 35) of canonical pathways of the curated gene sets (C2: CP) pointed to a strong enrichment of cell cycle as well as cell growth and metabolic pathways (Figure 8B and Supplemental Figure 10A). In particular, cell cycle–associated gene sets contained cell cycle phase transition and DNA replication signatures. Predominant cell growth–associated pathways included metabolism of RNA and proteins as well as nucleotides and amino acids. Moreover, genes involved in glucose transport, glucokinase regulation, pyruvate metabolism, and the citric acid cycle were enriched (for gene set lists, see Supplemental Information). Thus, c-Rel overexpression promotes key energy metabolism and cellular biosynthetic processes. As expected, the significantly enriched pathways also contained immune system signatures, including genes involved in activation of NF-κB in B cells.
The proto-oncogene Myc was among the top 10% of differentially upregulated genes. Assaying transcription factor target gene sets (C3: TFT) by GSEA also indicated enrichment of c-Myc–driven genes (see Supplemental Information). Of 46 gene sets containing the term MYC extracted from C2, 19 were significantly enriched at a family-wise error rate P value less than 0.05 (Figure 8C and Supplemental Figure 10A). Also in plasma cells GSEA showed enrichment of cell cycle–associated genes as well as upregulation of Igh genes (Supplemental Figure 10B).
GCB cells contain elevated c-Rel due to transcription-independent mechanisms.
Consistent with published data, Rel mRNA expression in CD19CreI/+ control mice (28, 29) was slightly lower in GCB cells and strongly reduced in plasma cells compared with naive B cells (Figure 8D and Supplemental Figure 10, C and D). Transgenic c-Rel overexpression increased mRNA 1.2-fold in GCB cells and 1.4-fold in plasma cells, but overall Rel mRNA expression in RelTG CD19CreI/+ terminal B cell differentiation still followed the same trend as in controls, which in GCB cells inversely correlated with c-Rel protein levels (compare with Figure 5A). Therefore, our data collectively demonstrate that c-Rel protein levels are specifically increased in GCB cells by transcription-independent mechanisms.
Enhanced proliferation contributes to the expansion of c-Rel–overexpressing GCB cells.
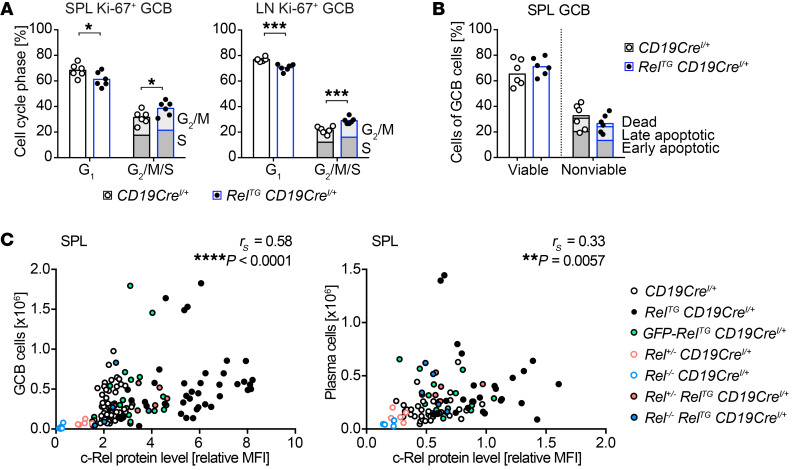
Our RNA sequencing data illustrate that genes controlling the central processes of cell division and cell growth are affected by c-Rel overexpression in GCB cells. In order to determine whether this translates into substantial alterations in proliferation, we performed flow cytometry–based cell cycle analyses in RelTG C19CreI/+ and control mice. Indeed, the percentage of Ki-67–positive c-Rel–overexpressing GCB cells in G2/M and S phase was significantly increased in LNs and spleen compared with controls (Figure 9A and Supplemental Figure 11A).
Figure 9. c-Rel protein abundance dose-dependently increases GCB cell and plasma cell numbers.
(A) Individual data points for percentages of cells in G1 and the sum of cells in G2/M and S phase obtained in 3 independent experiments are plotted. Bars represent mean values. *P ≤ 0.05, ***P ≤ 0.001, unpaired t test. (B) Assessment of apoptosis in GCB cells of mice immunized with SRBCs 12 days before analysis. Individual data points of all viable and the sum of all nonviable splenic GCB cells obtained in 2 independent experiments are plotted. Bars represent median values. Viable: caspase– (C–), annexin V– (AV–), 7-AAD–; early apoptotic: C+AV–/C–AV+/C+AV+, 7-AAD–; late apoptotic: C+AV–/C–AV+/C+AV+, 7-AAD+; non-apoptotic dead: C–, AV–, 7-AAD+; GCB cells: CD19+B220+CD95hiCD38lo. (C) Correlation of c-Rel protein abundance with cellular expansion. Spearman’s correlation coefficient (rS) and P values are given. c-Rel protein levels are normalized to non-GC or non-PC B cells of CD19CreI/+ mice. GCB cells (CD19+/B220+ CD95hiCD38lo): spleen, n = 140; plasma cells (B220loCD138+): spleen, n = 100. SPL, spleen; LN, lymph nodes. See Supplemental Figure 11, A–C.
In contrast, assessing the viability by staining for apoptotic markers annexin V and active caspases combined with a dead cell marker ex vivo did not reveal significant differences between c-Rel–overexpressing and control GCB cells (Figure 9B and Supplemental Figure 11B).
We conclude that c-Rel overexpression causes the expansion of GCB cells by enhancing proliferation.
c-Rel is a dose-dependent driver of terminal B cell differentiation over a wide dynamic range of protein levels.
