Abstract
Genetic polymorphisms have been shown to affect opioid requirement for pain relief. However, true genetic effect is often difficult to assess due to underlying pain conditions and placebo effects. The goal of this study was to understand how common polymorphisms affect opioid effects while controlling for these factors. A randomized, double-blind, placebo-controlled study was implemented to assess how opioid effects are modulated by COMT (rs6269, rs4633, rs4848, rs4680), OPRM1 (A118G) and OPRK1 (rs1051660, rs702764, rs16918875). 108 healthy subjects underwent experimental pain testing before and after morphine, butorphanol, and placebo (saline). Association analysis was performed between polymorphisms/haplotypes and opioid response, while correcting for race, gender, placebo effects and multiple comparisons. Pressure pain was significantly associated with rs6269 and rs4633 following butorphanol. The AA genotype of rs4680 or A_T_C_A/ A_T_C_A (rs6269_rs4633_ rs4818_rs4680) diplotype of COMT, combined with the AG genotype of OPRM1 A118G, showed significantly increased pressure pain threshold from butorphanol. Opioid effects on pressure, ischemic, heat pain and side effects were nominally associated with several SNPs and haplotypes. Effects were often present in one opioid but not the other. This indicates that these polymorphisms affect pain relief from opioids, and that their effects are opioid and pain modality specific.
Introduction:
Opioids are commonly used for not only surgical, but for non-surgical and often for chronic pain management. To date, there are over 30 opioids marketed worldwide for such purposes. Notably, there is tremendous inter-individual variation in response to opioid analgesia. For example, the morphine usage to achieve effective analgesia can vary five to tenfold for postoperative pain(1, 2). With such a wide interpersonal variability, the current practice of trial and error in opioid analgesia can result in insufficient analgesia, intolerable side effects(3), and prolonged patient suffering.
Variability in opioid analgesic requirement is thought to be, at least partially, secondary to genetic factors. Human genetic variants have been shown to affect pain perception and development(4–7). For opioid response, twin studies have suggested that genetic effects are responsible for 12–60% of variability(8). A recent genome-wide association study has identified a locus upstream of OPRM1 that increased oral methadone requirements for treating drug dependence in African Americans(9). Multiple candidate gene studies have also suggested that several genes involved in both the pharmacokinetics and pharmacodynamics of opioids can affect opioid response(10).
Two candidate genes, COMT and OPRM1, are widely studied in pain and opioid consumption. This is due to their presumed influences on both endogenous and exogenous analgesic responses(11). Despite the precedence of multiple studies exploring how polymorphism in COMT and OPRM1 can affect opioid requirement, it has been difficult to tease out whether these variants related to altered opioid responses based on the underlying pain conditions, affected the pharmacodynamics/kinetics of opioids, or even changed the placebo effect from opioid intake. This is because previous studies evaluated opioid requirement mostly in the setting of existing painful conditions(11). Placebo effects were also not corrected in most previous studies. It has been shown recently that the COMT Val158Met and OPRM1 A118G variants are powerful predictors of placebo analgesia(12). It is therefore possible that at least some of the effects observed by previous studies may reflect the influence of these genetic variants on placebo responses.
The goal of this study was to better understand how genetic variants can affect opioid response in a controlled manner. Non-specific effects, including the placebo effect, were corrected by using a randomized, double-blind, placebo-controlled study design. Healthy individuals without baseline pain conditions were recruited. This study also adopted experimental pain measurement as a more standardized readout for opioid effect. To evaluate if pharmacogenomic response is specific to one type of opioid, both morphine and butorphanol were tested. Both morphine and butorphanol exhibit binding at the μ- and κ-opioid receptors with varying efficacies(13). Morphine displays high efficacy at the μ-opioid receptor and low efficacy at the κ-opioid receptor(14), whereas butorphanol exhibits weak efficacy at both(15). OPRM1, OPRK1 and COMT were chosen as candidate genes given that OPRM1 encodes the μ-opioid receptor, OPRK1 encodes the κ-opioid receptor, and COMT encodes an important enzyme that contributes to pathways affecting dopaminergic, adrenergic, noradrenergic, and serotonin neurotransmission, and has been associated with morphine analgesia(16).
Material and Methods:
Detailed method has been published elsewhere(17–19). Study protocol is briefly mentioned below.
Study subjects:
Subjects were recruited through posted advertisements approved by local Institutional Review Board (IRB). This study recruited only healthy nonsmoking individuals between the ages of 18 and 45 without clinical pain, psychiatric disturbance, substance use disorder, or use of centrally acting medications assessed by a health history questionnaire. Participants with missing gender, age, race were excluded from the analysis. Participants with greater than 10% missing genotype were also excluded. After exclusion criteria were applied, 108 healthy participants were included in the analysis.
General Experimental Procedures
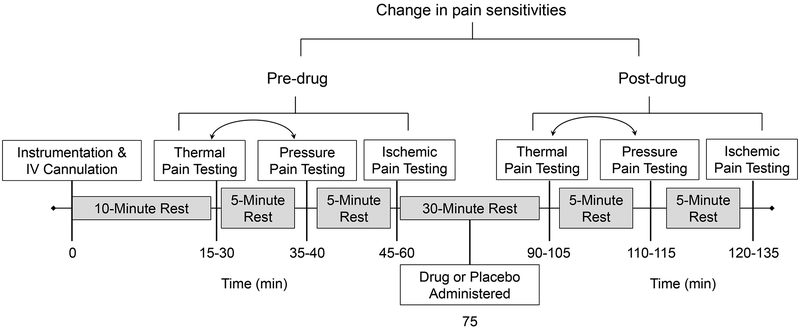
The pre-drug sensory testing protocol included thermal pain, pressure pain, and ischemic pain measures. Following pre-drug sensory testing, a 15-minute rest period was observed followed by the double-blind IV administration of 0.08 mg/kg of morphine, 0.016 mg/kg butorphanol, or saline over 5 minutes. These doses approximate a low–to-moderate clinical dose with estimated equianalgesia(20). Fifteen minutes following drug administration post-drug sensory testing was repeated in a manner identical to the pre-drug testing. A time line of the experimental session is presented in Figure 1.
Figure 1.
