Abstract
It remains unclear whether the increased risk of new-onset type 2 diabetes (T2D) seen in statin users is due to low LDL-C concentrations, or due to the statin-induced proportional change in LDL-C. In addition, genetic instruments have not been proposed before to examine whether liability to T2D might cause greater proportional statin-induced LDL-C lowering. Using summary level statistics from the Genomic Investigation of Statin Therapy (GIST, nmax=40,914) and DIAGRAM (nmax=159,208) consortia, we found a positive genetic correlation between LDL-C statin response and T2D using LD score regression (rgenetic=0.36, s.e.=0.13). However, mendelian randomization analyses did not provide support for statin response having a causal effect on T2D risk (OR 1.00 (95%CI: 0.97, 1.03) per 10% increase in statin response), nor that liability to T2D has a causal effect on statin-induced LDL-C response (0.20% increase in response (95%CI: −0.40, 0.80) per doubling of odds of liability to T2D). Although we found no evidence to suggest that proportional statin response influences T2D risk, a definitive assessment should be made in populations comprised exclusively of statin-users, as the presence of non-statin users in the DIAGRAM dataset may have substantially diluted our effect estimate.
Introduction
3-Hydroxy-3-methylglutaryl–coenzyme A (HMG-CoA) reductase inhibitors, also known as statins, have demonstrated consistent benefits to cardiovascular disease risk reduction, while being safe and well-tolerated for most people [1]. However, statin treatment has been linked to a modestly increased risk of new-onset type 2 diabetes (T2D), an observation first noted in the JUPITER trial [2] which has since been replicated in large-scale meta-analyses of randomized controlled trials [3–5]. As promising novel strategies for lowering low-density lipoprotein cholesterol (LDL-C) such as proprotein convertase subtilisin–kexin type 9 (PCSK9) inhibitors emerge, the safety of lipid-modifying treatments with regard to diabetes risk remains an important question.
In recent years, genetic epidemiology has started to untangle the complex link between LDL-C lowering and T2D risk. For example, analyses of patients with familial hypercholesterolemia have shown that the prevalence of T2D is significantly lower than among unaffected relatives, with variability by underlying mutation type [6]. Furthermore, apparent causal effects on T2D have been shown both for overall genetic predisposition to lower LDL-C concentrations [7–9] as well as for HMGCR-, NPC1L1-, and PCSK9-gene specific (i.e. on target) mechanisms of lowering LDL-C [9–13]. These findings, and recent reanalysis of statin trial data using Egger regression [14], suggest that statin-related dysglycaemia might be mediated largely through LDL-C lowering mechanisms rather than through proposed pleiotropic mechanisms of statins [15].
However, meta-regression approaches modeling heterogeneity among treatment effects from statin trials have produced conflicting results as to whether statin-induced proportional change of LDL-C influences T2D risk [3–5]. Previous Mendelian randomization (MR) studies have been unable to directly answer this question, as genetic instruments solely proxying lifelong lower levels of LDL-C have been utilized. In addition, genetic instruments have not been proposed before to examine whether the greater absolute cardiovascular disease (CVD) risk reduction conferred by statin therapy in individuals with T2D could result from greater proportional statin-induced LDL-C lowering. Findings from the largest pharmacogenomic meta-analysis for differential LDL-C response to statin therapy to date by the Genomic Investigation of Statin Therapy (GIST) consortium might be used to investigate these questions. We therefore aimed to use these data to examine the causal direction of the relationship between proportional statin response and T2D using a bidirectional two-sample MR approach.