Based on the dominant role for c-Rel in GCB cells, we aimed at deconstructing the relation between c-Rel protein levels and the extent of the GCB cell expansion. We integrated data obtained from c-Rel–overexpressing, control, and heterozygous and homozygous c-Rel–knockout mice. The highest c-Rel protein level was observed in c-Rel Tg–expressing GCB cells, while GFP–c-Rel fusion protein–expressing GCB cells were characterized by intermediate c-Rel levels. Remarkably, our data sets demonstrated a highly significant positive correlation of c-Rel protein levels and the expansion of GCB cell populations in spleen and LNs as well as the GALT over a large range of c-Rel levels (Figure 9C and Supplemental Figure 11C). In addition, also in plasma cells c-Rel protein abundance significantly correlated with population size (Figure 9C). We conclude that c-Rel transcription factor levels dictate GCB cell and plasma cell expansion in a strikingly dose-dependent manner.
B cell–specific c-Rel gain induces autoantibodies and renal immune complex deposition.
In humans, autoantibodies secreted by plasma cells are a hallmark of many autoimmune diseases. These autoantibodies are often class switched and are characterized by signs of somatic mutations and antigen selection indicating that the producing cells are of post-GC origin (36).
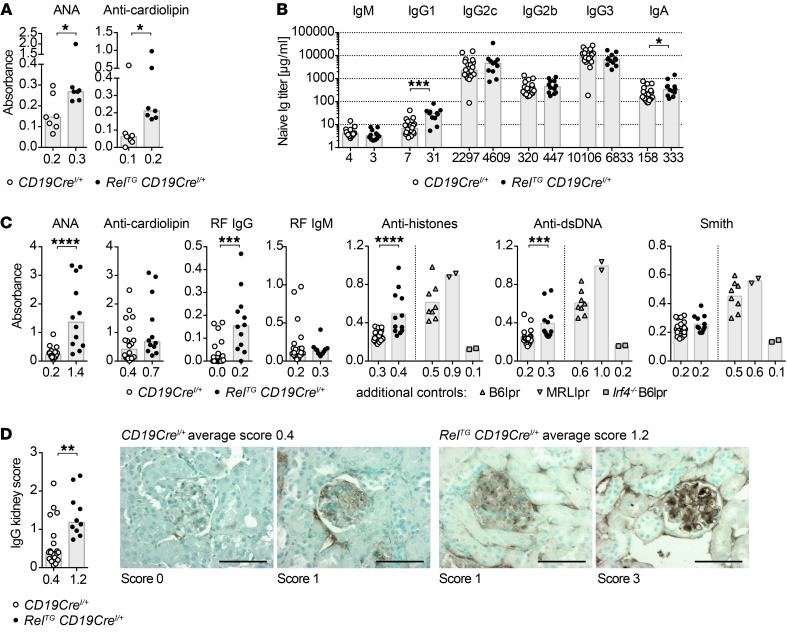
Remarkably, young RelTG CD19CreI/+ mice (8–13 weeks) already showed significantly elevated serum levels of anti-cardiolipin autoantibodies and anti-nuclear antibodies (ANAs) (Figure 10A). As many autoimmune diseases develop with time, we analyzed aged cohorts. Aged RelTG CD19CreI/+ mice (58–82 weeks) maintained expanded GCB cell and plasma cell populations, albeit not to the same extent as observed in young mice (Supplemental Figure 11, D and E). As in sera of young mice, titers of switched isotypes were elevated in aged cRelTG CD19CreI/+ mice, in particular IgG1 (Figure 10B), correlating with a higher percentage of IgG1-expressing plasma cells and a reduced fraction of unswitched IgM-positive plasma cells (Supplemental Figure 11F). These mice showed significantly elevated serum titers of ANAs and anti-dsDNA and anti-histone autoantibodies. In addition, anti-IgG rheumatoid factor autoantibodies significantly accumulated in aged RelTG CD19CreI/+ mice (Figure 10C). Finally, we detected significant deposition of IgG in the kidneys of aged RelTG CD19CreI/+ mice (Figure 10D).
Figure 10. c-Rel gain induces autoimmunity in mice.
(A) ANA and anti-cardiolipin autoantibody serum titers of 8- to 13-week-old mice determined by ELISA. Individual data points are plotted (n = 7). Bars and numbers below graphs are median values. *P ≤ 0.05, Mann-Whitney test. (B and C) ELISA for Ig subtypes (B) and autoantibodies (C) in serum of 58- to 82-week-old mice. Individual data points are plotted. Bars and numbers below graphs are median values (CD19CreI/+: median age 522 days, n = 20; RelTG CD19CreI/+: median age 511 days, n = 11–12). *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001, Mann-Whitney test. (D) IgG kidney histology. Kidney sections from mice aged 58–81 weeks (CD19CreI/+: median age 522 days, n = 19; RelTG CD19CreI/+: median age 513 days, n = 10) were stained with anti-mouse IgG to detect IgG deposits. For each mouse, 30 images were scored from 0 to 3 according to strength of positive IgG staining. The mean score for each mouse is shown as individual data points. Bars and numbers below bars are median values (scale bars: 50 μm). **P ≤ 0.01, Mann-Whitney test. ANA, anti-nuclear antibodies; RF, rheumatoid factor. See Supplemental Figure 11, D–F.
In conclusion, our results provide in vivo evidence for a functional relevance of c-Rel gain of function in B cells as a causative mechanism in autoimmunity.
Discussion
The c-Rel transcription factor is associated with human lymphomas through gain/amplification of its gene locus. In autoimmunity, not only are SNPs within the REL locus linked to human autoimmune diseases including rheumatoid arthritis (1, 2), but also c-Rel nuclear translocation was found to be increased in systemic lupus erythematosus (SLE) patients (37). These correlations raised the important question of the in vivo consequences of c-Rel gain, particularly in B cells. However, this issue remained unaddressed, largely owing to an absence “of transgenic mice that are capable of sustained B cell-specific expression of wild-type c-rel,” as noted over a decade ago (38). Here, we used cell type–specific overexpression to demonstrate that c-Rel gain causes strikingly dose-dependent hyperplasia of GCB cells and class-switched plasma cells followed by autoantibody production.
c-Rel plays essential roles in the effector functions of multiple cells implicated in GC reactions, including T cells and dendritic cells. Tfh cell–extrinsic (21) and –intrinsic roles for c-Rel were put forward to explain the lack of Tfh cells in c-Rel–knockout mice (39). The critical cell-intrinsic role of c-Rel in terminal B cell differentiation is underscored by our demonstration that c-Rel expression exclusively in the B cell lineage can restore GCs and plasma cells in otherwise completely c-Rel–deficient mice, including T and dendritic cells. The differentiation of c-Rel–containing GCB cells suffices to induce corresponding numbers of c-Rel–deficient Tfh cells, essentially ruling out a cell-intrinsic requirement for c-Rel in Tfh differentiation.