Time line representing the temporal structure of procedures during each experimental session. The boxed text represents the procedures (white) and rest breaks (gray) implemented during the experimental session. The numbers below the timeline reflect the approximate time in minutes at which experimental procedures were conducted. The bidirectional arrows between thermal pain and pressure pain indicate that these two procedures were conducted in counterbalanced order. Figure adapted from (19).
Pain Testing Procedures
Pressure Pain Threshold
Pressure pain threshold (PPT) was assessed with a handheld algometer (Pain Diagnostics and Therapeutics, Great Neck, NY, USA). Three sites were used to assess PPTs on the right side of the body: the center of the upper trapezius (posterior to the clavicle), the upper masseter (approximately midway between the ear opening and the corner of the mouth), and the ulna (dorsal forearm, approximately 8 cm distal to the elbow). The average of the three assessments for each site was calculated and used in subsequent analysis.
Thermal Pain
The thermal procedure involved assessment of heat pain threshold (HPTh), and heat pain tolerance (HPTo). Contact heat stimuli were delivered using a computer-controlled Medoc Thermal Sensory Analyzer. From a baseline of 32°C, probe temperature increased at a rate of 0.5°C/sec until the subject responded by pressing a button to indicate when they first felt pain (HPTh) and when they were no longer able to tolerate the pain (HPTo). For each measure, the average of all four trials was computed for use in subsequent analyses.
Ischemic Pain
Modified Submaximal Tourniquet Procedure
The right arm was exsanguinated by elevating it above heart level for 30 seconds, after which, the blood flow to the arm was occluded with a standard blood pressure cuff positioned proximal to the elbow and inflated to 240 mm Hg. Subjects performed 20 handgrip exercises of 2-second duration at 4-second intervals at 50% of their maximum grip strength. Subjects were instructed to report when they first felt pain (ischemic pain threshold), then to continue until the pain became intolerable (ischemic pain tolerance).
Side Effects
To assess side effects in response to morphine and butorphanol, subjects completed the Somatic Side Effects Questionnaire (SSE) and Cognitive and Affective Side Effects Questionnaire (CASE) 60 to 70 minutes after the administration of each opioid(21). For the purpose of analysis, SSE symptoms were collapsed into 7 symptom dimensions and CASE symptoms were collapsed into 6 symptom dimensions as determined by previous confirmatory factor analysis(22). A score of 3 (on a scale of 1 to 5) and above was defined as having significant side effect in that particular dimension.
Genotyping
Whole blood was collected by venipuncture from study participants who provided consent for genotyping. Genomic DNA was purified from leukocytes utilizing Qiagen PureGene Extraction Kits (Germantown, MD, USA). All individuals were genotyped using the Algynomics (Chapel Hill, NC) Pain Research Panel, a dedicated chip-based platform manufactured by Illumina. Eight SNPs within three genes were analyzed in this study: OPRM1 (rs1799971, or A118G), OPRK1 (rs1051660, rs702764 and rs16918875) and COMT (rs4633, rs6269, rs4818 and rs4680).
Data Analysis
The drug effect for each pain measure was determined by a change score calculated as the difference between the pre-drug value and the post-drug value. Thus, subjects had three separate change scores (morphine, saline, and butorphanol) for each experimental pain variable.
Associations between pain change score from opioids and each individual SNP were tested with PLINK(23), using the additive genetic model in linear regression, with race, gender and placebo response (saline change score) as covariates. Haplotype analysis of COMT was performed in a similar fashion. Analysis on the joint effects of COMT and OPRM1 was computed using a 2-tailed t-test with unequal variance. In this analysis, pain change score was corrected for placebo response by subtracting the pain change score from placebo. For side effect analysis, logistic regression was used with race and gender as covariates. A score of 3 and above was defined as having significant side effects in that particular side effect domain. Nominally significant association was set at P<0.05. Multiple testing correction was carried out with Bonferroni correction. Due to high linkage disequilibrium among tested SNPs, effective number of markers was set to the number of tested genes. Given that three genes were tested, the study-wide significant threshold was set to P<0.017 for SNP associations with opioid response and side effects. For COMT haplotype analysis, Bonferroni correction was used assuming four independent haplotypes. This would set haplotypic significant threshold at P<0.0125 after Bonferroni correction. Significant P value for joint effect of COMT and OPRM1 variants was set at P<0.05. The word “nominal association” refers to any association that has P<0.05 but does pass the multiple comparison P value threshold.
Result:
Demographic characteristics
The demographic characteristics of included subjects are presented in Table 1. The average age was 23.14 years old. Male to female ratio was about 1:1. Race was predominantly non-Hispanic white (87%), with an average body mass index of 24.39.
Table 1.
Subject characteristics (average ± standard deviation)
| Variables | Subjects (N=108) |
|---|---|
| Age | 23.14±4.55 |
| Male | 57 (52.78%) |
| Female | 51 (47.22%) |
| Race | |
| African American | 14 (12.96%) |
| Non-Hispanic White | 94 (87.04%) |
| Weight (kg) | 73.92 ±15.89 |
| Body mass index | 24.39 ±3.91 |
Genetic associations
The association results are listed in Table 2. rs6269 and rs4633 of COMT were found to be significantly associated altered pressure pain response following butorphanol. COMT rs4680, OPRM1 A118G(rs1799971), OPRK1 rs702764 and rs1051660 were nominally associated with different pain modalities and opioids. The AA genotype or A_T_C_A/ A_T_C_A (rs6269_rs4633_ rs4818_rs4680) diplotype of COMT rs4680, combined with the AG genotype of OPRM1 A118G, showed significantly increased pressure pain threshold from butorphanol. We detail the findings below.
Table 2.
