Abstract
The molecular mechanisms responsible for the high immunosuppressive capacity of CD4+ Tregs in tumors are not well known. High-dimensional single-cell profiling of T cells from chemotherapy-naive individuals with non–small-cell lung cancer identified the transcription factor IRF4 as specifically expressed by a subset of intratumoral CD4+ effector Tregs with superior suppressive activity. In contrast to the IRF4– counterparts, IRF4+ Tregs expressed a vast array of suppressive molecules, and their presence correlated with multiple exhausted subpopulations of T cells. Integration of transcriptomic and epigenomic data revealed that IRF4, either alone or in combination with its partner BATF, directly controlled a molecular program responsible for immunosuppression in tumors. Accordingly, deletion of Irf4 exclusively in Tregs resulted in delayed tumor growth in mice while the abundance of IRF4+ Tregs correlated with poor prognosis in patients with multiple human cancers. Thus, a common mechanism underlies immunosuppression in the tumor microenvironment irrespective of the tumor type.
Keywords: Immunology, Oncology
Keywords: Adaptive immunity, Cancer immunotherapy, T cells
Introduction
Despite recent clinical breakthroughs in adoptive T cell transfer approaches and checkpoint blockade in treating hematopoietic and solid tumors, suppression of the antitumor immune response in the tumor microenvironment (TME) is a major obstacle to tumor regression (1). Although antitumor immune cells, such as T and NK cells, normally infiltrate tumors, and their abundance correlates with better prognosis in certain cancers, the presence of inhibitory populations, such as regulatory CD4+ T cells (Tregs), counteract tumor rejection (2). The main role of Tregs is to maintain immune homeostasis in physiology by inhibiting effector T cells via different modes of action, including expression of cell-surface inhibitors, such as cytotoxic T lymphocyte–associated protein 4 (CTLA-4) (3); production of inhibitory cytokines, such as IL-10 (4), TGF-β (5), or IL-35 (6); depletion of IL-2 by overexpression of the high-affinity IL-2 receptor α chain (IL-2Rα, also known as CD25); purine-mediated suppression by CD39-dependent conversion of ATP (7); or direct cytotoxicity (8), thus preventing autoimmunity and immunopathology (reviewed in Vignali et al., ref. 9). Conversely, Tregs are aberrantly enriched in tumors and dampen antitumor immune responses (10). This detrimental effect on antitumor immunity is demonstrated by a plethora of preclinical data in which systemic Treg depletion or inhibition of their function promotes tumor regression (11–14). Unfortunately, depletion or inhibition of Tregs often results in severe autoimmunity, allergy, and immunopathology (11–14); therefore, new strategies are required to safely deplete Tregs specifically in the TME. The prerequisite to do so is the identification of a specific molecular program or molecules that are exclusively present in tumor-associated Tregs, which would therein allow us to spare circulating Tregs and maintain peripheral homeostasis.
Transcriptional profiling recently revealed that CD4+ Tregs isolated from colon, lung, and breast tumors are transcriptionally different from those isolated from the adjacent tissues and the blood (15, 16). In particular their transcriptional profile overlaps with that of effector Tregs described in mice, which show enhanced suppressive activity. This includes the increased expression of molecules, such as CTLA-4, 4-1BB, CCR8, ICOS, and others (17). Noteworthily, increased expression of CCR8 (15, 16), LAYN, or MAGEH1 (15) by the intratumoral Tregs correlated with poor prognosis, suggesting a pivotal role of these cells in inhibiting antitumor immune responses and/or favoring tumor growth.
The molecular mechanisms leading to increased Treg activity in tumors remain ill-defined. Studies performed over the last decade have elucidated in part the transcriptional network at the basis of murine effector Treg differentiation in lymphoid and nonlymphoid tissues. Once activated, Tregs undergo a program of effector differentiation that mirrors Th cell differentiation by expressing transcription factors (TFs) that regulate Th polarization, such as T-bet, GATA-3, or Bcl-6 (17). Additional TFs might be involved in regulating effector Tregs at specific tissue sites, such as PPARγ, which controls the unique transcriptional and metabolic signature of those cells residing in the visceral adipose tissue (18). Interferon regulatory factor 4 (IRF4), initially identified as the TF responsible for the generation of those Tregs specifically controlling Th2 responses (19), plays a central role in generating effector Tregs in peripheral organs (20). IRF4 is absent in quiescent T cells but is induced by T cell receptor (TCR) signaling. Mice with specific deletion of Irf4 in Tregs develop multiorgan autoimmunity due to exacerbated Th1, Th17, and Tfh responses and plasma cell infiltration (19). Notably, IRF4-deficient Tregs largely maintain a CD62Lhi naive-like phenotype and express reduced amounts of effector and suppressive molecules, such as ICOS, CTLA-4, and IL-10 (20). IRF4 cooperates with other TFs in DNA binding and regulating transcription in CD4+ T cells, namely the AP-1 family members BATF and JUN (21, 22). Notably, IRF4 was reported to be overexpressed by non–small-cell lung cancer–infiltrating (NSCLC-infiltrating) Tregs (23), but its role in determining the Treg transcriptional program, functionality, and regulation of antitumor immunity remains completely unexplored.
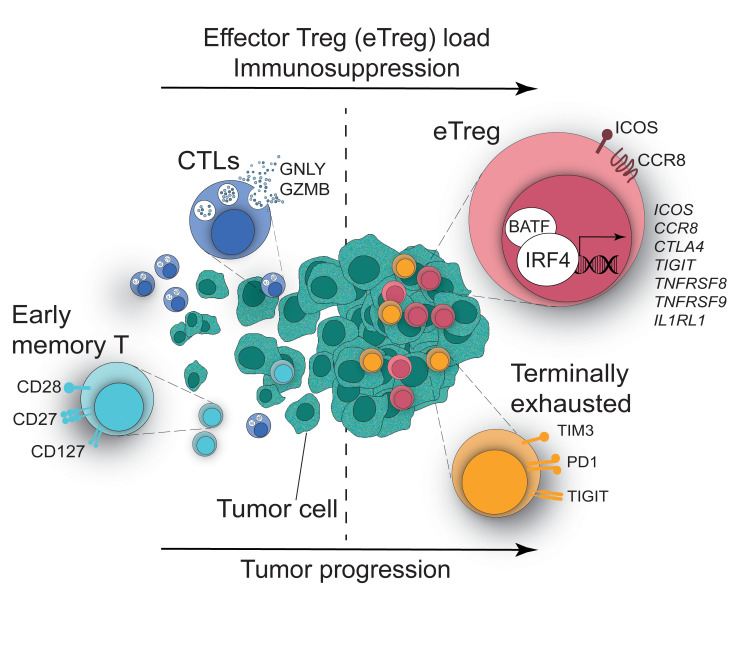
Here, we reported the existence of 2 subsets of tumor-infiltrating Tregs with differential expression of IRF4, as revealed by high-dimensional single-cell analysis of lung, liver, and melanoma-infiltrating T cells. IRF4 defined activated effector Tregs with enhanced suppressive activity, the abundance of which correlates with poor prognosis in multiple human cancers. Consistent with a direct requirement for IRF4 for the enhanced suppressive functions, deletion of Irf4 in Tregs enhanced antitumor immunity in a mouse model of cancer in vivo. We further identified a core network of immunosuppressive genes directly regulated by IRF4 in interaction with its molecular partner, BATF, thereby demonstrating that IRF4 instructs the suppressive activity of Tregs in human cancer.
Results
Treg heterogeneity in the TME and its relation to IRF4.
