Abstract
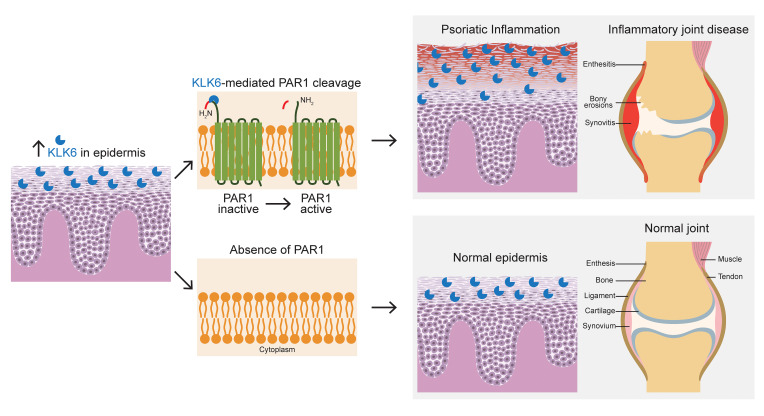
Kallikrein-related peptidase 6 (KLK6) is a secreted serine protease hypothesized to promote inflammation via cleavage of protease-activated receptor 1 (PAR1) and PAR2. KLK6 levels are elevated in multiple inflammatory and autoimmune conditions, but no definitive role in pathogenesis has been established. Here, we show that skin-targeted overexpression of KLK6 causes generalized, severe psoriasiform dermatitis with spontaneous development of debilitating psoriatic arthritis-like joint disease. The psoriatic skin and joint phenotypes are reversed by normalization of skin KLK6 levels and attenuated following genetic elimination of PAR1 but not PAR2. Conservation of this regulatory pathway was confirmed in human psoriasis using vorapaxar, an FDA-approved PAR1 antagonist, on explanted lesional skin from patients with psoriasis. Beyond defining a critical role for KLK6/PAR1 signaling in promoting psoriasis, our results demonstrate that KLK6/PAR1-mediated inflammation in the skin alone is sufficient to drive inflammatory joint disease. Further, we identify PAR1 as a promising cytokine-independent target in therapy of psoriasis and psoriatic arthritis.
Keywords: Dermatology, Inflammation
Keywords: Arthritis, Kallikrein, Skin
Introduction
Psoriasis is a chronic, incurable systemic inflammatory disease with complex and incompletely understood pathogenesis. Two percent to 3% of the population worldwide suffers from psoriasis, and affected individuals show higher rates of diverse comorbid conditions. The most classic among these is psoriatic arthritis (PsA), which is found in approximately one-third of patients and can cause severe, disabling joint disease in the peripheral and axial skeletons (1).
Kallikrein-related peptidase 6 (KLK6) belongs to a family of serine proteases that are emerging as prevalent biomarkers of inflammatory and malignant diseases. Cell culture and preclinical animal model studies suggest that KLK6 may promote inflammation and autoimmunity via cleavage of the G protein–coupled protease-activated receptor 1 (PAR1) and PAR2 (2, 3).
KLK6 levels are increased in multiple skin diseases including psoriasis and atopic dermatitis but normalize rapidly with efficacious therapy (4, 5). KLK6 is also found in joint synovium, urine, and serum of patients with PsA (6) and rheumatoid arthritis (7). Nonetheless, a pathogenic role for KLK6 remains to be identified. Furthermore, little is known about how KLK6/PAR1/2-mediated signaling influences autoimmunity.