Association of polymorphisms in OPRM1, OPRK1 and COMT genes with analgesic response from morphine and butorphanol in different pain modalities (*Nominal P<0.05, **study-wide significant P<0.017).
| Association with analgesic response from Morphine | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heat pain threshold | Heat pain tolerance | Ischemic pain threshold | Ischemic pain tolerance | Pressure pain, masseter | Pressure pain, trapezius | Pressure pain, ulna | |||||||||
| Gene | SNP | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P |
| OPRM1 | A118G | −0.075 | 0.87 | −0.18 | 0.52 | −3.80 | 0.81 | −26.37 | 0.41 | 0.23 | 0.030* | −0.15 | 0.52 | 0.25 | 0.34 |
| OPRK1 | rs1051660 | −0.41 | 0.28 | −0.46 | 0.042* | 11.85 | 0.37 | 33.68 | 0.19 | 0.015 | 0.86 | 0.20 | 0.32 | 0.36 | 0.081 |
| rs702764 | −0.12 | 0.68 | −0.28 | 0.10 | 14.23 | 0.15 | −8.32 | 0.67 | −0.028 | 0.68 | −0.03 | 0.84 | 0.0018 | 0.99 | |
| rs16918875 | −0.61 | 0.26 | −0.11 | 0.74 | 22.95 | 0.21 | 23.78 | 0.53 | 0.11 | 0.37 | −0.071 | 0.79 | 0.16 | 0.60 | |
| COMT | rs4633 | 0.079 | 0.73 | 0.028 | 0.84 | −11.42 | 0.16 | 21.67 | 0.19 | −0.0041 | 0.94 | 0.18 | 0.14 | −0.013 | 0.92 |
| rs4680 | 0.036 | 0.88 | −0.003 | 0.98 | −16.70 | 0.041* | 18.32 | 0.28 | −0.014 | 0.80 | 0.086 | 0.49 | −0.027 | 0.84 | |
| rs4818 | −0.18 | 0.49 | −0.16 | 0.31 | 12.99 | 0.15 | −22.64 | 0.20 | −0.044 | 0.46 | −0.25 | 0.067 | −0.12 | 0.42 | |
| rs6269 | −0.27 | 0.30 | −0.18 | 0.26 | 18.15 | 0.044* | −28.91 | 0.12 | −0.054 | 0.36 | −0.26 | 0.046* | −0.070 | 0.63 | |
| OPRM1 | A118G | 0.27 | 0.61 | 0.55 | 0.091 | −7.90 | 0.75 | 6.11 | 0.91 | 0.19 | 0.14 | −0.28 | 0.36 | 0.41 | 0.15 |
| OPRK1 | rs1051660 | −0.17 | 0.70 | −0.23 | 0.37 | 18.87 | 0.35 | 15.34 | 0.72 | −0.08 | 0.43 | −0.15 | 0.56 | 0.13 | 0.58 |
| rs702764 | 0.69 | 0.037* | −0.033 | 0.87 | 17.96 | 0.25 | 25.25 | 0.46 | 0.025 | 0.76 | 0.29 | 0.15 | −0.08 | 0.69 | |
| rs16918875 | 0.90 | 0.14 | −0.018 | 0.96 | 25.39 | 0.37 | 46.08 | 0.47 | −0.17 | 0.25 | −0.16 | 0.64 | −0.072 | 0.83 | |
| COMT | rs4633 | 0.012 | 0.96 | 0.039 | 0.81 | −9.99 | 0.43 | 20.42 | 0.47 | −0.015 | 0.81 | 0.085 | 0.60 | 0.37 | 0.015** |
| rs4680 | −0.040 | 0.88 | 0.015 | 0.92 | −13.04 | 0.31 | 11.05 | 0.70 | 0.0034 | 0.96 | 0.02 | 0.89 | 0.24 | 0.12 | |
| rs4818 | 0.086 | 0.77 | 0.081 | 0.65 | 3.18 | 0.82 | −27.14 | 0.39 | 0.021 | 0.77 | −0.25 | 0.16 | −0.35 | 0.025* | |
| rs6269 | 0.19 | 0.52 | 0.15 | 0.40 | −1.62 | 0.91 | −55.40 | 0.085 | 0.029 | 0.68 | −0.26 | 0.12 | −0.39 | 0.014** | |
COMT SNPs are associated with altered pressure pain threshold from butorphanol
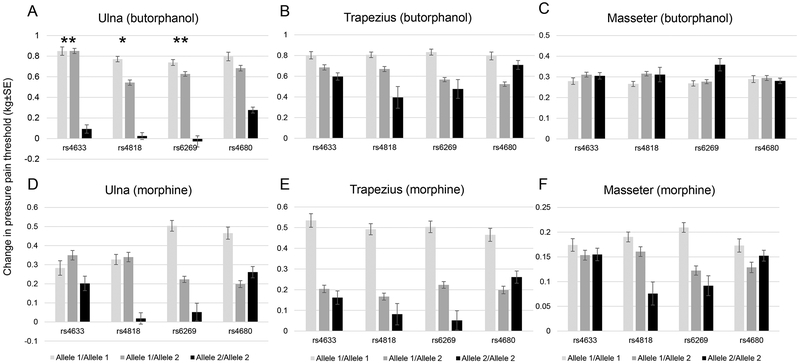
The COMT gene is spanned by three major haploblocks. rs6269, rs4633, rs4818 and rs4680 (Val158Met) reside on the second haploblock(24). There are two major forms of COMT enzyme: membrane bound (MB-COMT) and soluble (S-COMT). rs4633, rs4818 and rs4680 are located within the coding region for both S - and MB-COMT(25). rs6269 is located in the promoter region of S-COMT (26). In this study, rs6269, rs4633 of the COMT gene were associated with different changes in pressure pain threshold at the ulna following butorphanol, at study-wide significance (P=0.014, Beta= −0.39 and P=0.015, Beta= 0.37, respectively). COMT rs4818 was nominally associated with less change in pressure pain threshold at the ulna following butorphanol (P=0.025, Beta=−0.35). The pharmacogenomic effect of COMT polymorphism on changes in pressure pain threshold from each opioid is demonstrated in Figure 2. The trend is similar at the trapezius and with morphine, but associations were not statistically significant. For pressure pain assessed at the ulna, individuals with the GG genotype of rs6269 showed minimal butorphanol analgesia change compared to the AA or GA genotypes. Also, the CC (major allele) genotype of rs4633 was associated with much lower butorphanol analgesia. These associations were not present with morphine or for other pain modalities.
Figure 2.
