Abstract
Hypertension (HTN) is a significant risk factor for cardiovascular morbidity and mortality. Metabolic abnormalities, including adverse cholesterol and triglycerides (TG) profiles, are frequent comorbid findings with HTN and contribute to cardiovascular disease. Diuretics, which are used to treat HTN and heart failure, have been associated with worsening of fasting lipid concentrations. Genome-wide meta-analyses with 39,710 European-ancestry (EA) individuals and 9,925 African-ancestry (AA) individuals were performed to identify genetic variants that modify the effect of loop or thiazide diuretic use on blood lipid concentrations. Both longitudinal and cross-sectional data were used to compute cohort-specific interaction results, which were then combined through meta-analysis in each ancestry. These ancestry-specific results were further combined through trans-ancestry meta-analysis. Analysis of EA data identified two genome-wide significant (p < 5×10−8) loci with single nucleotide variant (SNV)-loop diuretic interaction on TG concentrations (including COL11A1). Analysis of AA data identified one genome-wide significant locus adjacent to BMP2 with SNV-loop diuretic interaction on TG concentrations. Trans-ancestry analysis strengthened evidence of association for SNV-loop diuretic interaction at two loci (KIAA1217 and BAALC). There were few significant SNV-thiazide diuretic interaction associations on TG concentrations and for either diuretic on cholesterol concentrations. Several promising loci were identified that may implicate biologic pathways that contribute to adverse metabolic side effects from diuretic therapy.
Introduction
Hypertension (HTN) is a significant risk factor for cardiovascular morbidity and mortality. However, even after accounting for the beneficial effects of blood pressure reduction on cardiovascular events, cardiovascular risk remains elevated despite intensive lowering of blood pressure.1 Metabolic abnormalities, including adverse cholesterol and triglycerides (TG) profiles, are frequent comorbid findings with HTN and may contribute to the residual cardiovascular disease (CVD) risk associated with antihypertensive therapy. Thiazide diuretics are recommended as first-line therapy for the treatment of HTN.2 Loop diuretics are a mainstay in the treatment of heart failure and are frequently used in patients with HTN complicated by renal insufficiency to control volume retention. However, there is little evidence to support a survival benefit with the use of loop diuretics.3 Both thiazide and loop diuretics have been associated with worsening fasting lipid concentrations,4–6 which may dampen the salutary effects of antihypertensive therapy in some patients. While there have been studies identifying genetic sources of inter-individual blood pressure response to primarily thiazide diuretic therapy,7–11 there is little literature regarding the interactions of diuretics with genetic variants on blood lipid concentrations.12
A meta-analysis of 56 randomized, placebo-controlled trials of diuretic monotherapy for the treatment of HTN, including both European-ancestry (EA) and African-ancestry (AA) individuals, showed that diuretic therapy was associated with average changes of 0.29 mmol/L for total cholesterol (TC), 0.24 mmol/L for low-density lipoprotein cholesterol (LDL), −0.02 mmol/L for high-density lipoprotein cholesterol (HDL), and 0.35 mmol/L for TG.13 Some long-term studies have suggested that effects of diuretics on lipid concentrations waned after one year.14,15 A retrospective analysis of the Systolic Hypertension in the Elderly Program (SHEP) showed persistent, albeit more modest, increases in lipid concentrations (three-year change in TG: 0.28 ± 0.86 mmol/L with chlorthalidone vs. 0.09 ± 0.71 mmol/L with placebo, P<0.001) when a cohort was randomized to a thiazide-like diuretic or a placebo;16 however, these results may have been affected by the use of non-diuretic antihypertensive drugs (atenolol and reserpine). Notably, the larger standard deviations suggest that an underlying genetic component may explain the high variability in individual’s lipid response to diuretic therapy.
Although the genetic underpinnings of fasting cholesterol and TG concentrations have been well-described, single nucleotide variants (SNVs) identified through genome-wide association studies (GWAS) explain only ~10–12% of heritability.17 Some studies have shown that the response of lipid concentrations to diuretics is significantly greater in AA than in EA individuals,13 further suggesting the modulating effect of genetic architecture. Unfortunately, most long-term studies were based on either almost exclusively EA subjects15 or studies including individuals of multiple ancestries where response by ancestry was not characterized.14 The purpose of this investigation was to determine whether common genetic variants modify the effect of diuretic use on blood lipid concentrations in persons of European and African ancestry. Exposure to thiazide and loop diuretics are considered separately since their mechanisms action differ; thiazide diuretics inhibit the sodium-chloride cotransporter (SLC12A3) in the renal distal convoluted tubule whereas loop diuretics inhibit the sodium-potassium-chloride cotransporter (SLC17A3) in the thick ascending loop of Henle.18 These analyses may identify biologic pathways that contribute to adverse metabolic side effects from diuretic therapy.
Materials and Methods
Study samples, phenotype, and genotype data
Data from 14 EA and 7 AA cohorts was used. Table 1 and Supplementary Table 1 provide summary characteristics of these cohorts; a detailed description is provided in the Supplementary Materials. Each study obtained informed consent from participants and approval from the appropriate institutional review boards. Genotyping was performed using Illumina (San Diego, CA, USA) or Affymetrix (Santa Clara, CA, USA) genotyping arrays. All studies performed imputation using HapMap Phase II data (release 22, build 36) except for the WHI cohorts which used the 1000 Genomes Project19 data. Imputation software packages used included MACH,20 Minimac,21 IMPUTE2,22 or BEAGLE23. Information on genotype and imputation for each study is presented in Supplementary Table 2.
Table 1.