Methods
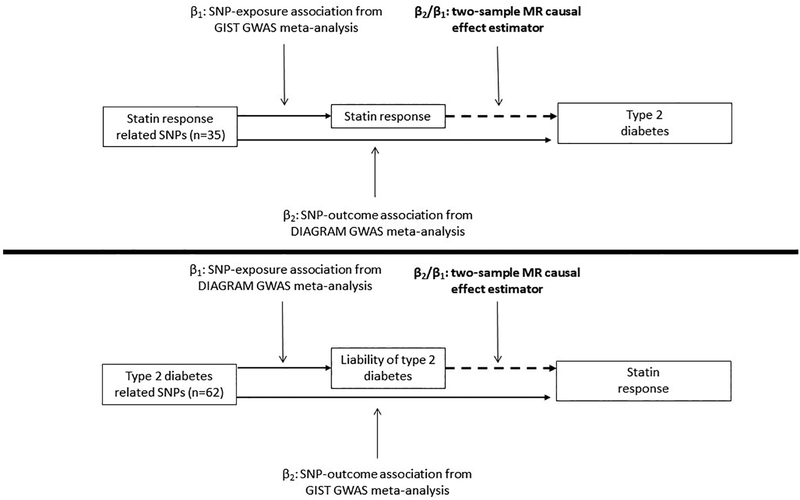
To assess the likelihood of a shared etiology between statin response and T2D, which may be the product of a causal relationship, we assessed their genetic correlation using cross-trait linkage disequilibrium (LD) score regression. To detect potential direct causality we performed a bidirectional two-sample MR-analysis, combining summary level statistics from the GIST [16] and DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) [17] consortia (Figure 1), to estimate: (1) the causal effect of statin-induced LDL-C response on T2D risk, and (2) the causal effect of liability to T2D on statin-induced LDL-C response. We refer to liability to T2D in this second analysis as it is not possible to determine whether individuals in the GIST dataset have been diagnosed with T2D.
Figure 1.
Overview of two-sample Mendelian randomization (MR) study on the bidirectional association between statin-induced LDL cholesterol response and type 2 diabetes (T2D). Top panel shows direction of statin response to T2D, bottom panel liability to T2D to statin response. Layout of figure based upon the work by Taylor et al. 2016 (PMID 27215954).
Causal effect of statin-induced LDL-C response on T2D
The GIST consortium’s 2014 meta-analysis on statin-induced LDL-C response included up to 18,596 statin-treated subjects in the discovery stage, of whom 9,064 (48.7%) were known to have a history of diabetes (type unspecified). The most promising signals (n=246) were taken forward for replication, to be validated in an additional 22,318 statin recipients [16]. Statin response had been defined as the difference between natural log-transformed on- and off-treatment LDL-C levels. Linear regression analyses using this statin response phenotype as dependent variable and genetic variant as independent variable were adjusted for natural log-transformed off-treatment LDL-C level, age, sex, and study-specific covariates including principal components of ancestry. Observational studies had additionally adjusted for statin type-specific equivalent dose in their regression models. The resulting regression coefficient thus approximates the fraction of differential LDL lowering in carriers versus non-carriers of the SNP. While lead variants for four independent loci (APOE, LPA, SLCO1B1, SORT1/CELSR2/PSRC1) were presented as the top genome-wide significant hits for statin response in the GIST paper [16], 63 correlated variants attained a p-value lower than 5×10−5 in a combined meta-analysis of the discovery and replication stage results.
To assess the effects of these instruments on T2D, we extracted discovery stage summary statistics for these 63 variants from DIAGRAM’s 2017 meta-analysis of genome-wide association data from 26,676 T2D case and 132,532 control subjects of European ancestry after imputation using the 1000 Genomes all ancestries reference panel (March, 2012 release) [17]. Contributing studies had performed logistic regression association analysis of T2D against each genetic variant, adjusted for age, sex, and principal components of ancestry. The summary statistics were extracted from the publicly available summary statistics dataset on the DIAGRAM website (http://www.diagram-consortium.org/). All variants were available in the DIAGRAM dataset, except two, for which we could not find suitable (i.e. high-LD) proxies. We subsequently LD clumped the set of variants using 0.001 as the maximum LD r2 value to ensure that the remaining instruments were essentially independent. This reduced the set of statin response instruments from 61 to 35 (Supplemental Table 1). We separately examine the effects of the full set of 35 statin response instruments and of the four top hits together. Possible dilution by the presence of non-statin users in DIAGRAM was explored through simple simulations (Supplementary Note).