COMT SNPs were associated with decreased response to opioids in pressure pain threshold. *Nominal P<0.05, **study-wide significant P<0.017. A. rs4633, rs4818, rs6269 were associated with decreased response to butorphanol in the ulna. B-F. There was a general trend in lower response to butorphanol and morphine in other body parts, but the associations with the COMT SNPs are were significant. Allele 1: rs4633: T, rs4818:C, rs6269: A, rs4680: A. Allele 2: rs4633: C, rs4818: G, rs6269:G, rs4680:G
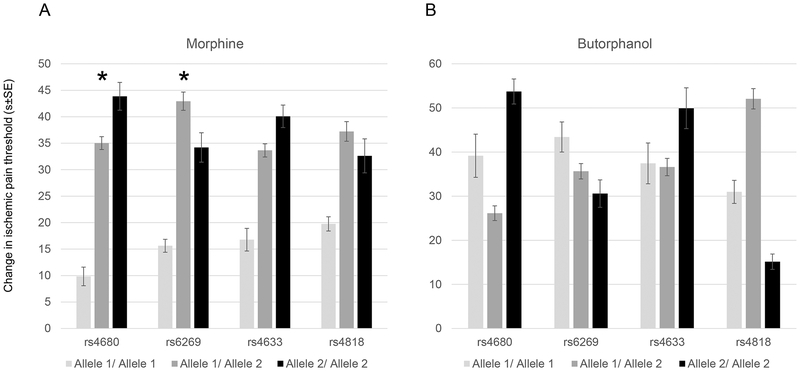
COMT Val158Met and rs6269 are associated with changes in ischemic pain threshold from morphine
rs4680, commonly referred to as Val158Met, was nominally associated with smaller morphine-induced changes in ischemic pain threshold (P=0.041, Beta=−16.70). The AA genotype, or the methionine substitution, showed considerably lower analgesia compared to the GG and GA genotypes. In addition to Val158Met, rs6269 was also nominally associated with higher change in ischemic pain threshold from morphine (P=0.044, Beta=18.15). The GA and GG genotypes showed greater increases in ischemic pain threshold following morphine compared to the AA genotype. Details of ischemic pain threshold are displayed in Figure 3.
Figure 3.
A. COMT SNPs were associated with increased response to morphine in ischemic pain threshold, but not with butorphanol (B). Allele 1: rs4633: T, rs4818:C, rs6269: A, rs4680: A. Allele 2: rs4633: C, rs4818: G, rs6269: G, rs4680: G. *Nominal P<0.05.
Haplotype analysis of COMT
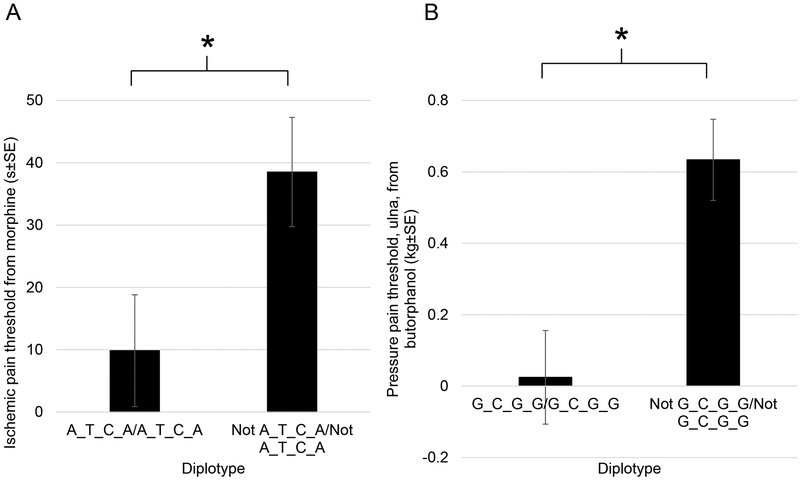
Given the association of rs6269, rs4633, and rs4680 with changes in response to opioids, we sought to evaluate the joint effect of these SNPs. Since rs4633, rs6269, rs4680 and rs4818 reside in the same haploblock, we performed a haplotype analysis for pressure pain and ischemic pain threshold. Frequency of the haplotype is shown in Table S1. Three haplotypes made up 96.64% of the included subjects similar to previously reported findings(24). The results of the haplotype analysis are shown in Table 3. Two haplotypes were found to be nominally associated with opioid response. The G _C _G_G (rs6269_rs4633_ rs4818_rs4680) haplotype was nominally associated with lower butorphanol effects on ulna pressure pain threshold (P=0.039, Beta=−0.34, haplotype frequency 39.6%). The A_T_ C_A haplotype was nominally associated with lower morphine effects on ischemic pain threshold (P=0.042, Beta=−16.5, haplotype frequency 45.3%). These findings are consistent with that from individuals SNPs. The effects of these haplotypes are demonstrated in Figure 4.
Table 3.
Association of COMT haplotypes with analgesic response from morphine and butorphanol in different pain modalities. SNP sequence in the haplotype: rs6269_rs4633_ rs4818_rs4680. *Nominal P<0.05.
| Morphine | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heat pain threshold | Heat pain tolerance | Ischemic pain threshold | Ischemic pain tolerance | Pressure pain, masseter | Pressure pain, trapezius | Pressure pain, ulna | ||||||||
| Haplotype | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P | BETA | P |
| A_T_C_A | 0.083 | 0.72 | 0.017 | 0.90 | −16.50 | 0.042* | 18.50 | 0.28 | −0.010 | 0.85 | 0.11 | 0.37 | −0.043 | 0.75 |
| G_C_G_G | −0.18 | 0.49 | −0.15 | 0.32 | 13.30 | 0.13 | −21.90 | 0.22 | −0.038 | 0.51 | −0.25 | 0.052 | −0.10 | 0.47 |
| A_C_C_G | 0.37 | 0.33 | 0.33 | 0.15 | 0.056 | 1.00 | 4.66 | 0.86 | 0.14 | 0.11 | 0.20 | 0.31 | 0.26 | 0.24 |
| G_C_C_G | −0.29 | 0.80 | 0.00 | 1.00 | 6.86 | 0.87 | −62.00 | 0.40 | −0.12 | 0.67 | −0.19 | 0.76 | −0.05 | 0.94 |
| A_T_C_A | −0.074 | 0.78 | 0.0090 | 0.96 | −15.60 | 0.22 | 11.30 | 0.69 | −0.0033 | 0.96 | 0.037 | 0.81 | 0.29 | 0.052 |
| G_C_G_G | 0.031 | 0.91 | 0.11 | 0.54 | −4.72 | 0.74 | −42.30 | 0.17 | 0.03 | 0.67 | −0.25 | 0.15 | −0.34 | 0.039* |
| A_C_C_G | −0.33 | 0.45 | −0.37 | 0.15 | 26.50 | 0.19 | 43.70 | 0.29 | −0.04 | 0.72 | 0.30 | 0.22 | −0.10 | 0.69 |
| G_C_C_G | 1.05 | 0.42 | 0.19 | 0.81 | 26.00 | 0.68 | −88.10 | 0.48 | 0.11 | 0.74 | 0.27 | 0.73 | 0.40 | 0.59 |
Figure 4.