Baseline characteristics of 21 participating cohorts of European and African ancestry. Seventeen cohorts were used for loop diuretic analysis.
| Ancestry | Study | N | Loop Exposed Subjects |
Thiazide Exposed Subjects |
TG (Mean & SD) |
|---|---|---|---|---|---|
| European | AGES | 1,677 | 153 | 466 | 1.17 (0.57) |
| ARIC | 8,881 | 179 | 1,179 | 1.55 (1.03) | |
| CHS | 3,174 | 218 | 838 | 1.58 (0.85) | |
| FHS | 7,068 | 163 | 567 | 1.48 (1.17) | |
| Health ABC | 1,828 | 130 | 329 | 1.73 (1.00) | |
| HVH1 | 440 | 51 | 167 | 2.25 (1.29) | |
| HVH2 | 192 | 21 | 80 | 2.36 (1.96) | |
| HyperGEN | 1,187 | 44 | 196 | 1.87 (1.31) | |
| MESA | 2,359 | NA | 563 | 1.49 (1.02) | |
| PROSPER | 4,592 | 618 | 1,402 | 1.52 (0.70) | |
| RS1 | 3,421 | 269 | 405 | 1.38 (0.61) | |
| RS2 | 2,096 | 86 | 147 | 1.40 (0.64) | |
| WHI CT GARNET Baseline | 1,222 | 24 | 142 | 1.65 (0.86) | |
| WHI CT GARNET Core | 861 | 15 | 78 | 1.66 (0.81) | |
| African | ARIC | 2,119 | 45 | 655 | 1.26 (0.86) |
| CHS | 709 | 104 | 250 | 1.30 (0.71) | |
| HyperGEN | 1,110 | 86 | 278 | 1.17 (0.77) | |
| JHS | 1,500 | 91 | 381 | 1.20 (0.96) | |
| WHI CT SHARe Baseline | 3,636 | 150 | 752 | 1.26 (0.76) | |
| WHI CT SHARe Core | 801 | 34 | 176 | 1.35 (0.64) | |
| WHI OS SHARe Baseline | 3,628 | 206 | 843 | 1.27 (0.77) |
BMI, body mass index (kg/m2); DBP, diastolic blood pressure (mmHg); SBP, systolic blood pressure (mmHg); TG, triglycerides (mmol/L).
In total, 49,635 subjects over 18 years of age with genotype, phenotype, and covariate information were available in this analysis. Both longitudinal and cross-sectional studies were used, and all available data for each subject was used. In longitudinal cohorts that had multiple clinic visits for each subject, those multiple measurements (obtained across clinic visits) were used in the analysis. In cross-sectional studies that had only a single visit for each subject, a single measurement was used. Phlebotomy was performed to measure blood lipids after a minimum 8-hour fast as per individual study protocols. Three fasting lipid traits were considered for analyses: TG, HDL, and LDL (all mmol/L). LDL was calculated via the Friedewald equation (LDL=TC-HDL- [TG/5]) for those with TG ≤4.52 mmol/L.24 If TG >4.52 mmol/L, LDL was set to missing unless directly assayed. LDL concentrations were adjusted for statin use as described elsewhere.25 TG concentrations were natural log-transformed for analysis.
Two patterns of diuretic exposure were used: 1) thiazide and thiazide-like diuretic use (yes/no) without use of loop; and 2) loop diuretic use (yes/no) without use of thiazide or thiazide-like diuretics. For each diuretic exposure, the unexposed group consisted of individuals taking neither diuretic. Drug use (yes/no) was defined as the use of any drug in the drug class, regardless of dosage, and with or without the use of a potassium supplement, as determined by self-report and/or inventory of medication bottles at each clinic visit. Examples of thiazide diuretic, loop diuretic, and statin medications are listed in Supplementary Table 3.
Statistical analyses
To determine the association of SNV-diuretic interactions on blood cholesterol and TG concentrations, the following regression model
was applied. Y is the fasting lipid value, D is the diuretic use (coded 0/1 for the absence/presence of the diuretic), G is the dosage of the imputed genetic variant coded additively (from 0 to 2), and C is the vector of all other covariates, which include age, sex, body mass index (BMI), principal components (to account for population stratification and admixture), and additional cohort-specific covariates (if any). Subjects with missing data for fasting (≥ 8 hours) lipid values, diuretic use, or any covariate were excluded from analysis. Subjects taking both thiazide and loop diuretics were also excluded from analysis.
Each study conducted GWAS analysis and provided the estimated SNV-diuretic interaction effect βGD and standard error. For longitudinal cohorts with repeated measures, generalized estimating equations (GEE) with an independent working correlation were used with an R package boss; for studies with cross-sectional data, linear regression was used. To account for relatedness in families, family studies used either GEE treating each family as a cluster using R software or the linear mixed effect model approach with a random polygenic component (for which the covariance matrix depends on the kinship matrix) using ProbABEL (Supplementary Table 2). When minor allele counts of SNVs among participants on the diuretic were small (< 10), the standardized interaction effect (βGD/SE) is not normally distributed. Note that, although a normal distribution is often appropriate in a large sample (and/or main effect analysis of GWAS), it is not appropriate in a GxE interaction study. Therefore, following our earlier work,26 cohort-specific P-values were computed based on a Student’s t-distribution. The degrees of freedom for the t-distribution depend on the number of drug-exposed participants (N_exposed), the SNV imputation quality, and the minor allele frequency. Before meta-analysis, SNVs were excluded if 2 • MAF • N_exposed • imputation quality measure < 10 to exclude unstable cohort-specific results that reflect small sample size, low MAF, or low imputation quality measures. As shown in Sitlani, et al.,27 we observed that this threshold provides a good balance between over-filtering and under-filtering.
Meta-analysis for each ancestry group was performed to combine these cohort-specific P-values with weighted Z-statistics using METAL,28 where weights were based on the number of drug-exposed subjects multiplied by the SNV imputation quality. As cohort-specific P-values were computed based on t distribution, inverse-variance weighted meta-analysis was not used. After meta-analysis, SNVs were excluded if heterogeneity P < 1×10−6 or they were available in fewer than three cohorts for EA results or two cohorts for AA results. The ancestry-specific results were further combined to perform trans-ancestry meta-analysis using MANTRA (Meta-ANalysis of TRansethnic Association studies).29 MANTRA accounts for similarity in allelic effects among closely-related populations, while allowing for heterogeneity across populations with more diverse ancestries. As MANTRA uses a Bayesian framework, a traditional fixed-effect meta-analysis with weighted Z-statistics was also performed using METAL.
