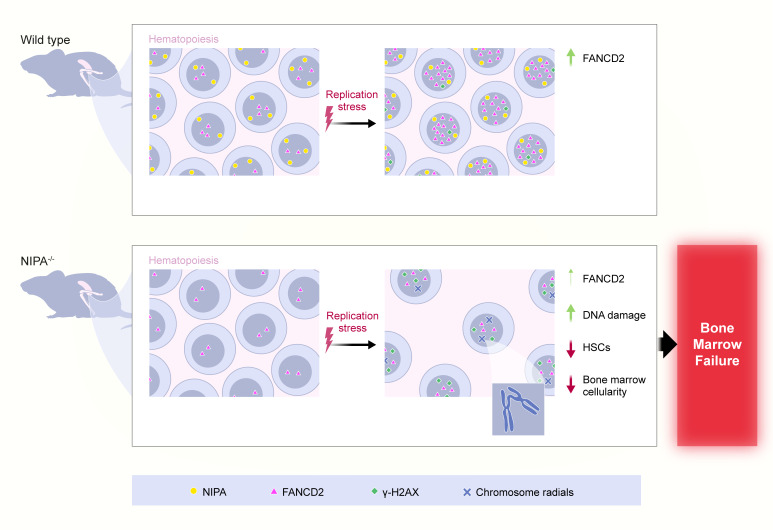
Abstract
Inherited bone marrow failure syndromes (IBMFSs) are a heterogeneous group of disorders characterized by defective hematopoiesis, impaired stem cell function, and cancer susceptibility. Diagnosis of IBMFS presents a major challenge due to the large variety of associated phenotypes, and novel, clinically relevant biomarkers are urgently needed. Our study identified nuclear interaction partner of ALK (NIPA) as an IBMFS gene, as it is significantly downregulated in a distinct subset of myelodysplastic syndrome–type (MDS-type) refractory cytopenia in children. Mechanistically, we showed that NIPA is major player in the Fanconi anemia (FA) pathway, which binds FANCD2 and regulates its nuclear abundance, making it essential for a functional DNA repair/FA/BRCA pathway. In a knockout mouse model, Nipa deficiency led to major cell-intrinsic defects, including a premature aging phenotype, with accumulation of DNA damage in hematopoietic stem cells (HSCs). Induction of replication stress triggered a reduction in and functional decline of murine HSCs, resulting in complete bone marrow failure and death of the knockout mice with 100% penetrance. Taken together, the results of our study add NIPA to the short list of FA-associated proteins, thereby highlighting its potential as a diagnostic marker and/or possible target in diseases characterized by hematopoietic failure.
Keywords: Hematology, Stem cells
Keywords: Bone marrow, DNA repair, Hematopoietic stem cells
Introduction
Inherited BM failure syndromes (IBMFSs) include a wide spectrum of hematological disorders characterized by hypocellular BM and peripheral blood (PB) cytopenia (1, 2). The best-characterized IBMFSs are Fanconi anemia (FA) and dyskeratosis congenita (DC), characterized by DNA damage repair deficiency and premature telomere shortening, respectively. It is a major diagnostic challenge to distinguish among IBMFS, myelodysplastic syndrome–type (MDS-type) refractory cytopenia of childhood (RCC), and severe aplastic anemia (SAA), due to overlapping phenotypes and pathological mechanisms. Furthermore, both the diagnostic tests and the number of cells available are limited (3–7). Therapeutic strategies for these entities differ, and challenging procedures including genetic analysis are required in order to verify the diagnosis. High-throughput analyses have identified new mutations and potential regulators of IBMFSs (8–13).
FA occurs worldwide and affects 1 in 100,000 births (14–16). However, due to the variety of phenotypes associated with FA, it is often underdiagnosed. Alterations in the FA/BRCA pathway are known to cause genomic instability and cancer in humans. For example, patients with FA have a several-100-fold-higher risk of acute myeloid leukemia (AML) induction than the non-FA population (17, 18). Interestingly, FA pathway aberrations can also contribute to cancer in individuals without FA (19–21). A total of 22 FANC genes have been characterized to date, and the elimination of any of these gene products results in disruption of the FA/BRCA pathway, a key regulator counteracting replication stress and promoting accurate DNA replication (22–24). Moreover, even heterozygous FANC mutations increase the risk of cancer, suggesting that the phenotypes resulting from disruption of these genes may be dose dependent. Among these proteins, FA complementation group D2 (FANCD2) is a crucial player, with a pivotal role in maintaining the integrity of stalled replication forks and processing interstrand crosslinks (ICLs) (25, 26). Hematopoietic stem cells (HSCs) cycle throughout their lifetime. Replication errors are associated with chromosome gaps or breaks, and accumulation of DNA damage is a hallmark of aging HSCs, leading to the induction of apoptosis in these cells (27–30). Because of this, FANCD2 has been shown to be an essential player in maintaining the HSC pool in both the steady state and under stress conditions (31–35). Although several studies highlight the impact of DNA damage on HSCs (27, 36–38), molecular mechanisms in the complex interplay of DNA repair/FA/BRCA pathway proteins and the aging of HSCs are not yet elucidated.
Nuclear interaction partner of ALK (NIPA; also called ZC3HC1) has been identified as a mammalian F-box protein that defines an SCF-type ubiquitin E3 ligase (SCFNIPA) and regulates the timing of mitotic entry (39). It also plays an important role in meiosis by promoting DNA double-strand break repair and formation of the synaptonemal complex, preventing a block of spermatogenesis during meiotic prophase and subsequent apoptosis (40).
In this work, we show that Nipa deficiency in mice mimics FA, with unresolved DNA damage in HSCs, BM failure, and mitomycin C (MMC) hypersensitivity defining NIPA as an FA-associated protein. Furthermore, the reduction in its expression in pediatric RCC patients indicates a potential function for NIPA as a biomarker and/or therapeutic target.
Results
Deletion of Nipa leads to progressive HSC reduction and BM aplasia in aged mice.
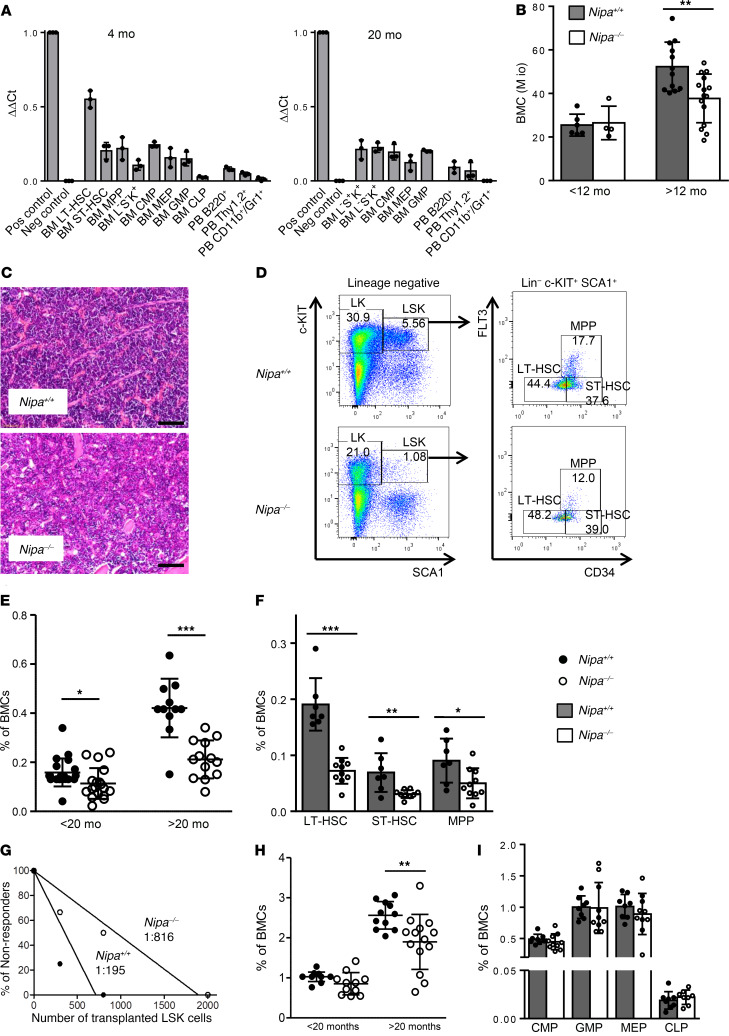
Nipa mRNA was highly expressed in young and old hematopoietic stem and progenitor cells, especially in the most primitive long-term repopulating HSCs (LT-HSCs) (Figure 1A). To analyze its function in these cells, we generated conditional Nipa–/– animals. Mice harboring 2 loxP sites spanning exons 1 and 2 of the Nipa locus (40) were bred with either a ubiquitously expressing CMV-Cre transgenic (41, 42) or a VAV-Cre transgenic mouse strain (43), leading to deletion of NIPA ubiquitously or only in hematopoietic tissues, respectively. Analysis of BM cellularity revealed significantly decreased cell numbers in old Nipa–/– mice compared with their WT littermates (Figure 1B), and histological examinations showed mild BM hypoplasia (Figure 1C). Immunophenotypic analysis of subpopulations revealed significant and progressive exhaustion of lineage marker–Sca-1+c-Kit+ (LSK) cells, with a 40% reduction in aged animals compared with their WT littermates (Figure 1, D–F). LT-HSCs (CD34–Flt3–LSK; P = 0.0005), short-term HSCs (ST-HSCs; CD34+Flt3–LSK; P = 0.006), and multipotent progenitors (MPPs; CD34+Flt3+LSK; P = 0.02) were reduced to similar degrees in Nipa–/– animals, with no age-related increase. In vivo limiting dilution transplantation (Tx) assays revealed a severe reduction in the number of functional HSCs within the LSK population, with a frequency of 1:816 Nipa–/– LSK cells compared with 1:195 WT LSK cells (Figure 1G). Analysis of more mature hematopoietic cells revealed a significant reduction in lineage marker–Sca-1–c–Kit+ (LK) frequencies in Nipa–/– mice older than 20 months (Figure 1H), whereas the common myeloid progenitor (CMP), megakaryocyte/erythrocyte progenitor (MEP), granulocyte/monocyte progenitor (GMP), and common lymphoid progenitor (CLP) populations were still within the normal ranges in aged mice (Figure 1I). Nipa–/– mice had normal RBC and WBC counts but mildly reduced numbers of platelets (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126215DS1). Nipa–/– mice older than 1 year showed mild B cell lymphopenia and monocytosis, pointing to a myeloid bias in aged Nipa–/– HSCs (Supplemental Figure 1, B–F).
Figure 1. Deletion of Nipa leads to progressive HSC reduction and BM aplasia with age.
(A) Expression levels of Nipa in immature and mature hematopoietic cell populations from 4- and 20-month-old WT mice, as assessed by qRT-PCR. Testis germ cells served as positive control, Nipa–/– BMCs as negative control. n = 3. (B) BM cellularity of Nipa+/+ and Nipa–/– mice younger and older than 12 months. n = 19 Nipa+/+; n = 18 Nipa–/–. Mio, 1 × 106. (C) Representative H&E staining of BM sections from 10-month-old mice. Original magnification, ×10. Scale bar: 100 μm. (D) Representative flow cytometry profiles of LSK cells, LT-HSCs, ST-HSCs, and MPPs in Nipa+/+ and Nipa–/– mice older than 20 months. (E) Quantification of LSK cells in Nipa+/+ and Nipa–/– BMCs from mice younger and older than 20 months. n = 32 Nipa+/+; n = 31 Nipa–/–. (F) Percentage of LSK subpopulations (LT-HSCs, ST-HSCs, and MPPs) in Nipa+/+ and Nipa–/– BMCs from mice older than 20 months. n = 7 Nipa+/+; n = 10 Nipa–/–. (G) Number of functional HSCs in vivo measured by limiting dilution Tx using different doses of LSK cells from 14- to 17-month-old mice. Nipa+/+, 1:195 (95% CI, 1:71–1:538); Nipa–/–, 1:816 (95% CI, 1:337-1:1973). (H) Quantification of LK cells in Nipa+/+ and Nipa–/– BMCs from mice younger and older than 20 months of age. n = 24 Nipa+/+; n = 26 Nipa–/–. (I) Percentage of hematopoietic progenitor populations (CMP, GMP, MEP, CLP) in Nipa+/+ and Nipa–/– BMCs of 20-month-old mice. n = 8 Nipa+/+; n = 10 Nipa–/–. *P < 0.05, **P < 0.01, ***P < 0.001. An unpaired 2-tailed Student’s t test (B, E, F, H, and I) or L-Calc software (G) was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figure 1.