A. COMT A_T_C_A haplotype was associated with decreased response to morphine in ischemic pain threshold. (B). COMT G_C_G_G haplotype was associated with decreased response to butorphanol in pressure pain threshold (ulna). *Nominal P<0.05.
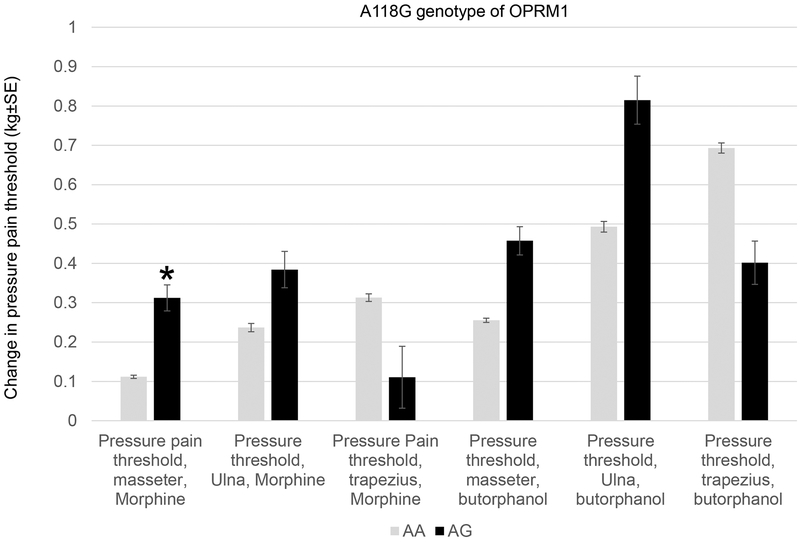
A118G polymorphism of OPRM1 is associated with morphine effects on pressure pain threshold.
A nominal association was found between the minor G allele of A118G polymorphism (rs1799971) of OPRM1 and morphine-induced change in pressure pain threshold at the masseter (P=0.030, Beta=0.23). The AG genotype showed greater increase in pressure pain threshold in response to morphine compared to the wild type AA genotype. Effect of the A118G polymorphism is illustrated in Figure 5.
Figure 5.
A118G of OPRM1 was associated with higher pressure pain threshold from morphine in the masseter. *Nominal P<0.05
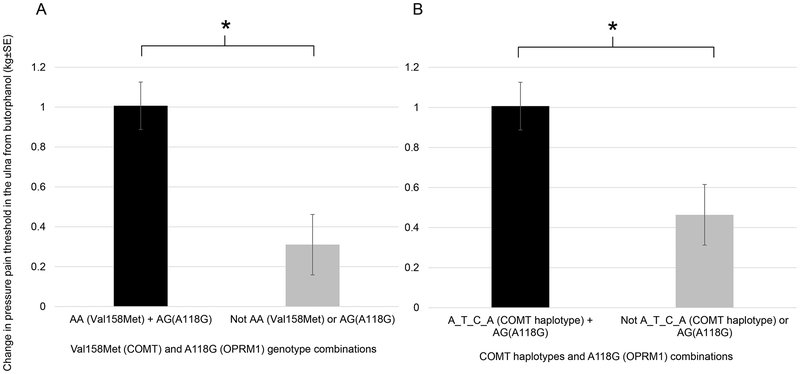
Joint effect of COMT and OPRM1 variants
The Val158Met variant of COMT (rs4680) and A118G (rs1799971) variant of OPRM1 have been observed to act in synergy to affect morphine requirement in patients with cancer pain(27). We sought to examine whether certain combinations of these genotypes are associated with opioid response in our study. Due to the association with minor alleles found with Val158Met and A118G, we first tested whether the combination of the Met/Met (AA) genotype of COMT Val158Met and AG genotype of A118G in OPRM1 changed the analgesic response from opioids. In subjects with this genotypic combination, we observed a statistically greater increase in pressure pain threshold at the ulna from butorphanol (P=0.016, corrected for placebo effect, compared to either Met/Met or AG genotype separately, 2-tailed t-test. See Supplementary Material Table S2 for other pain modalities). Because of the strong linkage disequilibrium between Val158Met and other SNPs within the COMT LD block, we also examined the A_T_C_A/ A_T_C_A (rs6269_rs4633_ rs4818_rs4680) diplotype that contains the AA genotype of Val158Met of COMT gene. We examined whether the combination of the A_T_C_A/ A_T_C_A diplotype and the AG genotype of A118G also affected pressure pain threshold. Again, we observed a greater increase in pressure pain threshold (ulna) from butorphanol in subjects with the A_T_C_A/A_T_C_A diplotype and the AG genotypes (P=0.009). The joint effects of COMT and OPRM1 variants/haplotypes are illustrated in Figure 6.
Figure 6.