The consequences of gain and loss of gene function approaches often do not inversely correlate. In the case of c-Rel, our findings reveal that overexpression compares reciprocally with ablation in the generation of GCB cells and class-switched antibodies, especially of the IgG1 isotype. Gain of c-Rel amplified both spontaneous and immunization-induced GC reactions to a similar extent, when c-Rel overexpression preceded (CD19Cre) and when it was initiated during GCB cell differentiation (Cγ1Cre). As Cγ1Cre-mediated recombination can be detected in 25%–50% of GCB cells at day 4 after immunization (27), c-Rel overexpression could affect the GC reaction from this stage onward in RelTG Cγ1CreI/+ mice. However, the strongly increased class switching and cellular division indicate that c-Rel exerts central functions during the proliferative phase of mature GCs. The enhanced isotype switching can be linked to increased cell division (40), as well as increased immunoglobulin germline transcription (16, 23). To what extent the 10-fold expansion of c-Rel–overexpressing IgG1-positive plasma cells is a consequence of the enlarged GCB cell population or is due to c-Rel–driven plasmablast and plasma cell expansion is a topic for future studies.
In accordance with the reduction in NF-κB–regulated gene expression in GCB cells, Rel mRNA (28, 29) was expressed at equal or lower levels in GC compared with follicular B cells. Unexpectedly, we uncovered that c-Rel protein levels were 2-fold increased in GCB cells compared with naive B cells. We detected the highest c-Rel protein level in LZ GCB cells, consistent with an enriched NF-κB signaling signature in this compartment (26), while in plasmablasts/plasma cells c-Rel protein levels were reduced. Our findings thus indicate that powerful post-transcriptional/-translational mechanisms determine c-Rel protein levels during terminal B cell differentiation. Enhanced degradation or lower translation rates of c-Rel in follicular B cells compared with GCB cells represent possible explanations. The existence of such mechanisms is supported by the strong B cell subset–specific difference in FLAG–c-Rel and GFP–c-Rel protein levels expressed from the BAC-Tg CAG promoter, which mediates similar transcription across all B cell subsets. Another, not mutually exclusive possibility is the preferential selection of cells with high c-Rel protein content in the GC. The more than 4-fold enrichment of FLAG–c-Rel–expressing cells in the GC compared with naive mature B cells clearly underscores this notion. Expression level dosage of transcription factors in immune cells can be a pivotal determinant of cell fate, as for instance described for IRF4 and IRF8 in GCB cell versus plasmablast decisions (41–43). Here, we demonstrate that posttranscriptional regulation of c-Rel protein levels linearly determines the size of the GCB cell pool and the descended plasma cell pool over a wide dynamic range of c-Rel protein levels (from 50% to over 8-fold enhanced compared with control naive B cells).
In GCB cells, enhanced c-Rel protein levels correlate with increased nuclear translocation and a profound change in gene expression. Notably, we did not find an enrichment of typical NF-κB target genes in c-Rel–overexpressing GCB cells. This suggests that c-Rel gain affects a unique set of genes in the GC. In overall agreement with the metabolic pathways affected by gene expression changes in c-Rel–knockout GCB cells (15), we find that c-Rel overexpression leads to an upregulation of genes enriched for mediators of cell cycle progression as well as cell growth and metabolism. In addition, the central cell cycle–associated transcription factor Myc was among the upregulated genes and c-Myc target gene signatures were positively enriched, indicating that c-Rel gain exerts some of its functions through c-Myc.
Our CAG promoter–controlled expression of c-Rel demonstrates that similar mRNA amounts can give rise to dramatically diverse c-Rel protein levels in different B cell subsets. This suggests that the autoimmunity-associated SNPs might cause detectable c-Rel protein changes only in selected lymphocyte subsets, such as GCB cells.
The strongly context-dependent regulation of c-Rel protein levels might explain why the relation of REL genomic status and c-Rel protein level and localization in lymphomas with REL gain/amplifications remains under debate (10, 44–47). Nevertheless, our data indicate that in GCB cells enhanced c-Rel mRNA results in an amplified increase in c-Rel protein levels and that cells containing high c-Rel levels have a tremendous advantage in the GC. Therefore, in the context of lymphomagenesis, c-Rel gain or locus amplification should result in a robust cellular expansion specifically in the GC, with an accumulation of additional mutations ultimately resulting in cellular transformation. This scenario, together with the GC-specific c-Rel gain–associated gene expression uncovered in our study, can also explain the puzzling preponderance of c-Rel gain/amplification in GCB cell–like versus activated B cell–like (ABC) diffuse large B cell lymphoma (DLBCL) (48). Based on an association of ABC-DLBCL with increased expression of general NF-κB target genes, NF-κB transcription factors are expected to rather play a role in ABC-DLBCL. However, in GCB cells c-Rel controls a different set of genes distinct from classical NF-κB target genes, which promote GCB cell expansion, in striking contrast to canonical NF-κB activation. We propose that transcription-independent upregulation of c-Rel protein levels and ensuing nuclear translocation allow GCB cells to execute a critical GC-specific c-Rel–mediated gene expression program while general NF-κB activity and consensus NF-κB target gene expression remain low. Whether c-Rel contributes through additional mechanisms to lymphomagenesis and how it cooperates with other relevant transforming events will be assessed in future studies.
Deregulated GC reactions are also associated with the development of autoimmunity (49), and in this line, we provide in vivo evidence for a connection of c-Rel gain of function and autoimmunity. Overexpression of c-Rel specifically in B cells was sufficient to cause spontaneous development of autoantibodies, most prominently anti-nuclear antibodies associated with SLE, but also rheumatoid arthritis, as well as rheumatoid factor, which is detected in most rheumatoid arthritis patients. Interestingly, detection of autoantibodies often precedes severe disease onset in humans (50–52), suggesting that enhanced c-Rel levels could drive early events in the pathogenesis of autoimmune diseases.