Results and Discussion
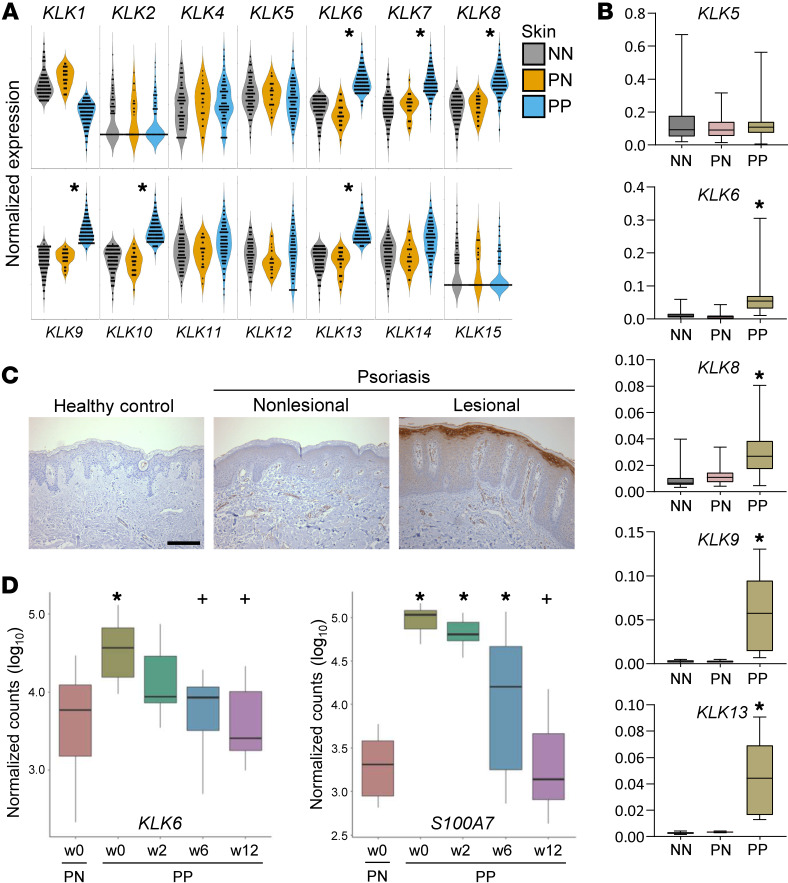
To investigate the role of KLK6 in inflammatory skin disease, we first compared expression of KLK transcripts in skin from healthy individuals to normal-appearing (nonlesional) and lesional skin from patients with psoriasis. KLK6 was significantly upregulated in lesional psoriatic skin, as were multiple other KLKs (Figure 1, A and B). Immunohistochemistry confirmed prominent KLK6 protein upregulation in infiltrating immune cells and keratinocytes of the superficial epidermis (Figure 1C). We further explored KLK6 dynamics during psoriasis treatment with etanercept, a soluble tumor necrosis factor (TNF) receptor that binds TNF to block its proinflammatory effects. KLK6 levels in the skin decreased rapidly in patients who were responsive to etanercept therapy, approximating levels in nonlesional skin by as early as 2 weeks (Figure 1D), sooner than observed for the traditional psoriasis biomarker psoriasin (S100A7). Thus, KLK6 shows disease-specific regulation, correlating closely with local disease activity.
Figure 1. KLK6 expression is significantly elevated in psoriatic lesions and parallels disease activity.
(A) Detection of KLK transcripts by RNA-seq in healthy control skin (NN, gray; N = 90), psoriatic patient nonlesional skin (PN, yellow; N = 26), and psoriatic patient lesional skin (PP, blue; N = 99). P values were computed using Wilcoxon’s rank-sum test; false discovery rate (FDR) was used to control the multiple testing. *P < 0.05 in PP vs. PN. (B) Relative expression of select KLK transcripts by qRT-PCR in a unique cohort of healthy controls and psoriatic patients (N ≥ 6 for all). Mean is indicated by the horizontal line. Box, 25th–75th percentile. Whiskers, minimum and maximum. *P < 0.005 in PP vs. both NN and PN by ordinary 1-way ANOVA with post hoc Tukey’s multiple-comparisons test. (C) Immunostaining of KLK6 in human skin. Results are representative of 4 biological replicates. Scale bar: 100 μm. (D) Detection of KLK6 and S100A7 transcripts by RNA-seq in PN and PP at the indicated weeks’ duration of etanercept therapy in responsive patients (N = 14). Mean is indicated by the horizontal line. Box, 25th–75th percentile. For definition of whiskers and additional details, see Supplemental Methods. *P < 0.05 vs. PN at w0; +P < 0.05 vs. PP at w0 by negative binomial test.