Genome-wide significance was defined as P < 5×10−8 from METAL with a fixed-effect meta-analysis or Bayes Factor >106 from MANTRA. Suggestive evidence of association was defined as P < 1×10−6 from METAL or Bayes Factor >105 from MANTRA. To assess type I error due to population stratification and other factors, quantile-quantile (QQ) plots were examined for all cohort-specific GWAS results for each pair of lipid and diuretic exposure. In addition, during meta-analysis, genomic control correction30 was applied to cohort-specific GWAS results if their genomic control lambda value was greater than 1. The gene locations referenced in the text and tables were obtained from the National Center for Biotechnology Information dbSNP database (reference assembly GRCh38.p2). Functional annotation information was sought using HaploReg31 and RegulomeDB.32
Results
The European-ancestry (EA) group included 39,710 subjects from 14 cohorts; the African-ancestry (AA) group included 9,925 subjects from 7 cohorts (Table 1). The number of subjects exposed to loop diuretics was 2,117 (5.3%) in EA and 784 (7.9%) in AA; the number exposed to thiazide and thiazide-like diuretics was 6,878 (17.3%) in EA and 3,923 (39.5%) in AA.
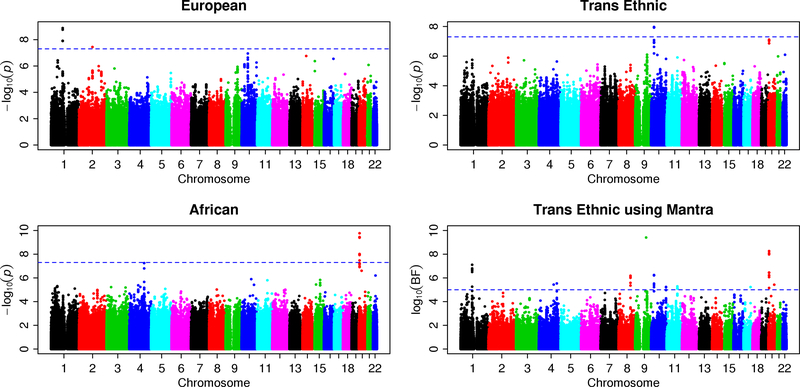
The QQ plots (Supplementary Figures 1–6) showed moderate inflation, in particular for the SNV-loop diuretic interaction terms for TG, LDL, and HDL analyses. The P-values based on a t-distribution (red crosses) that we used for our meta-analyses have better genomic control values than the P-values based on a standard normal distribution (black circles). Manhattan plots show the adjusted -log10(P) values for loop diuretics (Figures 1 and Supplementary Figures 7–8) and thiazide diuretics (Supplementary Figures 9–11) for TG, HDL, and LDL, respectively. Each figure includes plots of EA and AA separately, and for trans-ancestry meta-analysis using METAL and MANTRA. The SNVs reaching genome-wide significance (P < 5×10−8) for association with gene-medication interactions on TG concentration are shown in Table 2. Supplementary Table 4 shows the SNVs with suggestive evidence of SNV-diuretic interactions on each lipid trait (with P < 10−6 from METAL or Bayes Factor >105 from MANTRA).
Figure 1:
Manhattan plots for analysis of SNV-loop diuretic interaction on triglyceride concentrations. The ancestry-specific meta-analysis used 11 cohorts of European ancestry (upper-left panel) and 6 cohorts of African ancestry (lower-left panel). Trans-ancestry meta-analysis used fixed-effect weighted Z-statistics with METAL (upper-right panel) and a Bayesian framework with MANTRA (lower-right panel). The -log10(P) from METAL or log10(Bayes Factor) from MANTRA was plotted at the chromosomal location of each variant. Manhattan plots for the remaining lipid-diuretic pairs are shown in Supplementary Figures 7–11.
Table 2.
SNVs with genome-wide significant (with P < 5×10−8 from METAL or Bayes factor >106 from MANTRA) SNV-loop diuretic interactions on triglyceride concentrations identified from European-ancestry, African-ancestry, and trans-ancestry analyses.
| Marker | Chr | Position | Alleles | European Ancestry | African Ancestry | Trans Ancestry | Mantra | Neighboring Genes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | P | Effect | SE | Freq | P | Effect | SE | Freq | P | Effect | SE | log10BF | |||||
| rs1463034 | 1 | 103,227,805 | t/c | 0.92 | 1.91E-09 | 0.0126 | 0.0003 | 0.59 | 4.40E-01 | 0.0111 | 0.0003 | 0.83 | 2.27E-06 | 0.0121 | 0.0001 | 5.54 | COL11A1 |
| rs1870958 | 1 | 103,229,744 | t/g | 0.92 | 2.57E-09 | 0.0126 | 0.0003 | 0.59 | 5.00E-01 | 0.0111 | 0.0003 | 0.82 | 4.01E-06 | 0.0121 | 0.0001 | 6.53 | |
| rs2786125 | 1 | 103,235,061 | a/g | 0.06 | 1.94E-08 | 0.0100 | 0.0002 | 0.31 | 4.37E-01 | 0.0115 | 0.0003 | 0.14 | 2.18E-05 | 0.0105 | 0.0002 | 5.68 | |