Causal effect of liability to T2D on statin-induced LDL-C response
As candidate genetic instruments for liability of T2D we selected 128 genetic instruments at 113 loci. These 128 variants represent the established loci from the literature before the DIAGRAM’s 2017 publication as well as the novel signals detected therein, with 42 being genome-wide significant (p<5×10−8) in this DIAGRAM dataset. Discovery stage regression coefficients and standard errors for a total of 128 single nucleotide polymorphisms (SNPs) at 113 loci were extracted from the publicly available summary statistics dataset on the DIAGRAM website (http://www.diagram-consortium.org/).
Next, we extracted summary statistics for the identified T2D liability instruments from GIST’s 2014 genome-wide meta-analysis on statin-induced LDL-C response. Of note, none of the identified T2D liability instruments were among those SNPs carried forwards to the replication stage of the GIST meta-analysis. In total, 78 of the 128 instruments were available in the discovery GIST dataset. We subsequently LD clumped the set of variants, again using 0.001 as the maximum LD r2 value. This reduced the set of instruments from 78 to 62 (including 24 genome-wide significant instruments), which include 19 proxies with an r2≥0.8 with the original variant in 1000 Genomes European samples (Supplemental Table 2). To tease out possible bias from using weaker instruments, we aimed to examine the combined effects of the T2D liability instruments before and after restricting the analysis to the genome-wide significant instruments.
Sample overlap
Of the studies that contributed to the discovery-stage meta-analysis of GIST, four (Atherosclerosis Risk in Communities Study (ARIC), Framingham Heart Study, Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) I and II) also contributed to the DIAGRAM meta-analysis. Of the studies contributing to the replication-stage meta-analysis of GIST, one (Rotterdam Study) also contributed to the DIAGRAM meta-analysis. We were unable to precisely determine the overlap between the datasets. However, if we assume that all participants from these five studies contributed to both analyses, up to 5% (with respect to the larger dataset, i.e. DIAGRAM) of overlap may be present for our analyses.
Statistics - LD score regression
A causal relationship between two heritable traits should induce a genetic correlation between these traits. The use of LD score regression to quantify genetic correlation has been described in detail elsewhere [18, 19]. Briefly, under the assumption that biologically relevant variants are uniformly distributed across the genome, genetic variants in high LD with many nearby variants are more likely to tag causal variants, and therefore have larger squared effect size estimates. This can be quantified by each variant’s ‘LD score’: the sum of squared correlations with all nearby variants. Cross-trait LD score regression calculates the cross-product of test statistics from different GWAS meta-analyses, and then regresses the cross-product on that variant’s LD score. In our study, the genome-wide summary-level datasets of both GIST (discovery stage, including only variants with N>5000, and without genome control-correction) and DIAGRAM’s 2012 GWAS [20] were thus used to estimate the genetic correlation of statin-induced LDL-C proportional response with T2D using the LD Hub platform (http://ldsc.broadinstitute.org/ldhub/) [21]. After QC, 1,039,702 genetic variants which overlapped between the two GWAS datasets were included for this analysis.
Statistics – MR-analysis
Partial F-statistics were calculated per instrument as measure of instrument strength [22]. For each set of instruments separately (i. statin response-all (n=35), ii. statin response-restricted (n=4 genome-wide significant instruments), iii. T2D liability-all (n=62), iv. T2D liability-restricted (n=24 genome-wide significant instruments) a MR-analysis was performed using an inverse-variance weighted (IVW) linear regression, with instrument-outcome associations as dependent variable, instrument-exposure associations as independent variable, and with the intercept constrained to zero [23] Estimates of the causal effect of statin response on T2D are presented as odds ratio for T2D per 10% increase in statin response. For examining the effects of liability to T2D on statin response we rescaled effect estimates such that they represent increase in statin response (% extra lowering of LDL-C) per doubling of the odds of liability to T2D in the population, by multiplying the causal estimate by 0.693 (i.e. loge 2) prior to exponentiating.