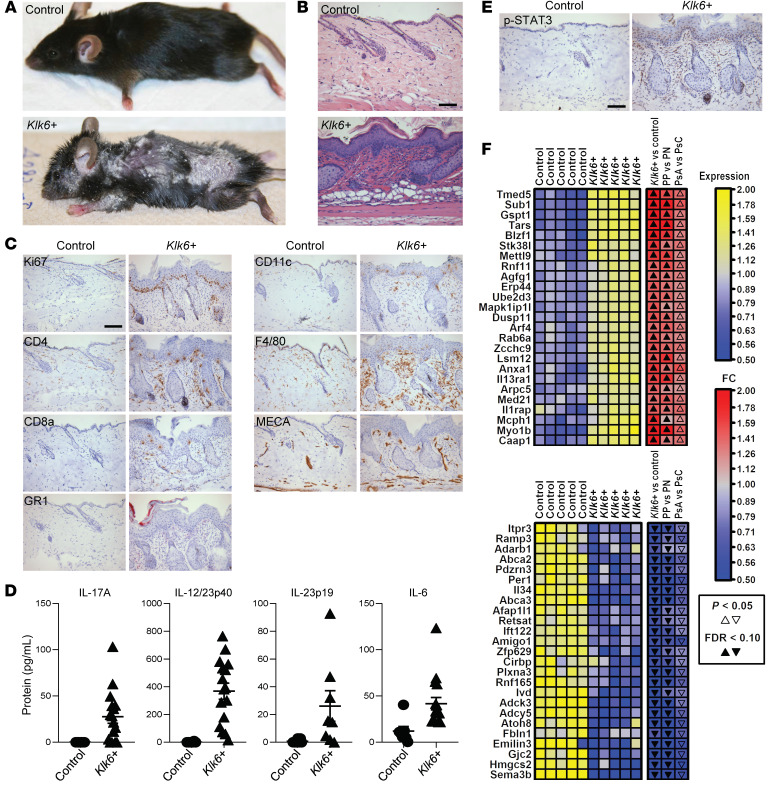
To investigate a pathogenic role for KLK6 in driving inflammatory disease in the skin, we developed Tet-off transgenic mice that overexpressed C-terminal Myc/His-tagged murine Klk6 in keratinocytes in the absence of doxycycline, hereafter referred to as Klk6+ transgenic mice. In these mice, transgene overexpression is under control of the bovine keratin 5 (K5) promoter, which drives expression in epidermal keratinocytes and some other stratified squamous epithelia (see Supplemental Methods and refs. 8, 9; supplemental material available online with this article; https://doi.org/10.1172/JCI133159DS1). By 2 months of age, all Klk6+ mice developed diffuse scaling and redness of the skin (Figure 2A). Histology revealed features of psoriasis including hyperplasia and focal parakeratosis, an indicator of abnormal keratinocyte maturation (Figure 2B). Ki67 staining demonstrated epidermal hyperproliferation (Figure 2C). Immunohistochemistry for inflammatory cell markers showed a brisk mixed infiltrate in the superficial dermis as well as collections of intraepidermal neutrophils (Figure 2C, GR1) comparable to the hallmark Munro’s microabscesses of psoriasis. Increased dermal vascularity was observed (Figure 2C, MECA), consistent with the hypervascularity of psoriatic plaques. Thus, epidermal overexpression of Klk6 causes a severe, generalized rash with histological features of psoriasis.
Figure 2. Epidermal Klk6 overexpression causes a severe, generalized skin rash with histologic, transcriptional, and immunological features of psoriasis.
(A) Gross images of age- and sex-matched control and Klk6+ transgenic mice. (B) H&E staining of control (N = 19) and Klk6+ (N = 38) dorsal skin. (C) Immunostaining in control and Klk6+ dorsal skin. Ki67, cell proliferation; CD4 and CD8a, T lymphocyte; GR1, granulocyte (including neutrophil); CD11c, dendritic cell; F4/80, macrophage; MECA, vascular endothelial cell. (D) Detection of IL-17A, the IL-12/23p40 subunit, the IL-23p19 subunit, and IL-6 by ELISA in control and Klk6+ skin. N ≥ 7 for all groups. P = 0.0027, P < 0.0001, P = 0.0056, and P = 0.0015, respectively, by Mann-Whitney test. Mean and SEM are indicated. (E) Phospho-STAT3 (p-STAT3) immunostaining of control and Klk6+ dorsal skin. (F) Blue-yellow heatmap showing relative expression of transcripts enriched (top panel) and depleted (bottom panel) in Klk6+ (N = 5) vs. control (N = 5) dorsal skin. Adjacent blue-red heatmaps show fold change (FC) estimates from the following 3 comparisons: Klk6+ vs. control mice, psoriatic lesional (PP) vs. nonlesional (PN) skin, and lesional skin from psoriatic arthritis patients (PsA) vs. cutaneous-only psoriatic patients (PsC). Significance by P value or false discovery rate (FDR) is indicated by solid or open arrowheads, respectively. Panels depict the 25 transcripts with the highest minimum (top) or lowest maximum (bottom) FC estimates across the 3 comparisons. For additional details, see Supplemental Methods. Scale bars: 100 μm (B, C, and E).