Joint effect of COMT and OPRM1 polymorphisms on opioid response. The Met/Met genotype of Val158Met combined with the AG genotype of A118G increased pressure pain threshold response in the ulna from butorphanol (A). The A_T_C_A (rs6269_rs4633_ rs4818_rs4680) haplotype of COMT combined with the AG genotype of A118G increased pressure pain threshold response in the ulna from butorphanol (B). *Study-wide significant P<0.05
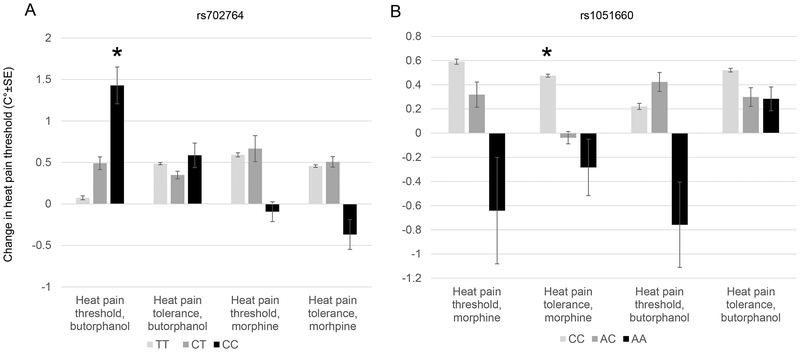
Pharmacogenomics effects of OPRK1 variants
We found that the CC genotype of rs702764 was nominally associated with greater butorphanol-induced increase in heat pain threshold (P=0.037, Beta=0.69). On the other hand, the AA genotype of rs1051660 was nominally associated with reduced morphine effects on heat pain tolerance (P=0.042, Beta=−0.46). The complete results for OPRK1 and heat pain analgesia are shown in Figure 7.
Figure 7.
OPRK1 SNPs associated with altered heat pain tolerance and threshold from opioids. rs702764 of OPRK1 was associated with increased heat pain threshold to butorphanol (A). rs1051660 is associated with decreased heat pain tolerance from morphine (B). *Nominal P<0.05.
Genetic association with side effects
We sought to evaluate whether variants in COMT, OPRM1 and OPRK1 were associated with side effects from opioids. COMT rs4633 and rs4680 were found to be nominally associated with the side effect of “Feeling In Control” from morphine (OR=1.90, P=0.028; OR=2.00, P=0.020, respectively). Subjects with these variants also showed a trend toward association with this side effect for butorphanol, but it did not reach statistical significance (OR=2.01, P=0.099; OR=2.06, P=0.091, respectively). Full association results of all side effects are shown in Supplementary Material Table S3.
Discussion:
To our knowledge, this is the first randomized, double-blind, placebo-controlled study to explore the pharmacogenomic effect of COMT, OPRM1 and OPRK1 variants on opioid responses in healthy adults. We report that variants and haplotypes have at least nominal association with altered opioid responses, and their effects are opioid and pain modality specific. Pressure pain response was found to be significantly associated with rs6269 and rs4633 following butorphanol. The AA genotype of rs4680 or A_T_C_A/ A_T_C_A (rs6269_rs4633_ rs4818_rs4680) diplotype of COMT, combined with the AG genotype of OPRM1 A118G, showed significantly increased pressure pain threshold from butorphanol. Opioid effects on pressure, ischemic, heat pain and side effects were nominally associated with several SNPs and haplotypes. The effects of these polymorphisms were often present in one opioid but not the other.
Experimental pain studies using healthy individuals provide controlled evaluations of pharmacogenomic effects while controlling for baseline pain variations. Earlier experimental pain studies exploring OPRM1 and COMT associations with opioid analgesia included only 10 to 50 healthy subjects(28–31). Further, none of these studies corrected for placebo effects, which have been reported to be influenced by OPRM1 and COMT polymorphisms(12). OPRK1 genotype has not previously been evaluated in this fashion. Our present study, including 108 subjects with correction for non-specific effects by using a randomized-controlled design, is the largest controlled study to date.
This work showed that homozygosity for the COMT Val158Met variant (AA) was found to have less morphine-induced decrease in ischemic pain threshold compared to the GG and GA genotypes. Its corresponding diplotype, A_T_C_A/A_T_C_A, was also found to have with similar effects following morphine. These findings are consistent with previous studies that the methionine substitution at this amino acid diminishes the regional μ-opioid system responses to pain(16).
In a previous haplotypic study of COMT and pain sensitivity in healthy individuals, the G_C_ G_G (rs6269_rs4633_ rs4818_rs4680) haplotype was found to relate to low pain sensitivity and the A_T_C_A haplotype was correlated to average pain sensitivity(24). In our study, the G _C _G_G (rs6269_rs4633_ rs4818_rs4680) haplotype was nominally associated with lower butorphanol effects on pressure threshold, and the A_T_C_A haplotype was associated with lower morphine effects on ischemic pain threshold. These effects are consistent between haplotypes and individuals SNPs. The G_C_G_G haplotype replicates the pressure pain threshold findings from butorphanol in individual SNP association (rs6269, rs4633, rs4818). The A_T_C_A haplotype also replicates the ischemic pain effect from morphine in individual SNP association (rs6269, rs4680). Our study suggests that these haplotypes not only affect baseline pain sensitivities, as suggested in previous studies, but may also affect opioid analgesic responses.
The results from this study suggest that the minor (G) allele of the OPRM1 A118G polymorphism is associated with increases in pressure pain threshold in response to morphine. This is consistent with previous findings that the G allele was associated with increased pressure pain threshold in healthy individuals(32). In cancer and acute pain, the relationship of the A118G variant with opioid consumption has been investigated with mixed results. Some studies have shown that this variant increases opioid consumption(33–36), some have shown decrease in consumption(37), and some have shown no difference(38, 39). If pressure pain threshold was positively correlated with pain suppression in cancer or postoperative pain, our study would support decreased consumption of opioids in these painful conditions.
Combined effects of COMT Val158Met and OPRM1 A118G have been explored in several studies with cancer pain and postoperative pain. The Val158Met variant of COMT and A118G variant of OPRM1 have been observed to act in synergy to affect morphine usage in patients with cancer pain(27). Heterozygous patients with A118G and Val158Met consumed significantly less morphine in the postoperative period compared to homozygous patients with A118G(39). In the current study, the combination of the COMT Met/Met (AA) genotype and AG genotype of A118G in OPRM1 were observed in relation to increases in pressure pain threshold following butorphanol administration. The association remained significant when it was expanded to the combined effect of A_T_C_A/ A_T_C_A (rs6269_rs4633_ rs4818_rs4680) haplotype in COMT and the AG genotype of A118G in OPRM1. These observations are consistent with previous observations that the Val158Met genotype of COMT and A118G of OPRM1 could be potentially used as joint biomarkers for opioid response.