Fittingly, transgenic mice overexpressing Bruton’s tyrosine kinase (Btk) in B cells display spontaneous GC formation with autoantibody production, and Btk-overexpressing B cells show elevated nuclear translocation of c-Rel in unstimulated or anti-IgM–stimulated conditions (53). Indeed, Btk plays an important role in c-Rel activation, as Btk-deficient B cells have strongly reduced c-Rel DNA binding upon stimulation (54).
More than 30 years after the discovery of v-Rel as a transcription factor capable of transforming chicken B cells, our cell type–specific c-Rel gain-of-function mouse models presented in this study reveal fundamental aspects of c-Rel regulation and function in terminal B cell differentiation. We uncovered that GCB cells employ nontranscriptional mechanisms to produce elevated amounts of c-Rel protein from less mRNA, which partially translocate to the nucleus. GCB cells, especially centroblasts, are generally characterized by comparatively low expression of NF-κB target genes, including NF-κB negative feedback inhibitory proteins. IκBβ, especially, is expressed at a much lower level in GCB cells compared with follicular or marginal zone B cells. Therefore, it seems plausible that the increased presence of c-Rel protein in GCB cells in the context of unchanged or even reduced levels of inhibitory proteins leads to a shift of the balance toward nuclear c-Rel. In our RNA sequencing–based gene expression analysis, we do not detect elevated expression of inhibitory proteins that are classical NF-κB targets, including IκBα or IκBβ, in c-Rel–Tg GCB cells compared with their control counterparts. Therefore, the dramatically elevated c-Rel protein levels in the transgenic GCB cells would lead to an even higher tendency toward nuclear translocation. This notion fits with the general idea that enhanced c-Rel stability in GCB cells allows these cells to modulate a c-Rel–specific gene expression program that does not rely on classical NF-κB–inducing stimuli and does not reflect classical NF-κB target genes. An additional factor reported to influence c-Rel nuclear translocation is Pin1, which when coexpressed with c-Rel in chicken spleen cells induces nuclear localization of c-Rel in the absence of stimulation (55). Pin1 is expressed at higher levels in GCB cells compared with follicular B cells or marginal zone B cells (29) and could contribute to the enhanced nuclear c-Rel levels in the former. The specific mechanisms regulating the dynamics of c-Rel nuclear translocation in GCB cells represent intriguing goals for future studies. These yet unknown mechanisms translate enhanced c-Rel protein levels into selective dose-dependent expansion of both GCB cells and highly class-switched plasma cells. The culmination of these effects in the production of class-switched autoantibodies deposited in kidneys of aged mice provides direct in vivo evidence for a functional connection of enhanced c-Rel function in B cells with autoimmunity.
Methods
Genetically modified mice.
All described mouse lines were maintained on a C57BL/6 background and housed in specific pathogen–free animal facilities of the Max Planck Institute of Biochemistry and the School of Medicine, Technical University of Munich. All animal procedures were approved by the Regierung of Oberbayern.
The conditional c-Rel–Tg mouse lines were generated for this study. Young mice were sacrificed for experiments at 8 weeks to a maximum of 16 weeks of age. For the total cohort of aged mice, the age ranged between 409 and 571 days at day of analysis, with a median age of 511 or 522 days for all 12 RelTG CD19CreI/+ experimental and all 20 CD19CreI/+ control mice, respectively.
Generation of c-Rel BAC-Tg mouse lines.
Using recombination-mediated genetic engineering of a bacterial artificial chromosome (BAC) containing the mouse Rel genetic locus but no other genes (BAC RP23-259L5), the following segments were inserted: 3×FLAG–IRES(internal ribosomal entry site)–hygromycin B–resistance at the Rel carboxy terminus; flippase (Flp) recognition target (Frt) sites flanking Rel exon 9; CAG(CMV early enhancer/chicken β-actin promoter)-loxP-STOP-neomycin/kanamycin-resistance-loxP combined with an HA tag or GFP to the first translated Rel exon (see scheme in Supplemental Figure 1).
Modified c-Rel/GFP–c-Rel BAC constructs were electroporated into C57BL/6 ES cells (Artemis Pharmaceuticals), and selection, isolation, expansion, and storage of ES cells were performed as previously described (56).
Following screening by PCR and Southern blot, selected clones were Cre-transduced with 1 μM TAT-Cre/HTNC (Protein Production Core Facility, Max Planck Institute of Biochemistry) in ADCF-MAb without l-glutamine (HyClone, Thermo Fisher Scientific) for 4–5 hours and functionally verified by hygromycin B (Sigma-Aldrich) selection as well as flow cytometry and Western blot to assess c-Rel/GFP–c-Rel protein expression. ES clones carrying single BAC integrants identified by Southern blot were selected (Supplemental Figure 1) for blastocyst injection.
Flow cytometry and imaging flow cytometry.
Single-cell suspensions were prepared from mouse lymphoid organs followed by red blood cell lysis (Gey’s solution). Fc receptors were blocked by incubation with anti–mouse CD16/CD32 (93, eBioscience) before stainings with the antibodies listed in Supplemental Methods. Biotinylated primary antibodies were visualized with streptavidin fluorochrome conjugates (eBioscience, BioLegend). For intracellular stainings the FoxP3 kit (eBioscience) was applied. 7-AAD staining solution (7-amino-actinomycin D, eBioscience) or LIVE/DEAD Fixable Near-IR (infrared) Dead Cell Stain Kit (Invitrogen) was used for discrimination of viable cells. Doublets were excluded based on FSC-H/A and SSC-W/A characteristics. DRAQ5 (Abcam) was used to visualize DNA content for cell cycle analysis or nuclear staining in imaging flow cytometry.