| rs2622870 | 1 | 103,236,564 | t/g | 0.94 | 1.98E-08 | 0.0127 | 0.0003 | 0.69 | 3.40E-01 | 0.0110 | 0.0003 | 0.87 | 2.14E-05 | 0.0122 | 0.0002 | 6.14 | |
| rs2622874 | 1 | 103,239,505 | a/c | 0.06 | 1.93E-08 | 0.0100 | 0.0002 | 0.31 | 3.26E-01 | 0.0116 | 0.0003 | 0.13 | 2.31E-05 | 0.0104 | 0.0002 | 6.77 | |
| rs7607797 | 2 | 117,944,672 | t/c | 0.97 | 3.60E-08 | 0.0127 | 0.0003 | 0.66 | 4.62E-01 | 0.0111 | 0.0003 | 0.86 | 6.88E-05 | 0.0121 | 0.0002 | 3.51 | DDX18 |
| rs7002454 | 8 | 104,347,955 | t/c | 0.95 | 8.78E-05 | 0.0124 | 0.0002 | 0.87 | 3.58E-02 | 0.0122 | 0.0005 | 0.93 | 1.10E-05 | 0.0124 | 0.0002 | 6.10 | BAALC / FZD6 |
| rs6985929 | 8 | 104,348,009 | a/g | 0.95 | 8.86E-05 | 0.0124 | 0.0002 | 0.87 | 3.29E-02 | 0.0124 | 0.0005 | 0.93 | 1.02E-05 | 0.0124 | 0.0002 | 6.17 | |
| rs10508671 | 10 | 24,574,062 | t/c | 0.03 | 9.43E-07 | 0.0102 | 0.0002 | 0.25 | 2.12E-03 | 0.0103 | 0.0003 | 0.11 | 1.07E-08 | 0.0102 | 0.0002 | 6.23 | KIAA1217 / MIR603 / ARHGAP21 |
| rs10508672 | 10 | 24,574,441 | a/g | 0.03 | 9.52E-07 | 0.0102 | 0.0002 | 0.24 | 2.32E-03 | 0.0103 | 0.0003 | 0.11 | 1.18E-08 | 0.0102 | 0.0002 | 6.20 | |
| rs3852940 | 20 | 6,165,600 | a/g | 0.79 | 6.78E-01 | 0.0112 | 0.0002 | 0.70 | 1.18E-08 | 0.0132 | 0.0004 | 0.77 | 1.09E-02 | 0.0116 | 0.0001 | 5.73 | CRLS1 / LRRN4 / FERMT1 / CASC20 / BMP2 |
| rs6054016 | 20 | 6,174,368 | t/c | 0.06 | 6.06E-01 | 0.0114 | 0.0003 | 0.19 | 4.06E-10 | 0.0092 | 0.0003 | 0.10 | 2.15E-03 | 0.0103 | 0.0002 | 7.84 | |
| rs6054018 | 20 | 6,175,236 | t/g | 0.94 | 6.12E-01 | 0.0112 | 0.0003 | 0.80 | 1.70E-10 | 0.0139 | 0.0004 | 0.90 | 1.83E-03 | 0.0124 | 0.0002 | 7.60 | |
| rs8120588 | 20 | 6,178,849 | t/c | 0.94 | 6.24E-01 | 0.0112 | 0.0003 | 0.81 | 3.75E-10 | 0.0139 | 0.0004 | 0.90 | 1.88E-03 | 0.0124 | 0.0002 | 7.99 | |
| rs7348828 | 20 | 6,182,498 | a/g | 0.94 | 6.38E-01 | 0.0112 | 0.0003 | 0.81 | 3.56E-10 | 0.0139 | 0.0004 | 0.90 | 1.75E-03 | 0.0124 | 0.0002 | 6.60 | |
| rs8122198 | 20 | 6,186,686 | t/c | 0.94 | 8.99E-01 | 0.0113 | 0.0003 | 0.85 | 9.53E-09 | 0.0141 | 0.0006 | 0.91 | 2.46E-03 | 0.0122 | 0.0002 | 6.29 | |
| rs6054037 | 20 | 6,187,877 | c/g | 0.94 | 8.69E-01 | 0.0113 | 0.0003 | 0.85 | 9.94E-09 | 0.0141 | 0.0006 | 0.91 | 2.73E-03 | 0.0122 | 0.0002 | 6.43 | |
| rs6038400 | 20 | 6,188,201 | a/c | 0.06 | 8.66E-01 | 0.0113 | 0.0003 | 0.15 | 1.03E-08 | 0.0091 | 0.0004 | 0.09 | 2.78E-03 | 0.0104 | 0.0002 | 5.08 | |
| rs3852942 | 20 | 6,192,559 | a/g | 0.06 | 8.71E-01 | 0.0113 | 0.0003 | 0.17 | 3.36E-08 | 0.0093 | 0.0003 | 0.10 | 3.46E-03 | 0.0104 | 0.0002 | 4.53 | |
Covariates included in the analysis: age, sex, BMI, principal components, and additional cohort-specific covariates (if any). Abbreviations: Chr, chromosome, Freq, frequency of allele 1. Bolded genes include intragenic SNVs. Bolded P-values and MANTRA log10BF identify values that are genome-wide significant. Units for Effect (SE) is mmol/L.
Loop diuretics with blood triglyceride (TG)
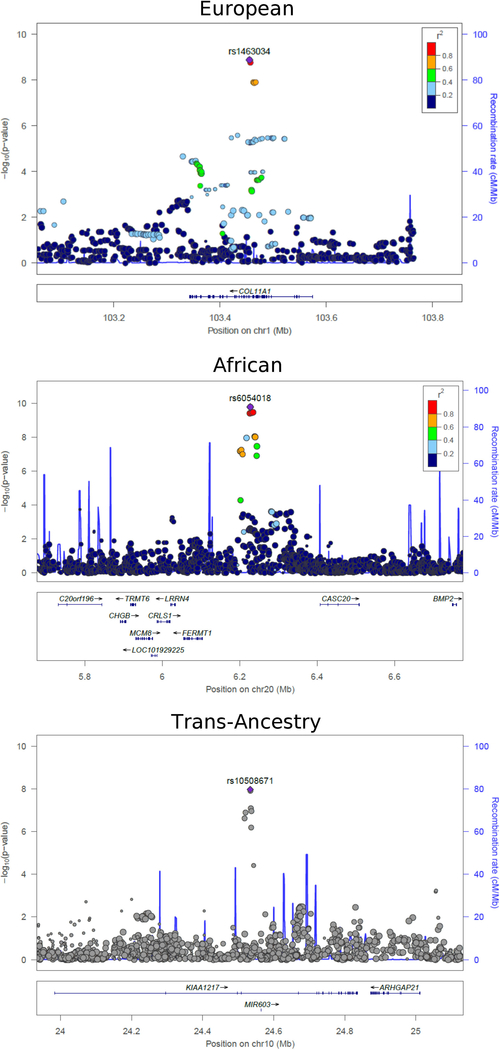
Analysis of the EA data identified two loci with genome-wide significant SNV-loop diuretic interaction effects (P < 5×10−8) on log-transformed TG concentrations (Figure 1). Another 8 loci demonstrated a suggestive association (P < 1×10−6). The locus with the strongest evidence of association included a six-SNV cluster (most significant rs1463034, P = 1.91×10−9, βGD = 0.0012 ± 0.0002 mmol/L) spanning four introns (7,256 bp) in COL11A1 on chromosome 1 (Figure 2A). A suggestive locus included a six-SNV cluster spanning a single intron (18,804 bp) on chromosome 10 in KIAA1217 (most significant rs7077598, P = 7.48×10−7). Another suggestive locus of 11 SNVs was found on chromosome 10 which is approximately 145 kb downstream of DKK1 (most significant rs10762762, P = 1.12×10−7). Within this locus, rs1441122 (P = 9.86×10−7) showed moderate evidence of altering the binding motif for the transcription factor TRIM28 in a human embryonic kidney cell line (RegulomeDB Score 3a, http://www.regulomedb.org/).