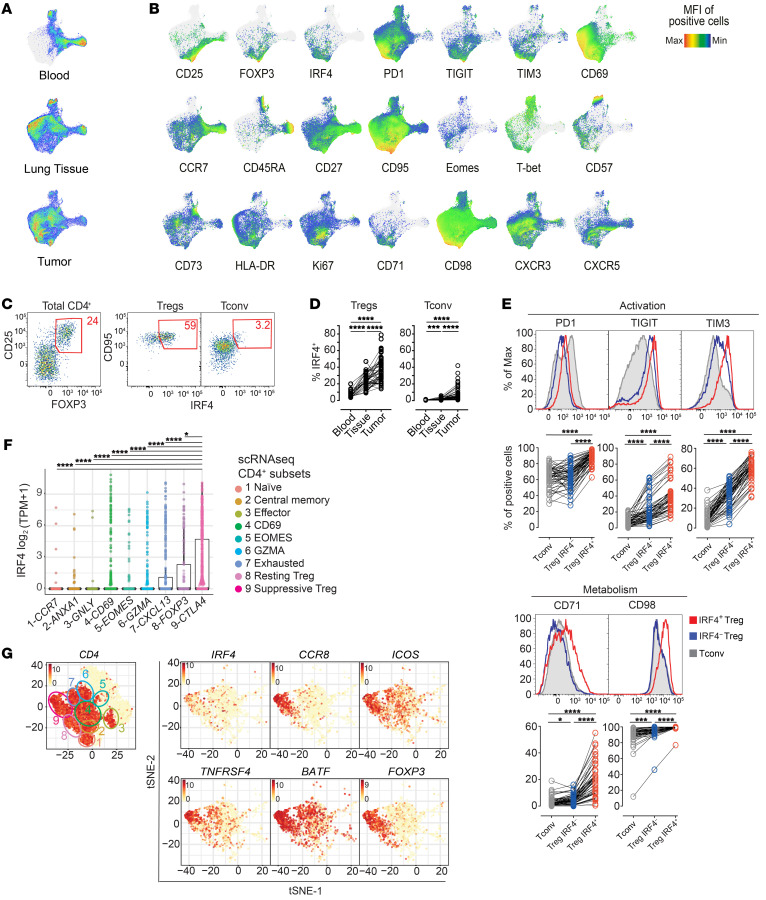
To gain insight into CD4+ T cell phenotypes at the tumor site, we initially investigated T cells in a cohort of 53 patients with NSCLC (Supplemental Table 1 and ref. 24; supplemental material available online with this article; https://doi.org/10.1172/JCI130426DS1) with a 27-parameter polychromatic flow cytometry panel encompassing markers of memory and effector T cell differentiation, activation, metabolic activity, and exhaustion as well as Treg markers (Supplemental Table 2). We profiled tumors (n = 53), paired adjacent cancer-free lung tissues (n = 45), and peripheral blood samples (PB; n = 22) of treatment-naive patients (Supplemental Table 1). We next used Uniform Manifold Approximation and Projection (UMAP), a dimensionality reduction visualization approach that preserves the local and the global structure of single-cell data, to simplify the visualization of marker coexpression in a 2D space (Figure 1, A and B). Overall, CD4+ T cells from the 3 body sites displayed a different single-cell profile (Figure 1A). Previously described dynamics aside (24), such as the loss of CD45RA+, CCR7+, and CD27+ (identifying naive and early memory cells) and the accumulation of HLA-DR+ activated and PD1+ T cells, we also noticed the accumulation of CD25+Foxp3+ Tregs expressing the TF IRF4 in tumor versus adjacent lung and blood samples (Figure 1, B–D). Manual gating of flow cytometry data indicated that approximately 40% of CD25+Foxp3+ Tregs in tumors expressed IRF4, while the rest were IRF4– (Figure 1, C and D), suggesting Treg heterogeneity at the tumor site. In addition, the majority of conventional CD4+ T (Tconv) cells lacked IRF4 expression (Figure 1, C and D). Flow cytometric analysis also revealed that IRF4+ Tregs retain increased expression of PD1, TIGIT, and TIM3 receptors and CD71 and CD98 metabolic markers compared with both IRF4– Tregs and Tconv cells in tumors (Figure 1E). These markers are generally upregulated with activation. In line with our results, reanalysis of published single-cell RNA-sequencing (scRNA-seq) data (25) of CD4+ tumor-infiltrating lymphocytes from patients with NSCLC identified IRF4 expression as largely confined to a subpopulation of CTLAhi Tregs (subset 9-CTLA4; Figure 1F), while it was relatively lower in other cell subsets, including in nonactivated Tregs (8-FOXP3). CTLA4hi Tregs, which also express FOXP3, displayed an effector signature, including expression of CCR8, ICOS, TNFRSF4 (encoding OX-40), TNFRSF9 (encoding CD137/4-1BB), and the IRF4 transcriptional partner BATF (Figure 1G). We identified a similar subset of IRF4-expressing Tregs by scRNA-seq analysis of CD45+ cells infiltrating hepatocellular carcinoma (Supplemental Figure 1A and ref. 26). Notably, IRF4 expression correlated with multiple Treg genes but not with TFs related to other Th cell subsets in single CD4+ T cells isolated from melanomas (Supplemental Figure 1B and ref. 27). Thus, a common phenotypic and gene expression architecture defines tumor-infiltrating Tregs in multiple human cancers.
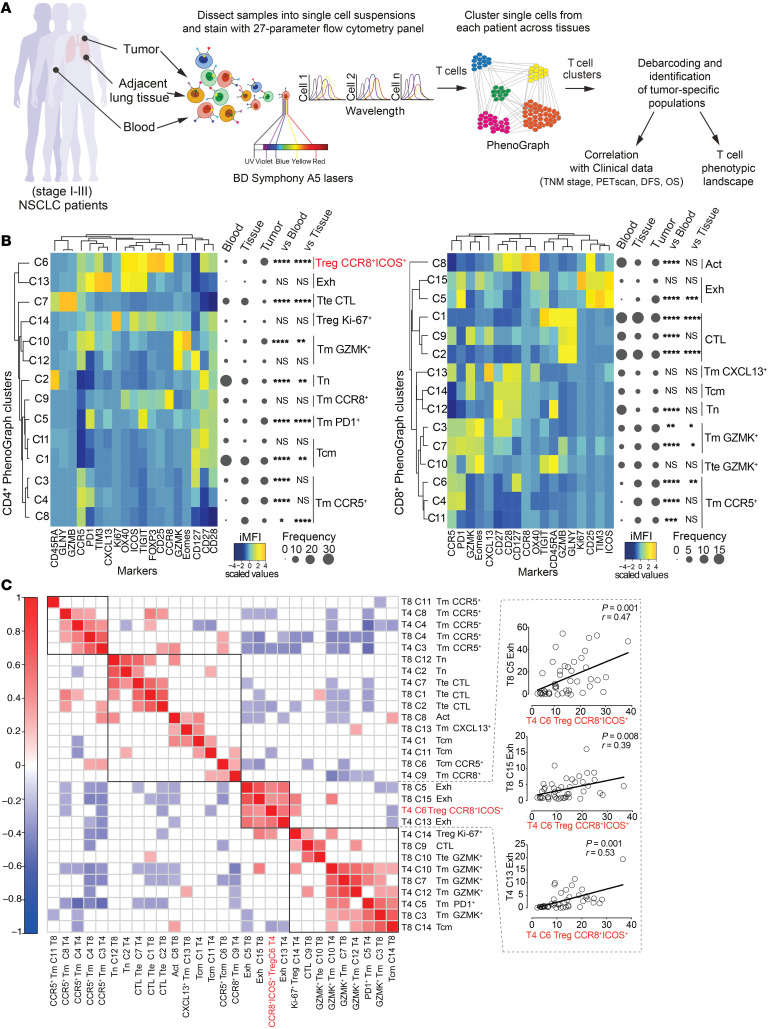
Figure 1. IRF4 identifies effector Tregs enriched in human tumors.
(A) UMAP analysis of concatenated CD4+ T cells (1,500 cells/sample) from tumor (n = 53), adjacent lung tissue (n = 45), and blood (n = 22) samples from patients with NSCLC. (B) UMAP of relative marker expression by concatenated CD4+ T cells from the same samples in A. (C) Manual gating analysis of CD4+CD25+FOXP3+ Tregs expressing IRF4+ by flow cytometry. Numbers indicate the percentage of positive cells. (D) Summary plot representing the IRF4 expression in CD4+ Tregs and conventional T (Tconv cells; defined as CD25loFoxP3–) cells from the same patients as in A (***P < 0.0005, ****P < 0.0001, Kruskal-Wallis test). (E) Representative distribution by flow cytometry (top) and summary of the percentage of expression of selected markers (bottom) in tumor-infiltrating IRF4+ and IRF4– Tregs and Tconv cells (*P < 0.01, ***P < 0.0005, ****P < 0.0001, nonparametric Friedman test). (F) Box plot showing the log2(TPM + 1) expression of IRF4 transcript across 9 CD4+ T cells clusters as identified by single-cell RNA-seq (25). Each dot represents a single cell (*P ≤ 0.01, ****P ≤ 0.0001, Wilcoxon test). (G) t-SNE plots illustrating the expression of selected genes in single CD4+ T cells from lung tumor lesions. Cell clusters, depicted on the left, were identified as in F.
IRF4 expression defines effector Tregs with enhanced suppressive potential capable of promoting tumor growth in vivo.