For staining of apoptotic cells, the CaspGlow active caspase staining kit (fluorescein, BioVision) was combined with 7-AAD and the Annexin V apoptosis detection kit (APC, eBioscience).
Samples were acquired on FACSCanto II (BD Biosciences), Cytoflex S, or Cytoflex LX (Beckman Coulter) flow cytometers or the ImageStream MKII (Luminex Corp.) imaging flow cytometer. Cell sorting was performed on a FACSAria II (BD Biosciences).
For imaging flow cytometry data analysis, the integrated module for the nuclear localization score of the IDEAS analysis software wizard was used to assess the degree of nuclear translocation (32). To determine the proportion of GCB cells containing significant amounts of nuclear c-Rel, the percentage of cells with a nuclear translocation score of ≥2.7 was calculated based on the average nuclear localization score of 2.7 of anti-CD40–stimulated control B cells, which demonstrate clear localization of c-Rel in the nucleus (see results shown in Figure 6D).
Cell separation.
For cell separation by magnetic-activated cell sorting (MACS), cell suspensions were prestained with anti-IgD–biotin (11- 26c, eBioscience) or anti-CD138–PE (281-2, BioLegend) antibodies and subsequently labeled with anti-biotin or anti-PE magnetic microbeads (Miltenyi Biotec), respectively, or directly labeled with anti-CD43 magnetic microbeads (Miltenyi Biotec). Sample separation was performed on an autoMACS Pro Separator (Miltenyi Biotec).
Capillary Western blot.
Splenocytes were enriched for GCB cells using a Germinal Center B (PNA) MicroBeads Kit (Miltenyi Biotec). Subsequently, GCB cells (CD3–CD11b–CD11c–CD19+IgDloCD38–FAS+) were FACS-sorted from the positive fraction and non-GCB cells (CD3–CD11b–CD11c–CD19+IgDhiCD38+FAS–) from the negative fraction. Cells from 2–3 mice were combined for fractionation of 1 × 106 GCB cells or B cells using an NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce Biotechnology). Nuclear and cytoplasmic fractions were analyzed by Simple Western system (Protein Simple). The following antibodies were used: c-Rel (polyclonal AF2699, R&D Systems), lamin B2 (E1S1Q, Cell Signaling Technology), α/β-tubulin (polyclonal 2148, Cell Signaling Technology), PTEN (138G6, Cell Signaling Technology), HRP-conjugated anti–goat IgG and anti–rabbit IgG (Protein Simple Detection Modules). The obtained data were processed for quantification with Compass software (Protein Simple, v4.0.0). Quantified c-Rel protein expression values were normalized to lamin B2 (nuclear fraction) or PTEN (cytoplasmic fraction).
Immunizations.
For immunizations, sheep red blood cells (SRBCs; Oxoid Ltd., Thermo Fisher Scientific) were washed 3 times in PBS, and the cell number was adjusted to 1 × 109 cells/mL, of which 100 μL per mouse was injected intraperitoneally. Mice were analyzed 10–12 days after immunization unless noted otherwise. Mice were immunized with 50 μg 4-hydroxy-3-nitrophenyl-acetyl chicken γ-globulin (NP-CG; Biosearch Technologies) in alum (Thermo Fisher Scientific) and analyzed 14 days later or bled at the indicated time points.
Somatic hypermutation and affinity maturation analysis.
Amplification of V186.2 IgG1-specific transcripts from PNA-positive MACS-enriched FACS-sorted GCB cells (CD3–CD11b–CD19+CD38–FAS+IgD–) from spleens of NP-CG–immunized 8-week-old mice was performed as previously described (57). Briefly, RNA was extracted using the AllPrep DNA/RNA Micro kit (Qiagen). IgG1-specific cDNA was generated using SMARTScribe Reverse Transcriptase (Takara Bio) followed by a semi-nested PCR to amplify the V186.2-specific IgG1 transcripts using Q5 Hot Start High-Fidelity DNA polymerase (New England Biolabs) (57). Final PCR amplicons were cloned and sequenced (Eurofins Genomics). Sequencing results were compared with the VBASE2 database for sequence analysis using Lasergene SeqMan Pro (DNASTAR). Additionally, VDJ sequences were analyzed using IMGT/HighV-Quest (ImMunoGeneTics).
RNA sequencing and gene expression analysis.
Desired splenocyte populations were pre-separated by MACS followed by FACS sorting of 1000 cells per sample into 96-well plates containing TCL buffer (Qiagen) (naive B cells: pre-MACS IgD+CD43+, sort IgDhiIgMlo; GCB cells: pre-MACS IgD–CD43–, sort B220+IgD–CD38loCD95hiGL7+; plasma cells: pre-MACS CD138+, sort B220loCD138+).
Bulk SCRB-seq (58, 59) RNA sequencing libraries were generated as previously described (60), with minor modifications. First, RNA was purified using magnetic beads (GC Biotech) and eluted in reverse transcription master mix containing barcoded oligo-dT primers. cDNA products were pooled and unincorporated primers digested by ExonucleaseI (New England Biolabs). Next, the cDNA pool was preamplified using KAPA HiFi HotStart polymerase (KAPA Biosystems). Sequencing libraries were generated using the Nextera XT kit (Illumina) and sequenced on a HiSeq1500 in High Output mode. Sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE143156).
Raw data were processed with Drop-seq tools v1.12 (Broad Institute) and mapped to the GRCm38p5 reference genome. Transcripts were annotated according to Ensembl release GRCm38.87. Differential gene expression between groups of interest was determined with DESeq2. A gene was considered to be differentially expressed with an FDR adjusted P value of 0.05 or less. Gene set enrichment analysis was conducted with GSEA (34, 35) using the pre-ranked list approach.
Vectra Polaris immunohistochemistry.