To further characterize the inflammation in Klk6+ transgenic mouse skin, we assessed activation of key psoriasis inflammatory pathways. IL-17A, the p40 and p19 subunits of IL-23, and IL-6 protein were highly upregulated in transgenic skin (Figure 2D), and phosphorylated STAT3 was diffusely increased in keratinocytes and inflammatory infiltrate of transgenic skin (Figure 2E). For a broader examination of the transcriptional effects of Klk6 overexpression, we performed RNA sequencing (RNA-seq) of Klk6+ transgenic and control mouse skin and identified differentially expressed genes (DEGs). Upregulated DEGs showed enrichment for Gene Ontology and KEGG terms related to keratinization, epidermal development, cellular proliferation, and steroid biosynthesis (Supplemental Figure 1). Transgenic DEGs showed prominent overlap with genes differentially enriched in lesional versus nonlesional skin of patients with psoriasis (Figure 2F), whereas similar comparison to skin of patients with atopic dermatitis showed only a modest overlap (Supplemental Figure 2). Of note, KLK6 itself has been defined as a gene that differentiates between psoriasis and eczema (10). Strongly upregulated transcripts supporting a psoriasis-like phenotype in Klk6+ transgenic mouse skin include Il17a (580-fold, false discovery rate [FDR] = 3.66 × 10–42), Il17f (380-fold, FDR = 7.27 × 10–61), Il22 (1123-fold, FDR = 1.04 × 10–37), Il24 (293-fold, FDR = 9.92 × 10–32), Il36a (203-fold, FDR = 3.16 × 10–241), Klk13 (216-fold, FDR = 1.24 × 10–193), and Il19 (374-fold, FDR = 1.96 × 10–148), in comparison with relatively modest changes in key atopic dermatitis transcripts Il4 (5-fold, FDR = 0.00209), Il13 (5-fold, FDR = 0.00426), Ccr7 (0.92-fold, FDR = 0.816), and Ccl17 (1.4-fold, FDR = 0.282). These are consistent with the classification criteria suggested by Quaranta et al. (10), validating the dermatitis of Klk6+ transgenic mice as immunologically and transcriptionally analogous to human psoriasis.
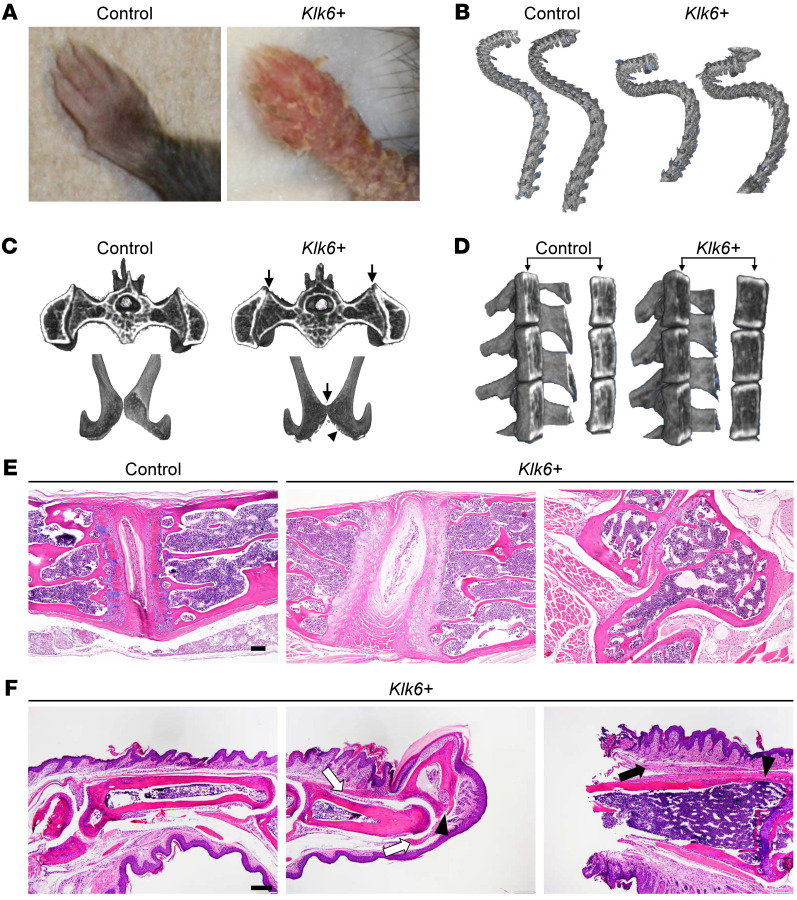
Intriguingly, Klk6+ transgenic mouse skin is more transcriptionally similar to lesions of patients with psoriasis who have PsA than those who do not (PsC) (Figure 2F and Supplemental Figure 3). We therefore examined Klk6+ transgenic mice for manifestations of inflammatory arthritis. Transgenic forelimbs showed severe erythema and scaling (Figure 3A), with variable nail dystrophy and digital swelling akin to the characteristic dactylitis, or sausage digit, of PsA. Additionally, transgenic mice developed excessive cervico-thoracic spinal curvature, termed kyphosis, by 10 weeks of age that led to a hunched posture and impaired mobility (Supplemental Video 1). To better characterize the bony abnormalities, we performed micro–computed tomography of transgenic mice. This revealed kyphosis (Figure 3B), decreased joint space in the sacroiliac joints and pubic symphysis, and erosions in the pubic symphysis in some, but not all, Klk6+ transgenic mice (Figure 3C and Figure 4B). Transgenic mice also showed significantly (P = 1.4 × 10–6) decreased vertebral bone mineral density (Figure 3D), which is commonly seen in patients with inflammatory arthritis, including those with PsA (11). H&E-stained spine sections revealed thinning of the trabecular bone and subchondral plate, impaired calcification of the vertebral cartilage, and abnormalities of the vertebral disc and surrounding fibrous tissue in Klk6+ transgenic mice (Figure 3E), supporting our gross and radiographic observations of axial skeleton disease. As peripheral joints, in particular the digits, are the most common site of inflammation in PsA, we examined transgenic paws for evidence of inflammatory arthritis. Klk6+ paws showed features of spondyloarthritis including entheseal inflammation and synovitis in approximately 80% of the mice examined, with severely affected mice showing signs of bone erosion (Figure 3F). Together, these gross, radiographic, and histologic findings demonstrate that overexpression of Klk6 in the skin alone is sufficient to cause progressive axial skeletal bony changes and peripheral joint inflammatory arthritis consistent with PsA.