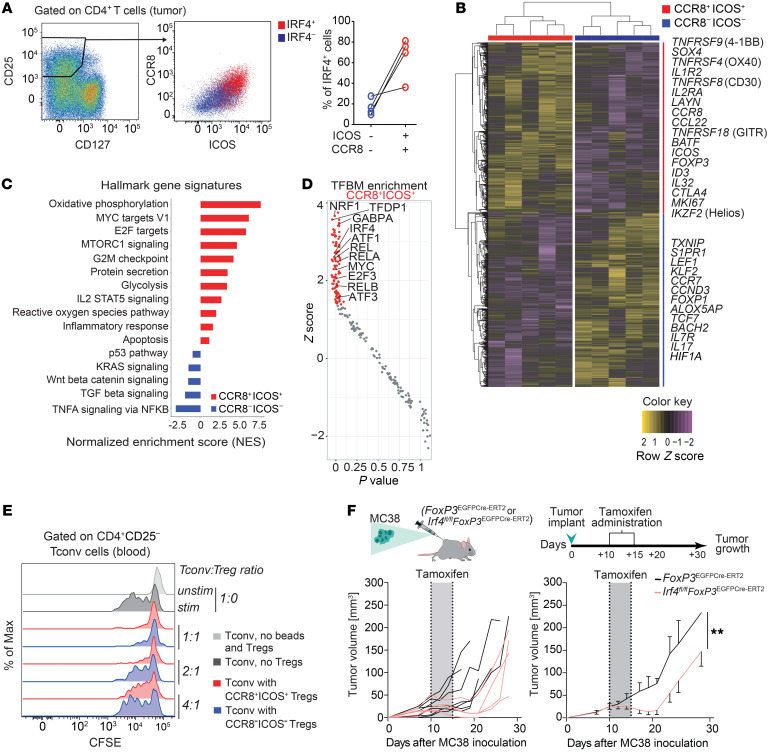
We further characterized IRF4+ and IRF4– Tregs by bulk RNA-seq. As the intranuclear localization of TFs precluded the isolation of viable cells based on IRF4 expression levels, we used a surrogate surface staining strategy for FACS-based isolation of Treg subsets. Bulk Tregs from tumors, defined as CD25hiCD127lo, were further separated according to CCR8 and ICOS to identify IRF4+ and IRF4– Tregs (Figure 2A, Supplemental Figure 2A, and Methods). As expected, both of these subsets expressed high levels of FOXP3 compared with Tconv cells (Supplemental Figure 2B) and their gene expression significantly overlapped with a tumor-infiltrating Treg signature, as obtained from a NSCLC scRNA-seq data set (ref. 25 and Supplemental Figure 2C), confirming the Treg identity of these subsets.
Figure 2. Transcriptional and functional profiling defines the effector and enhanced suppressive nature of IRF4+ Tregs.
(A) Representative CCR8 and ICOS expression in tumor-infiltrating CD25hiCD127lo Treg subsets defined by IRF4 expression and t percentage of IRF4 expression in tumor-infiltrating Tregs gated as CCR8–ICOS– or CCR8+ICOS+. (B) Heatmap of differentially expressed genes (DEGs) in the FACS-sorted CCR8+ICOS+ versus ICOS–CCR8– tumor-infiltrating Tregs, as obtained by RNA-seq (FDR < 0.05). Selected DEGs are indicated. For some genes, protein names are indicated. (C) Hallmark gene sets (MsigDB; as obtained by GSEA) significantly enriched in cells sorted as in B. (D) Transcription factor binding motif (TFBM) enrichment analysis by pScan of RNA-seq data obtained as in B. Colored dots indicate significant hits. (E) CFSE-labeled CD4+ CD25– T (Tconv) cells dilution from a representative blood sample. Tconv cells were cocultured with Suppression Inspector MACSiBead beads and different ratios of intratumoral Treg subsets for 5 days. Data are representative of 5 independent experiments. (F) Tumor volumes in FoxP3EGFP-cre-ERT2(control) (n = 7) or Irf4fl/flFoxP3EGFP-cre-ERT2 (n = 5) mice following the administration of tamoxifen. Tumor curves in individual mice and mean ± SEM of the same cohort are shown. **P < 0.01, paired Student’s t test.
A multidimensional scaling plot of gene expression profiles showed that sorted intratumoral CCR8+ICOS+ (IRF4+) and CCR8–ICOS– (IRF4–) clustered separately, indicating substantial differences at the transcriptional level (Supplemental Figure 2D). Overall, we identified 2674 differentially expressed genes (DEGs; FDR < 0.05; Supplemental Table 3). CCR8+ICOS+ Tregs expressed high amounts of transcripts encoding molecules involved in effector differentiation (e.g., TNFRSF9, TNFRSF4, TNFRSF18, IL2RA), transcriptional regulation of effector Tregs (BATF), proliferation (MKI67), Treg identity (IKZF2, encoding the TF Helios [ref. 28]), costimulation (ICOS) and suppressive function (CTLA4). Additional transcripts included IL32, CCL22, and CX3CR1. By contrast, CCR8– ICOS– Tregs expressed high amounts of transcripts encoding molecules associated with early differentiation or quiescence, including CCR7, KLF2, LEF1, TCF7 (29), BACH2 (repressing effector programs to stabilize Treg-mediated immune homeostasis, ref. 30), and TXNIP (a negative regulator of AKT signaling and glycolytic metabolism) (Figure 2B and Supplemental Table 3). Gene set enrichment analysis (GSEA) further revealed that CCR8–ICOS– Tregs were largely quiescent and displayed transcriptional signatures of Wnt/β-catenin and TGF-β signaling. In contrast, CCR8+ICOS+ Tregs displayed enhanced signatures of metabolic activity, including oxidative phosphorylation and glycolysis; mTORC1-dependent activity, previously shown to favor murine Treg activation and prevention of autoimmunity (31); and reactive oxygen species metabolism, consistent with enhanced activation and/or mitochondrial respiration (Figure 2C, Supplemental Table 4, and ref. 32). To identify transcriptional regulators of CCR8+ICOS+ intratumoral Tregs, we performed computational analysis of TF-binding motif enrichment at the promoters (–950, +50 bp from the TSS) of DEGs obtained from bulk RNA-seq data. This analysis showed enrichment of binding motifs of TFs, including IRF4, predicted to be preferentially active in the CCR8+ICOS+ Treg subset (Figure 2D). We additionally identified NRF1, involved in antioxidant defense, lipid metabolism, and mitochondrial respiratory function (33); MYC, a master regulator of metabolic reprogramming in activated T cells (34); and the NF-κB family members REL, RELA, and RELB. In particular, RelA has been previously shown to regulate effector Treg activity in nonlymphoid tissues downstream of TNFRSFs (35). Thus, our transcriptional profiling revealed that intratumoral IRF4+ Tregs are highly activated compared with IRF4– Tregs and supported the notion that they have enhanced suppressive potential. To confirm this hypothesis, FACS-sorted CCR8+ICOS+ (IRF4+) and CCR8–ICOS– (IRF4–) intratumoral Tregs were further tested for their capacity to suppress proliferation of autologous CD4+CD25– Tconv cells in vitro (Figure 2E). Both Treg subsets were effective in this regard at a 1:1 Tconv cell/Treg ratio, while only CCR8+ICOS+ Tregs maintained substantial suppressive capacity at a 2:1 ratio (Figure 2E).
IRF4 has been shown to promote effector Treg differentiation in peripheral tissues and to limit autoimmunity (20), but its importance in suppressing antitumor immune responses is unknown. To test the functional role of IRF4 in tumor-infiltrating Tregs, we used mice that allow tamoxifen-inducible deletion of IRF4 specifically in Tregs (Irf4fl/flFoxp3EGFP-cre-ERT2) and controls (Foxp3EGFP-cre-ERT2) (Figure 2F). Induced deletion of Irf4 in FoxP3+ cells in MC38 tumor-bearing mice resulted in a significant delay in tumor growth (Figure 2F), indicating that IRF4+ Tregs suppress antitumor immunity. To further test the Treg-intrinsic role of IRF4, we reconstituted lethally irradiated mice with a mix of congenically marked bone marrow from WT mice and mice with a T cell–specific deletion of IRF4 (Irf4fl/flCd4Cre). We also generated mixed control chimeras containing WT and Cd4Cre control bone marrow. Flow cytometric analysis of tumor-infiltrating Tregs in these chimeric mice showed severely impaired representation of IRF4-deficient Tregs at the tumor site but not in the spleen and lack of ICOS expression, a direct target of IRF4. In contrast, control chimeras showed similar contribution of Tregs at both sites and robust ICOS expression (Supplemental Figure 2, E and F).
IRF4 and its partner BATF control a molecular program of effector Treg differentiation and suppression in tumors.