Tonsils were fixed for 12 hours in 4% formalin and embedded in paraffin. For immunohistochemistry, 1.5-μm sections were used. Multispectral immunohistochemistry was performed sequentially after antigen retrieval with epitope retrieval buffer (PerkinElmer). Slides were incubated for 1 hour with the following antibodies: c-Rel (4727, Cell Signaling Technology), Bcl6 (EP278, Cell Marque), CD20 (L26, PerkinElmer), Ki-67 (SP6, Cell Marque), and B220 (RA30B2, Thermo Fisher Scientific). TSA-Opal520, TSA-Opal570, TSA-Opal620, and TSA-Opal650 (PerkinElmer) were applied for each antibody. Nuclei were stained with DAPI (Vectashield). Slides were imaged using the PerkinElmer Vectra Polaris platform. Images were analyzed using PerkinElmer inForm software, and the nuclear intensity of c-Rel in CD20+Bcl6+ (human) or B220+Bcl6+ (mouse) versus CD20+Bcl6– (human) or B220+Bcl6– (mouse) cells was quantified.
Immunofluorescence.
Spleens were fixed in 4% PFA in PBS for 2 hours, washed in PBS, and then incubated in 15% sucrose in PBS for 4 hours. The spleens were transferred into 30% sucrose, incubated overnight at 4°C, and snap-frozen in OCT. For c-Rel quantification, 10-μm sections were air-dried for 1 hour at room temperature and fixed in ice-cold acetone for 6 minutes. Fixed sections were air-dried for 30 minutes and rehydrated for 10 minutes in PBS followed by 1 hour of incubation in blocking/permeabilizing buffer (3% BSA and 1% Triton X-100 in PBS). Thirty-micrometer sections were permeabilized, blocked, and stained in 0.1 M Tris with 1% BSA, 0.3% Triton X-100, and normal mouse serum (MilliporeSigma). Serial sections were prepared and visually inspected by epifluorescence light microscopy before acquisition of representative areas by confocal microscopy. Sections were stained by incubation at 4°C for 2 days with goat anti–c-Rel (AF2699, R&D Systems) and rabbit anti-Bcl6 antibodies (N-3, Santa Cruz Biotechnology) in dilution buffer (1% BSA and 0.5% Triton X-100 in PBS). The sections were then incubated at 4°C overnight with donkey anti–goat IgG–Alexa Fluor 405 (ab175665, Abcam), chicken anti–rabbit IgG–Alexa Fluor 488 (A21441, Invitrogen), rat anti-B220–Alexa Fluor 594 (RA3-6B2, BD Biosciences), and DRAQ5 (Abcam). Images were captured with a Leica TSC SP8 confocal microscope with ×63 optic. For quantification of c-Rel intensities, see Supplemental Methods.
IgG immunostainings on kidney sections.
Kidneys were fixed in 4% buffered formalin, processed, and embedded in paraffin. Immunostaining was performed on 2-μm sections using biotinylated anti–mouse IgG (BA-9200, Vector Laboratories). IgG deposition in kidneys was scored semiquantitatively (0 to 3) by assessment of the extent of a positive signal.
ELISA.
ELISA to detect mouse serum titers of IgA, IgG2b, IgG2c, and IgG3 antibody subclasses was performed using kits (Bethyl Laboratories). For quantification of IgM and IgG1 serum titers, 96-well ELISA plates (Nunc MaxiSorp) were coated with anti–mouse λ/κ (Goat Anti-Mouse Lambda-UNLB and Kappa-UNLB, Southern Biotech) combined with sample detection based on anti–mouse IgG1–biotin (biotin rat anti-mouse IgG1, A85-1, BD Pharmingen) or IgM-biotin conjugate (biotin rat anti–mouse IgM, R6-60.2, BD Pharmingen) and streptavidin-HRP (Invitrogen). Serial dilutions of mouse IgM/κ or IgG/κ isotype controls served as a reference to calculate absolute concentrations (BD Pharmingen). Presence of anti-nuclear antibodies (ANAs) and cardiolipin antibodies was assessed based on kits (Varelisa Phadia) combined with detection by anti–mouse IgG–HRP conjugate (goat anti–mouse IgG–HRP, Southern Biotech). To capture rheumatoid factor, 96-well ELISA plates (Nunc MaxiSorp) were coated with rabbit IgG (AB2337118, Jackson ImmunoResearch Laboratories), and mouse serum Igs were detected using IgG kit components (Bethyl Laboratories) or as described for IgM above. Tetramethylbenzidine (TMB) was used as an HRP substrate (BD OptEIA, BD Biosciences).
For NP2 and NP23 ELISAs, plates were coated with 12.5 μg/mL NP2-BSA or NP23-BSA (Biosearch Technologies), combined with sample detected based on anti-mouse IgG1–biotin, similarly to the method previously described (57). Serial dilutions (six 1:4 steps starting with a 1:400 dilution) of the samples were measured together with an IgG1 standard, and the titer was calculated using the absorbance summation method (61).
For detection of antibodies against dsDNA, Nunc MaxiSorp ELISA plates (Thermo Fisher Scientific) were coated with poly-l-lysine (Trevigen) and PBS (ratio 1:1) for 1 hour. Plates were washed with Tris-NaCl (50 mM Tris and 0.14 M NaCl, pH 7.5), and dsDNA from mouse embryonic stem cells was coated in SSC (pH 7.0) buffer overnight. For anti-nucleosome antibodies, MaxiSorp ELISA plates were precoated with poly-l-lysine and PBS (1:1) for 1 hour. Plates were washed with Tris-NaCl buffer and incubated with mouse DNA-histones (2 μg/mL) in SSC buffer overnight at 4°C. For anti-Smith antibodies, MaxiSorp ELISA plates were coated with Smith antigen (Immunovision) in 0.05 M carbonate-bicarbonate buffer overnight at 4°C. Sera from 24-week C57BL/6lpr/lpr and 24-week MRLlpr/lpr mice, as well as IgG-poor serum, were used as controls. One percent BSA in Tris-NaCl was used for blocking. HRP-conjugated antibody against mouse IgG (A90-131P, Bethyl Laboratories) was used as secondary antibody. Absorbance was measured at 450 nm with a Sunrise plate reader (TECAN).
Statistics.