Instrument-outcome associations were plotted against instrument-exposure associations to visualize the resulting regression line from the IVW analysis using the full and restricted sets of instruments. Furthermore, causal effect estimates for the individual instruments (i.e. Wald ratios) were plotted against the inverse of their standard error to facilitate visual detection of possible horizontal pleiotropy (i.e. a direct effect on the outcome rather than via the exposure). We subsequently performed three complementary sensitivity analyses which relax the assumption of no horizontal pleiotropy amongst the genetic variants. First, MR-Egger regression, of which the intercept formally tests for the presence of unbalanced horizontal pleiotropy, and the slope reflects the causal effect estimate after adjusting for this pleiotropy by adding an intercept to the IVW method [24]. Additional approaches that are similarly more robust to potential violations of the instrumental variable assumptions than the conventional (i.e. IVW) MR-analysis were the weighted median-[25] and the weighted mode-based estimator [26], which respectively use the weighted median of, and the highest density of, the ratio estimates across the individual instruments as estimate of the true causal effect. We additionally tested for heterogeneity in effects estimates by means of Cochran’s Q and Rucker’s Q’ [27, 28]. First-order weights were utilized for this calculation. Finally, as several of the proposed instruments for statin-induced LDL-C response are known to independently associate with fasting LDL-C levels, we performed a multivariable MR-analysis for the analysis of statin-response to T2D, adjusted for effects on fasting LDL-C levels [29]. This multivariable analysis included 32 statin response instruments, and 64 instruments for fasting LDL-C concentrations from the Global Lipids Genetics Consortium 2013 GWAS on blood lipid levels [30]. The number of instruments used for the multivariable MR-analysis differ from the other MR-analyses due to not all instruments being available in all three GWAS datasets. All MR-analyses were carried out in R version 3.4.2 [31], without correction for multiple testing, using the TwoSampleMR R-package which accompanies the MR-base analytical platform, and the sample code provided by the methodology paper on multivariable MR [29, 32] (code available upon request).
Results
We found a statistically significant positive genetic correlation of statin-induced LDL-C response with type 2 diabetes (rgenetic (s.e.) = 0.36 (0.13), p = 0.0071) using LD score regression.
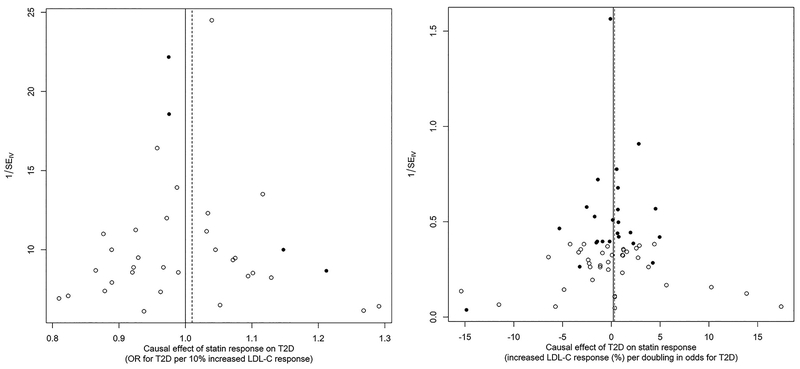
The median F-statistic (25, 75th percentile) was 23.2 (16.3, 37.8) for the full set of statin response instruments, and 18.1 (16.2, 20.9) for the full set of T2D liability instruments. These respectively increased to 44.8 (35.5, 70.7) and 73.1 (38.8, 120.3) for the restricted sets of instruments. As shown in Figure 2 and the Table, we did not find statistical evidence that statin-induced differential LDL-C response has a causal effect on T2D risk, nor that liability to T2D has a causal effect on statin-induced differential LDL-C response. This held true for both the full and restricted sets of instruments, and results from all sensitivity analyses were consistent with these findings. More specifically, our results for the full sets of instruments indicate the OR of T2D is 1.00 (95% CI: 0.97, 1.03) per 10% increase in statin-induced LDL-C response, and that statin-induced LDL-C response is increased by 0.20% (95% CI: −0.40, 0.80) per doubling of the odds of T2D liability. Evidence of unbalanced horizontal pleiotropy was present only for the restricted set of statin response instruments, as indicated by the MR-Egger intercept (intercept (95% CI): 1.04 (1.01, 1.07)) and Figure 3, but this is likely an artefact of including such a small number of instruments (n=4). Neither Cochran’s Q nor Rucker’s Q’ were suggestive of substantial heterogeneity in the effect estimates. Finally, a multivariable MR-analysis where we adjust for effects of all SNPs on fasted LDL-C did not lead to different conclusions regarding the effect of statin response on T2D risk (OR 1.02 (95% CI: 0.97, 1.07 per 10% increase in statin-induced LDL-C response).