Together, these data indicate that Nipa is expressed throughout life in hematopoietic cells, particularly in LT-HSCs, and that deletion of Nipa causes progressive, age-related loss of HSCs in mice.
Nipa–/– HSCs show reduced repopulating ability and a bias toward myeloid differentiation.
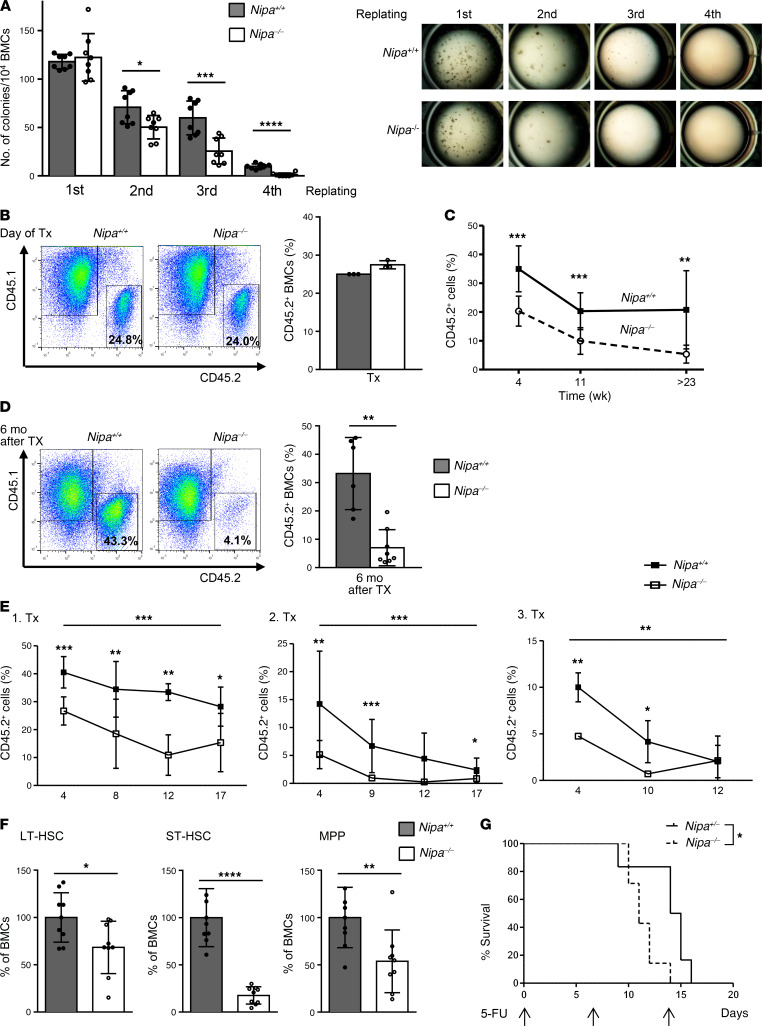
To further examine the role of NIPA in HSC reconstitution capacity and exhaustion, we performed in vitro and in vivo repopulation experiments. Although Nipa–/– BM cells (BMCs) gave rise to colony numbers similar to those in WT cells, their serial repopulation revealed greatly reduced ability to form colonies (Figure 2A). To test the competitive reconstitution potential of Nipa-deficient BMCs, we transplanted CD45.2+ Nipa+/+ or Nipa–/– BMCs together with competing CD45.1+ WT BMCs at a ratio of 1:3 into lethally irradiated recipient mice (Figure 2B and Supplemental Figure 2A). Chimerism was measured at the indicated time points, showing a severe and sustained reduction in CD45.2+ WBCs in mice transplanted with Nipa–/– BMCs (Figure 2C). The reduced competitiveness of Nipa–/– BMCs affected all hematopoietic organs and lineages, with less than 10% BM chimerism 6 months after Tx compared with 33% for WT BMCs (Figure 2D; P = 0.003). To further determine whether the HSC pool was reduced in transplanted Nipa–/– BMCs or whether Nipa–/– HSCs exhibited functional defects, we performed serial Tx of LSK cells (Supplemental Figure 2, B and C). Indeed, Tx of equal numbers of sorted WT or Nipa–/– LSK cells showed that the repopulation capacity of Nipa–/– LSK cells was greatly impaired, with reduced WBC chimerism in all 3 serial Tx assays, suggesting a functional loss of repopulating Nipa–/– cells (Figure 2E). The reduced repopulation ability was also reflected by splenic as well as LT-HSC chimerism (Supplemental Figure 2, D and E). Nevertheless, long-term reconstitution seemed to be at least partially maintained, as we were able to detect rare donor cells in the recipients of tertiary Tx of Nipa–/– LSK cells (Figure 2E). In Tx of Nipa–/– LSK cells, myeloid repopulation was favored over lymphocytic reconstitution, with greater stability of granulocytes and monocytes seen in the first course of Tx (Supplemental Figure 2F).
Figure 2. Nipa–/– HSCs show reduced repopulating ability, limited self-renewal potential, and bias toward myeloid differentiation.
(A) In vitro CFU assay of Nipa+/+ and Nipa–/– BMCs replated 4 times. Quantification of colonies and representative images. Data are from 4 independent experiments. n = 8 Nipa+/+; n = 8 Nipa–/–. (B) Representative flow cytometry profiles of BM chimerism of transplanted BMC mixture (day of Tx) injected into recipient mice in a competitive BM Tx assay. Quantified percentage of donor-derived BMCs normalized to WT, set 25%. n = 3. (C) Percentages of donor-derived PB cells analyzed by flow cytometry at the indicated time points in competitive BM Tx assay. Data from 3 independent Tx assays with at least 5 mice per group and genotype. (D) Representative flow cytometry profiles of CD45.1/CD45.2 BM chimerism after 6 months in recipient mice in competitive BM Tx assay. Quantified percentage of donor-derived BMCs. n = 8 Nipa+/+; n = 6 Nipa–/–. (E) Percentages of donor-derived PB cells analyzed by flow cytometry at the indicated time points in 3 serial LSK Tx assays. Data are from 2 independent Tx assays with 3–7 mice per group and genotype. (F) Percentages of LT-HSCs, ST-HSCs, and MPPs in 6- to 8-month-old Nipa+/+ and Nipa–/– BMCs after 5-FU injection on day –4, normalized to WT, set 100%. n = 9 Nipa+/+; n = 9 Nipa–/–. (G) Kaplan-Meier survival curve of 11-month-old Nipa+/x and Nipa–/– mice following regular 5-FU administration. n = 6 Nipa+/x; n = 7 Nipa–/–. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. An unpaired 2-tailed Student’s t test (A–F) or log-rank (Mantel-Cox) test (G) was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figures 2–4.
Taken together, the results indicate that Nipa deletion both reduces the HSC pool and causes a cell-intrinsic long-term repopulation defect, with impaired self-renewal in serial transplantations and a bias toward myeloid differentiation.
Nipa deficiency accelerates 5-fluorouracil–mediated hematopoietic exhaustion.
To evaluate the influence of Nipa deficiency on the regenerative potential of HSCs under stress conditions, we analyzed BM cellularity and the frequency of early hematopoietic cell populations in young mice treated with 5-fluorouracil (5-FU). Nipa–/– mice showed BM aplasia 10 days after weekly 5-FU injection (Supplemental Figure 3A) and a significant reduction in LT-HSCs (P = 0.04), ST-HSCs (P < 0.0001), and MPPs (P = 0.008) 96 hours after a single 5-FU injection compared with their littermate controls (Figure 2F). When mice were treated weekly with 5-FU, we observed reduced survival of Nipa–/– animals, indicating reduced regenerative potential under the replication stress of Nipa-deficient HSCs (Figure 2G). To determine the extent of HSC proliferation after 5-FU treatment, we performed BrdU incorporation assays and observed significantly higher amounts of Nipa–/– LSK cells in S phase 5 days after drug administration, compared with WT LSK cells (Supplemental Figure 3B). This implies a role for NIPA in maintaining HSC quiescence. Furthermore, in cell trace assays, which were combined with annexin staining, we demonstrated loss of Nipa–/– HSCs after induction of replication stress by a reduction in cell numbers (Supplemental Figure 3C) and significantly higher rates of apoptosis (Supplemental Figure 3E). However, rare surviving cells were able to perform adequate rounds of cell cycling, as in cell trace assays both Nipa+/+ and Nipa–/– LSK cells cycled 4 times after in vivo 5-FU treatment and 3 days of ex vivo culture (Supplemental Figure 3D). Steady-state cell cycle analyses revealed no differences in aged, nonstimulated HSCs (Supplemental Figure 4, A and B).
These results suggest an important role for NIPA in retaining HSC maintenance under 5-FU–induced stress conditions. Moreover, by inducing replication stress through application of 5-FU, we were able to trigger the phenotypic reduction and functional decline of aged Nipa–/– HSCs in young animals.
Nipa–/– HSCs are unable to repair DNA damage and prone to cell death.
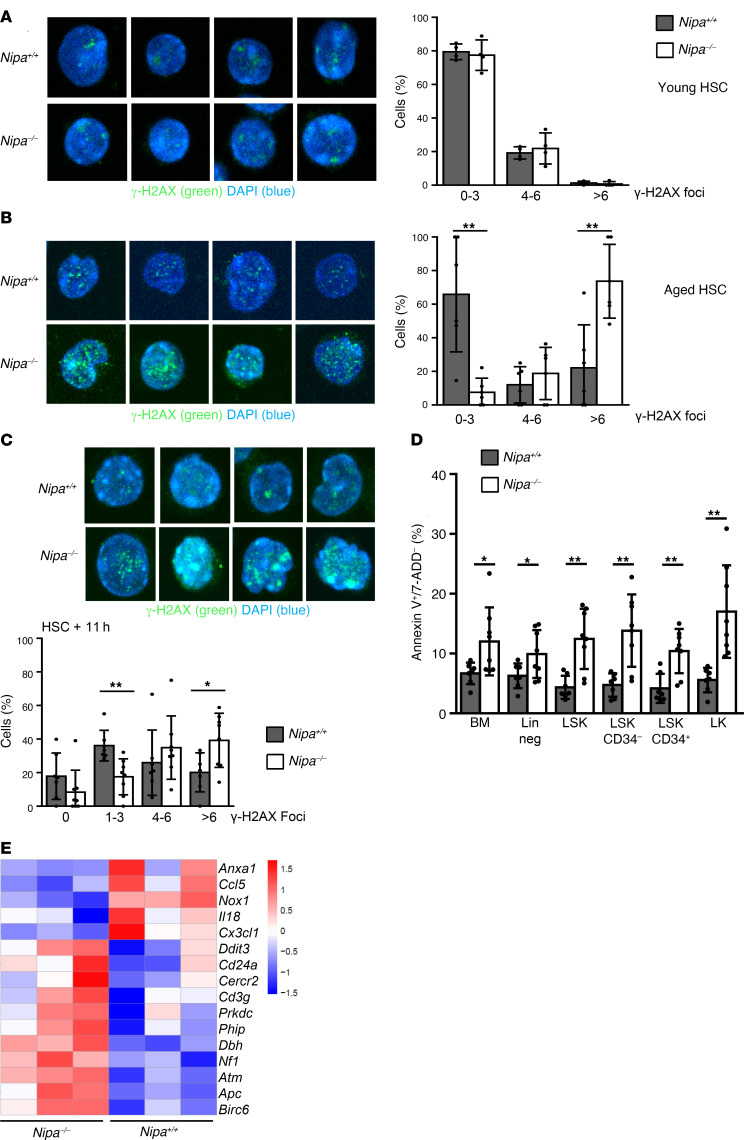
To clarify whether the phenotype of aged or stressed Nipa–/– LSK cells is associated with increased levels of DNA damage, we examined the accumulation of foci of the phosphorylated histone H2A variant H2AX at Ser139 (γ-H2AX), which is indicative of unresolved DNA damage. No difference in DNA damage foci between Nipa+/+ and Nipa–/– LSK cells was found at 5 months of age (Figure 3A), whereas the amount of γ-H2AX foci was markedly enhanced in aged Nipa–/– LSK cells (Figure 3B; P = 0.008). The accumulation of γ-H2AX foci in aged Nipa–/– mice was restricted to the primitive hematopoietic stem and progenitor cell compartment, since we did not detect such differences in mature, lineage-positive cells (Supplemental Figure 5A).