Flow cytometry data were analyzed with FlowJo (Tree Star). The FlowJo cell cycle platform based on the Watson pragmatic model was applied for cell cycle analysis. To quantify nuclear translocation of imaging flow cytometry data, the nuclear translocation wizard of the IDEAS (Luminex Corp.) software was used. ImageJ (NIH) (62) was used for quantification of Southern blot images. Graphical representation and statistical analysis were performed in Prism (GraphPad Software) and R (63, 64). Where statistically significant results are indicated, the respective statistical test and P values are provided in the figure legends. P less than or equal to 0.05 was considered statistically significant. All performed t tests were 2-tailed.
Study approval.
Written informed consent was received from participants before inclusion in this study. Material was double-anonymized before usage. All animal procedures were approved by the Regierung of Oberbayern (Munich, Germany).
Author contributions
MKH designed, performed, and analyzed most experiments and wrote the manuscript. HOS, VS, and CD performed and analyzed experiments. DK generated the modified BACs. ML performed and analyzed ELISAs and histology. EK performed and analyzed capillary Western blot experiments. CZ performed the RNA sequencing experiment. TE performed analysis of RNA sequencing experiments. GP and MR performed and analyzed Vectra Polaris experiments. HL, BM, JPB, VFS, WE, RR, and HJA contributed to experimental design and data analysis. MSS conceptualized the work, directed the study, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We are grateful to M. Mann and D. Krappmann for advice and to R. Fässler and C. Peschel for support. We thank M.J. Weißenhorn from the transgenic core facility at the Max Planck Institute of Biochemistry as well as M. Utzt of the DKTK cell sorting core facility. We acknowledge the Ludwig-Maximilians-Universität Biomedical Center Core Facility Flow Cytometry for providing equipment and expertise. We thank S. Klein and D. Saur for help with immunofluorescence microscopy and J. Knogler, M. Mugler, and B. Habermehl for technical assistance. This project received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement 682435) to MSS; from the DFG through SCHM220 2.1 to MSS, CRC1335 project P05 to MSS and VFS, and CRC1243 project A12 to MSS; and from an FCI Kekulé PhD fellowship to MKH. JPB is supported by an International Junior Research Group grant (N-LW-2016-370, Elite Network of Bavaria).
Version 1. 03/19/2020
In-Press Preview
Version 2. 05/18/2020
Electronic publication
Version 3. 06/01/2020
Print issue publication
Footnotes
MKH’s present address is: Merck KGaA, Darmstadt, Germany.
DK’s present address is: Singapore Immunology Network (SIgN), Singapore.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):3270–3286.https://doi.org/10.1172/JCI124382.
Contributor Information
Maike Kober-Hasslacher, Email: maikekober@gmx.de.
Hyunju Oh-Strauß, Email: hyunju.oh@tum.de.
Dilip Kumar, Email: dilip_kumar@immunol.a-star.edu.sg.
Carina Diehl, Email: carina.steinecke@tum.de.
Maciej Lech, Email: m_lechu@yahoo.com.
Thomas Engleitner, Email: Thomas.Engleitner@tum.de.
Eslam Katab, Email: eslam.katab@tum.de.
Guido Piontek, Email: Guido.Piontek@med.uni-muenchen.de.
Hongwei Li, Email: hongwei.li@tum.de.
Björn Menze, Email: bjoern.menze@tum.de.
Christoph Ziegenhain, Email: cziegenhain@gmail.com.
Wolfgang Enard, Email: enard@biologie.uni-muenchen.de.
Roland Rad, Email: roland.rad@tum.de.
Jan P. Böttcher, Email: j.boettcher@tum.de.
Hans-Joachim Anders, Email: hjanders@med.uni-muenchen.de.
Martina Rudelius, Email: Martina.Rudelius@med.uni-muenchen.de.
Marc Schmidt-Supprian, Email: supprian@gmail.com.
References
- 1.Gregersen PK, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41(7):820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre S, et al. Confirmation of association of the REL locus with rheumatoid arthritis susceptibility in the UK population. Ann Rheum Dis. 2010;69(8):1572–1573. doi: 10.1136/ard.2009.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genetic Analysis of Psoriasis Consortium the Wellcome Trust Case Control Consortium 2. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42(11):985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGovern DP, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42(4):332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trynka G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-κB signalling. Gut. 2009;58(8):1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 6.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinghaus E, et al. Genome-wide meta-analysis of psoriatic arthritis identifies susceptibility locus at REL. J Invest Dermatol. 2012;132(4):1133–1140. doi: 10.1038/jid.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmore TD. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 1999;18(49):6925–6937. doi: 10.1038/sj.onc.1203222. [DOI] [PubMed] [Google Scholar]
- 10.Gilmore TD, Gerondakis S. The c-Rel transcription factor in development and disease. Genes Cancer. 2011;2(7):695–711. doi: 10.1177/1947601911421925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerondakis S, et al. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25(51):6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 12.Carrasco D, Weih F, Bravo R. Developmental expression of the mouse c-rel proto-oncogene in hematopoietic organs. Development. 1994;120(10):2991–3004. doi: 10.1242/dev.120.10.2991. [DOI] [PubMed] [Google Scholar]
- 13.Kaileh M, Sen R. NF-κB function in B lymphocytes. Immunol Rev. 2012;246(1):254–271. doi: 10.1111/j.1600-065X.2012.01106.x. [DOI] [PubMed] [Google Scholar]
- 14.Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol. 2009;27(1):693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 15.Heise N, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J Exp Med. 2014;211(10):2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köntgen F, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9(16):1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 17.Grumont RJ, et al. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-κB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med. 1998;187(5):663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tumang JR, et al. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur J Immunol. 1998;28(12):4299–4312. doi: 10.1002/(SICI)1521-4141(199812)28:12<4299::AID-IMMU4299>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Hsia CY, Cheng S, Owyang AM, Dowdy SF, Liou HC. c-Rel regulation of the cell cycle in primary mouse B lymphocytes. Int Immunol. 2002;14(8):905–916. doi: 10.1093/intimm/dxf055. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, Hsia CY, Leone G, Liou HC. Cyclin E and Bcl-xL cooperatively induce cell cycle progression in c-Rel–/– B cells. Oncogene. 2003;22(52):8472–8486. doi: 10.1038/sj.onc.1206917. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Hardy K, Bunting K, Daley S, Ma L, Shannon MF. Regulation of the IL-21 gene by the NF-κB transcription factor c-Rel. J Immunol. 2010;185(4):2350–2359. doi: 10.4049/jimmunol.1000317. [DOI] [PubMed] [Google Scholar]