Figure 3. Skin-directed Klk6 overexpression leads to debilitating joint inflammation and bony abnormalities consistent with psoriatic arthritis (PsA).
(A) Gross images of control and Klk6+ transgenic forelimbs. (B) Curvature of control and Klk6+ spines visualized by micro-computed tomography (micro-CT). Data are representative of 6 animals per group. (C) Pelvic joints of control and Klk6+ mice visualized by micro-CT. Upper panels, sacroiliac (SI) joint; lower panels, pubic symphysis. Arrows, distortion of joint space in SI joints and bony erosions in pubic symphysis. Data are representative of 6 animals per group. SI joints are magnified in Figure 4B. (D) Vertebral body bone density as visualized by brightness (higher density) on micro-CT. (E) H&E staining of control and Klk6+ spine sections. Rightmost panel, misshapen vertebra at level of gross kyphosis. Data are representative of 3 animals. Scale bar: 100 μm. (F) H&E staining of Klk6+ paw sections showing synovial inflammation (black arrows) and enthesitis (white arrows). Arrowheads, bony erosions. Data are representative of 5 animals. Scale bar: 200 μm.
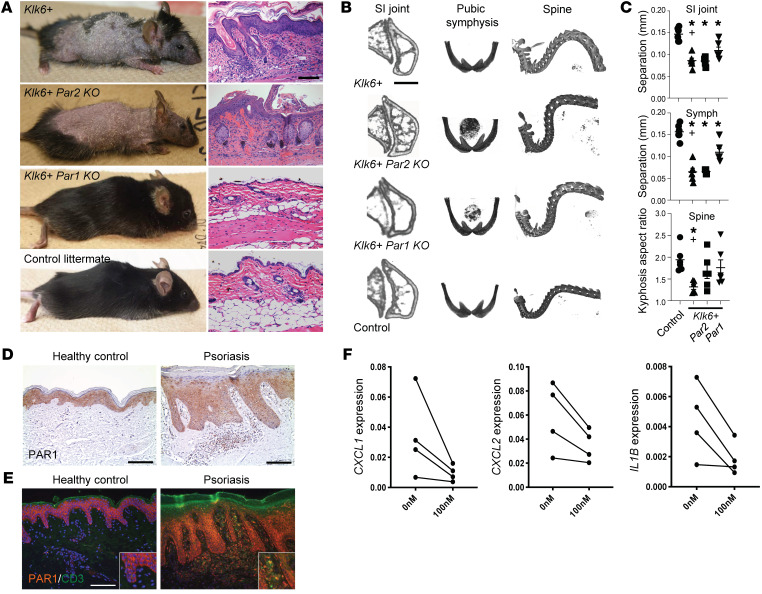
Figure 4. KLK6 induces psoriatic skin and joint disease through PAR1, but not PAR2.
(A) Gross images and H&E dorsal skin staining of 10-week-old mice of the indicated genotypes. (B) Micro-CT images of the sacroiliac (SI) joint, pubic symphysis, and cervico-thoracic kyphosis (spine) in mice of the indicated genotypes. (C) Quantitative analysis of micro-CT images of the SI joint, pubic symphysis (Symph), and cervico-thoracic kyphosis in mice of the indicated genotypes. Mean and SEM are indicated. N ≥ 5 per group. *P < 0.05 vs. control; +P < 0.05 for Klk6+ vs. Klk6+ Par1KO by ordinary 1-way ANOVA with post hoc Tukey’s multiple-comparisons test (SI joint and Symph) or a post hoc uncorrected Fisher’s LSD test (Spine). Control and Klk6+ data are reproduced in Supplemental Figure 4E. (D) PAR1 immunostaining in human healthy control and lesional psoriatic skin. (E) PAR1 (orange) and CD3 (green) immunofluorescence in human healthy control and lesional psoriatic skin. (F) Detection of indicated proinflammatory marker transcripts by qRT-PCR of lesional psoriatic skin explants (N = 4) treated with 0 or 100 nM of the PAR1 antagonist vorapaxar. P < 0.05 by paired t test for all. Scale bars: 100 μm (A, D, and E).