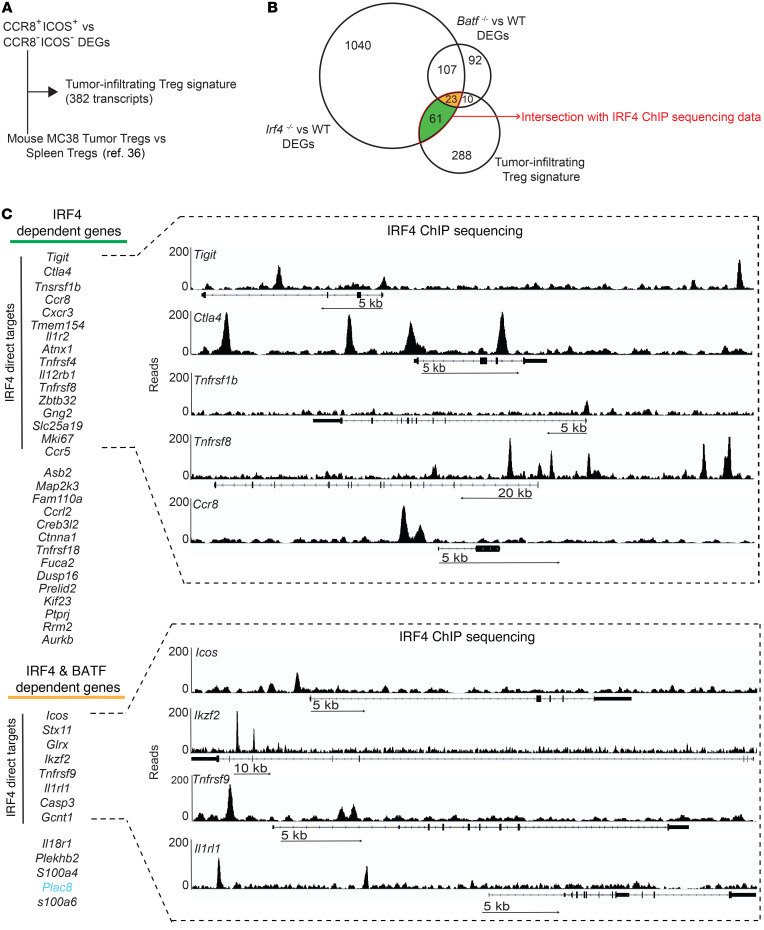
Next, we formally tested the role of IRF4 and its transcriptional partner BATF in determining the features of tumor-infiltrating Tregs by integrating our data from human Tregs with gene expression profiles and epigenomic data from mouse Tregs (Supplemental Table 5). As mouse versus human gene regulation is difficult to infer due to species differences in noncoding regions, we first defined a conserved tumor-infiltrating Treg signature by integrating DEGs of CCR8+ICOS+ versus CCR8– ICOS– Tregs from Figure 2B and Supplemental Table 3 and DEGs of tumor-infiltrating Tregs versus spleen Tregs from a murine model (36), as outlined in Figure 3A. In total, we identified 382 transcripts that were specifically upregulated or downregulated in mouse and human tumor-infiltrating Tregs (Figure 3A and Supplemental Table 6). Next, we defined the transcriptional program that is dependent on IRF4 or BATF expression by performing RNA-seq and analysis of gene expression profiles of splenic Tregs from WT versus Irf4–/– mice. We also reanalyzed a published data set comparing WT versus Batf–/– Tregs (37). Overall, this identified 1241 and 232 genes that were controlled by IRF4 and BATF, respectively (Figure 3B and Supplemental Table 5). GSEA revealed that many genes involved in lymphocyte activation, proliferation, and differentiation were under the joint control of IRF4 and BATF; those involved in apoptosis were controlled by BATF alone; and those related to immunosuppression, i.e., IL-37– and IL-10–dependent signaling pathways, were under the control of IRF4 alone (Supplemental Figure 3B). A relevant proportion (94 genes; 24.6%) of the conserved tumor Treg signature between humans and mice was controlled by IRF4 and/or BATF (Figure 3B; IRF4 and BATF DEGs vs. tumor-infiltrating Treg signature: P = 1.1 × 10–14 and P = 3.3 × 10–18, respectively; hypergeometric test; data not shown). We further made use of our ChIP deep-sequencing (ChIP-seq) data to assess IRF4 occupancy at the loci of interest (Supplemental Table 7) and thus define a direct role of this TF in controlling gene expression in tumor-infiltrating Tregs. Supplemental Figure 3B shows the distribution of regions in the genome that are bound directly by IRF4 according to their distance from transcriptional start sites (TSSs) (see also Supplemental Table 7). Despite a small fraction mapped in promoter regions (0–1 Kb), the majority of IRF4-bound sites mapped at 10 to 100 Kb distance from the TSS, suggesting regulation of gene expression at enhancer regions. Tumor-infiltrating Treg genes dependent on IRF4 included several Tnfr family members involved in effector Treg differentiation, such as Tnfrsf1b, Tnfrsf8, Tnfrsf18, and Tnfrsf9 (the latter also dependent on Batf expression) (35); chemokine receptors, such as Ccr8, Cxcr3, and Ccr5, likely involved in the localization of Tregs to tumors or to the lung and expressed by murine effector Tregs (38); and Zbtb32, Mki67, Map2k3, Kif23, Rrm2, and Aurkb, previously linked to cell cycle in other cell types and likely contributing to Treg proliferation (Figure 3C). Instead, Icos, Ikzf2 (encoding Helios), and Il1rl1 (also known as ST2 or IL-33 receptor) were dependent on both Irf4 and Batf. Thus, Irf4 and Batf directly and indirectly control a program of effector Treg differentiation and immunosuppression in cancer.
Figure 3. Irf4 and its partner Baft directly and indirectly control a molecular program of effector Treg differentiation and suppression.
(A) Identification of a shared gene expression signature between tumor-infiltrating human CCR8+ICOS+ Tregs versus CCR8–ICOS– Tregs and murine Tregs (36). (B) Venn diagram of the number of genes of the tumor-specific Treg signature obtained as in A that are differentially expressed in splenic Tregs from Batf–/– and Irf4–/– mice. Genes controlled only by Batf (n = 10) were of limited interest and thus not further investigated. (C) List of tumor-infiltrating Treg genes that are dependent on the expression of Irf4 or Irf4 and Batf. All genes are induced, except for Plac8, which is repressed (indicated in light blue). Those genes directly controlled by IRF4 binding to the genome, as obtained from ChIP-seq analysis of murine Tregs, are highlighted. Genomic binding of Irf4 to the DNA for selected genes is depicted.
scRNA-seq–guided high-dimensional flow cytometry profiling reveals that CCR8+ICOS+ (IRF4+) effector Tregs associate with multiple exhaustion traits of T cells.
To further characterize the T cell phenotypic landscape associated with Treg subsets in patients with NSCLC cancer in an unbiased manner, we designed a second high-dimensional flow cytometry panel guided by results obtained with scRNA-seq (25) and bulk RNA-seq (Figure 2 and Supplemental Table 2). We preferred markers identified by fluorochrome-conjugated antibodies providing a high signal-to-noise ratio (e.g., CCR8 and ICOS instead of IRF4) so as to enhance the identification of subsets by the clustering algorithm, as recently suggested (39). We profiled millions of single cells from tumors (n = 45), paired adjacent cancer-free lung tissues (n = 23), and peripheral blood samples (n = 23) of treatment-naive patients (Supplemental Table 1). A schematic representation of the analysis pipeline is shown in Figure 4A. By applying PhenoGraph, a computational algorithm capable of clustering single cells without bias according to the relative expression of these molecules in the multidimensional space (40), we identified 14 different CD4+ and 15 different CD8+ T cell clusters and defined their abundance as a percentage of total CD4+ or CD8+ T cells in each sample type (Figure 4B). In addition, for this panel of markers, UMAP of single-cell distributions and principal component analysis (PCA) of cluster abundance in the different samples clearly distinguished T cells from the peripheral blood, lung tissues, and tumors (Supplemental Figure 4, A and B), indicating that different sites have highly distinct T cell profiles. We next calculated the integrated median fluorescence intensity (iMFI) values of each marker in each PhenoGraph cluster so as to obtain information on cluster identity (Figure 4B, the Methods, and ref. 39). Metaclustering of PhenoGraph clusters and markers grouped subpopulations with similar immunophenotypes. Similar to that in previous findings (24), we documented the loss of subsets of naive (CD4+, C2; CD8+, C12) and cytotoxic (CD4+, C7; CD8+, C1, C9, C2) T cells and the accumulation of exhausted T cells (CD8, C5) in tumors compared with the blood or the adjacent lung tissue (Figure 4B). Moreover, in line with data shown in Figure 1, we revealed the increased presence of CCR8+ICOS+ (IRF4+) activated Tregs (CD4+, C6) in tumors. Next, we investigated the association of these cells with specific T cell subsets in the TME by performing a Pearson correlation analysis of the abundance of CD4+ and CD8+ T cell clusters as identified by PhenoGraph in 45 patients (Figure 4C). Notably, intratumoral CCR8+ICOS+ Tregs correlated with CD4+ (C13) and CD8+ (C5 and C15) T cells with features of exhaustion (i.e., expressing PD1, TIM3, and TIGIT in different combinations and intensities). At the same time, CCR8+ICOS+ Tregs negatively correlated with cytotoxic CD8+ T cells (C2, coexpressing GZMB and GNLY and thus armed for rapid effector functions) and with clusters of T cells bearing a central memory T phenotype (CD4+, C11 and CD8+, C14). In summary, high-dimensional single-cell profiling identifies a T cell signature with increased frequencies of CCR8+ICOS+ effector Tregs associated with T cell exhaustion.
Figure 4. Abundance of CCR8+ICOS+ intratumoral Tregs is associated with multiple features of T cell exhaustion.
(A) Experimental workflow. (B) Heatmaps of the relative expression, depicted as integrated MFI (iMFI: MFI × percentage of antigen expression) of markers (columns) in discrete CD4+ (left) and CD8+ (right) Phenograph clusters (rows). Tm, memory; Tcm, central memory; Tn, naive; Exh, exhausted; Act, activated; CTL, cytotoxic T lymphocyte. Tte, terminal effector. Data are further metaclustered to group subpopulations with similar immune-phenotypes. The median frequency of each PhenoGraph cluster in the different compartments is depicted by using Balloon plots. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, tumor versus blood or versus normal tissue samples; 2-way ANOVA with Bonferroni’s post hoc test. (C) Correlogram showing Pearson correlation between frequencies of CD4+ (T4) and CD8+ (T8) PhenoGraph clusters in tumor samples from 45 patients with early-stage (I–III) non–small-cell lung cancer (NSCLC). Nonsignificant correlations (P value > 0.05) were left blank.