Figure 2.
Scatter plots of instrument-outcomes (y-axis) against individual instrument-exposure (x-axis) per-allele effects, shown separately for statin response (left panel) and liability of type 2 diabetes instruments (T2D, right panel). The filled dots correspond to the restricted lists of variants (see text), while the full set included all dots. The lines correspond to the inverse-variance weighted combined MR estimator, for the restricted (dashed line) and full set of instruments.
Table.
Mendelian randomization (MR) estimators for the bidirectional association between type 2 diabetes (T2D) and statin-induced LDL-C response
| Number of instruments | F-statistic, median (25th, 75th percentile) | Estimate (IVW method, 95% CI) | Sensitivity analyses | Tests of heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|
| Weighted mode estimate (95% CI) | Weighted median estimate (95% CI) | MR-Egger estimate (95% CI) | MR-Egger intercept (95% CI) | Cochrane’s Q (p-value) | Rucker’s Q’ (p-value) | ||||
| Statin response ➔ T2D1 | 35 | 18.1 (16.2, 20.9) | 1.00 (0.97, 1.03) | 0.99 (0.93, 1.04) | 0.98 (0.94, 1.02) | 0.99 (0.94, 1.04) | 1.02 (0.92, 1.14) | 31.0 (0.61) | 31.0 (0.57) |
| 4 | 73.1 (38.8, 120.3) | 1.01 (0.93, 1.09) | 0.98 (0.91, 1.05) | 0.98 (0.91, 1.05) | 0.91 (0.81, 1.01) | 1.04 (1.01, 1.07) | 3.4 (0.33) | 0.2 (0.93) | |
| T2D liability ➔ Statin response2 | 62 | 23.2 (16.3, 37.8) | 0.20 (−0.40, 0.80) | 0.11 (−0.95, 1.18) | 0.03 (−0.97, 1.02) | 0.62 (−0.55, 1.80) | −0.04 (−0.15, 0.06) | 65.5 (0.32) | 65.3 (0.30) |
| 24 | 44.8 (35.5, 70.7) | 0.32 (−0.47, 1.12) | 0.04 (−1.07, 1.14) | 0.08 (−0.99, 1,15) | 1.24 (−0.67, 3.15) | −0.12 (−0.36, 0.11) | 32.4 (0.09) | 32.3 (0.07) | |
IVW denotes inverse-variance weighted. The different MR estimators can be interpreted as
odds ratio for T2D per 10% increase in proportional statin response, and
the effect on proportional statin response (%) per doubling in the odds of liability to T2D, respectively. A positive statin response value corresponds to an increased LDL cholesterol lowering effect of statin therapy.
Figure 3.