Figure 3. Nipa–/– HSCs are unable to repair DNA damage and prone to cell death.
(A) Immunofluorescence for γ-H2AX foci in young (5 months) Nipa+/+ and Nipa–/– HSCs. Representative confocal microscopy images and quantitative graph are shown. Data from 4 independent experiments. n = 53 Nipa+/+; n =62 Nipa–/–. Original magnification, ×63. (B) Immunofluorescence for γ-H2AX foci in aged (11–18 months) Nipa+/+ and Nipa–/– HSCs. Representative confocal microscopy images and quantitative graph are shown. Data are from 6 independent experiments. n = 136 Nipa+/+; n = 78 Nipa–/–. Original magnification, ×63. (C) Representative confocal microscopy images and quantitative results for 8-month-old HSCs stained for γ-H2AX 11 hours after 4-Gy irradiation. Data are from 7 independent experiments. n = 183 Nipa+/+; n = 230 Nipa–/–. Original magnification, ×63. (D) Percentages of early apoptotic cells (annexin V+7-AAD–) within hematopoietic subpopulations from Nipa+/+ and Nipa–/– mice older than 20 months. n = 8 Nipa+/+; n = 8 Nipa–/–. Lin neg, lineage-negative. (E) Heatmap of expression levels of apoptosis-related genes between aged (>20 months) untreated Nipa+/+ and Nipa–/– HSCs analyzed by microarray. Color scale represents row Z-score mRNA intensity values. *P < 0.05, **P < 0.01. An unpaired 2-tailed Student’s t test was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figure 5.
To clarify whether the accumulation of DNA damage sites in aged Nipa–/– animals was due to defective DNA repair, we irradiated sorted young LSK cells (4 Gy) and monitored for DNA damage repair by measuring γ-H2AX kinetics. We were not able to find gross differences in early DNA damage induction in young LSK cells 3 hours after irradiation (Supplemental Figure 5B). However, 11 hours after irradiation, the majority of Nipa+/+ HSCs showed successful DNA damage repair, as illustrated by resolution of the γ-H2AX foci. Nipa–/– LSK cells showed prolonged DNA damage signals, with a significantly higher percentage of cells with more than 6 γ-H2AX foci (39.3%, vs. 19.6% in WT cells; P = 0.02; Figure 3C). This suggests that the DNA repair pathway is defective in Nipa–/– HSCs. Of note, we did not find any differences in resolution of γ-H2AX foci in more differentiated cell populations (Supplemental Figure 5C), indicating that Nipa deficiency impairs the irradiation-induced DNA damage response predominantly in LSK cells.
In addition, we observed a significant increase in annexin V+7AAD– cells, especially in primitive CD34– Nipa–/– LSK cells (13.8%, compared with 4.7% in WT LSK cells) in mice older than 20 months (Figure 3D and Supplemental Figure 5D). The increased susceptibility of Nipa–/– LSK cells to cell death was further enhanced in the absence of protective BM niche–derived signals (30%, vs. 3.8% annexin V+7AAD– cells; Supplemental Figure 5E; P < 0.001). Cell death could also be triggered by Tx stress, as young Nipa–/– BMCs showed an increase in annexin V+7AAD– cells 17 weeks after a second LSK Tx compared with Nipa+/+ BMCs (Supplemental Figure 5F).
To further delineate which pathways are deregulated in Nipa-deficient aged or DNA-damaged HSCs, we performed global gene expression analysis on untreated and irradiated (4 Gy) cells of aged mice (Supplemental Figure 5G). In agreement with the above-described experiments, the microarray data showed significantly upregulated expression of apoptotic genes in both aged and DNA-damaged Nipa–/– LSK cells (Figure 3E and Supplemental Figure 5H). Untreated Nipa–/– LSK cells showed aberrations in their DNA repair expression profile, with overexpression of genes involved in DNA damage repair (Supplemental Figure 5I).
Taken together, our results highlight increased sensitivity to cell death in aged and DNA-damaged Nipa–/– LSK cells, which was associated with an accumulation of DNA damage that may be due to alterations in the DNA repair pathway.
NIPA interacts with FANCD2.
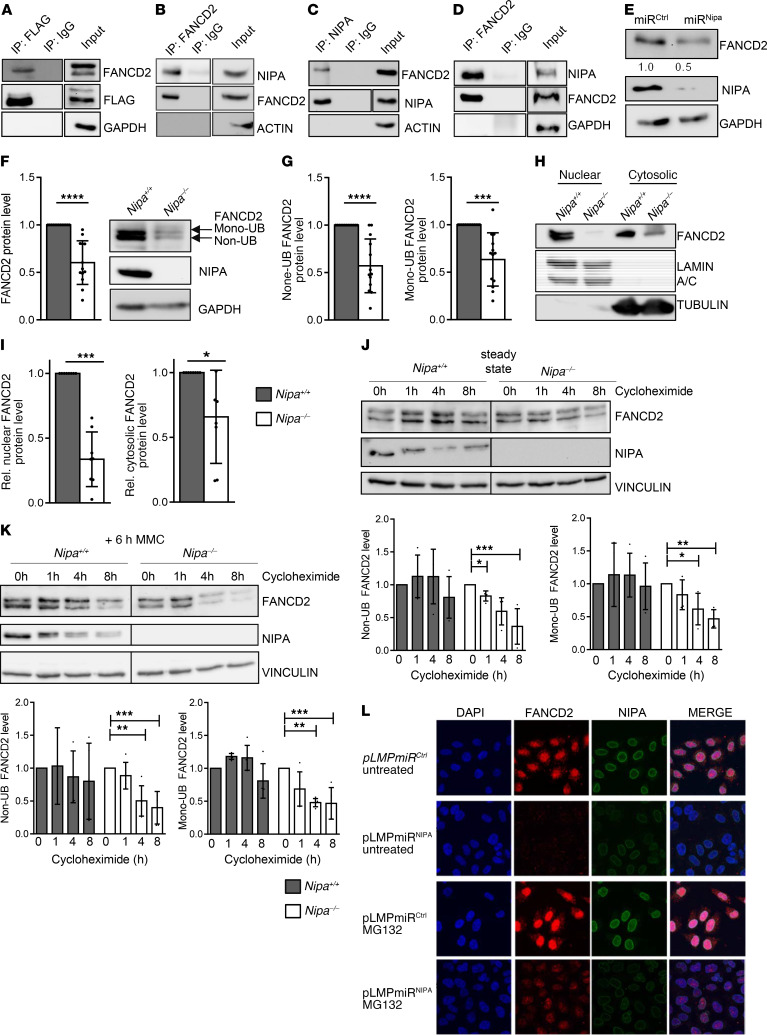
To further investigate the underlying mechanisms and identify NIPA-interacting proteins, we performed mass spectrometry analysis of NIPA-overexpressing HEK293T cells. Through this analysis, we found that FANCD2 interacts with NIPA and confirmed the binding by coimmunoprecipitation experiments in Phoenix and HeLa cells, as well as in hematopoietic primary cells and cell lines (Figure 4, A and B, and Supplemental Figure 6, A–D). Coimmunoprecipitation of transiently overexpressed FLAG-hNIPA in Phoenix cells and endogenous NIPA in HeLa cells through FANCD2 and vice versa confirmed the robust NIPA-FANCD2 interaction (Figure 4, C and D). Although FancD2 mRNA levels were unaffected in the absence of NIPA (Supplemental Figure 7A), non- and monoubiquitinated FANCD2 protein levels were significantly reduced in Nipa-deficient cells, indicating that NIPA is important for FANCD2 protein stabilization (Figure 4, E–G; P < 0.0001). Localization analyses of FANCD2 by immunofluorescence in HeLa cells retrovirally transfected with pLMP miRCtrl or miRNipa showed significantly reduced FANCD2 levels predominantly in the nucleus of Nipa-downregulated cells (Supplemental Figure 7B). Nuclear and cytosolic extracts of Nipa–/– primary mouse embryonic fibroblasts (MEFs) reinforced this result, showing greatly reduced FANCD2 protein levels, especially in the nuclear fraction (Figure 4, H and I). Cycloheximide treatment of primary MEFs resulted in a reduced half-life of FANCD2 in Nipa–/– cells (Figure 4, J and K), suggesting that NIPA is important for FANCD2 protein stability. Inhibition of the proteasomal pathway by MG132 led to partial rescue of FANCD2 protein levels (Figure 4L and Supplemental Figure 8, A and B), once again suggesting that NIPA prevents FANCD2 from untimely (proteasomal) degradation. In order to investigate steady-state levels of other FANC family members in Nipa-deficient animals, we examined FANCG, FANCE, FANCF, and FANCI protein levels, with only slightly reduced amounts of FANCI in Nipa–/– primary MEFs (Supplemental Figure 9, A and B).
Figure 4. NIPA interacts with FANCD2.
(A) FLAG coimmunoprecipitation of Phoenix E cells transiently transfected with FLAG-hNIPA. (B) FANCD2 coimmunoprecipitation of Phoenix E cells transiently transfected with FLAG-hNIPA. (C) NIPA coimmunoprecipitation of HeLa cells with endogenous levels of NIPA. (D) FANCD2 coimmunoprecipitation of HeLa cells with endogenous levels of NIPA. (E) Western blot analysis of HeLa cells retrovirally transfected with pLMP miRCtrl or miRNipa for FANCD2, normalized to GAPDH. (F) Western blot analysis of Nipa+/+ and Nipa–/– primary MEFs for FANCD2, NIPA, and GAPDH. Quantification of relative FANCD2 protein levels normalized to GAPDH of Nipa+/+ and Nipa–/– primary MEFs. n = 13 Nipa+/+; n = 13 Nipa–/–. (G) Quantification of relative nonubiquitinated (Non-UB) and monoubiquitinated (Mono-UB) FANCD2 protein levels of Nipa+/+ and Nipa–/– primary MEFs analyzed by Western blot. n = 13 Nipa+/+; n = 13 Nipa–/–. (H) Western blot analysis of nuclear and cytosolic extracts of Nipa+/+ and Nipa–/– primary MEFs for FANCD2, lamin A/C, and tubulin. (I) Quantification of relative (rel.) FANCD2 protein levels normalized to lamin A/C (nuclear) or tubulin (cytosolic) of Nipa+/+ and Nipa–/– primary MEFs. n = 8 Nipa+/+; n = 8 Nipa–/–. (J) Western blot analysis of non- and mono-UB FANCD2 levels of Nipa+/+ and Nipa–/– primary MEFs (steady state) treated with cycloheximide for the indicated times. n =4 Nipa+/+; n = 4 Nipa–/–. (K) Western blot analysis of non- and mono-UB FANCD2 levels of Nipa+/+ and Nipa–/– primary MEFs (6 hours, 0.5 μM MMC) treated with cycloheximide for the indicated times. n = 5 Nipa+/+; n = 5 Nipa–/–. (L) Immunofluorescence for FANCD2, NIPA, and DAPI in untreated and 4-hour MG132–treated (5 μM) in HeLa cells retrovirally transfected with pLMP miRCtrl or miRNipa. Representative confocal microscopy images are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A paired 2-tailed Student’s t test Original magnification, ×63. (F, G, and I) or Dunnett’s test (J and K) was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figures 6–9.
Nipa-deficient cells show MMC hypersensitivity as a hallmark of FA.