Figure 2:
Regional plots of significant SNV-loop diuretic interaction effects on triglyceride concentrations on chromosome 1 in European ancestry (top), chromosome 20 in African ancestry (middle), and chromosome 10 in trans-ancestry analyses of European and African ancestries (bottom). Plots were created using LocusZoom software (http://csg.sph.umich.edu/locuszoom/). Linkage disequilibrium (LD, r2) was based on hg19/1000 Genomes Nov 2014 EUR for EA and AFR hg19/1000 Genomes Nov 2014 for AA. Because no LD information was available for trans-ancestry results combining EA and AA results, the bottom plot does not show LD.
Analysis of the AA data identified one locus with genome-wide significant interaction effects on TG concentrations, with two additional loci with suggestive P-values (Figure 1). This 9-SNV locus (most significant rs6054018, P = 1.70×10−10, βGD = 0.0024 ± 0.0003 mmol/L; Figure 2B) is in an intergenic region on chromosome 20 approximately 500 kb upstream of BMP2 and approximately 114 kb downstream of FERMT1.
Trans-ancestry association tests strengthened evidence of association for the KIAA1217 locus, elevating two of the six SNVs in this locus from suggestive in EA to genome-wide significance. In particular, at rs10508671, the SNV-loop diuretic interaction effect (βGD) was consistent in both ancestries (−0.0011 vs −0.0010 mmol/L, Supplementary Table 4), and therefore trans-ancestry test provided stronger evidence of association (EA P = 9.43×10−7, AA P = 0.002, trans-ancestry P = 1.08×10−8, MANTRA log10[BF] = 6.23, Figure 2C). MANTRA also strengthened the association for a locus on chromosome 8 (most significant rs6985929, EA P = 8.86×10−5, trans-ancestry P = 1.02×10−5, MANTRA log10[BF] = 6.17) that overlaps an intron in BAALC and that is approximately 32 kb upstream of FZD6. Notably, the effect sizes of SNV-loop diuretic interactions is small (ranging from 0.0010–0.0025 mmol/L, Table 2), and were greater for AA than for EA or trans-ancestry analyses.
Loop diuretics with blood HDL and LDL
No loci reached genome-wide significance for SNV-loop diuretic interactions on HDL or LDL concentrations in the analyses of EA data. Suggestive association was noted with 11 SNVs in U6 (rs203202, P = 1.65×10−7) for LDL. For LDL analyses in AA (Supplementary Figure 8), one SNV on chromosome 6 approximately 35 kb 5’ of PHIP reached genome-wide significance (rs12208017, P = 3.73×10−8, βGD = 0.3538 ± 0.0638 mmol/L); this SNV is in an expression quantitative trait locus (eQTL) for both PHIP and the adjacent gene, IRAK1BP1. An additional SNV with a suggestive association (rs1312663, P = 4.06×10−7) is located approximately 100 kb upstream of SLC24A4. Trans-ancestry analyses newly identified two SNVs in two loci with suggestive interaction associations on HDL, including a SNV on chromosome 11 in TEAD1 (rs7924536, P = 5.79×10−7, MANTRA log10[BF] = 5.04).
Thiazide diuretics with blood TG, HDL and LDL
For analyses of SNV-thiazide diuretic interactions in EA and AA, no loci reached genome-wide significance for TG, HDL, or LDL (Supplementary Figures 9–11). In EA, suggestive association was noted with two loci including MYO16 (rs1926511, P = 1.01×10−7) for TG and one locus for LDL. In AA, three loci yielded suggestive associations with HDL including a seven-SNV locus on chromosome 14 in TRIP11 (most significant rs10083510, P = 3.20×10−7). SNVs in this locus are eQTLs for FBLN5.
In trans-ancestry analyses, a single intronic SNV on chromosome 20 in MACROD2 (rs6043629, P = 1.12×10−8, βGD = 0.0382 ± 0.0068 mmol/L, MANTRA log10[BF] = 6.62) reached genome-wide significance for a thiazide diuretic interaction association on HDL; the strength of the association in EA (P = 1.25×10−5) and AA (P = 1.82×10−4) cohorts analyzed separately were much lower. With analysis of SNV-thiazide interactions on LDL, four SNVs on chromosome 17 in KIAA0753 showed suggestive association in trans-ancestry (most significant rs2304976, P = 1.31×10−7, MANTRA log10[BF] = 5.34) although neither SNV approached significance in separate EA and AA analyses.
Discussion
Genome-wide meta-analyses with 39,710 European-ancestry (EA) individuals and 9,925 African-ancestry (AA) individuals were performed to identify genetic variants that modify the effect of loop or thiazide diuretic use on blood lipid concentrations. As the effects of loop and thiazide diuretic classes on lipids may differ,5,6 each with different effects on renal sodium excretion,18 analyses were performed on each of diuretic classes separately. Ancestry-specific analyses (of EA and AA data) identified genome-wide significant variants in three loci with SNV-loop diuretic interaction on TG concentrations; trans-ancestry analysis strengthened evidence of association at two additional loci.
The most significant associations with TG concentrations in EA were observed at intronic SNVs clustered in COL11A1 (collagen, type XI, alpha 1) on chromosome 1. COL11A1 produces a structural/adhesion protein secreted by primary rat adipocytes.33 The expression of this gene is up-regulated during human adipogenesis,34 suggesting a role in the modulation of lipid traits. Although it remains unclear how diuretics modulate the effect of COL11A1 variants on lipid traits, a COL11A1 mutation has been shown to be associated with nephrogenic diabetes insipidus,35 which is characterized by an inability of the kidney to concentrate urine, thus providing a possible mechanistic link between COL11A1, diuretics, and lipids.