CCR8+ICOS+ (IRF4+) effector Tregs define a signature of disease progression in NSCLC.
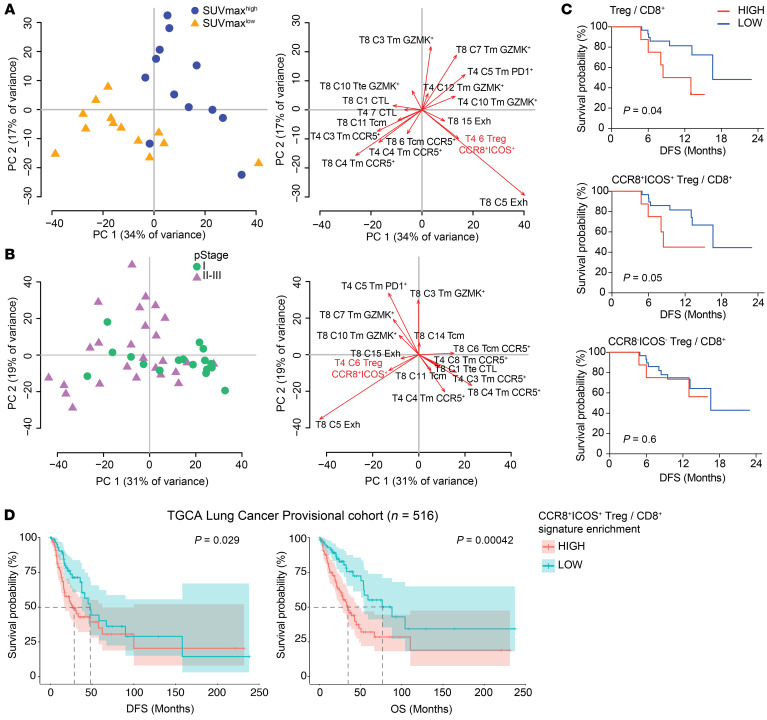
We finally tested whether a phenotypic landscape of T cells involving IRF4+ effector Tregs could be found to be different in patients with cancer with different prognoses. First, a subset of our cohort of patients (n = 25) was subdivided in 2 groups according to the median distribution of the maximum standardized value of fluorodeoxyglucose uptake (SUVmax), a PET indicator of tumor glycolysis and aggressiveness. PCA revealed a bimodal separation of the SUVmaxlo and SUVmaxhi groups of patients according to the relative frequencies of CD4+ and CD8+ PhenoGraph clusters (Figure 5A, left), indicating that they display a different T cell profile as a whole. Analysis of PCA loadings identified clusters contributing the most to such distribution (Figure 5A, right). Specifically, SUVmaxhi patients harbored increased frequencies of CCR8+ICOS+ Tregs (C6), subsets of CD8+ (C5 and C15), and CD4+ T cells (C5) with traits of exhaustion or memory T cells expressing GZMK, EOMES (contributing to T cell dysfunction in humans [ref. 41]), and PD1 (CD8+, C3, C7), while SUVmaxlo patients harbored increased frequencies of CCR5+ clusters (CD8+, C4, C6 and CD4+, C3, C4) as well as terminal effector, cytotoxic T cells (CD8+, C1 and CD4+, C7) (Figure 5A). We observed similar trends when considering a parameter of disease progression, i.e., the international TNM classification, according to which CCR8+ICOS+ Tregs (CD4+, C6) as well as T cells featuring exhaustion/activation markers (CD8+, C5 and C15) were more abundant in patients with advanced pathological stage II and III (Figure 5B).
Figure 5. CCR8+ICOS+ Tregs define a signature of disease progression in NSCLC.
(A) Left: Principal component analysis (PCA) plot showing the distribution of patients (n = 48) according to the frequency of CD4+ and CD8+ PhenoGraph clusters in each patient (Tm, memory; Tcm, central memory). Patients were classified according to pathological stage (pStage) I, II, or III of the International TNM classification. Right: PCA loading plot of PhenoGraph clusters most contributing to the PCA output on the left. (B) Left: PCA plot showing the distribution of patients (n = 26) according to the frequency of CD4+ and CD8+ PhenoGraph clusters in each patient. The cohort was subdivided in 2 groups according to the median distribution of maximum standardized uptake value (SUVmax). Right: PCA loading plot as in A. (C) Kaplan-Meier progression-free survival curves according to the intratumoral frequencies of Tregs subsets over CD8+ T cells in each patient (n = 38). The cohort was subdivided in 2 groups according to the percentile rank (set at 0.8). The P value was calculated by Gehan Breslow-Wilcoxon test. (D) Kaplan-Meier disease-free survival (DFS) and overall survival (OS) curves in the TCGA lung adenocarcinoma (LUAD) lung cancer cohort (n = 516). Patients were grouped by percentile rank (set at 0.8) according to the enrichment of the CCR8+ICOS+ bulk Treg signature (as obtained in Figure 2B) as relative to the CD8+ T cell signature. + indicates censored observations. P values were calculated by multivariate Cox regression. Dotted lines indicate the time at which 50% of the cohort was still free of the event.
Infiltration of CD8+ T cells is a predictor of good prognosis in multiple types of cancer (42). We thus investigated disease-free survival (DFS) of our patients according to the Treg/CD8+ T cell ratio (as determined by their frequency among CD3+) at the tumors site and found that a higher ratio was significantly associated with early tumor relapse (Figure 5C). As predicted by their superior suppressive activity, this was due to the contribution of CCR8+ICOS+, but not of CCR8– ICOS–, Tregs (Figure 5C). We next validated our findings in larger cohorts of individuals, such as those from The Cancer Genome Atlas (TCGA). In these, the degree of infiltration of specific subpopulations was determined by enrichment of transcriptional signatures (see Methods). Similar to results from our NSCLC cohort, we found that a higher CCR8+ICOS+ Treg/CD8+ T cell signature in bulk RNA-seq data from primary biopsies was associated both with worse DFS and overall survival in lung adenocarcinoma (Figure 5D and ref. 43), hepatocellular carcinoma (Supplemental Figure 5A and ref. 44), and melanoma (Supplemental Figure 5B), thereby underlying a common biology of Treg-mediated suppression of antitumor immunity in multiple human cancers.
Discussion
Human tumors are often infiltrated by large numbers of CD4+ Tregs that display a highly activated phenotype and enhanced suppressive capacity compared with those present in the peripheral and the adjacent tumor-free tissues (15, 16). We have now shown that the transcriptional program associated with such activation and suppression in the TME is driven by the TF IRF4 in combination with its molecular partner BATF. Signals downstream of the TCR are likely involved in IRF4 induction (45), possibly suggesting that human Tregs suppress antitumor immunity in an antigen-specific way.
IRF4, previously linked to effector Treg differentiation in murine tissues in physiology (20), is now shown to control the formation of ICOS+ effector Tregs in the TME that, in turn, favor tumor growth in a mouse model of cancer. IRF4 controls gene expression of tumor-infiltrating Tregs both directly, by binding gene promoters or distal regulatory regions, and indirectly, by inducing the expression of additional transcriptional regulators, such as IKZF2 (Helios), which is required for Treg stability (28). IRF4 alone binds DNA poorly; however, binding is increased when IRF4 is part of a macromolecular complex involving AP-1 family members BATF, JUN, JUNB, or JUND, which recognize DNA motifs known as AP-1/IRF composite elements (AICEs) (21, 46). Although the regions bound by these TFs are largely overlapping, IRF4 and BATF also bind unique regions (21), which possibly explains why their deficiency has differential, specific effects on gene expression. We did not formally test whether BATF is responsible for the expression of specific genes by direct binding; however, additional computational investigations identified AICE motifs in the proximity of genes that are linked to effector Treg differentiation and function, including Ccr8, Icos, Ctla4, Cxcr3, Il12rb1, Tnfrsf1b, and Tnfrsf8 (data not shown). In relation to this, JunB has recently been shown to control murine effector Treg differentiation in the lung and colon (47), while additional TFs, such as STAT3, may physically interact with c-Jun (48). STAT3 mRNA is upregulated and the STAT3 binding motif is enriched in IRF4+ Tregs compared with IRF4– Tregs (data not shown), which overall suggests that a complex range of molecular interactions is cooperating to shape the effector differentiation and enhanced suppressive activity of intratumoral Tregs.