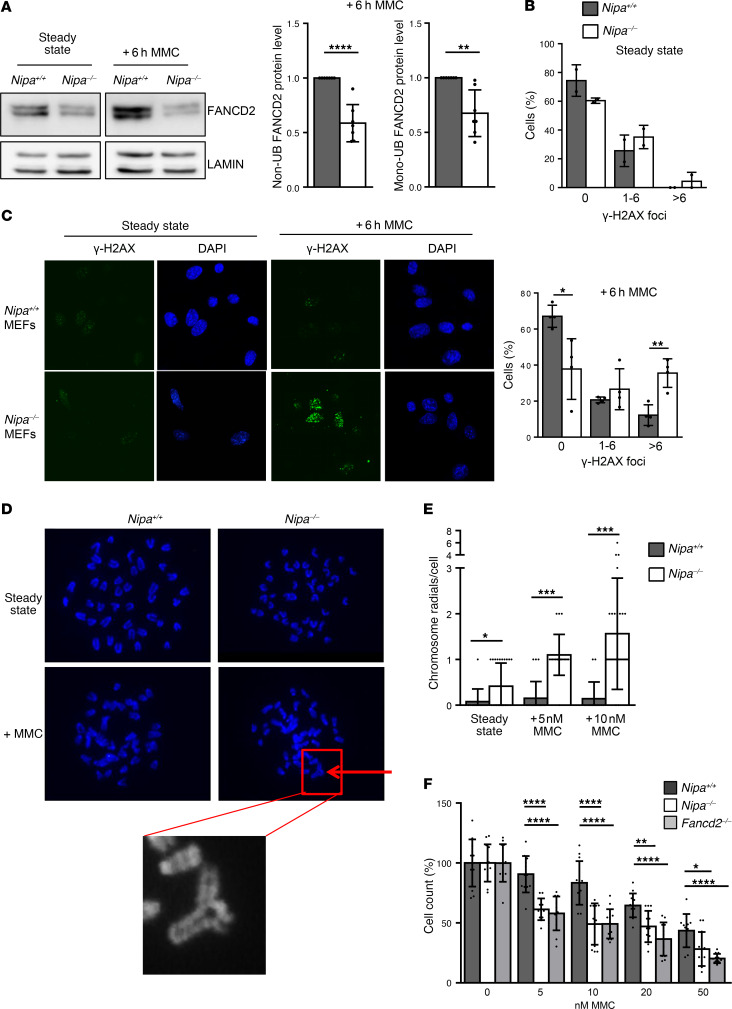
To test the activation ability of the FA/BRCA pathway in the absence and presence of NIPA, we treated primary MEFs with MMC, which strongly activates the FA pathway via DNA interstrand cross-linkage. We examined mRNA and protein levels of FANCD2, and found that while FancD2 transcription was comparable in Nipa+/+ and Nipa–/– primary MEFs (Supplemental Figure 10A), non- and monoubiquitinated FANCD2 protein levels were dramatically reduced, particularly after MMC treatment in Nipa–/– compared with WT MEFs (Figure 5A and Supplemental Figure 10B; P < 0.0001). Immunofluorescence of untreated and MMC-treated primary MEFs demonstrated the MMC hypersensitivity of Nipa–/– cells, shown by significantly increased numbers of γ-H2AX foci after 6 hours of MMC treatment (Figure 5, B and C, and Supplemental Figure 10C). Importantly, we showed a significant increase in radial chromosomes, which is a hallmark of FA, in MMC-treated Nipa-deficient primary splenocytes (Figure 5, D and E). Nipa–/– cells further displayed high sensitivity to MMC, as confirmed by reduced survival rates. Interestingly, the MMC hypersensitivity was seen in both Nipa–/– and Fancd2-deficient cells, with Fancd2-deficient cells showing a trend to even higher MMC sensitivity after treatment with 20 or 50 nM MMC (Figure 5F).
Figure 5. Nipa-deficient cells display MMC hypersensitivity as a hallmark of FA.
(A) Western blot analysis of Nipa+/+ and Nipa–/– primary MEFs (untreated and treated +6 hours with 0.5 μM MMC) for FANCD2 and lamin. Quantification of relative non- and monoubiquitinated FANCD2 protein levels of Nipa+/+ and Nipa–/– primary MEFs is shown. n = 7 Nipa+/+; n = 7 Nipa–/–. (B) Quantification of γ-H2AX foci in untreated and 0.5 μM MMC–treated Nipa+/+ and Nipa–/– primary MEFs. No MMC: data from 2 independent experiments; n = 74 Nipa+/+, n = 112 Nipa–/–. +6 hours MMC: data from 4 independent experiments; n = 121 Nipa+/+, n = 93 Nipa–/–. (C) Immunofluorescence for γ-H2AX foci in untreated and 6-hour MMC-treated (0.5 μM) Nipa+/+ and Nipa–/– primary MEFs. Representative confocal microscopy images are shown. Original magnification, ×63. (D) Representative images of DAPI-stained metaphase spreads of untreated and MMC-treated (5 nM) Nipa+/+ and Nipa–/– spleen cells after 48 hours of in vitro growth. Arrow indicates chromosome radials. Original magnification, ×100. (E) Quantification of chromosome radials per cell of untreated and MMC-treated (5 and 10 nM) Nipa+/+ and Nipa–/– spleen cells after 48 hours of in vitro growth. No MMC: n =40 Nipa+/+; n = 40 Nipa–/–. +MMC: n = 20 Nipa+/+; n = 20 Nipa–/–. (F) Cell survival assay of Nipa+/+, Nipa–/–, and Fancd2–/– MEFs measured after 5 days of culture with the indicated concentrations of MMC. Data from 2 independent experiments are shown. n = 12 Nipa+/+; n = 12 Nipa–/–; n =12 Fancd2–/–. One-way ANOVA, P = 6.4 × 10–7 (5 nM); P = 4.9 × 10–6 (10 nM); P = 1.7 × 10–5 (20 nM); P = 1.0 × 10–4 (50 nM). Reported P values in the figure from unpaired 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A paired (A) or unpaired (B and E) 2-tailed Student’s t test was used for statistical analyses. Data are represented as mean ± SD. See also Supplemental Figure 10.
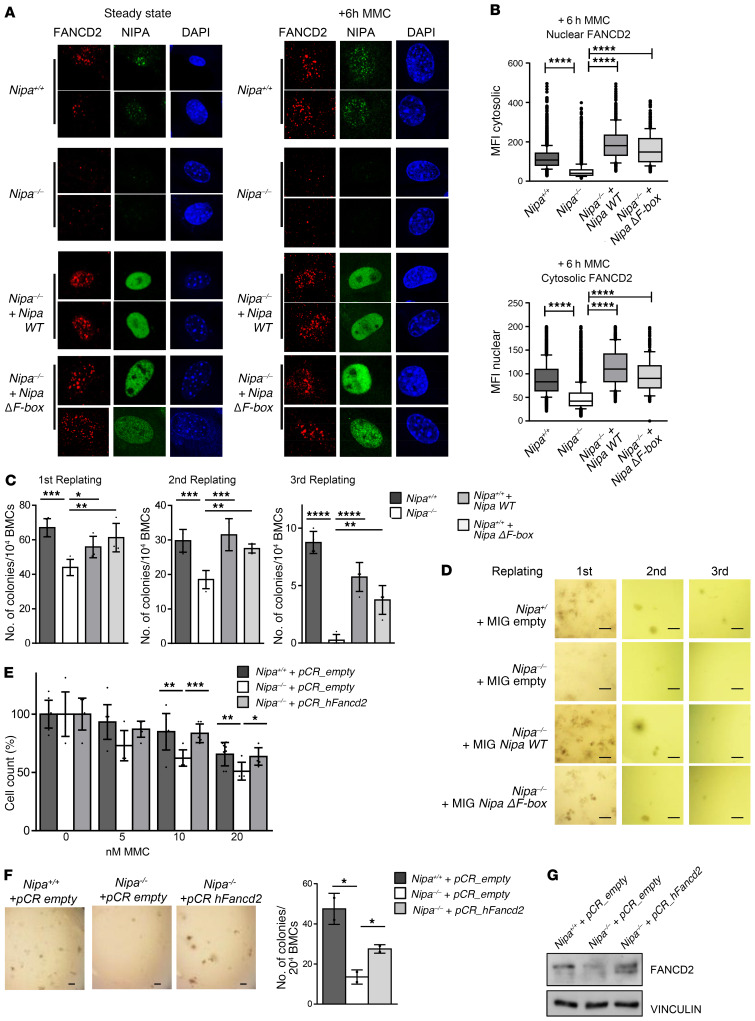
At this point, we sought to examine whether reexpression of Nipa, Nipa mutants, or Fancd2 can rescue the observed phenotype. Indeed, reexpression of Nipa (WT) was able to increase FANCD2 levels (Figure 6, A and B, and Supplemental Figure 11A) and also to functionally reverse the observed repopulating defects in Nipa–/– HSCs (Figure 6, C and D). Reexpression of a Nipa mutant with a deleted and thereby inactive F-box (referred to here as NipaΔF-box), was also able to rescue the functional repopulation defects of Nipa–/– HSCs and also showed strong binding to FANCD2 (Figure 6, A–D, and Supplemental Figure 11, A–E). Reexpression of Fancd2 in primary Nipa–/– BMCs restored the repopulation capacity in CFU assays, further emphasizing that the Nipa–/– phenotype in HSCs is caused by impaired FANCD2 levels with a disruption of the FA pathway (Figure 6, F and G). We were also able to restore the MMC sensitivity of Nipa–/– MEFs by Fancd2 reexpression in those cells (Figure 6E). To investigate whether NIPA and FANCD2 are epistatic, we downregulated Fancd2 in Nipa-deficient spleen cells and examined chromosomal radials after MMC treatment. As shown in Supplemental Figure 12, A–C, NIPA was epistatic to FANCD2. However, we found a slight increase in chromosomal radials after Fancd2 knockdown in Nipa–/– spleen cells (Supplemental Figure 12, A–C). In agreement with these results, Nipa–/– primary MEFs with downregulated Fancd2 showed slightly increased MMC hypersensitivity compared with solely Nipa-deficient MEFs (Supplemental Figure 12, D and E; 10, 20, and 50 nM MMC; P = 0.1). Therefore, we suggest that NIPA is epistatic to FANCD2, with slight additional FANCD2-dependent effects on MMC sensitivity in Nipa-deficient cells.
Figure 6. Fancd2 restoration can overcome the defects of Nipa-deficient cells.
(A) Immunofluorescence for FANCD2, NIPA and DAPI, in untreated and 6-hour MMC–treated (0.5 μM) Nipa+/+ and Nipa–/– primary MEFs retrovirally transfected with pBABENipaWT or pBABENipaΔF-box. Representative confocal microscopy images are shown. Original magnification, ×63. (B) Average fluorescence intensity of FANCD2 staining in the nucleus and the cytosol of the Nipa+/+ and Nipa–/– primary MEFs (+6 hour MMC) shown in A analyzed by image cytometry. Box-and-whisker plots: 10th to 90th percentile, with outliers aligned. Nuclear FANCD2: n = 4622 Nipa+/+; n = 5035 Nipa–/–; n = 722 Nipa–/– + Nipa WT; n = 315 Nipa–/– + Nipa ΔF-box. Cytosolic FANCD2: n = 3929 Nipa+/+; n = 4414 Nipa–/–; n = 593 Nipa–/– + Nipa WT; n =287 Nipa–/– + Nipa ΔF-box. Reported P values in the figure from Dunnett’s test. (C) Nipa+/+ or Nipa–/– BMCs were retrovirally infected with pMIGempty, pMIGNipaWT, or pMIGNipaΔF-box and used for in vitro CFU assay. Quantification of colonies is shown. n = 4 Nipa+/+; n = 4 Nipa–/–; n = 4 Nipa–/– + Nipa WT; n = 4 Nipa–/– + Nipa ΔF-box. Reported P values in the figure from Dunnett’s test. (D) Representative images of CFU assay described in C. Scale bars: 1000 μm. (E) Cell survival assay of Nipa+/+ and Nipa–/– primary MEFs lentivirally infected with pCRempty or pCRFancd2 measured after 5 days of culture with the indicated concentrations of MMC. Data from 2 independent tests are shown. n = 9 Nipa+/+ + pCRempty; n = 6 Nipa–/– + pCRempty; n = 6 Nipa–/– + pCRFancd2. One-way ANOVA, P = 0.004 (10 nM); P = 0.014 (20 nM). Reported P values in the figure from unpaired 2-tailed Student’s t test. (F) Nipa+/+ or Nipa–/– BMCs were lentivirally infected with pCRempty or pCRFancd2 and used for in vitro CFU assay. Representative images and quantification of colonies are shown. Scale bar: 1000 μm. n = 2 Nipa+/+ + pCRempty; n =2 Nipa–/– + pCRempty; n = 2 Nipa–/– +pCRFancd2. One-way ANOVA, P = 0.015. Reported P values in the figure from unpaired 2-tailed Student’s t test. (G) Representative Western blot of BMCs used for CFU assay shown in A. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are represented as mean ± SD. See also Supplemental Figures 11 and 12.