Trans-ancestry analysis combining European and African results provided evidence of association at intronic SNVs clustered in KIAA1217 (an uncharacterized long coding DNA) which are just 5’ to an intronic microRNA (MIR603) on chromosome 10. KIAA1217 variants have been associated with obesity.36 MIR603 has been associated with benign and malignant tumors.37–40 Of potential interest, SNVs in both KIAA1217 and COL11A1 have been associated with lumbar disk herniation which, in turn, has been associated with blood lipid concentrations41 and cardiovascular risk traits and disease.42
Loci with genome-wide significant associations found in intragenic regions may impact the transcription of genes tens or hundreds of kilobases away from their target genes.43 The cluster of SNVs on chromosome 20 reaching genome-wide significance for TG in AA lies between two genes. FERMT1 (fermitin family member 1) is a membrane-associated protein that links intracellular structural proteins to the extracellular matrix. While mutations in this gene have been implicated in a heritable skin disorder,44 connections to cardiovascular traits are sparse. Perhaps more relevant is this locus’ proximity to BMP2 (bone morphogenetic protein 2), the expression of which is regulated by angiotensin II,45 a key regulator of blood pressure and fluid/electrolyte balance. Furthermore, BMP2, a member of the transforming growth factor (TGF)-β superfamily of cytokines, is expressed in endothelial cells, and has been shown to stimulate adjacent smooth muscle and endothelial cells to proliferate, differentiate, and deposit extra-cellular matrix, thus affecting blood pressure and contributing to atherosclerosis by processes that also includes Wnt signaling.46 BMP2 also participates in the differentiation of mesenchymal stem cells to mature adipocytes,47 thus potentially affecting blood lipid concentrations. Variants in the BMP2 receptor gene (BMPR2) have also been implicated in familial forms of pulmonary arterial hypertension.48
A cluster of SNVs spanning the 3’ end and intragenic region adjacent to BAALC and 5’ of FZD6 was nominally associated with TG response to loop diuretic therapy in both races; however, trans-ancestry meta-analysis by METAL and MANTRA strengthened the association. There is little evidence to link BAALC (brain and acute leukemia, cytoplasmic) with TG concentrations. However, both FZD6 (frizzled class receptor 6), which is approximately 32 kb downstream from this locus on chromosome 8, and DKK1 (dickkopf WNT signaling pathway inhibitor 1), included in a suggestive locus on chromosome 10, are negative regulars of the β-catenin/Wnt signaling cascade49,50 that not only alters the expression of genes which regulate renal sodium, chloride and potassium handling,51 but is inhibited by the loop diuretic ethacrynic acid.52 Both FZD6 and DKK1 have also been associated with renal development,53,54 glomerular damage, and proteinuria.55,56
Trans-ancestry analyses identified SNVs in TEAD1 (TEA domain family member 1 [SV40 transcriptional enhancer factor]) as significantly associated with HDL concentrations. This transcription factor is another β-catenin/Wnt pathway member57 that has been shown to modulate adipocyte differentiation and proliferation58 and play a role in regulating insulin sensitivity via skeletal muscle fiber type switching.59,60
In the analyses of SNV-loop diuretic interactions in AA data with LDL, a SNV reaching genome-wide significance adjacent to PHIP (pleckstrin homology domain interacting protein) is within 270 kb of another SNV (rs16890334) previously identified as having a genome-wide significant association with blood pressure response to a high-sodium diet intervention in Han Chinese.61
No loci from analyses of thiazide and thiazide-like diuretics with lipid traits reached genome-wide significance in EA or AA. However, trans-ancestry analyses identified a MACROD2 (MACRO domain containing 2) with genome-wide significance for a diuretic interaction effect on HDL. This gene was found to have a significant association with coronary artery disease and hypertension using a two-marker testing approach in a reanalysis of data from the Wellcome Trust Case Control Consortium,62 and a suggestive association with brain infarcts in another meta-analysis;63 however, the role of this gene in determining cardiovascular traits remain uncertain.
Limitations
Although this study identified interesting SNV-loop diuretic interactions as modulators of TG concentrations, there were limitations in this study. First, the study design assumed therapeutic class effects although individual drugs may have different effects on lipids.13,64,65 Information regarding the dosage, duration, or adequacy of diuretic therapy may have also affected the traits but were not available in most studies. Second, because potassium-sparing diuretics were frequently co-administered with thiazide and thiazide-like diuretics, it was not feasible to pursue the sole contributions of potassium-sparing diuretics in this study. Third, the effects of other classes of anti-hypertensive medications that may affect lipid levels, such as calcium, β-adrenergic, and α-adrenergic receptor blockers,66 were not assessed in this study. Fourth, the potential contribution of co-morbidities (e.g. diabetes, renal dysfunction) and lifestyle choices (e.g. alcohol consumption, physical activity) on lipid traits was not considered. Fifth, the relatively smaller number of AA subjects exposed to loop diuretics may have reduced the power to detect significant association or increased the risk for spurious findings in this ancestry group. Finally, given the number of tests performed and the relatively small effect sizes for the interactions, this effort should be construed as a discovery analysis that warrants replication.
Conclusions
This genome-wide SNV-diuretic interaction meta-analysis on blood lipid concentrations used data from 39,710 European-ancestry individuals and 9,925 African-ancestry individuals. The results of the present study suggest stronger interaction effects for loop versus thiazide diuretics, identifying several genome-wide significant loci of small effect sizes. The results of this study are typical of most GWAS studies wherein loci of small-to-modest effects are identified.67–77 Although these loci are likely not action able in terms of genotype-guided therapy, some may nonetheless be biologically and clinically relevant,78 perhaps by identifying biologic pathways that contribute to adverse metabolic side effects from diuretic therapy. These findings may also explain, at least in part, some of the residual CVD risk following reduction of blood pressure in patients treated with anti-hypertensive medications.
Supplementary Material
Acknowledgements
This work was partially supported by National Institutes of Health National Heart, Lung, Blood Institute grants R01HL103612 (BMP) and K25HL121091 (YJS). Study-specific support included in the Supplementary Material.