We next investigated whether the Klk6+ transgenic phenotype is dependent on continued Klk6 overexpression. We allowed transgenic mice to develop robust skin and joint manifestations and then repressed transgene expression with doxycycline for 4 weeks to restore Klk6 to wild-type levels (Supplemental Figure 4A). This fully ameliorated the cutaneous phenotype (Supplemental Figure 4, B and C), indicating that KLK6-induced skin inflammation is not self-perpetuating. Unexpectedly, this also led to partial and in some cases complete reversal of bony abnormalities (Supplemental Figure 4, D and E), suggesting that persistence of inflammatory arthritis requires ongoing KLK6-driven skin inflammation.
Finally, we sought to define the mechanism through which KLK6 induces psoriatic skin and joint disease. We crossed the Klk6+ Tet-off transgenic mice with Par1- and Par2-knockout lines and assessed skin and joint phenotypes. Although the Klk6+ Par2KO phenotype was essentially indistinguishable from that of the Klk6+ transgenic mice, Klk6+ Par1KO mice showed rescue of the cutaneous abnormalities and significant amelioration of the bony abnormalities (Figure 4, A–C). This indicates that KLK6/PAR1 signaling is imperative for the skin and joint inflammatory phenotypes in these mice, although the persistence of subtle residual bony disease in Klk6+ Par1KO mice suggests the possible existence of other minor mediators of KLK6 signaling acting in parallel to PAR1.
To determine whether the KLK6/PAR1 signaling underlying the mouse inflammatory phenotypes is conserved in human psoriasis, we assessed colocalization of KLK6 and PAR1 in human skin. PAR1 was expressed throughout the epidermis, with nuclear staining more prominent in the superficial epidermis, where KLK6 is most abundant (Figure 4, D and E). Many infiltrating dermal inflammatory cells also showed PAR1 staining (Figure 4E). As a proof of concept, we then applied vorapaxar, an FDA-approved PAR1 antagonist, to cultured explanted lesional skin from psoriatic patients. Vorapaxar therapy decreased psoriasis-associated proinflammatory markers CXCL1, CXCL2, and IL1B (Figure 4F), confirming that KLK6/PAR1 signaling drives inflammation in psoriasis in humans.
Despite intensive investigation, the mechanistic connection between PsA and psoriasis remains unclear. Although many mouse psoriasis models have been developed through epidermal genetic alterations, very few show spontaneous evolution of joint disease (12, 13), challenging the hypothesis that PsA reflects spillover of cutaneous inflammation. Here, we have shown that Klk6 overexpression limited to the epidermis is sufficient to drive not only psoriasis-like cutaneous disease but also inflammatory arthritis via signaling through PAR1. Whether joint disease requires intermediary inflammation in other compartments such as the gut is of considerable interest and remains to be investigated.
The mechanisms by which KLK6/PAR1 signaling promotes psoriasis and joint inflammation remain to be determined. STAT3 was diffusely increased in Klk6+ mouse skin (Figure 2E) and has previously been linked with KLK6/PAR1 signaling in injured spinal cord (14). STAT3 activation is a hallmark of psoriatic epidermis, and STAT3 strongly correlates with KLK6 in human psoriatic lesional skin (15) (Spearman’s correlation coefficient ρ of 0.71; P = 3.3 × 10–5), providing a mechanistic link between KLK6/PAR1 and STAT3 activation. STAT3 plays a critical role in the feedback loop between keratinocytes and immune cells, driving production of TNF, IL-23, and IL-17, which in turn feed back onto keratinocytes to drive further generation of KLK6 and downstream activation of STAT3. Recently, expression of hyperactive STAT3 in mouse CD4+ T cells was shown to promote psoriasis-like skin disease, enthesitis/tendonitis, and periarticular bone erosion accompanied by osteopenia (16). Klk6+ transgenic mice have high levels of Il22 (>1000-fold relative to WT), Il17a (569-fold), Il17f (378-fold), Il19 (372-fold), and Il24 (292-fold) (Supplemental Figure 1), cytokines that contribute to STAT3-mediated inflammation between keratinocytes and immune cells in psoriatic lesional skin and joints, suggesting KLK6-mediated PAR1 cleavage occurs upstream of these pathways.