Based on these results, we conclude that NIPA, independent of its F-box function, is essential for the nuclear abundance of FANCD2 and correct initiation of the FA/BRCA pathway.
Loss of NIPA leads to a BM failure phenotype in mice resembling that of functional Fanc deficiency.
Mice with a dysfunctional FA pathway such as FancA–/–, FancD2–/–, or FancG–/– showed overall mild hematopoietic anomalies under steady-state conditions, but application of chronic stress induced complete hematopoietic collapse in some strains (38, 44). Based on the defective activation of the FA/BRCA pathway, we hypothesized that Nipa-deficient mice have an FA-like phenotype. To provoke this distinct phenotype, we treated Nipa–/– mice repeatedly with poly(I:C) (Supplemental Figure 13A and ref. 38). To exclude Nipa-deficient niche–dependent effects in the experimental setup, we performed the long-term assay using a hematopoietic cell–specific VavTg/WT NipaCKO/CKO mouse strain. After short-term poly(I:C) treatment of young Nipa–/– mice, we showed significantly reduced blood counts (Supplemental Figure 13B), diminished BM cellularity (Supplemental Figure 13C), and lower levels of LSK and LK cells (Supplemental Figure 13D). An 5-ethynyl-2′-deoxyuridine (EdU) assay did not identify cell cycle defects in Nipa-deficient BMCs (Supplemental Figure 13E).
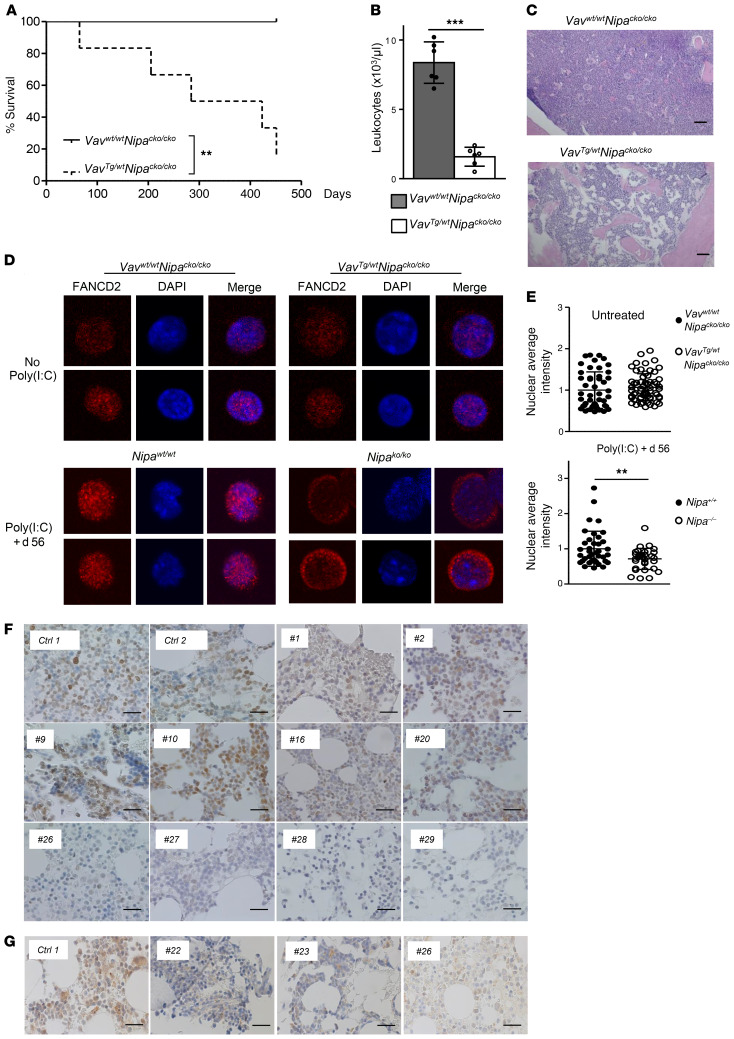
Even more remarkable were the results of the chronic stress assay: while control mice survived repeated poly(I:C) injections for more than 400 days, all of the treated VavTg/WT NipaCKO/CKO animals died due to BM failure, with a median latency of 354 days (Figure 7A; P = 0.001). WBC counts were greatly reduced at the time of death (Figure 7B; P < 0.0001), and BM sections showed substantial aplasia (Figure 7C and Supplemental Figure 13F), with a slight increase in fibrosis as measured by reticulin staining of VavTg/WT NipaCKO/CKO BM sections (Supplemental Figure 13G). To further test our hypothesis that this dramatic phenotype was caused by a functional Fancd2 deficiency in Nipa–/– animals, we analyzed FANCD2 in young untreated and poly(I:C)-treated HSCs via immunofluorescence. As expected, untreated young HSCs showed low FANCD2 levels independent of NIPA (Figure 7, D and E, and Supplemental Figure 13H). In contrast, poly(I:C)-treated WT HSCs exhibited a strong FANCD2 signal, whereas Nipa-deficient HSCs showed significantly reduced FANCD2 levels. The FANCD2 reduction occurred predominantly in the nucleus, with significantly reduced nuclear FANCD2 staining in Nipa–/– HSCs on day 56 of repetitive poly(I:C) treatment (Figure 7, D and E, and Supplemental Figure 13H; P = 0.006). This was most likely the cause for the aberrant DNA repair/FA/BRCA pathway in Nipa-deficient HSCs.
Figure 7. Loss of NIPA leads to a BM failure phenotype resembling functional Fanc deficiency.
(A) Kaplan-Meier survival curve of VavWT/WT NipaCKO/CKO and VavTg/WT NipaCKO/CKO mice following regular poly(I:C) treatment dying from BM failure. n = 8 VavWT/WT NipaCKO/CKO, n = 6 VavTg/WT NipaCKO/CKO. (B) Leukocyte counts of VavWT/WT NipaCKO/CKO and VavTg/WT NipaCKO/CKO mice following regular poly(I:C) treatment at death compared with control mice at the same time points. n = 6 Nipa+/+; n = 6 Nipa–/–. (C) Representative H&E staining of BM sections of VavWT/WT NipaCKO/CKO and VavTg/WT NipaCKO/CKO mice following regular poly(I:C) treatment sacrificed when moribund or at end of experiment. Original magnification, ×10. Scale bars: 100 μm. (D) Representative confocal microscopy images of LSK cells isolated from untreated or poly(I:C)-treated (day 56) mice (aged 3–5 months). Cells are stained for FANCD2 (red) and DAPI (blue). Original magnification, ×63. (E) Average fluorescence intensity of FANCD2 staining in the nuclei of untreated and poly(I:C)-treated HSCs. Results were normalized to control cells set to 1. Untreated: n = 48 VavWT/WT NipaCKO/CKO; n = 62 VavTg/WT NipaCKO/CKO. Poly(I:C) +d56: n = 40 Nipa+/+; n = 32 Nipa–/–. (F) Representative IHC staining of NIPA (brown) in BM sections from patients with FA (patients 1 and 2) and RCC with (9 and 10) and without (16, 20, and 26–29) known germline predisposition demonstrates distinct NIPA downregulation (patients 26–29) in approximately 45% of analyzed RCC cases. Control samples (Ctrl 1 and 2) represent IHC NIPA BM sections from adult healthy donors. Original magnification, ×63. Scale bar: 20 μm. (G) Representative immunohistochemical staining of FANCD2 (brown) in BM sections from NIPA-downregulated RCC patients (22, 23, and 26). Control sample (Ctrl 1) represents IHC FANCD2 BM section from an adult healthy donor. Original magnification, ×63. Scale bars: 20 μm. **P < 0.01, ***P < 0.001. A log-rank (Mantel-Cox) test (A) or unpaired 2-tailed Student’s t test (B and E) was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figures 13 and 14.
Taken together, these data indicate that loss of NIPA leads to a BM failure phenotype in mice under chronic stress conditions resembling that of animals with a dysfunctional FA/BRCA signaling cascade. This points to a functional FA pathway defect in LSK cells due to reduced nuclear FANCD2 levels in the case of Nipa deficiency and defines NIPA as an FA-associated protein.
NIPA is downregulated in BM cells from a subset of patients with hypocellular RCC.
Based on the pathological MMC sensitivity in vitro and the FA-like phenotype of Nipa-deficient mice, we postulated that Nipa deficiency might cause FA in humans or play a role in other human disorders characterized by hypocellularity of the BM. First, we sequenced the NIPA gene in 5 patients with confirmed but unclassified FA who were negative for all FA genes described to date, but found no NIPA mutations when using whole exome sequencing (WES). We next focused on BM samples from children with MDS-type RCC, a disease characterized by pancytopenia, BM dysplasia, normal blast percentages, and often BM hypocellularity. We also included patients with genetically defined FA (Supplemental Figure 14). NIPA expression was determined by IHC on trephine biopsy slides. As expected, FA patients with documented germline FANC gene mutations showed physiological NIPA expression in BM cells (Supplemental Figure 14, patients 1–6, and Figure 7F, patients 1 and 2). In contrast, only 13 of 24 RCC patients showed normal NIPA expression (Supplemental Figure 14, patients 8–20), while 11 showed greatly reduced NIPA levels (Figure 7F and Supplemental Figure 14, patients 21–31). The presence of an underlying germline syndrome predisposing to MDS (including FA) was investigated in all RCC patients. Surprisingly, in the patients with a defined germline mutation (i.e., in GATA2, SAMD9L, or RUNX1), we found normal levels of NIPA expression (Supplemental Figure 14, patients 8–13, and Figure 7F, patients 9 and 10). Of the 18 patients without a known genetic predisposition and having a normal karyotype, 11 showed a marked reduction in NIPA expression (Supplemental Figure 14, patients 21–31, and Figure 7F, patients 26–29). Interestingly, the RCC samples with downregulated NIPA expression also showed reduced FANCD2 levels (Figure 7G). Together with the findings in our mouse model, this result indicates that a reduction in NIPA levels might contribute to hematopoietic failure in these patients.
To investigate whether NIPA mutations or deletions account for its reduced expression in RCC patients, we performed WES of 8 of the samples with reduced NIPA expression. We did not detect any NIPA mutations, suggesting epigenetic and/or (post)transcriptional regulation as the most likely reasons for NIPA downregulation in these patients.
In summary, we found reduced NIPA expression in a subset of patients with hypocellular BM and peripheral cytopenia. Further studies will be required in order to uncover a causal link between NIPA expression and hematopoietic failure in RCC patients, and to evaluate NIPA expression as a novel biomarker and potential therapeutic target in patients with RCC and FA.
Discussion
Nipa is expressed in the murine hematopoietic system throughout life, with particularly high expression levels in the primitive HSC pool, indicating a potential role for NIPA in these cells. Indeed, within our study, we were able to identify a crucial function of the protein within the DNA repair/FA/BRCA network.