Footnotes
Supplementary information is available at The Pharmacogenomics Journal’s website.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Zanchetti A, Hansson L, Menard J, Leonetti G, Rahn KH, Warnold I et al. Risk assessment and treatment benefit in intensively treated hypertensive patients of the Hypertension Optimal Treatment (HOT) study. J Hypertens 2001; 19: 819–825. [DOI] [PubMed] [Google Scholar]
- 2.James PA, Oparil S, Carter BL, Cushman WC, Dennison-Himmelfarb C, Handler J et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014; 311: 507–520. [DOI] [PubMed] [Google Scholar]
- 3.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr. et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA 2003; 289: 2560–2572. [DOI] [PubMed] [Google Scholar]
- 4.Brook RD. Mechanism of differential effects of antihypertensive agents on serum lipids. Curr Hypertens Rep 2000; 2: 370–377. [DOI] [PubMed] [Google Scholar]
- 5.Cutler R. Effect of antihypertensive agents on lipid metabolism. Am J Cardiol 1983; 51: 628–631. [DOI] [PubMed] [Google Scholar]
- 6.Ames RP. The influence of non-beta-blocking drugs on the lipid profile: are diuretics outclassed as initial therapy for hypertension? Am Heart J 1987; 114: 998–1006. [DOI] [PubMed] [Google Scholar]
- 7.Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M et al. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet 1997; 349: 1353–1357. [DOI] [PubMed] [Google Scholar]
- 8.Citterio L, Lanzani C, Manunta P. Polymorphisms, hypertension and thiazide diuretics. Pharmacogenomics 2011; 12: 1587–1604. [DOI] [PubMed] [Google Scholar]
- 9.Gong Y, McDonough CW, Wang Z, Hou W, Cooper-DeHoff RM, Langaee TY et al. Hypertension susceptibility loci and blood pressure response to antihypertensives: results from the pharmacogenomic evaluation of antihypertensive responses study. Circ Cardiovasc Genet 2012; 5: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonough CW, Burbage SE, Duarte JD, Gong Y, Langaee TY, Turner ST et al. Association of variants in NEDD4L with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J Hypertens 2013; 31: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner ST, Boerwinkle E, O’Connell JR, Bailey KR, Gong Y, Chapman AB et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension 2013; 62: 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeks E, Janssen RG, Kroon AA, Keulen ET, Geurts JM, de Leeuw PW et al. Association between the alpha-adducin Gly460Trp polymorphism and systolic blood pressure in familial combined hyperlipidemia. Am J Hypertens 2001; 14: 1185–1190. [DOI] [PubMed] [Google Scholar]
- 13.Kasiske BL, Ma JZ, Kalil RS, Louis TA. Effects of antihypertensive therapy on serum lipids. Ann Intern Med 1995; 122: 133–141. [DOI] [PubMed] [Google Scholar]
- 14.Lakshman MR, Reda DJ, Materson BJ, Cushman WC, Freis ED. Diuretics and beta-blockers do not have adverse effects at 1 year on plasma lipid and lipoprotein profiles in men with hypertension. Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. Arch Intern Med 1999; 159: 551–558. [DOI] [PubMed] [Google Scholar]
- 15.Ott SM, LaCroix AZ, Ichikawa LE, Scholes D, Barlow WE. Effect of low-dose thiazide diuretics on plasma lipids: results from a double-blind, randomized clinical trial in older men and women. J Am Geriatr Soc 2003; 51: 340–347. [DOI] [PubMed] [Google Scholar]
- 16.Savage PJ, Pressel SL, Curb JD, Schron EB, Applegate WB, Black HR et al. Influence of long-term, low-dose, diuretic-based, antihypertensive therapy on glucose, lipid, uric acid, and potassium levels in older men and women with isolated systolic hypertension: The Systolic Hypertension in the Elderly Program. SHEP Cooperative Research Group. Arch Intern Med 1998; 158: 741–751. [DOI] [PubMed] [Google Scholar]
- 17.Maitland-van der Zee AH, Turner ST, Schwartz GL, Chapman AB, Klungel OH, Boerwinkle E. Demographic, environmental, and genetic predictors of metabolic side effects of hydrochlorothiazide treatment in hypertensive subjects. Am J Hypertens 2005; 18: 1077–1083. [DOI] [PubMed] [Google Scholar]
- 18.Jentzer JC, DeWald TA, Hernandez AF. Combination of loop diuretics with thiazide-type diuretics in heart failure. J Am Coll Cardiol 2010; 56: 1527–1534. [DOI] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium, Abecasis GR, Auton A Brooks LD, DePristo MA Durbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009; 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972; 18: 499–502. [PubMed] [Google Scholar]
- 25.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet 2014; 94: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avery CL, Sitlani CM, Arking DE, Arnett DK, Bis JC, Boerwinkle E et al. Drug-gene interactions and the search for missing heritability: a cross-sectional pharmacogenomics study of the QT interval. Pharmacogenomics J 2014; 14: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitlani CM, Rice KM, Lumley T, McKnight B, Cupples LA, Avery CL et al. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat Med 2015; 34: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 2011; 35: 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999; 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 31.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res 2008; 7: 1251–1263. [DOI] [PubMed] [Google Scholar]
- 34.Hung SC, Chang CF, Ma HL, Chen TH, Low-Tone Ho L. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene 2004; 340: 141–150. [DOI] [PubMed] [Google Scholar]
- 35.Kamsteeg EJ, Wormhoudt TA, Rijss JP, van Os CH, Deen PM. An impaired routing of wild-type aquaporin-2 after tetramerization with an aquaporin-2 mutant explains dominant nephrogenic diabetes insipidus. EMBO J 1999; 18: 2394–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang KS, Liu X, Zheng S, Zeng M, Pan Y, Callahan K. A novel locus for body mass index on 5p15.2: a meta-analysis of two genome-wide association studies. Gene 2012; 500: 80–84. [DOI] [PubMed] [Google Scholar]
- 37.Kushwaha D, Ramakrishnan V, Ng K, Steed T, Nguyen T, Futalan D et al. A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget 2014; 5: 4026–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mussnich P, D’Angelo D, Leone V, Croce CM, Fusco A. The High Mobility Group A proteins contribute to thyroid cell transformation by regulating miR-603 and miR-10b expression. Mol Oncol 2013; 7: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, Raverot G et al. Altered microRNA expression profile in human pituitary GH adenomas: down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab 2012; 97: E1128–1138. [DOI] [PubMed] [Google Scholar]