Funnel plots of individual causal effect estimates (Wald ratios) for statin response on type 2 diabetes (T2D, left panel), and liability to T2D on statin response (right panel). The black dots correspond to the restricted lists of variants (see text), while the full set included all dots. The lines correspond to the inverse-variance weighted combined MR estimator, for the restricted (dashed) and full set of instruments.
Discussion
Using LD score regression we found a positive genetic correlation of proportional statin response with T2D using genome-wide data, pointing to shared genetic determinants between these traits. However, our bi-directional MR analyses did not provide evidence of direct causal mechanisms of either statin-induced LDL-lowering on risk for T2D, nor liability to T2D on statin response. Sensitivity analyses of these MR analyses showed consistent results, suggesting that the issue of horizontal pleiotropy is unlikely to substantially influence our results.
The findings from the MR analyses suggest that statin-induced proportional change of LDL-C is unlikely to influence T2D risk. If true, this would indicate that it is not the degree of proportional lowering of LDL-C levels in response to statin therapy which increases the risk for diabetes, but the low levels in an absolute sense which may result from this transition. Indeed, results from the JUPITER trial have shown that achieving LDL-C concentrations <30 mg/dl with high-intensity statin therapy was associated with more physician-reported diabetes [33], which was not observed when a threshold of <50 mg/dl was considered [34]. Of further note here is a recent comparison of the risk of T2D in a large electronic health record database between individuals with low and normal LDL-C levels, which showed that LDL-C levels below 60 mg/dl occurring in absence of statin therapy are also associated with higher T2D risk [35]. Moreover, also in line with our results, researchers using data from 129,170 participants free from T2D at baseline from 20 statin trials did not observe evidence of a clinically relevant association between LDL-C proportional lowering at 1 year and within-trial odds ratios for new-onset type 2 diabetes (log-odds per 1% reduction in LDL-C: 0.004 (95% CI −0.001, 0.009)) [10].
However, we cannot exclude the possibility that a direct causal effect of statin-induced proportional LDL-C response on T2D may exist, but was (substantially) diluted by the presence of non-statin users in the DIAGRAM dataset. This is because proposed genetic instruments for pharmacological response phenotypes can only exert their effect in the presence of (i.e. are conditional on) drug usage. More intuitively, if this type of instrument were to associate with an outcome in a population which includes no relevant drug users, this must reflect an alternative pathway unrelated to that specific drug response (i.e. horizontal pleiotropy), or at least a shared genetic etiology between the two traits. Therefore, assessment of any causal effect of instruments derived from pharmacogenetic studies should ideally also be examined in populations composed solely of individuals using the drug of interest. An analogous dilemma has been described in the context of smoking heaviness, where a SNP which strongly predicts cigarettes per day was detected in a GWAS sample including daily smokers, and found to exert an effect only after a person has become an established smoker [36, 37]. Due to our use of summary statistics, we were unable to stratify our analyses on statin use. In addition, it was not possible to weight for the prevalence of statin use, as this is unknown for the DIAGRAM consortium, where statin use is additionally likely to be differential by case/control status. Therefore, given our null results, it is more appropriate to conclude that our MR analyses did not provide evidence for a shared genetic etiology between statin-induced proportional LDL-C response and T2D.
Furthermore, our observation that liability to T2D does not associate with LDL-C response resulting from statin treatment is consistent with previous studies showing that, while individuals with type 2 diabetes are likely to gain greater clinical benefit from statin therapy in terms of absolute CVD risk reduction, this does not result from differential lowering of their LDL-C concentrations when compared with non-diabetics [38].
An alternative non-causal explanation for the genetic correlation, which does suggest the presence of (some form of) directionally consistent pleiotropy, may be that both traits are independently influenced by the same underlying biological pathway [39]. Similar observations have been made in the field of neuropsychiatric diseases, where patterns of genetic correlation often reflect shared biological processes rather than causal relationships [40]. More generally, caution has been advised against treating LD score regression results as a singular assessment of causality, amongst others due to its lack of validation against some known causal relationships [41].