In the absence of NIPA, HSCs show characteristic features of premature aging (45) in aged and challenged mice: the finding of impaired reconstitution efficiency and repopulation capacity in competitive and serial Tx, respectively, suggests that Nipa deficiency leads to cell-intrinsic HSC defects. Reduced reconstitution efficiency is a classical feature of aged HSCs, as has been shown previously by Liang et al., who correlated BM reconstitution efficiency indirectly with age, finding 2- to 3-fold-lower engraftment of old compared with young HSCs (46, 47). Consistent with other reports showing a switch from lymphoid to myeloid differentiation in aged HSCs (48, 49), Nipa–/– HSCs tended to differentiate toward the myeloid lineage. One hallmark of aged HSCs is the accumulation of γ-H2AX foci (50, 51). The accumulation of a large number of γ-H2AX foci in aged Nipa–/– HSCs suggests that unresolved DNA damage might be causative for premature aging, as seen in mice defective for DNA repair pathway genes. Recent findings implicate replication stress as a major cause of DNA damage induction in HSCs (31, 52), and it has been shown that physiological stresses (such as infections and blood loss) trigger quiescent HSCs to enter the cell cycle, with accumulation of DNA damage as a direct consequence (38). Interestingly, we demonstrated that NIPA regulates the nuclear abundance of FANCD2, a key regulator of the FA/BRCA pathway directly involved in counteracting replication stress by maintaining the integrity of stalled replication forks and processing ICLs (22, 25, 26); and it has been shown that the nuclear localization of FANCD2 is essential for its function (53). Although we cannot exclude that NIPA has several interaction partners involved in DNA damage repair, the interaction of NIPA with FANCD2 and its interplay with the FA/BRCA pathway define NIPA as an FA-associated protein that is essential for preventing the accumulation of DNA damage. Like Fanc-mutated cells, Nipa deficiency results in increased MMC sensitivity, which is a substantial hallmark of a nonfunctional FA/BRCA pathway (34, 35).
Based on these observations, we hypothesized that Nipa-deficient mice resemble Fanc gene–knockout animals. Indeed, Nipa–/– mice have gross hematopoietic and nonhematopoietic similarities to those strains. Most existing FA mouse models show functional defects of HSCs, hypersensitivity to MMC, and increased γ-H2AX levels, all of which are features of the Nipa–/– phenotype (44, 54–57). FancB+/– and FancA–/– mice show reduced platelet counts, as was found in the Nipa–/– animals. However, most Fanc-knockout mouse strains (Fancc–/–, Fancg–/–, Fancd1–/–, Fancd2–/–, and Usp1–/–) show only limited alterations in PB counts, comparable to the Nipa-knockout mouse. Most importantly, exposure of Nipa–/– mice to chronic stress conditions induced full BM failure with peripheral cytopenia and death of the animals, which closely mirrors the phenotype of FancA–/– mice (38). Along with the hematopoietic phenotype, Nipa–/– mice showed a reduced Mendelian birth rate, similar to Fancd2- and other Fanc-knockout animals. Furthermore, in both Nipa- and Fanc-knockout strains, mice show growth retardation and germ cell defects with hypogonadism. The germ cell defects in our Nipa–/– mice were caused by a block of spermatogenesis during meiotic prophase due to altered DNA damage repair followed by apoptosis (40). Regarding cancer susceptibility, Fanc-knockout mice mostly develop cancer after 15 months of age or under stress conditions (44).
In our study, we showed that NIPA and FANCD2 interact which each other and identified a profound and essential regulation of FANCD2 by NIPA. A central remaining question is how NIPA influences FANCD2 abundance and its localization. In previous studies, NIPA was identified as an F-box protein that is part of an SCF-type E3-ubiquitin ligase (39). We show that the F-box function of NIPA seems not to play a crucial role in the regulation of FANCD2 or the murine BM failure phenotype: First, FANCD2 levels were downregulated in NIPA-knockdown and -knockout cells, but should accumulate if FANCD2 would be an SCFNIPA target. Second, we generated an F-box–deleted Nipa mutant and showed that it was still able to bind and interact with FANCD2. Therefore, we believe that the F-box function of NIPA plays a subordinate role in regulating the FA/BRCA axis. Alternatively, NIPA may act as a scaffold protein for FANCD2, thereby stabilizing the nuclear abundance of FANCD2. Another plausible possibility could be that NIPA, which is located at the nuclear envelope of the cell, is directly involved in the nuclear export of the protein. In addition, it is most likely that NIPA has pleiotropic effects after DNA damage, as damaged Nipa–/– cells show some features (γ-H2AX foci after irradiation) that do not essentially correspond to the FANCD2 pathway. Further biochemical and in vivo analysis with different Nipa and FancD2 mutants will unravel the precise mode of the functional NIPA-FANCD2 interaction.
Based on the FA-like phenotype of Nipa-deficient mice, we predict that there is also an FA phenotype in humans. Along with the very variable clinical picture, 90% of FA patients develop BM failure within the first 40 years of life, and the incidence of hematopoietic and solid malignancies in those patients is high (58). Only a very few of the patients diagnosed with FA by the MMC hypersensitivity assay have no mutation in 1 of the 22 known homozygous or compound heterozygous FANC genes. Despite the rarity of those unclassified FA patients, we were able to sequence 5 patient samples collected at the German reference diagnostics center for FA (Institute of Human Genetics, Biozentrum), but did not identify any NIPA mutations. However, we intend to continue to screen genetically undefined FA cases for NIPA deletions and include expression analyses of the protein. As classical IBMFSs such as FA and DC cannot be diagnosed by BM morphology alone, and differentiation of IBMFS and RCC remains a major diagnostic challenge (5, 59), we also evaluated NIPA status in patients with hematopoietic failure. In recent years, it became evident that other forms of BM failure or familial MDS (GATA2 haploinsufficiency, SAMD9/9L syndromes, MYSM1 deficiency, etc.) can also appear as hypocellular RCC. Still, in approximately 85% of RCC patients, no predisposing germline mutation can be identified (our unpublished observation, European Working Group of MDS in Childhood [EWOG-MDS] study). In addition, biomarkers that predict disease progression (i.e., transfusion dependency, severe neutropenia, and/or progression to advanced MDS) are missing. Within our study, we provide evidence that NIPA expression is relevant in diseases characterized by hematopoietic failure. In a subset of patients with MDS-type RCC, we found a distinct and significant downregulation of NIPA. None of the 11 RCC patients with reduced NIPA expression had mutations in genes known to be involved in BM failure or myeloid malignancies (i.e., GATA2, SAMD9/9L, or RUNX1). We also did not find mutations in the NIPA gene in 8 of those samples, suggesting epigenetic or posttranslational mechanisms as a cause for the reduction in NIPA protein levels. Interestingly, NIPA is located at chromosome 7q32.2, a region frequently found to be deregulated in methylation analyses of myelodysplastic syndromes (60–63), suggesting the possibility of epigenetic regulation of NIPA in RCC. In contrast, all of the 20 patients with genetically defined FA or GATA2, SAMD9L, or RUNX1 syndromes examined showed normal NIPA expression. This might indicate that NIPA is an additional regulator of hematopoietic function and that its loss can be the cause of hematopoietic failure in the absence of other known reasons. Further studies will help to understand how NIPA expression is regulated, and larger patient cohorts will be required in order to determine whether NIPA expression levels can be used to distinguish between familial MDS/IBMFS cases (GATA2, SAMD9/9L, etc.) and sporadic disease, or to predict disease outcome. Finally, the putative causal relationship between NIPA expression and RCC pathogenesis could open the way for its use as a therapeutic target.
Taken together, our results add NIPA to the short list of FA-associated proteins and identify a NIPA/FANCD2 axis in which the nuclear abundance of FANCD2 is regulated by NIPA. Further analyses of NIPA in hypoplastic BM disorders will expand the molecular and clinical landscape of IBMFS and RCC, and uncover the clinical potential and relevance of NIPA as a diagnostic tool and possible target in affected patients.
Methods
Mice.
NipaCKO/CKO and NipaKO/KO mice were generated using a conditional knockout strategy (40). To achieve tissue-specific Nipa deletion, we crossed NipaCKO/CKO mice with Vav-Cre (VavTg/WT) transgenic mice (43). Littermates or age- and sex-matched mice were used as controls. Recipient mice and donor mice for support BM were either B6.SJL-PtprcaPep3b/Boy (CD45.1) mice or CD45.1/CD45.2 heterozygotes that were of the F1 generation of C57BL/6 and B6.SJL-PtprcaPep3b/Boy mice (The Jackson Laboratory).
Plasmids and siRNAs.
pLMPmiRCtrl and pLMPmiRNipa were generated by subcloning the target-specific hairpin sequence from Eurofins according to the manufacturer’s instructions. siRNANIPA corresponded to sense CAGAUUGAAUCGUCCAUGA•d(TT) (39). A firefly luciferase siRNA served as a control. FANCD2 Silencer Select siRNA and control Silencer Select siRNA were purchased from Thermo Fisher Scientific and used according to the manufacturer’s protocol. MigR1 FLAG-hNipaWT, MigR1 FLAG-hNipaΔF-box, pBABE hNipaWT, and pBABE hNipaΔF-box were provided by F. Bassermann (Technische Universität München, Munich, Germany). pCR1317 (pCR hFancd2; Addgene plasmid 111127) and pCR1265 (pCR empty; Addgene plasmid 111094) were a gift from Jacob Corn (64). VSVG and CMV vectors were produced by a member of our research team.
Cell lines.
Primary Nipa+/+ or Nipa–/– MEFs were prepared from embryos on E13.5, but only used at early passage and cultured in DMEM (PAA Laboratories) supplemented with 15% FCS under low oxygen conditions. Phoenix E helper virus-free ecotropic packaging cells (G. Nolan, Stanford University, Stanford, California, USA) and HeLa cells (ATCC) were maintained in DMEM supplemented with 10% FCS. The mouse pro-B cell line Ba/F3 (DSMZ) and the BM-derived cell line 32D were cultured in RPMI 1640 (Life Technologies) supplemented with 10% FCS, 2 ng/mL IL-3, and penicillin/streptomycin (65). HPC7 cells (RRID:CVCL_RB19) were cultured in IMDM (Gibco) supplemented with 10% FCS, 200 mM l-glutamine, 1-thyioglycerol, stem cell factor, and penicillin/streptomycin. All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
To inhibit protein synthesis, cells were cultured in the presence of 25 μg/mL cycloheximide, and to inhibit proteasomal degradation, cells were cultured in the presence of 10 μM MG132 (66).
Transfection, and retroviral and lentiviral transduction.
Retrovirus preparation and generation of transiently transfected cell lines were performed using Lipofectamine 2000 (Invitrogen) (67). To generate stable retrovirally infected cell lines of HeLa cell line origin, cells were transduced by 2–4 rounds of spin infection (1200 g, 32°C, 90 minutes) every 12 hours in retroviral supernatant supplemented with 4 g/mL Polybrene (Sigma-Aldrich) and 10% DMEM. Retroviral and lentiviral infection of BMCs and primary MEFs was performed as previously described (67–69). MMC was used in a final concentration of 0.1 or 0.5 μM.
MMC sensitivity assay.
Primary MEFs were plated in triplicate for each drug dose and treated with different concentrations of freshly made MMC. After 5 days in culture, cells were harvested and counted. Cell numbers of the MMC-treated samples were normalized to the cell numbers in the untreated sample (70).
Immunoprecipitation, Western blotting, and cytosolic/nuclear extracts.
For Immunoprecipitation and Western blotting, the following antibodies were used (40, 71, 72): NIPA (B-10), lamin A/C (E-1), and tubulin (TU-02) antibodies were obtained from Santa Cruz Biotechnology. NIPA (HPA 024023), FLAG (M2), and β-actin (A5441) antibodies were purchased from Sigma-Aldrich; GAPDH (OSG-00033G) from Osenses; FANCD2 (EPR2302) from Abcam; and (NB100-182) from Novus Biologicals. Cytosolic and nuclear extracts were generated according to the manufacturer’s protocol using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific).
Flow cytometric analysis and cell sorting.