- 40.Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J et al. Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One 2012; 7: e31241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longo UG, Denaro L, Spiezia F, Forriol F, Maffulli N, Denaro V. Symptomatic disc herniation and serum lipid levels. Eur Spine J 2011; 20: 1658–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jhawar BS, Fuchs CS, Colditz GA, Stampfer MJ. Cardiovascular risk factors for physician-diagnosed lumbar disc herniation. Spine J 2006; 6: 684–691. [DOI] [PubMed] [Google Scholar]
- 43.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev 2009; 19: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel DH, Ashton GH, Penagos HG, Lee JV, Feiler HS, Wilhelmsen KC et al. Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellular-matrix linker protein UNC-112, causes Kindler syndrome. Am J Hum Genet 2003; 73: 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaffe IZ, Mendelsohn ME. Angiotensin II and aldosterone regulate gene transcription via functional mineralocortocoid receptors in human coronary artery smooth muscle cells. Circ Res 2005; 96: 643–650. [DOI] [PubMed] [Google Scholar]
- 46.Al-Aly Z. Arterial calcification: a tumor necrosis factor-alpha mediated vascular Wnt-opathy. Transl Res 2008; 151: 233–239. [DOI] [PubMed] [Google Scholar]
- 47.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 2008; 454: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Rivas G, Jerjes-Sanchez C, Rodriguez D, Garcia-Pelaez J, Trevino V. A systematic review of genetic mutations in pulmonary arterial hypertension. BMC Med Genet 2017; 18: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golan T, Yaniv A, Bafico A, Liu G, Gazit A. The human Frizzled 6 (HFz6) acts as a negative regulator of the canonical Wnt. beta-catenin signaling cascade. J Biol Chem 2004; 279: 14879–14888. [DOI] [PubMed] [Google Scholar]
- 50.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 2001; 411: 321–325. [DOI] [PubMed] [Google Scholar]
- 51.Kahle KT, Wilson FH, Leng Q, Lalioti MD, O’Connell AD, Dong K et al. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet 2003; 35: 372–376. [DOI] [PubMed] [Google Scholar]
- 52.Lu D, Liu JX, Endo T, Zhou H, Yao S, Willert K et al. Ethacrynic acid exhibits selective toxicity to chronic lymphocytic leukemia cells by inhibition of the Wnt/beta-catenin pathway. PLoS One 2009; 4: e8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyons JP, Miller RK, Zhou X, Weidinger G, Deroo T, Denayer T et al. Requirement of Wnt/beta-catenin signaling in pronephric kidney development. Mech Dev 2009; 126: 142–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pietila I, Ellwanger K, Railo A, Jokela T, Barrantes Idel B, Shan J et al. Secreted Wnt antagonist Dickkopf-1 controls kidney papilla development coordinated by Wnt-7b signalling. Dev Biol 2011; 353: 50–60. [DOI] [PubMed] [Google Scholar]
- 55.Dai C, Stolz DB, Kiss LP, Monga SP, Holzman LB, Liu Y. Wnt/beta-catenin signaling promotes podocyte dysfunction and albuminuria. J Am Soc Nephrol 2009; 20: 1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato H, Susztak K. Repair problems in podocytes: Wnt, Notch, and glomerulosclerosis. Semin Nephrol 2012; 32: 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 2014; 158: 157–170. [DOI] [PubMed] [Google Scholar]
- 58.An Y, Kang Q, Zhao Y, Hu X, Li N. Lats2 modulates adipocyte proliferation and differentiation via hippo signaling. PLoS One 2013; 8: e72042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou D, Strakovsky RS, Zhang X, Pan YX. The skeletal muscle Wnt pathway may modulate insulin resistance and muscle development in a diet-induced obese rat model. Obesity (Silver Spring) 2012; 20: 1577–1584. [DOI] [PubMed] [Google Scholar]
- 60.Tsika RW, Schramm C, Simmer G, Fitzsimons DP, Moss RL, Ji J. Overexpression of TEAD-1 in transgenic mouse striated muscles produces a slower skeletal muscle contractile phenotype. J Biol Chem 2008; 283: 36154–36167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He J, Kelly TN, Zhao Q, Li H, Huang J, Wang L et al. Genome-wide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet 2013; 6: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slavin TP, Feng T, Schnell A, Zhu X, Elston RC. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet 2011; 130: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Debette S, Bis JC, Fornage M, Schmidt H, Ikram MA, Sigurdsson S et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke 2010; 41: 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weidmann P, Uehlinger DE, Gerber A. Antihypertensive treatment and serum lipoproteins. J Hypertens 1985; 3: 297–306. [DOI] [PubMed] [Google Scholar]
- 65.Ferrari P, Weidmann P. Lipoproteins during antihypertensive therapy. Study supported by the Swiss National Science Foundation. S Afr Med J 1989; Suppl: 13–17. [PubMed] [Google Scholar]
- 66.Krone W, Nagele H. Effects of antihypertensives on plasma lipids and lipoprotein metabolism. Am Heart J 1988; 116: 1729–1734. [DOI] [PubMed] [Google Scholar]
- 67.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008; 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 2009; 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A et al. Genome-wide association study of blood pressure and hypertension. Nat Genet 2009; 41: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L et al. Genome-wide association study identifies eight loci associated with blood pressure. Nature genetics 2009; 41: 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Human molecular genetics 2011; 20: 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet 2011; 43: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics 2011; 43: 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo Y, Tomlinson B, Chu T, Fang YJ, Gui H, Tang CS et al. A genome-wide linkage and association scan reveals novel loci for hypertension and blood pressure traits. PLoS One 2012; 7: e31489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet 2017; 49: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nature genetics 2016; 48: 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andaleon A, Mogil LS, Wheeler HE. Gene-based association study for lipid traits in diverse cohorts implicates BACE1 and SIDT2 regulation in triglyceride levels. PeerJ 2018; 6: e4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.