The pharmacogenomic effect of OPRK1 on oxycodone was previously explored in a small study, but no association was found(40). In another study, OPRK1 polymorphisms was found to be associated with pressure pain tolerance(41). Our study suggests that variants in OPRK1 are nominally associated with greater opioid-induced changes in heat pain perception. This supports that notion that the OPRK1 gene plays a role in opioid responsiveness.
Side effects from medications can be a powerful predictor of addiction propensity. One such example is polymorphisms in alcohol aldehyde dehydrogenases resulting in flushing and nausea when alcohol is consumed, which in turn becomes protective against alcoholism(42). The COMT enzyme catalyzes the breakdown of dopamine, which plays a prominent role in drug reward. In this study, the minor alleles of COMT rs4633 and rs4680 were nominally associated with the morphine-induced side effect of “Feeling In Control”. This “Feeling In Control” sensation could be one of the mechanisms through which opioid addiction develops. Indeed, rs4680 (Val158Met) has been associated with opiate addiction(43).
There are several limitations to this study. First, although this is the largest study of its kind, analysis of more participants would provide higher confidence in the current findings. Second, while the use of experimental pain models provides advantages, the clinical application of the current findings is unclear, as the directions of effects varied greatly for different pain modalities. Third, this is a hypothesis-driven study and other variants within the genome were not evaluated. Effects of other variants in the genome could confound the results through gene-gene interactions.
Despite these limitations, this study provides important insights into the complexity of opioid pharmacogenomics and validates some smaller studies in the literature. Our results suggest that the pharmacogenomic effects of COMT, OPRM1 and OPRK1 variants are specific to each experimental pain modality and opioid. This should not be surprising, as we have previously shown that analgesic responses are themselves modality- and drug-specific(21). This suggests that generalization of pharmacogenomic studies using different measurement tools and opioids may be difficult due the specificities of these associations. Further studies with larger sample sizes and standardized pain measurement will be necessary to evaluate the genetic contributions to opioid analgesia.
Supplementary Material
Acknowledgments:
This work was supported by NIH/NINDS grant R01NS041670.
Footnotes
Conflict of interests: Authors declared no conflict of interests.
References:
- 1.Coulbault L, Beaussier M, Verstuyft C, Weickmans H, Dubert L, Trégouet D, et al. Environmental and genetic factors associated with morphine response in the postoperative period. Clin Pharmacol Ther 2006;79(4):316–24. [DOI] [PubMed] [Google Scholar]
- 2.Pico L, Hernot S, Nègre I, Samii K, Fletcher D. Peroperative titration of morphine improves immediate postoperative analgesia after total hip arthroplasty. Can J Anaesth 2000;47(4):309–14. [DOI] [PubMed] [Google Scholar]
- 3.Hunold KM, Esserman DA, Isaacs CG, Dickey RM, Pereira GF, Fillingim RB, et al. Side effects from oral opioids in older adults during the first week of treatment for acute musculoskeletal pain. Acad Emerg Med 2013;20(9):872–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fillingim R, Wallace M, Herbstman D, Ribeiro-Dasilva M, Staud R. Genetic contributions to pain: a review of findings in humans. Oral Diseases 2008;14(8):673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho KWD, Wallace MR, Sibille KT, Bartley EJ, Cruz-Almeida Y, Glover TL, et al. Single Nucleotide Polymorphism in the COL11A2 Gene Associated with Heat Pain Sensitivity in Knee Osteoarthritis. Mol Pain 2017;13:1744806917724259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho KWD, Jerath NU. V144D Mutation of SPTLC1 Can Present with Both Painful and Painless Phenotypes in Hereditary Sensory and Autonomic Neuropathies Type I. Case Reports in Genetics. 2018;2018:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho KWD, Jerath NU. T118M Variant of PMP22 Gene Presents with Painful Peripheral Neuropathy and Varying Charcot-Marie-Tooth Features: A Case Series and Review of the Literature. Case Reports in Genetics. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angst MS, Phillips NG, Drover DR, Tingle M, Ray A, Swan GE, et al. Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain 2012;153(7):1397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith AH, Jensen KP, Li J, Nunez Y, Farrer LA, Hakonarson H, et al. Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Mol Psychiatry 2017;22(3):346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd RA, Hotham E, Hall C, Williams M, Suppiah V. Pharmacogenomics and Patient Treatment Parameters to Opioid Treatment in Chronic Pain: A Focus on Morphine, Oxycodone, Tramadol, and Fentanyl. Pain Med 2017;18(12):2369–87. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen LM, Olesen AE, Branford R, Christrup LL, Sato H, Drewes AM. Association Between Human Pain-Related Genotypes and Variability in Opioid Analgesia: An Updated Review. Pain Pract 2015;15(6):580–94. [DOI] [PubMed] [Google Scholar]
- 12.Aslaksen PM, Forsberg JT, Gjerstad J. The opioid receptor mu 1 (OPRM1) rs1799971 and catechol-O-methyltransferase (COMT) rs4680 as genetic markers for placebo analgesia. Pain 2018. [DOI] [PubMed] [Google Scholar]
- 13.Emmerson PJ, Clark MJ, Mansour A, Akil H, Woods JH, Medzihradsky F. Characterization of opioid agonist efficacy in a C6 glioma cell line expressing the mu opioid receptor. J Pharmacol Exp Ther 1996;278(3):1121–7. [PubMed] [Google Scholar]
- 14.Yamada H, Shimoyama N, Sora I, Uhl GR, Fukuda Y, Moriya H, et al. Morphine can produce analgesia via spinal kappa opioid receptors in the absence of mu opioid receptors. Brain Res 2006;1083(1):61–9. [DOI] [PubMed] [Google Scholar]
- 15.Walsh SL, Chausmer AE, Strain EC, Bigelow GE. Evaluation of the mu and kappa opioid actions of butorphanol in humans through differential naltrexone blockade. Psychopharmacology (Berl) 2008;196(1):143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science 2003;299(5610):1240–3. [DOI] [PubMed] [Google Scholar]