Flow cytometric analysis was performed using single-cell suspensions incubated with the indicated antibodies (73). A CyAn ADP Analyzer (Beckman Coulter), FACSCanto II, LSRFortessa, FACSAria III, FACSAria Fusion (BD Biosciences), and Beckman Coulter MoFlo XDP were used for analyzing and sorting. Antibodies used to stain cell surface markers were: anti–mouse CD11b (Mac-1, M1/70), CD16/32 (clone 93), CD34 (RAM34), CD45.1 (LY-5.1, A20), CD45.2 (LY-5.2, 104), CD45R/B220 (RA3-6B2), CD45 (30-F11), CD90.2 (Thy1.2, 53-2.1), CD117 (c-KIT, 2B8), CD127 (IL-7Ra, A7R34), CD135 (FLT3, A2F10), Gr-1 (Ly-6G, RB6-8C5), Sca-1 (D7), and Ter-119 (TER-119), all obtained from BD Biosciences or eBioscience. Lineage-positive cells were removed using the Lineage Cell Depletion Kit (Miltenyi Biotec) before sorting.
Apoptosis measurement was performed as previously described using annexin V/7-AAD and cell cycle analysis by BrdU/propidium iodide (PI) (67, 74).
Immunofluorescence and microscopy.
For γ-H2AX, NIPA, and FANCD2 measurements, HeLa cells, primary MEFs, or HSCs were fixed (4%PFA), permeabilized (100%methanol), and after blocking stained for primary and secondary antibodies (75). Anti–γ-H2AX (JBW301; Merck Millipore), anti-NIPA (B-10; Santa Cruz Biotechnology), and anti-FANCD2 (NB100-182, Novus Biologicals) antibodies were used. Image acquisition was performed using a Leica TCS SP2 AOBS confocal microscope equipped with a ×63 NA 1.4 HCX PL APO objective or an Olympus scanR Screening Station. Image analysis was performed using MetaMorph v.7.6.3.0 or Olympus scanR acquisition software.
CFU methylcellulose assay.
To analyze stem cell potential with serial replating, 10,000 BMCs were plated in MethoCult 3434 (StemCell Technologies) (67). After 6–10 days, colonies were counted, and 10,000 BMCs were replated in new MethoCult medium.
Transplantation assays.
For competitive BM Tx, Nipa+/+ or Nipa–/– BMCs (CD45.2+) were isolated from donor mice, mixed with CD45.1+ competitor BMCs, and transplanted via tail vein injection into lethally irradiated (10 Gy fractionated) WT hosts (CD45.2+). For serial LSK cell Tx, 2000 sorted Nipa+/+ or Nipa–/– LSK cells were transplanted together with support BMCs using CD45.1+CD45.2+ double-positive, lethally irradiated recipient mice. For second and third serial Tx, donor LSK cells were isolated and transplanted at 17-week intervals. For the limiting dilution Tx assay, we transplanted 300, 800, or 2000 Nipa+/+ or Nipa–/– LSK cells together with 0.5 × 106 CD45.1+ support BMCs into lethally irradiated recipient mice. Engraftment was assessed as positive if chimerism was greater than 5% when all lineages were included. To monitor hematopoietic recovery in all Tx experiments, blood counts were measured using a scil Vet ABC Blood Counter. Donor cell engraftment was determined using CD45.1+/CD45.2+ discrimination.
Treatment with 5-FU and poly(I:C).
To induce hematopoietic stress, 5-FU (150 mg/kg body weight, Ribosepharm GmbH) was injected intraperitoneally at the indicated time points. Mice were analyzed after the first injection or monitored for hematopoietic exhaustion during weekly application. For poly(I:C) treatment (5 mg/kg body weight, i.p.), mice were injected twice a week for 4 weeks, followed by 4 weeks of recovery. For the long-term poly(I:C) assay, this treatment cycle was repeated 9 times (504 days).
Chromosome preparation.
Spleen cells were cultured for 48 hours in RPMI medium supplemented with 10% FCS, 1% penicillin/streptomycin (10,000 U/mL), 1% nonessential amino acids (×1000), 1% sodium pyruvate, 50 μM β-mercaptoethanol, 5 ng/mL murine IL-4, and 25 μg/mL LPS. Reagents were purchased from Sigma-Aldrich or Gibco. Cells were exposed to either no drug or MMC in a final concentration of 5 nM. After incubation with Colcemid (0.5 μg/mL) for 3 hours at 37°C, the mitotic metaphases were prepared by resuspending the cells in hypotonic 0.075 M KCl in H2O for 30 minutes, followed by fixing in methanol/acetic acid solution (3:1), dropping on cold, wet slides, and air-drying. Chromosomes were stained with DAPI (1.0 × 10–6 g/mL in PBS buffer, pH 7.0), mounted with antifade (Vector Laboratories), and scaled with a coverslip. Digital images were recorded with a digital camera (Sensys, Photometrics) on an Axioplan II fluorescence microscope (Zeiss) with Plan Apochromat 63/40 or 100/1.30 objectives (room temperature) using the Vysis workstation QUIPS (76).
Patients.
Diagnosis of FA was established using the MMC hypersensitivity assay. Diagnosis of MDS-type RCC was established according to WHO criteria (77, 78). In all RCC patients, FA was excluded, and GATA2, RUNX1, SAMD9, and SAMD9L genes were sequenced in 23 of 24 patients. RCC patients were enrolled in the EWOG-MDS study (ClinicalTrials.gov NCT00047268).
Histology and IHC.
For histology, tissues were dissected, fixed (4% formalin), and paraffin embedded. Sections (3–5 μm) were cut and stained with H&E (40). Immunohistochemistry was performed using NIPA (HPA024023, Sigma-Aldrich), FANCD2 (NB100-182, Novus Biologicals), and reticulin antibodies.
Gene expression analysis.
RNA from primary BM/PB cells and MEFs was isolated with the RNeasy Mini Kit (QIAGEN). cDNA was generated using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed using LightCycler 480 SYBR Green I Master (Roche Life Science) (73). Primer sequences used for quantitative qRT-PCR were as follows: mNipa primer RT1S (5′-GGGAGCTGATAGATGAAGGAATT-3′), mNipa primer RT1AS (5′-GCCCACTTCAAAGAAGAAAAGGTTT-3′), mGapdh primer S (5′-AAGGTCATCCCAGAGCTGAA-3′), mGapdh primer AS (5′-CTGCTTCACCACCTTCTTGA-3′), m18S primer S (5′-GCAATTATTCCCCATGAACG-3′), and m18S primer AS (5′-GGCCTCACTAAACCATCCAA-3′). For microarray analysis, RNA integrity was analyzed by capillary electrophoresis using a Fragment Analyzer (Advanced Analytical Technologies). RNA samples were further processed with the Affymetrix WT Pico Kit as described by the manufacturer. Labeled fragments were hybridized to GeneChip Mouse Gene ST 2.0 arrays for 16 hours at 45°C at 60 rpm in an Affymetrix Hybridization Oven 640. After washing and staining, the arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G. CEL files were produced from the raw data with Affymetrix GeneChip Command Console Software (75). Regulated genes were identified using a linear-based model, limma R package (http://bioinf.wehi.edu.au/limma). The threshold for significance was set to a P value of less than 0.01.
Nipa sequencing.
Sufficient amounts of DNA were available for exome sequencing from 17 patients. Sequencing libraries were prepared at the DKFZ Genomics and Proteomics Core Facility using the Agilent SureSelectXT Human All Exon V64 kit and subsequently sequenced on a HiSeq 4000 instrument using the 100-bp paired-end mode. First, bad quality reads were removed using Trimmomatic 0.36 (79). The selected reads were then aligned to the human reference genome hg19 using BWA-MEM 0.7.15 (80). Postalignment processing including base recalibration and insertion/deletion (INDEL) realignment was performed with GATK 3.6 (81). Finally, we called single nucleotide variants (SNVs) and INDELs with VarScan2 2.4.3 (82). Synonymous and nonexonic mutations were filtered out, as well as mutations found in more than 0.1% of the healthy population (gnomAD) (83). Calling of both SNVs and INDELs was performed without using paired germline control samples.
Accession number.
The microarray data reported herein were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE104735).
Statistics.
A paired or unpaired 2-tailed Student’s t test was used for statistical analyses unless otherwise stated. Data were plotted as mean ± SD as indicated. A closed testing procedure was applied to control for the error rate in multiple comparisons of 3 groups; i.e., tests of the global hypothesis of no differences between 3 groups were performed first, and 2-group comparisons were performed only if the 3-group comparisons yielded a significant result. To compare several groups with 1 control group, Dunnett’s test was applied. The survival curves were analyzed using a log-rank (Mantel-Cox) test; for limiting dilution Tx assays, L-Calc software (StemCell Technologies) was used. A P value less than 0.05 was considered significant.
Study approval.
All procedures were performed in accordance with national and institutional guidelines for animal care and experiments. Animal studies were approved by the institutional review board of Regierungspräsidium Freiburg, Freiburg, Germany. The EWOG-MDS study (ClinicalTrials.gov NCT00047268) was approved by the ethics committee of the University of Freiburg (no. 430/16). Written informed consent for participation was obtained from patients and parents.
Author contributions
SK performed and designed experiments, analyzed data, and wrote the manuscript. ME, CMN, MW, and IB provided and analyzed human patient samples and data collected within the EWOG-MDS study. GA, DP, and MB performed and analyzed array and WES data. RI, TP, and CK helped with mouse experiments. AMR and MZ performed immunoprecipitation analyses. TR helped with biochemical experiments. MF supported flow cytometry, sorting, and microscopy analyses. HK, KS, TAM, and NCW provided critical material. MK, SL, and BK performed mass spectrometry and data analysis. MP helped with the isolation and preparation of chromosomes for radial identification. MR and ASG performed and analyzed histology samples. ME, MB, CD, JDA, MLC, DS, RZ, and CP contributed to the development of the manuscript. ME, RAJO, and JD developed the concept and analyzed the data. ALI developed the overall concept, supervised the experiments, analyzed data, and wrote the manuscript. All authors edited and reviewed the manuscript.
Supplementary Material
Acknowledgments
We are very grateful to Sabina Schaberg for excellent technical assistance; M. Auer and M. Sickinger for helping with mouse experiments; and A. Fischer and W. Truckenmüller (Coordinating Study Center, EWOG-MDS) for assistance with patient samples. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to ALI and JD (DU227-3-1), CD and JD (FOR2032/2B1), and RAJO (OO8/16 and FOR2032/2B3). ALI is supported by a European Training Network grant (MSCA-ITN-2015-ETN-ALKATRAS), the DKTK (L693), and a BSL grant from the Government of Baden-Württemberg. ME is funded by European Research Council starting grant ApoptoMDS (ERC-STG-638145). GA and MB are supported by the DFG within the CRC850. MB is funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (DeCaRe, FKZ01ZX1409B). TAM is supported by a research grant from Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS, 02FN/2017). Michal Roessler illustrated the graphical abstract. G. Ihorst reviewed statistical analyses.
Version 1. 04/27/2020
Electronic publication
Version 2. 06/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):2827–2844.https://doi.org/10.1172/JCI126215.
Contributor Information
Stefanie Kreutmair, Email: stefanie.kreutmair@uniklinik-freiburg.de.
Miriam Erlacher, Email: miriam.erlacher@uniklinik-freiburg.de.
Geoffroy Andrieux, Email: geoffroy.andrieux@mol-med.uni-freiburg.de.
Melissa Zwick, Email: melissa.zwick@web.de.
Tamina Rückert, Email: tamina.rueckert@uniklinik-freiburg.de.
Milena Pantic, Email: milena.pantic@uniklinik-freiburg.de.
Teresa Poggio, Email: teresa.poggio@uniklinik-freiburg.de.
Khalid Shoumariyeh, Email: khalid.shoumariyeh@uniklinik-freiburg.de.
Tony A. Mueller, Email: tony.mueller@uniklinik-freiburg.de.
Hiroyuki Kawaguchi, Email: hiroyuki.kawaguchi@gmail.com.
Marie Follo, Email: marie.follo@uniklinik-freiburg.de.
Cathrin Klingeberg, Email: cathy020781@gmx.de.
Marcin Wlodarski, Email: marcin.wlodarski@uniklinik-freiburg.de.
Irith Baumann, Email: iri.baumann@gmail.com.
Dietmar Pfeifer, Email: dietmar.pfeifer@uniklinik-freiburg.de.
Michal Kulinski, Email: michal.kulinski@hotmail.de.