Neutrophils and other innate immune cells are important in the pathogenesis of PsA, and IL-23/IL17A signaling and IL-22 expression are increased in PsA patient synovial membranes and entheses contributing to joint inflammation and bone remodeling (17). Klk6+ transgenic mice have high levels of IL-23, IL-17A/F, and IL-22 (Figure 2D and Supplemental Figure 1), and IL-23/IL-17 signaling promotes production of CXCL1, CXCL2, and CXCL5, which promote neutrophil recruitment and migration into joint spaces. CXCL1 strongly correlates with KLK6 (ρ of 0.75; P = 9.6 × 10–6) in psoriatic patient lesional skin, and voropaxar treatment of psoriatic skin explants decreased CXCL1 expression (Figure 4F). Other transcripts strongly associated with KLK6 in psoriatic lesional skin include IL36A (ρ of 0.7; P = 6 × 10–5) and PI3 (ρ of 0.81; P = 1.7 × 10–6), which are also critically involved in keratinocyte–immune cell hyperactivation and inflammation and may link skin inflammation with joint and bone disease.
Using the Klk6+ transgenic mouse, we have identified a critical role for KLK6 in promoting psoriasis-like inflammation via PAR1 signaling, suggesting that targeting PAR1 may offer a cytokine-independent approach for treating psoriasis. The unexpected discovery that normalization of murine KLK6 levels largely reverses KLK6-driven bony disease may reflect the plasticity of the skeletal and articular system of young mice; however, there is precedent for reversal of PsA with therapy in adult patients (18). Ongoing investigation of development and regression of PsA-like joint and bone disease in the Klk6+ mouse and, perhaps, future evaluation of the effects of KLK6 normalization in patients with psoriasis will provide further insight.
Methods
Refer to Supplemental Methods for details.
Data availability.
The RNA-seq data sets have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE144312.
Statistics.
Experimental data are presented as mean ± SEM unless otherwise indicated. P values for experiments comparing 2 groups were calculated using either a 2-tailed Student’s t test or a Mann-Whitney test. For experiments comparing more than 2 groups, an ordinary 1-way ANOVA was used with post hoc Tukey’s multiple-comparisons test, unless otherwise noted. P less than 0.05 was considered statistically significant.
Study approval.
Human samples were obtained from volunteer patients with psoriasis and healthy controls with informed written consent before inclusion in the study in accordance with Declaration of Helsinki principles. All protocols were approved by the University of Michigan institutional review board. All animal experiments were approved by the Case Western Reserve University institutional animal care and use committee and conformed to the American Association for Accreditation of Laboratory Animal Care guidelines.
Author contributions
JEG, RJM, and NLW conceived the study and designed experiments. WRS, LCT, RR, RJM, JEG, and NLW analyzed the data. JEL, YF, DD, RR, XX, PAK, DG, RU, MIC, MKS, and NLW conducted the experiments. DG, RR, RJM, MEH, CM, SAR, JUS, JMK, JEG, ACB, and NLW contributed to the phenotyping and understanding of the arthritic phenotype. ACB, JEG, and NLW wrote the manuscript with input from all authors.
Assigning of order of shared first author position was done as follows: ACB interpreted the data, generated the figures, and wrote the manuscript and was assigned first position; JEL conducted the majority of the animal analyses and was placed second; and YF designed the mouse cloning and completed critical experiments and analyses in the initial years of the study and was placed third.
Supplementary Material
Acknowledgments
This work was supported by the Lozick Discovery Grant and a Bridge Grant from the National Psoriasis Foundation (to NLW), the Babcock Endowment Fund (to JEG), the A. Alfred Taubman Medical Research Institute (to JEG), NIH awards R01-AR069071 and R01-AR073196 (to JEG and NLW); P30-AR075043, R01-AI130025 (to JEG); P30-AR39750, P50-AR055508, P50-AR070590, R01-AR063437, R01-AR062546, and R21-AR063852 (to NLW); T32-AR007197 (to ACB), and a core utilization grant from the Clinical and Translational Science Collaborative of Cleveland (4UL1TR0002548), from the National Center for Advancing Translational Sciences (NCATS) component of the NIH and NIH Roadmap for Medical Research. The contents of this paper are the responsibility of the authors and do not necessarily represent the official views of the NIH.
Version 1. 03/10/2020
In-Press Preview
Version 2. 05/11/2020
Electronic publication
Version 3. 06/01/2020
Print issue publication
Funding Statement
to NLW
to JEG
to JEG
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):3151–3157.https://doi.org/10.1172/JCI133159.
Contributor Information
Allison C. Billi, Email: acbilli@med.umich.edu.
Jessica E. Ludwig, Email: jel151@case.edu.
Yi Fritz, Email: yxf107@case.edu.
Richard Rozic, Email: rickrozic@gmail.com.
William R. Swindell, Email: ws277814@ohio.edu.
Lam C. Tsoi, Email: alextsoi@umich.edu.