Our study reveals an additional important aspect, namely that effector differentiation is not a feature of all intratumoral Tregs. Rather, a subset of these cells with increased expression of IRF4 is preferentially expanded in lung, liver, and melanoma tumors compared with the adjacent tumor-free tissues and peripheral blood, as revealed by scRNA-seq. These IRF4+ Tregs express high amounts of molecules associated with enhanced immunosuppression, have increased metabolic demand, and are phenotypically and transcriptionally distinct from the more quiescent IRF4– Tregs. Whereas in humans approximately 40% of Tregs express IRF4, as detected by flow cytometry, deletion of Irf4 in mice results in the near-complete loss of Tregs in tumors, which possibly indicates that low levels of IRF4 are also present in CCR8–ICOS– Tregs. An alternative hypothesis is that slow tumor development, as seen in humans, results in a balanced infiltration of fewer and more activated Tregs. In line with this, approximately 40% of Tregs infiltrating lung adenocarcinoma forming in the K-rasG12D autochthonous model (which better recapitulates of the physiology of lung adenocarcinoma) express the IL-33 receptor ST2 (encoded by Il1rl1) (49), a direct genomic target of IRF4 (Figure 3C). Similarly, a recent human study found that approximately 50% of breast cancer-infiltrating Tregs express CCR8 (50), thereby corroborating our results of Treg heterogeneity in the TME. Despite not reaching the capability of scRNA-seq in terms of number of parameters being measured, the high-dimensional flow cytometry used here investigates T cell phenotypes as a whole in a large cohort of patients compared with those recently investigated by scRNA-seq (25) and is thus capable of further defining the relationships between Treg subsets and other T cell populations within the TME. In this way, we revealed that IRF4+ Tregs positively correlate with multiple subsets of exhausted and activated CD4+ and CD8+ T cells and their abundance is associated both with relapse and poor overall survival in multiple human cancers. Therefore, deep immunophenotyping with improved and high-throughput single-cell technologies can pinpoint those subsets associated with slower tumor growth and favorable antitumor responses with enhanced precision. We anticipate that the simple addition of an effector Treg marker (e.g., IRF4, ICOS, or CCR8 or a TNFR superfamily member) to FoxP3 staining, as done by immunohistochemistry in immunoscore approaches (51), will improve the definition of patients with improved prognosis.
The identification of CD4+ Treg heterogeneity in the TME and, as a consequence, of a molecular program mastering the differentiation of those Tregs with enhanced suppressive capacity offers potentially novel opportunities to reverse immunosuppression while favoring antitumor immune responses. Depletion of Tregs has been tested in a number of preclinical approaches to promote antitumor responses (11–14); however, novel strategies interfering selectively with the activated effector Treg state are emerging as promising tools to boost effective antitumor immunity without resulting in overt autoimmunity due to the loss of peripheral tolerance (14, 52). We expect that targeting those signals leading to IRF4 activation or downstream IRF4-dependent Treg activation would result in a similar scenario. This approach may be widely applicable, as our data show that IRF4-driven effector Treg differentiation is common to at least 3 human tumor types, i.e., lung cancer, hepatocellular carcinoma, and melanoma. Further definition of the molecular network orchestrating the suppressive capacity of intratumoral Tregs and, most importantly, the identification of specific players that are not active in antitumor-infiltrating lymphocytes is anticipated to benefit cancer immunotherapy strategies.
Methods
Study design.
The characteristics of the patients and of the samples used in this study as well as the procedures of cell isolation have been described previously (24). Details on the patients’ characteristics included in this manuscript are further indicated in Supplemental Table 1. Information on the pathological stage, determined by an institutional pathologist (some patients were restaged as III following examination of the tumor), was available for all patients, while results of the preoperative FDG PET-CT scan were available for 25 patients. Details on obtainment of PET scans have been described previously (24).
Polychromatic flow cytometry and cell sorting.
Antibodies used in the study are listed in Supplemental Table 2. Flow cytometry procedures for high-dimensional single-cell panel development have been described previously (24, 39). Additional panels used for further characterizing the IRF4+ CD4+ cell subset are listed in Supplemental Table 2. All data were acquired on a FACS Symphony A5 flow cytometer (BD Biosciences) equipped with 5 lasers (UV, 350 nm; violet, 405 nm; blue, 488 nm; yellow/green, 561 nm; red, 640 nm; all tuned at 100 mW, except for UV, which tuned at 60 mW) and the capability of detecting 30 parameters. Flow cytometry data were compensated in FlowJo by using single stained controls (BD Compbeads incubated with fluorescently conjugated antibodies), as described previously (53). CCR8+ICOS+ and CCR8–ICOS– Tregs, pregated as CD4+ Aqua– CD25+CD127–, were isolated from tumor samples with a FACSAria cell sorter (BD Biosciences). CCR8 and ICOS proved to be the best combination over other markers to isolate Treg subsets with differential IRF4 expression (data not shown).
Suppression of T cell proliferation by Tregs.
Live (Aqua–) CD4+ CD25– Tconv cells were isolated from patients’ blood samples with a FACSAria cell sorter and stained with a CellTrace CFSE kit (final concentration: 2 μM; Thermo Fisher Scientific) according to the manufacturer’s protocol. Subsequently, cells were plated in R10 U-bottom 96-well plates (10,000 cells/well) and stimulated with human Treg Suppression Inspector beads (Miltenyi Biotec) for 5 days at 37°C. Tconv cells cultured alone, in the absence of bead stimulation, were used as nonproliferating, negative control. CCR8+ICOS+ and CCR8–ICOS– Tregs were FACS sorted from tumors and were added to autologous Tconv cell cultures at different ratios (Tconv cells/Treg ratio = 1:1, 2:1, and 4:1). CFSE dilution was evaluated at day 5.
High-dimensional flow cytometry data analysis.
Flow Cytometry Standard (FCS) 3.0 files were imported into FlowJo software (version 9) and analyzed by standard gating to remove aggregates and dead cells and identify CD3+CD4+ or CD8+ T cells. Three thousand CD4+ and CD8+ T cells per sample were subsequently imported in FlowJo (version 10), biexponentially transformed, and exported for further analysis in Python (version 3.7.3) by a custom-made pipeline of PhenoGraph (available here http://github.com/luglilab/Cytophenograph, where we modified the linux-community and the core.py script of PhenoGraph package in order to fix the seed to “123456”). Blood, adjacent lung tissue, and tumor samples were labeled with a unique computational barcode for further identification and converted in comma separated (CSV) files and concatenated in a single matrix by using the merge function of pandas package. The K value, indicating the number of nearest neighbors identified in the first iteration of the algorithm, was set at 40 and 60 for CD4+ and CD8+ T cells clustering, respectively. Clusters representing less than 1% of total CD4+ or CD8+ T cells were removed in subsequent analysis. The data were then reorganized and saved as new CSV files, one for each cluster, that were further analyzed in FlowJo to determine the frequency of positive cells for each marker and the corresponding median fluorescent intensity (MFI). These values were multiplied to derive the iMFI (rescaled to values from 0 to 100; Figure 4B and refs. 24, 39) and then visualized in a heatmap. Subsequent metaclustering of iMFI values was performed using the gplots R package. Hierarchical metaclustering of all samples, based on the frequency of PhenoGraph clusters (Figure 4B), was performed in R according to the Ward minimum variance method. UMAP was obtained by UMAP Python package and visualized in FlowJo 10.
Melanoma data set.
Normalized scRNA-seq counts were retrieved from the Gene Expression Omnibus (GEO GSE72056). Analysis was restricted to the cells labeled as “T cells,” as previously defined by Tirosh et al. (27). T cells were divided into CD4+ and CD8+, based on the normalized expression levels (E) of CD4 (E > 4) and CD8 (average of CD8A and CD8B, E > 4). Among all samples composing the data set, 466 were identified as CD4+. To asses which genes have the expression profile most correlated with that of IRF4, a Pearson correlation coefficient, indicated as score in Supplemental Figure 1B, and a corresponding P value were computed between each gene expression profile in the matrix (consisting of 23,686 genes) and IRF4. In order to obtain the CD8 signature used in Supplemental Figure 5B, DEGs in the pairwise comparison between CD8+ and CD4+ T cell subsets were determined by the “FindAllMarkers” function coded in the Seurat R package (version 2.3.4) with default parameters (54). In this way, we obtained 225 specific genes for the CD8+ subpopulation respect to the CD4+ T cells.
Lung and liver data sets.
We took advantage of the web server for exploration of NSCLC (http://lung.cancer-pku.cn) and hepatocellular carcinoma (http://hcc.cancer-pku.cn) single T cell RNA-seq data to assess the expression levels of markers of interest within the CD4+ intratumoral subpopulations. Lung and liver tumor-infiltrating CD8+ signatures (Figure 5D and Supplemental Figure 5A) were obtained by combining all the DEGs from the CD8+ tissue-specific clusters and by further excluding the circulating clusters labeled as CD8-C1-LEF1 (26, 27).
Mice.
Irf4–/– mice (55) were originally provided by Tak Mak (Princess Margaret Cancer Centre, University Health Network, Toronto, Canada). Irf4–/– mice were crossed to Foxp3RFP mice. Foxp3EGFP-Cre-ERT2 mice (JAX 016961) (56) were crossed to Irf4-floxed mice (JAX 009380) (57) to allow for specific deletion of Irf4 in Foxp3+ cells following tamoxifen treatment.