While an important strength of our analyses is the use of large-scale GWAS data, increasing the power of our investigations for both directions of causality, we purposely included instruments which did not attain genome-wide significance in their corresponding GWAS to increase the number of instruments. Though instrument-exposure associations will have been estimated with less precision for the weaker (i.e. sub-threshold) instruments, we considered this issue analogous to possible misspecification of weights in allele scores, which causal estimates have been shown to be generally robust to [42]. However, we cannot exclude the possibility of weak instrument bias, particularly for the full set of T2D liability instruments, which included several instruments with an individual F-statistic below 10 [22]. Given the relatively small overlap between the GWAS datasets, it is likely that any weak instrument bias would be towards the null [43]. However, the analysis using the restricted list of strong instruments reassuringly showed similar results.
In conclusion, our results suggest that liability to T2D is unlikely to influence LDL-C response to a statin, but provided some evidence of a shared genetic etiology between statin-induced LDL-C response and T2D. Future studies should make a definitive assessment of direct causal effects of statin-induced proportional LDL-C response on T2D in populations of statin users.
Supplementary Material
Acknowledgements:
The authors gratefully acknowledge the DIAGRAM consortium for making their GWAS summary data publicly available. In addition, we wish to express our gratitude to all studies participating in the GIST consortium. Full study-specific acknowledgments for the GIST consortium are given in ref 16.
Funding: JWJ is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). This work is also supported in part by the National Heart, Lung, and Blood Institute grants HL105756 (infrastructure grant for the Cohorts for Heart and Ageing Research in Genetic Epidemiology (CHARGE) consortium, to BMP), GM109145 (to CMS), and GM120523 (to QF).
Disclosures: BMP serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. DIC received research support for independent genetic analysis in JUPITER from AstraZeneca. RMK serves on the Merck Global Atherosclerosis Advisory Board. The remaining authors declare no competing financial interests.
Footnotes
Supplementary material: Supplementary information is available at The Pharmacogenomics Journal’s website.
References
- 1.Adhyaru BB, Jacobson TA. Safety and efficacy of statin therapy. Nat Rev Cardiol 2018; 15: 757–769. [DOI] [PubMed] [Google Scholar]
- 2.Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM Jr., Kastelein JJ, et al. Rosuvastatin to Prevent Vascular Events in Men and Women with Elevated C-Reactive Protein. N Engl J Med 2008; 359: 2195–2207. [DOI] [PubMed] [Google Scholar]
- 3.Sattar N, Preiss D, Murray HM, Welsh P, Buckley BM, de Craen AJM, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet 2010; 375: 735–742. [DOI] [PubMed] [Google Scholar]
- 4.Preiss D, Seshasai SR, Welsh P, Murphy SA, Ho JE, Waters DD, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA 2011; 305: 2556–2564. [DOI] [PubMed] [Google Scholar]
- 5.Navarese EP, Buffon A, Andreotti F, Kozinski M, Welton N, Fabiszak T, et al. Meta-analysis of impact of different types and doses of statins on new-onset diabetes mellitus. Am J Cardiol 2013; 111: 1123–1130. [DOI] [PubMed] [Google Scholar]
- 6.Besseling J, Kastelein JJ, Defesche JC, Hutten BA, Hovingh GK. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA 2015; 313: 1029–1036. [DOI] [PubMed] [Google Scholar]
- 7.Fall T, Xie W, Poon W, Yaghootkar H, Mägi R, GENESIS consortium, et al. Using Genetic Variants to Assess the Relationship Between Circulating Lipids and Type 2 Diabetes. Diabetes 2015; 64: 2676–2684. [DOI] [PubMed] [Google Scholar]