- 17.Fillingim RB, Ness TJ, Glover TL, Campbell CM, Hastie BA, Price DD, et al. Morphine responses and experimental pain: sex differences in side effects and cardiovascular responses but not analgesia. J Pain 2005;6(2):116–24. [DOI] [PubMed] [Google Scholar]
- 18.Fillingim RB, Ness TJ, Glover TL, Campbell CM, Price DD, Staud R. Experimental pain models reveal no sex differences in pentazocine analgesia in humans. Anesthesiology 2004;100(5):1263–70. [DOI] [PubMed] [Google Scholar]
- 19.Sibille KT, Kindler LL, Glover TL, Gonzalez RD, Staud R, Riley JL, et al. Individual differences in morphine and butorphanol analgesia: a laboratory pain study. Pain Med 2011;12(7):1076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunton L, Chabner B, Knollman B. Goodman & Gilman’s The Pharmacological Basis of Therapeutics 12th ed: McGraw-Hill Education; 2011. [Google Scholar]
- 21.Kindler LL, Sibille KT, Glover TL, Staud R, Riley JL, Fillingim RB. Drug response profiles to experimental pain are opioid and pain modality specific. J Pain 2011;12(3):340–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley JL, Hastie BA, Glover TL, Fillingim RB, Staud R, Campbell CM. Cognitive-affective and somatic side effects of morphine and pentazocine: side-effect profiles in healthy adults. Pain Med 2010;11(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet 2005;14(1):135–43. [DOI] [PubMed] [Google Scholar]
- 25.Li T, Ball D, Zhao J, Murray RM, Liu X, Sham PC, et al. Family-based linkage disequilibrium mapping using SNP marker haplotypes: application to a potential locus for schizophrenia at chromosome 22q11. Mol Psychiatry 2000;5(4):452. [DOI] [PubMed] [Google Scholar]
- 26.Shifman S, Bronstein M, Sternfeld M, Pisanté-Shalom A, Lev-Lehman E, Weizman A, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet 2002;71(6):1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes-Gibby CC, Shete S, Rakvåg T, Bhat SV, Skorpen F, Bruera E, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain 2007;130(1–2):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwisler ST, Enggaard TP, Noehr-Jensen L, Mikkelsen S, Verstuyft C, Becquemont L, et al. The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam Clin Pharmacol 2010;24(4):517–24. [DOI] [PubMed] [Google Scholar]
- 29.Oertel BG, Schmidt R, Schneider A, Geisslinger G, Lötsch J. The mu-opioid receptor gene polymorphism 118A>G depletes alfentanil-induced analgesia and protects against respiratory depression in homozygous carriers. Pharmacogenet Genomics 2006;16(9):625–36. [DOI] [PubMed] [Google Scholar]
- 30.Romberg RR, Olofsen E, Bijl H, Taschner PE, Teppema LJ, Sarton EY, et al. Polymorphism of mu-opioid receptor gene (OPRM1:c.118A>G) does not protect against opioid-induced respiratory depression despite reduced analgesic response. Anesthesiology 2005;102(3):522–30. [DOI] [PubMed] [Google Scholar]
- 31.Jensen KB, Lonsdorf TB, Schalling M, Kosek E, Ingvar M. Increased sensitivity to thermal pain following a single opiate dose is influenced by the COMT val(158)met polymorphism. PLoS One 2009;4(6):e6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillingim RB, Kaplan L, Staud R, Ness TJ, Glover TL, Campbell CM, et al. The A118G single nucleotide polymorphism of the mu-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain 2005;6(3):159–67. [DOI] [PubMed] [Google Scholar]
- 33.Klepstad P, Rakvåg TT, Kaasa S, Holthe M, Dale O, Borchgrevink PC, et al. The 118 A > G polymorphism in the human mu-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol Scand 2004;48(10):1232–9. [DOI] [PubMed] [Google Scholar]
- 34.Chou WY, Yang LC, Lu HF, Ko JY, Wang CH, Lin SH, et al. Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand 2006;50(7):787–92. [DOI] [PubMed] [Google Scholar]
- 35.Chou WY, Wang CH, Liu PH, Liu CC, Tseng CC, Jawan B. Human opioid receptor A118G polymorphism affects intravenous patient-controlled analgesia morphine consumption after total abdominal hysterectomy. Anesthesiology 2006;105(2):334–7. [DOI] [PubMed] [Google Scholar]
- 36.Sia AT, Lim Y, Lim EC, Ocampo CE, Lim WY, Cheong P, et al. Influence of mu-opioid receptor variant on morphine use and self-rated pain following abdominal hysterectomy. J Pain 2013;14(10):1045–52. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda K, Hayashida M, Ide S, Saita N, Kokita Y, Kasai S, et al. Association between OPRM1 gene polymorphisms and fentanyl sensitivity in patients undergoing painful cosmetic surgery. Pain 2009;147(1–3):194–201. [DOI] [PubMed] [Google Scholar]
- 38.Janicki PK, Schuler G, Francis D, Bohr A, Gordin V, Jarzembowski T, et al. A genetic association study of the functional A118G polymorphism of the human μ-opioid receptor gene in patients with acute and chronic pain. Anesthesia & Analgesia 2006;103(4):1011–7. [DOI] [PubMed] [Google Scholar]
- 39.Kolesnikov Y, Gabovits B, Levin A, Voiko E, Veske A. Combined catechol-O-methyltransferase and mu-opioid receptor gene polymorphisms affect morphine postoperative analgesia and central side effects. Anesth Analg 2011;112(2):448–53. [DOI] [PubMed] [Google Scholar]
- 40.Olesen AE, Sato H, Nielsen LM, Staahl C, Droney J, Gretton S, et al. The genetic influences on oxycodone response characteristics in human experimental pain. Fundam Clin Pharmacol 2015;29(4):417–25. [DOI] [PubMed] [Google Scholar]
- 41.Sato H, Droney J, Ross J, Olesen AE, Staahl C, Andresen T, et al. Gender, variation in opioid receptor genes and sensitivity to experimental pain. Mol Pain 2013;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tawa EA, Hall SD, Lohoff FW. Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol 2016;51(5):507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oosterhuis BE, LaForge KS, Proudnikov D, Ho A, Nielsen DA, Gianotti R, et al. Catechol‐O‐methyltransferase (COMT) gene variants: Possible association of the Val158Met variant with opiate addiction in hispanic women. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2008;147(6):793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.