Martina Rudelius, Email: Martina.Rudelius@med.uni-muenchen.de.
Simone Lemeer, Email: S.M.Lemeer@uu.nl.
Bernhard Kuster, Email: kuster@tum.de.
Christine Dierks, Email: christine.dierks@uniklinik-freiburg.de.
Christian Peschel, Email: christian.peschel@tum.de.
Nina Cabezas-Wallscheid, Email: cabezas@ie-freiburg.mpg.de.
Jesus Duque-Afonso, Email: jesus.duque.afonso@uniklinik-freiburg.de.
Robert Zeiser, Email: robert.zeiser@uniklinik-freiburg.de.
Michael L. Cleary, Email: mcleary@stanford.edu.
Detlev Schindler, Email: schindler@biozentrum.uni-wuerzburg.de.
Annette Schmitt-Graeff, Email: annette.schmitt-graeff@uniklinik-freiburg.de.
Melanie Boerries, Email: melanie.boerries@uniklinik-freiburg.de.
Charlotte M. Niemeyer, Email: charlotte.niemeyer@uniklinik-freiburg.de.
Robert A.J. Oostendorp, Email: robert.oostendorp@tum.de.
Justus Duyster, Email: justus.duyster@uniklinik-freiburg.de.
Anna Lena Illert, Email: lena.illert@uniklinik-freiburg.de.
References
- 1.Parikh S, Bessler M. Recent insights into inherited bone marrow failure syndromes. Curr Opin Pediatr. 2012;24(1):23–32. doi: 10.1097/MOP.0b013e32834eca77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24(3):101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann I, et al. Morphological differentiation of severe aplastic anaemia from hypocellular refractory cytopenia of childhood: reproducibility of histopathological diagnostic criteria. Histopathology. 2012;61(1):10–17. doi: 10.1111/j.1365-2559.2011.04156.x. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimi A, et al. High incidence of Fanconi anaemia in patients with a morphological picture consistent with refractory cytopenia of childhood. Br J Haematol. 2013;160(1):109–111. doi: 10.1111/bjh.12083. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer CM, Baumann I. Classification of childhood aplastic anemia and myelodysplastic syndrome. Hematology Am Soc Hematol Educ Program. 2011;2011:84–89. doi: 10.1182/asheducation-2011.1.84. [DOI] [PubMed] [Google Scholar]
- 6.Erlacher M, Strahm B. Missing cells: pathophysiology, diagnosis, and management of (pan)cytopenia in childhood. Front Pediatr. 2015;3:64. doi: 10.3389/fped.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurre P. Hematopoietic development: a gap in our understanding of inherited bone marrow failure. Exp Hematol. 2018;59:1–8. doi: 10.1016/j.exphem.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Ripperger T, et al. Childhood cancer predisposition syndromes-A concise review and recommendations by the Cancer Predisposition Working Group of the Society for Pediatric Oncology and Hematology. Am J Med Genet A. 2017;173(4):1017–1037. doi: 10.1002/ajmg.a.38142. [DOI] [PubMed] [Google Scholar]
- 9.Karow A, Flotho C, Schneider M, Fliegauf M, Niemeyer CM, European Working Group of Myelodysplastic Syndromes in Childhood Mutations of the Shwachman-Bodian-Diamond syndrome gene in patients presenting with refractory cytopenia — do we have to screen? Haematologica. 2010;95(4):689–690. doi: 10.3324/haematol.2009.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locatelli F, Strahm B. How I treat myelodysplastic syndromes of childhood. Blood. 2018;131(13):1406–1414. doi: 10.1182/blood-2017-09-765214. [DOI] [PubMed] [Google Scholar]
- 11.Yao CY, et al. Distinct mutation profile and prognostic relevance in patients with hypoplastic myelodysplastic syndromes (h-MDS) Oncotarget. 2016;7(39):63177–63188. doi: 10.18632/oncotarget.11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bluteau O, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131(7):717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 13.Valli R, et al. Bone marrow failure may be caused by chromosome anomalies exerting effects on RUNX1T1 gene. Mol Cytogenet. 2018;11:2. doi: 10.1186/s13039-017-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamary H, et al. Frequency and natural history of inherited bone marrow failure syndromes: the Israeli Inherited Bone Marrow Failure Registry. Haematologica. 2010;95(8):1300–1307. doi: 10.3324/haematol.2009.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011;155A(8):1877–1883. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callén E, et al. A common founder mutation in FANCA underlies the world’s highest prevalence of Fanconi anemia in Gypsy families from Spain. Blood. 2005;105(5):1946–1949. doi: 10.1182/blood-2004-07-2588. [DOI] [PubMed] [Google Scholar]
- 17.Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27(3-4):214–221. doi: 10.1016/j.beha.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alter BP, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 21.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37(9):931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 22.van Twest S, et al. Mechanism of ubiquitination and deubiquitination in the Fanconi anemia pathway. Mol Cell. 2017;65(2):247–259. doi: 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107(11):4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 24.Bagby GC. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10(1):68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 25.Panneerselvam J, et al. Basal level of FANCD2 monoubiquitination is required for the maintenance of a sufficient number of licensed-replication origins to fire at a normal rate. Oncotarget. 2014;5(5):1326–1337. doi: 10.18632/oncotarget.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Che R, Zhang J, Nepal M, Han B, Fei P. Multifaceted Fanconi anemia signaling. Trends Genet. 2018;34(3):171–183. doi: 10.1016/j.tig.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 28.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annu Rev Med. 2005;56:495–508. doi: 10.1146/annurev.med.56.082103.104546. [DOI] [PubMed] [Google Scholar]
- 30.Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15(1):37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 34.Nepal M, Che R, Ma C, Zhang J, Fei P. FANCD2 and DNA Damage. Int J Mol Sci. 2017;18(8):E1804. doi: 10.3390/ijms18081804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y, et al. Constitutive role of the Fanconi anemia D2 gene in the replication stress response. J Biol Chem. 2017;292(49):20184–20195. doi: 10.1074/jbc.M117.814780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viale A, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457(7225):51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 37.Parmar K, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28(7):1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 39.Bassermann F, et al. NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell. 2005;122(1):45–57. doi: 10.1016/j.cell.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 40.Illert AL, et al. Targeted inactivation of nuclear interaction partner of ALK disrupts meiotic prophase. Development. 2012;139(14):2523–2534. doi: 10.1242/dev.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajewsky K, et al. Conditional gene targeting. J Clin Invest. 1996;98(3):600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23(24):5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132(1):203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- 44.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668(1-2):133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dykstra B, de Haan G. Hematopoietic stem cell aging and self-renewal. Cell Tissue Res. 2008;331(1):91–101. doi: 10.1007/s00441-007-0529-9. [DOI] [PubMed] [Google Scholar]
- 46.Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208(13):2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Y, Van Zant G, Szilvassy SJ. Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood. 2005;106(4):1479–1487. doi: 10.1182/blood-2004-11-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akunuru S, Geiger H. Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol Med. 2016;22(8):701–712. doi: 10.1016/j.molmed.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benz C, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Watters GP, Smart DJ, Harvey JS, Austin CA. H2AX phosphorylation as a genotoxicity endpoint. Mutat Res. 2009;679(1-2):50–58. doi: 10.1016/j.mrgentox.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 52.Beerman I, Rossi DJ. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16(6):613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boisvert RA, Rego MA, Azzinaro PA, Mauro M, Howlett NG. Coordinate nuclear targeting of the FANCD2 and FANCI proteins via a FANCD2 nuclear localization signal. PLoS ONE. 2013;8(11):e81387. doi: 10.1371/journal.pone.0081387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du W, et al. Fancb deficiency impairs hematopoietic stem cell function. Sci Rep. 2015;5:18127. doi: 10.1038/srep18127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mgbemena VE, Signer RAJ, Wijayatunge R, Laxson T, Morrison SJ, Ross TS. Distinct Brca1 mutations differentially reduce hematopoietic stem cell function. Cell Rep. 2017;18(4):947–960. doi: 10.1016/j.celrep.2016.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerabona D, Sun Z, Nalepa G. Leukemia and chromosomal instability in aged Fancc-/- mice. Exp Hematol. 2016;44(5):352–357. doi: 10.1016/j.exphem.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasanthakumar A, et al. Brca1 deficiency causes bone marrow failure and spontaneous hematologic malignancies in mice. Blood. 2016;127(3):310–313. doi: 10.1182/blood-2015-03-635599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kutler DI, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101(4):1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 59.Iwafuchi H, Ito M. Differences in the bone marrow histology between childhood myelodysplastic syndrome with multilineage dysplasia and refractory cytopenia of childhood without multilineage dysplasia. Histopathology. 2019;74(2):239–247. doi: 10.1111/his.13721. [DOI] [PubMed] [Google Scholar]
- 60.Shallis RM, Ahmad R, Zeidan AM. The genetic and molecular pathogenesis of myelodysplastic syndromes. Eur J Haematol. 2018;101(3):260–271. doi: 10.1111/ejh.13092. [DOI] [PubMed] [Google Scholar]
- 61.Will B, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120(10):2076–2086. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou L, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286(28):25211–25223. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang Y, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113(6):1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson CD, et al. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet. 2018;50(8):1132–1139. doi: 10.1038/s41588-018-0174-0. [DOI] [PubMed] [Google Scholar]
- 65. doi: 10.3324/haematol.2018.212027. Yu C, et al. Phosphorylation of Beclin-1 by BCR-ABL suppresses autophagy in chronic myeloid leukemia [published online August 8, 2019]. Haematologica. https://doi.org/10.3324/haematol.2018.212027. [DOI] [PMC free article] [PubMed]
- 66.Klitzing Cv, et al. APC/C(Cdh1)-mediated degradation of the F-box protein NIPA is regulated by its association with Skp1. PLoS ONE. 2011;6(12):e28998. doi: 10.1371/journal.pone.0028998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Illert AL, et al. Grb10 is involved in BCR-ABL-positive leukemia in mice. Leukemia. 2015;29(4):858–868. doi: 10.1038/leu.2014.283. [DOI] [PubMed] [Google Scholar]
- 68.Grundler R, Miething C, Thiede C, Peschel C, Duyster J. FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood. 2005;105(12):4792–4799. doi: 10.1182/blood-2004-11-4430. [DOI] [PubMed] [Google Scholar]
- 69.Duque-Afonso J, et al. E2A-PBX1 remodels oncogenic signaling networks in B-cell precursor acute lymphoid leukemia. Cancer Res. 2016;76(23):6937–6949. doi: 10.1158/0008-5472.CAN-16-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Illert AL, et al. Extracellular signal-regulated kinase 2 (ERK2) mediates phosphorylation and inactivation of nuclear interaction partner of anaplastic lymphoma kinase (NIPA) at G2/M. J Biol Chem. 2012;287(45):37997–38005. doi: 10.1074/jbc.M112.373464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102(2):646–651. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- 73.Müller TA, et al. Lineage-specific STAT5 target gene activation in hematopoietic progenitor cells predicts the FLT3(+)-mediated leukemic phenotype. Leukemia. 2016;30(8):1725–1733. doi: 10.1038/leu.2016.72. [DOI] [PubMed] [Google Scholar]
- 74.Tomiatti V, et al. Cks1 is a critical regulator of hematopoietic stem cell quiescence and cycling, operating upstream of Cdk inhibitors. Oncogene. 2015;34(33):4347–4357. doi: 10.1038/onc.2014.364. [DOI] [PubMed] [Google Scholar]
- 75.Klein C, et al. Ptch2 loss drives myeloproliferation and myeloproliferative neoplasm progression. J Exp Med. 2016;213(2):273–290. doi: 10.1084/jem.20150556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voelxen N, et al. B-cell signaling in persistent polyclonal B lymphocytosis (PPBL) Immunol Cell Biol. 2016;94(9):830–837. doi: 10.1038/icb.2016.46. [DOI] [PubMed] [Google Scholar]
- 77.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 78. Baumann I, Niemeyer C, Shannon K. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008:104–107. [Google Scholar]
- 79.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 (q-bio.GN). Revised May 26, 2013. Accessed April 15, 2020. [Google Scholar]
- 81.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koboldt D, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.