Shahla Abdollahi-Roodsaz, Email: sabdollahi@celgene.com.
Xianying Xing, Email: xingx@med.umich.edu.
Doina Diaconu, Email: doina.diaconu@case.edu.
Ranjitha Uppala, Email: uranjith@med.umich.edu.
Maya I. Camhi, Email: mcamhi@outlook.com.
Philip A. Klenotic, Email: pxk258@case.edu.
Mrinal K. Sarkar, Email: mksarkar@umich.edu.
M. Elaine Husni, Email: HUSNIE@ccf.org.
Jose U. Scher, Email: Jose.Scher@nyumc.org.
Christine McDonald, Email: mcdonac2@ccf.org.
J. Michelle Kahlenberg, Email: mkahlenb@umich.edu.
Ronald J. Midura, Email: midurar@ccf.org.
Johann E. Gudjonsson, Email: johanng@med.umich.edu.
Nicole L. Ward, Email: nlw4@cwru.edu.
References
- 1.Takeshita J, et al. Psoriasis and comorbid diseases: Epidemiology. J Am Acad Dermatol. 2017;76(3):377–390. doi: 10.1016/j.jaad.2016.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oikonomopoulou K, et al. Kallikrein-mediated cell signalling: targeting proteinase-activated receptors (PARs) Biol Chem. 2006;387(6):817–824. doi: 10.1515/BC.2006.104. [DOI] [PubMed] [Google Scholar]
- 3.Radulovic M, Yoon H, Wu J, Mustafa K, Fehlings MG, Scarisbrick IA. Genetic targeting of protease activated receptor 2 reduces inflammatory astrogliosis and improves recovery of function after spinal cord injury. Neurobiol Dis. 2015;83:75–89. doi: 10.1016/j.nbd.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khattri S, et al. Cyclosporine in patients with atopic dermatitis modulates activated inflammatory pathways and reverses epidermal pathology. J Allergy Clin Immunol. 2014;133(6):1626–1634. doi: 10.1016/j.jaci.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krueger JG, et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. J Allergy Clin Immunol. 2012;130(1):145–54.e9. doi: 10.1016/j.jaci.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eissa A, et al. Serum kallikrein-8 correlates with skin activity, but not psoriatic arthritis, in patients with psoriatic disease. Clin Chem Lab Med. 2013;51(2):317–325. doi: 10.1515/cclm-2012-0251. [DOI] [PubMed] [Google Scholar]
- 7.Stalmach A, et al. Identification of urinary peptide biomarkers associated with rheumatoid arthritis. PLoS One. 2014;9(8):e104625. doi: 10.1371/journal.pone.0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramírez A, Bravo A, Jorcano JL, Vidal M. Sequences 5’ of the bovine keratin 5 gene direct tissue- and cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation. 1994;58(1):53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- 9.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115(5):788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 10.Quaranta M, et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med. 2014;6(244):244ra90. doi: 10.1126/scitranslmed.3008946. [DOI] [PubMed] [Google Scholar]
- 11.Kathuria P, Gordon KB, Silverberg JI. Association of psoriasis and psoriatic arthritis with osteoporosis and pathological fractures. J Am Acad Dermatol. 2017;76(6):1045–1053.e3. doi: 10.1016/j.jaad.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 12.Winge MC, et al. RAC1 activation drives pathologic interactions between the epidermis and immune cells. J Clin Invest. 2016;126(7):2661–2677. doi: 10.1172/JCI85738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenz R, et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 2005;437(7057):369–375. doi: 10.1038/nature03963. [DOI] [PubMed] [Google Scholar]
- 14.Radulovic M, Yoon H, Wu J, Mustafa K, Scarisbrick IA. Targeting the thrombin receptor modulates inflammation and astrogliosis to improve recovery after spinal cord injury. Neurobiol Dis. 2016;93:226–242. doi: 10.1016/j.nbd.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsoi LC, et al. Atopic dermatitis is an IL-13-dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol. 2019;139(7):1480–1489. doi: 10.1016/j.jid.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, et al. Augmented Th17 differentiation leads to cutaneous and synovio-entheseal inflammation in a novel model of psoriatic arthritis. Arthritis Rheumatol. 2018;70(6):855–867. doi: 10.1002/art.40447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blauvelt A, Chiricozzi A. The immunologic role of IL-17 in psoriasis and psoriatic arthritis pathogenesis. Clin Rev Allergy Immunol. 2018;55(3):379–390. doi: 10.1007/s12016-018-8702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eder L, Chandran V, Gladman DD. Repair of radiographic joint damage following treatment with etanercept in psoriatic arthritis is demonstrable by 3 radiographic methods. J Rheumatol. 2011;38(6):1066–1070. doi: 10.3899/jrheum.100919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data sets have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE144312.