Mouse procedures.
Mice were irradiated (2 × 5.5 Gy) and reconstituted by i.v. injection of 200 μL bone marrow isolated from femurs and tibias of donor mice. Following irradiation mice were treated with neomycin in drinking water for 4 weeks and allowed to recover for at least 8 weeks. MC38 tumor cells (provided by Stephen Nutt, Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia) were grown in DMEM with 10% FCS and 1% PenStrep (Gibco) in 37°C, 5% CO2 incubator and passaged every 2 to 3 days. For tumor inoculation, 1 × 106 cells were resuspended in 100 μl PBS and injected subcutaneously into the right flank of the mice. Tumor growth was assessed with LCD Digital Vernier Calipers (Protech) every 2 to 3 days. Tamoxifen (2 mg/mouse) was administered in 200 μL sunflower oil i.p. for 5 consecutive days. Upon the experimental endpoint, mice were euthanized with CO2 and cervical dislocation, and tumors were excised with forceps and scissors. Tumors were mechanically dissociated and digested in 1 mg/mL Collagenase IV (Gibco) in RPMI medium for 45 minutes at 37°C, with constant agitation.
Treg isolation from mice.
Single-cell suspensions from spleens were enriched for CD4+ T cells by depleting CD8+ T cells and B cells using anti-CD8 and B220 antibodies. The CD4+ T cell–enriched cell suspension was stained with CD4 (RM 4-5) and TCRβ (H57-597) antibodies. CD4+TCRβ+Foxp3RFP+ cells were sorted from Foxp3RFP (WT) and Irf4–/–Foxp3RFP mice.
Mouse RNA isolation and sequencing.
RNA purification was performed following the manufacturer’s protocol using the RNAeasy Plus Mini Kit (Qiagen). RNA from WT and Irf4–/– Tregs was sequenced using Illumina platform (75-bp paired-end reads).
RNA-seq and bioinformatic analysis.
RNA isolation of the FACS-purified CCR8+ICOS+ and CCR8–ICOS– Tregs was performed following the manufacturer’s protocol using the Quick-RNA Microprep kit (Zymo Research). RNA quality control was performed with the Agilent 2200 Tape Station system, and only RNAs having a RIN >8 were used for library preparation. Libraries for mRNA sequencing were prepared starting from 1.5 ng total RNA for each sample by using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech-Takara). All samples were sequenced on an Illumina NextSeq 500 at an average of 32.9 million 75-bp single-end reads. After quality control, raw reads were aligned to the human genome (GRCh38.p12) using the STAR aligner with default parameters (version 2.7.0) (58). Gene-based read counts were then obtained using HTSeq count (59) module (version 0.11) and GENCODE v29.gtf annotation (60). The read counts were imported into R statistical software, and differential gene expression analysis was performed using the edgeR package (version 3.22) (61). For pair-wise comparisons, raw read counts were normalized using the TMM method (trimmed mean of log-ratio values) (62), and genes that failed to achieve a counts per million (CPM) mapped reads value greater than 1 in at least 2 libraries were not considered. P values were adjusted using the Benjamini-Hochberg method. Genes were considered differentially expressed when FDR < 0.05 and when they had an expression change of more than 1 log2 fold change. The heatmap representing the log2 of CPM was obtained using pheatmap R package (version 1.0.12) with the distance method “correlation” for both rows and columns.
Gene signature identification (RNA-seq).
GSEA was performed using GSEA (version 3.0) software (Broad Institute of MIT) and gene list ranked based on log2 fold changes. The gene set enrichment analysis was conducted in preranked mode with scoring scheme “classic” and 1,000 permutations. The maximum gene set size was fixed at 5,000 genes, and the minimum size was fixed at 10 genes. The gene signature was retrieved from the H collection (h.all.v6.2.symbols.gmt) of the Molecular Signatures Database (MSigDB v6.2). The GSEA in Supplemental Figure 3B was performed with custom gene sets relevant to immunological signatures (https://github.com/luglilab/ProjectScripts_Treg_Irf4/blob/master/Figure3S/Mouse_ImmunePath_February_01_2019_symbol.gmt). The dot plot was generated with a custom Rscript (https://github.com/luglilab/ProjectScripts_Treg_Irf4/blob/master/Figure3S/ImmunoSignatureDotPlot.r).
Motif enrichment analysis.
The PScan software tool (version 1.5) was used to perform the in silico computational analysis of overrepresented TF binding sites within the 5′-promoter regions of DEGs (63). PScan was run on [–950, +50] bp upstream regions onto the Homo Sapiens JASPAR 2018_NR database (64). Results were summarized with a scatter plot, where P values were plotted against Z score on vertical axis by using the Python Matplotlib package (version 3.0.3).
Bioinformatic analysis of microarray data.
Microarray probe fluorescence signals downloaded from the Gene Expression Omnibus (GEO GSE89656 and GSE61077, samples GSM1496276, GSM1496277, GSM1496274, GSM1496275) were converted to expression values using robust multiarray average procedure RMA (65) of the Bioconductor Affy package. Fluorescence intensities were background adjusted and normalized using quantile normalization, and expression values were calculated using median polish summarization and custom chip definition files for a total of 18,075 custom probe sets for Mouse Genome 430 2.0 Array based on Entrez genes (Mouse4302_Mm_ENTREZG version 21.0.0) and 12,426 custom probe sets for Mouse Genome 430A 2.0 Array based on Entrez genes (Mouse430A2_Mm_ENTREZG version 21.0.0). To identify genes that are differentially expressed, we compared the expression profiles of Batf−/− Tregs and WT Tregs, using limma algorithm coded in the same R package (66). All data analyses were performed in R (version 3.4.4) using Bioconductor libraries and R statistical packages.
Bioinformatic analysis of ChIP-seq data.
Raw data were downloaded from the Gene Expression Omnibus (GEO GSE98264). Reads were aligned to mouse genome GRCm38.p6/mm10 using Bowtie2 (version 2.1.0) (67) in local alignment mode. After alignment to the reference genome, mitochondrial and ambiguously mapped reads were discarded with Samtools (68) and further used for sorting and indexing mapping files. Bigwig files for IGV (69) genome coverage visualization were generated with the multiBamSummary module from the deepTools suite (version 3.2.0) (70). To call peaks, we used MACS2 (version 2.1.2) (71) with these parameters: callpeak gsize mm nomodel extsize 147 and Q value 1 × 10–3. Peaks within 30 kb upstream and 10 kb downstream of the TSS or within intragenic regions were annotated with the closest TSS using ChIPSeeker (version 1.18.0) (72) and GENCODE M20.gtf gene annotation.
Survival analysis.
Transcriptomic and clinical data of human lung adenocarcinoma, hepatocellular carcinoma, and melanoma from TCGA database (provisional cohorts) were obtained from the cBioPortal platform. Signatures of CCR8+ICOS+ Tregs (Figure 2B) and CD8+ T cells (see “Gene signature identification”) were used to calculate patient-specific enrichment scores from specimens of lung adenocarcinoma (LUAD), hepatocellular carcinoma (LIHC) and melanoma (SKCM) data sets (“GSVA” R package) (73). Survival curves were calculated between groups of patients subdivided according to the percentile rank (set at 0.8) of the resulting scores. The R packages “survival” (http://cran.rproject.org/web/packages/survival/index.html) and “survminer” (http://cran.r-project.org/web/packages/survminer/index.html) were used to assess statistics and obtain survival curves.
Data availability.
The gene expression data generated in this study are available in the GEO database (GSE128822).
Statistics.
Statistical analyses were performed using GraphPad Prism (version 7), unless specified otherwise. Data were first tested for normal distribution with D’Agostino-Pearson, Shapiro-Wilk, or Kolmogorov-Smirnov normality tests and then analyzed with Student’s t test (paired, 2 tailed), Wilcoxon rank test (paired nonparametric data), or Mann-Whitney (unpaired nonparametric data) when comparing 2 groups or Friedman test or 2-way ANOVA with Bonferroni’s post hoc test for multiple comparisons when comparing 3 groups (specific tests are specified in the figure legends).
Correlation of gene expression from scRNA-seq data was determined by calculating the Pearson correlation coefficient. P values of less than or equal to 0.05 were considered significant. PCA was performed by the prcomp function of the R stats package. Hypergeometric tests between Irf4 or Batf DEGs or scRNA-seq signature (25) and the murine tumor-infiltrating Treg signature (Figure 3B) were run with GeneOverlap R package.
Study approval.
All human experiments were approved by the Humanitas Clinical and Research Center Internal Review Board (approval 1501). All patients provided written informed consent. All mice were maintained and used in accordance with the guidelines of the University of Melbourne Animal Ethics Committee.