- 22.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15(3):137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harling-McNabb L, Deliyannis G, Jackson DC, Gerondakis S, Grigoriadis G, Brown LE. Mice lacking the transcription factor subunit Rel can clear an influenza infection and have functional anti-viral cytotoxic T cells but do not develop an optimal antibody response. Int Immunol. 1999;11(9):1431–1439. doi: 10.1093/intimm/11.9.1431. [DOI] [PubMed] [Google Scholar]
- 24.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki Y, et al. Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24(6):729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casola S, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103(19):7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi W, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16(6):663–673. doi: 10.1038/ni.3154. [DOI] [PubMed] [Google Scholar]
- 29.Heng TS, Painter MW, Immunological Genome Project Consortium The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 30.Ferch U, et al. MALT1 directs B cell receptor-induced canonical nuclear factor-kappaB signaling selectively to the c-Rel subunit. Nat Immunol. 2007;8(9):984–991. doi: 10.1038/ni1493. [DOI] [PubMed] [Google Scholar]
- 31.Wuerzberger-Davis SM, et al. Nuclear export of the NF-κB inhibitor IκBα is required for proper B cell and secondary lymphoid tissue formation. Immunity. 2011;34(2):188–200. doi: 10.1016/j.immuni.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.George TC, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311(1-2):117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 33. Sommer C, Straehle C, Hamprecht FA, Köthe U. ilastik: Interactive Learning and Segmentation Toolkit. In: IEEE. 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. New York, New York, USA: IEEE; 2011:230–233. [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28(1):18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, et al. Aberrant CD40-induced NF-κB activation in human lupus B lymphocytes. PLoS One. 2012;7(8):e41644. doi: 10.1371/journal.pone.0041644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23(13):2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- 39.Liu WH, et al. A miR-155-Peli1-c-Rel pathway controls the generation and function of T follicular helper cells. J Exp Med. 2016;213(9):1901–1919. doi: 10.1084/jem.20160204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp Med. 1996;184(1):277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 42.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25(2):225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Ochiai K, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38(5):918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilmore TD, Starczynowski DT, Kalaitzidis D. RELevant gene amplification in B-cell lymphomas? Blood. 2004;103(8):3243–3244; author reply 3244. doi: 10.1182/blood-2003-11-4019. [DOI] [PubMed] [Google Scholar]
- 45.Houldsworth J, et al. Relationship between REL amplification, REL function, and clinical and biologic features in diffuse large B-cell lymphomas. Blood. 2004;103(5):1862–1868. doi: 10.1182/blood-2003-04-1359. [DOI] [PubMed] [Google Scholar]
- 46.Weniger MA, et al. Gains of REL in primary mediastinal B-cell lymphoma coincide with nuclear accumulation of REL protein. Genes Chromosomes Cancer. 2007;46(4):406–415. doi: 10.1002/gcc.20420. [DOI] [PubMed] [Google Scholar]
- 47.Li L, et al. Prognostic impact of c-Rel nuclear expression and REL amplification and crosstalk between c-Rel and the p53 pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6(27):23157–23180. doi: 10.18632/oncotarget.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9(12):845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 50.Arbuckle MR, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 51.Scofield RH. Autoantibodies as predictors of disease. Lancet. 2004;363(9420):1544–1546. doi: 10.1016/S0140-6736(04)16154-0. [DOI] [PubMed] [Google Scholar]
- 52.Bizzaro N. Autoantibodies as predictors of disease: the clinical and experimental evidence. Autoimmun Rev. 2007;6(6):325–333. doi: 10.1016/j.autrev.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Kil LP, et al. Btk levels set the threshold for B-cell activation and negative selection of autoreactive B cells in mice. Blood. 2012;119(16):3744–3756. doi: 10.1182/blood-2011-12-397919. [DOI] [PubMed] [Google Scholar]
- 54.Shinners NP, et al. Bruton’s tyrosine kinase mediates NF-kappa B activation and B cell survival by B cell-activating factor receptor of the TNF-R family. J Immunol. 2007;179(6):3872–3880. doi: 10.4049/jimmunol.179.6.3872. [DOI] [PubMed] [Google Scholar]
- 55.Fan G, et al. Peptidyl-prolyl isomerase Pin1 markedly enhances the oncogenic activity of the rel proteins in the nuclear factor-κB family. Cancer Res. 2009;69(11):4589–4597. doi: 10.1158/0008-5472.CAN-08-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt-Supprian M, et al. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5(6):981–992. doi: 10.1016/S1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 57.Heise N, Klein U. Somatic hypermutation and affinity maturation analysis using the 4-hydroxy-3-nitrophenyl-acetyl (NP) system. Methods Mol Biol. 2017;1623:191–208. doi: 10.1007/978-1-4939-7095-7_16. [DOI] [PubMed] [Google Scholar]
- 58. doi: 10.1101/543066. Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, Mikkelsen TS. Characterization of directed differentiation by high-throughput single-cell RNA-Seq [preprint]. Posted on bioRxiv March 5, 2014. [DOI]
- 59.Ziegenhain C, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65(4):631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 60.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. The impact of amplification on differential expression analyses by RNA-seq. Sci Rep. 2016;6:25533. doi: 10.1038/srep25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartman H, Wang Y, Schroeder HW, Cui X. Absorbance summation: a novel approach for analyzing high-throughput ELISA data in the absence of a standard. PLoS One. 2018;13(6):e0198528. doi: 10.1371/journal.pone.0198528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raschka S. Instant Heat Maps in R How-To. Birmingham, United Kingdom: Packt Publishing Ltd; 2013. [Google Scholar]
- 64. R Core Team. R: A Language and Environment for Statistical Computing. http://www.r-project.org Accessed March 25, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.