Author contributions
GA, J. Brummelman, SP, EMCM, EPT, CP, AV, and J. Blume performed experiments. FSC provided expertise with cell sorting. VZ, CP, and AS provided technical support. MA, PN, and GV performed surgery. AL, E. Lopci, and GV provided clinical information and helped with clinical interpretation of the data. RR, MK, and MP provided expertise in data analysis. GA, J. Brummelman, SP, AK, and E. Lugli conceived the study. E. Lugli supervised the study. GA and E. Lugli wrote the paper. All authors edited the paper. GA, J. Brummelman, and SP share first authorship. GA and J. Brummelman provided equal contribution in conceiving the overall study and in performing the vast majority of the experiments, while SP conceived and performed all the bioinformatic analyses. Given the importance of bioinformatics in this manuscript, these authors deserve equal contribution. The order of the first authors reflects the leadership exerted in the study.
Supplementary Material
Acknowledgments
We wish to thank Elisa Dieci (Humanitas Clinical and Research Center) for helping with patients’ information, Gianluca Rotta and Jens Fleischer (BD Biosciences) for helping with FACSymphony A5 instrument setup, as well as Silvia Piconese (University of Rome La Sapienza), Gioacchino Natoli (European Institute of Oncology), and the members of the Laboratory of Translational Immunology for critical discussion. This work has been supported by grants from the Associazione Italiana per la Ricerca sul Cancro (investigator grant 20676 to E. Lugli and My First AIRC Grant 189323 to E. Lopci), Bristol-Myers Squibb (preclinical grant CA209-9YM to E. Lugli), and the Humanitas Clinical and Research Center. J. Brummelman is a recipient of the “Fondo di beneficenza Intesa San Paolo” fellowship from AIRC. EMCM and PN are recipients of the 2017 Fondazione Umberto Veronesi postdoctoral fellowship. Purchase of the BD FACSymphony A5 has been in part defrayed by a grant from Italian Ministry of Health (agreement 82/2015). AK is supported by grants and a fellowship from the National Health and Medical Research Council of Australia. J. Blume was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
Version 1. 03/03/2020
In-Press Preview
Version 2. 05/11/2020
Electronic publication
Version 3. 06/01/2020
Print issue publication
Version 4. 10/28/2020
Affiliation corrected for Agnese Losurdo
Footnotes
Conflict of interest: The Laboratory of Translational Immunology receives reagents in-kind from BD Biosciences Italy as part of a collaborative research agreement.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(6):3137–3150.https://doi.org/10.1172/JCI130426.
Contributor Information
Giorgia Alvisi, Email: giorgia.alvisi@humanitasresearch.it.
Jolanda Brummelman, Email: Jolanda.Brummelman@radboudumc.nl.
Simone Puccio, Email: simone.puccio@humanitasresearch.it.
Elisa Paoluzzi Tomada, Email: elisa.paoluzzitomada@st.hunimed.eu.
Agnese Losurdo, Email: agnese.losurdo@cancercenter.humanitas.it.
Veronica Zanon, Email: Veronica.Zanon@humanitasresearch.it.
Clelia Peano, Email: clelia.peano@humanitasresearch.it.
Federico S. Colombo, Email: Federico_Simone.Colombo@humanitasresearch.it.
Alice Scarpa, Email: alice.scarpa@humanitasresearch.it.
Marco Alloisio, Email: marco.alloisio@hunimed.eu.
Ajithkumar Vasanthakumar, Email: vasanthakumar@wehi.edu.au.
Rahul Roychoudhuri, Email: rahul.roychoudhuri@babraham.ac.uk.
Marinos Kallikourdis, Email: marinos.kallikourdis@gmail.com.
Massimiliano Pagani, Email: pagani@ingm.org.
Egesta Lopci, Email: egesta.lopci@cancercenter.humanitas.it.
Pierluigi Novellis, Email: Novellis.pierluigi@hsr.it.
Jonas Blume, Email: jonas.blume@unimelb.edu.au.
Axel Kallies, Email: kallies@wehi.edu.au.
Giulia Veronesi, Email: veronesi.giulia@hsr.it.
Enrico Lugli, Email: enrico.lugli@humanitasresearch.it.
References
- 1.Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16(10):599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27(1):109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 4.Larmonier N, et al. Tumor-derived CD4(+)CD25(+) regulatory T cell suppression of dendritic cell function involves TGF-beta and IL-10. Cancer Immunol Immunother. 2007;56(1):48–59. doi: 10.1007/s00262-006-0160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ML, et al. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci USA. 2005;102(2):419–424. doi: 10.1073/pnas.0408197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450(7169):566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 7.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204(6):1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman WJ, Verbsky JW, Barchet W, Colonna M, Atkinson JP, Ley TJ. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21(4):589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8(7):523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol. 2015;33:101–111. doi: 10.1016/j.coi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Wei WZ, et al. Concurrent induction of antitumor immunity and autoimmune thyroiditis in CD4+ CD25+ regulatory T cell-depleted mice. Cancer Res. 2005;65(18):8471–8478. doi: 10.1158/0008-5472.CAN-05-0934. [DOI] [PubMed] [Google Scholar]
- 12.Dannull J, et al. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest. 2005;115(12):3623–3633. doi: 10.1172/JCI25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colombo MP, Piconese S. Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat Rev Cancer. 2007;7(11):880–887. doi: 10.1038/nrc2250. [DOI] [PubMed] [Google Scholar]
- 14.Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529(7587):532–536. doi: 10.1038/nature16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Simone M, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45(5):1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol. 2013;34(2):74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12(4):304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 21.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490(7421):543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tussiwand R, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490(7421):502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akimova T, et al. Human lung tumor FOXP3+ Tregs upregulate four “Treg-locking” transcription factors. JCI Insight. 2017;2(16):94075. doi: 10.1172/jci.insight.94075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med. 2018;215(10):2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HJ, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350(6258):334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei X, et al. Reciprocal expression of IL-35 and IL-10 defines two distinct effector Treg subsets that are required for maintenance of immune tolerance. Cell Rep. 2017;21(7):1853–1869. doi: 10.1016/j.celrep.2017.10.090. [DOI] [PubMed] [Google Scholar]
- 30.Roychoudhuri R, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498(7455):506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman NM, et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat Commun. 2018;9(1):2095. doi: 10.1038/s41467-018-04392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilipow K, et al. Antioxidant metabolism regulates CD8+ T memory stem cell formation and antitumor immunity. JCI Insight. 2018;3(18):122299. doi: 10.1172/jci.insight.122299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirotsu Y, Hataya N, Katsuoka F, Yamamoto M. NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1β genes. Mol Cell Biol. 2012;32(14):2760–2770. doi: 10.1128/MCB.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasanthakumar A, et al. The TNF receptor superfamily-NF-κB axis is critical to maintain effector regulatory T cells in lymphoid and non-lymphoid tissues. Cell Rep. 2017;20(12):2906–2920. doi: 10.1016/j.celrep.2017.08.068. [DOI] [PubMed] [Google Scholar]
- 36.Magnuson AM, et al. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci USA. 2018;115(45):E10672–E10681. doi: 10.1073/pnas.1810580115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayatsu N, et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity. 2017;47(2):268–283.e9. doi: 10.1016/j.immuni.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Kallikourdis M, Andersen KG, Welch KA, Betz AG. Alloantigen-enhanced accumulation of CCR5+ ‘effector’ regulatory T cells in the gravid uterus. Proc Natl Acad Sci USA. 2007;104(2):594–599. doi: 10.1073/pnas.0604268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brummelman J, et al. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat Protoc. 2019;14(7):1946–1969. doi: 10.1038/s41596-019-0166-2. [DOI] [PubMed] [Google Scholar]
- 40.Levine JH, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, He Y, Hao J, Ni L, Dong C. High levels of eomes promote exhaustion of anti-tumor CD8+ T cells. Front Immunol. 2018;9:2981. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39(1):11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15(11):1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glasmacher E, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338(6109):975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koizumi SI, et al. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat Commun. 2018;9(1):5344. doi: 10.1038/s41467-018-07735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer TS, Sanders LK, Nathans D. Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92(20):9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A, et al. IL-33 signaling alters regulatory T cell diversity in support of tumor development. Cell Rep. 2019;29(10):2998–3008.e8. doi: 10.1016/j.celrep.2019.10.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, et al. Connecting blood and intratumoral Treg cell activity in predicting future relapse in breast cancer. Nat Immunol. 2019;20(9):1220–1230. doi: 10.1038/s41590-019-0429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavalleri T, et al. Combined low densities of FoxP3+ and CD3+ tumor-infiltrating lymphocytes identify stage II colorectal cancer at high risk of progression. Cancer Immunol Res. 2019;7(5):751–758. doi: 10.1158/2326-6066.CIR-18-0661. [DOI] [PubMed] [Google Scholar]
- 52.Di Pilato M, et al. Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature. 2019;570(7759):112–116. doi: 10.1038/s41586-019-1215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lugli E, Zanon V, Mavilio D, Roberto A. FACS analysis of memory T lymphocytes. Methods Mol Biol. 2017;1514:31–47. doi: 10.1007/978-1-4939-6548-9_3. [DOI] [PubMed] [Google Scholar]
- 54.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittrücker HW, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275(5299):540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 56.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37(Web Server issue):W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 66.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Wilks C, Antonescu V, Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019;35(3):421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 73.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data generated in this study are available in the GEO database (GSE128822).