- 8.White J, Swerdlow DI, Preiss D, Fairhurst-Hunter Z, Keating BJ, Asselbergs FW, et al. Association of Lipid Fractions With Risks for Coronary Artery Disease and Diabetes. JAMA Cardiol 2016; 1: 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017; 49: 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JEL, Shah T, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. The Lancet 2015; 385: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt AF, Swerdlow DI, Holmes MV, Patel RS, Fairhurst-Hunter Z, Lyall DM, et al. PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. The Lancet Diabetes & Endocrinology 2017; 5: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotta LA, Sharp SJ, Burgess S, Perry JRB, Stewart ID, Willems SM, et al. Association Between Low-Density Lipoprotein Cholesterol-Lowering Genetic Variants and Risk of Type 2 Diabetes: A Meta-analysis. JAMA 2016; 316: 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med 2016; 375: 2144–2153. [DOI] [PubMed] [Google Scholar]
- 14.Labos C, Brophy JM, Smith GD, Sniderman AD, Thanassoulis G. Evaluation of the Pleiotropic Effects of Statins: A Reanalysis of the Randomized Trial Evidence Using Egger Regression-Brief Report. Arterioscler Thromb Vasc Biol 2018; 38: 262–265. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Liao JK. Pleiotropic Effects of Statins. Circulation Journal 2010; 74: 818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postmus I, Trompet S, Deshmukh HA, Barnes MR, Li X, Warren HR, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun 2014; 5: 5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 2017; 66: 2888–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012; 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017; 33: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock J, Yogo M. Testing for Weak Instruments in Linear IV Regression, Cambridge, New York, 2005. [Google Scholar]
- 23.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013; 37: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015; 44: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016; 40: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017; 46: 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rucker G, Schwarzer G, Carpenter JR, Binder H, Schumacher M. Treatment-effect estimates adjusted for small-study effects via a limit meta-analysis. Biostatistics 2011; 12: 122–142. [DOI] [PubMed] [Google Scholar]
- 28.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 2017; 36: 1783–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Thompson SG. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 2015; 181: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: 2017. [Google Scholar]
- 32.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018; 7: pii: e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Everett BM, Mora S, Glynn RJ, MacFadyen J, Ridker PM. Safety profile of subjects treated to very low low-density lipoprotein cholesterol levels (<30 mg/dl) with rosuvastatin 20 mg daily (from JUPITER). Am J Cardiol 2014; 114: 1682–1689. [DOI] [PubMed] [Google Scholar]
- 34.Hsia J, MacFadyen JG, Monyak J, Ridker PM. Cardiovascular event reduction and adverse events among subjects attaining low-density lipoprotein cholesterol <50 mg/dl with rosuvastatin. The JUPITER trial (Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin). J Am Coll Cardiol 2011; 57: 1666–1675. [DOI] [PubMed] [Google Scholar]
- 35.Feng Q, Wei WQ, Chung CP, Levinson RT, Sundermann AC, Mosley JD, et al. Relationship between very low low-density lipoprotein cholesterol concentrations not due to statin therapy and risk of type 2 diabetes: A US-based cross-sectional observational study using electronic health records. PLoS Med 2018; 15: e1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gage SH, Jones HJ, Taylor AE, Burgess S, Zammit S, Munafo MR. Investigating causality in associations between smoking initiation and schizophrenia using Mendelian randomization. Sci Rep 2017; 7: 40653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ware JJ, van den Bree MB, Munafo MR. Association of the CHRNA5-A3-B4 gene cluster with heaviness of smoking: a meta-analysis. Nicotine Tob Res 2011; 13: 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cholesterol Treatment Trialist’ (CTT) Collaborators Kearney PM, Blackwell L, Collins R, Keech A, Simes J, et al. Efficacy of cholesterol-lowering therapy in 18 686 people with diabetes in 14 randomised trials of statins: a meta-analysis. Lancet 2008; 371: 117–125. [DOI] [PubMed] [Google Scholar]
- 39.van Rheenen W, Peyrot WJ, Schork AJ, Lee SH, Wray NR. Genetic correlations of polygenic disease traits: from theory to practice. Nat Rev Genet 2019; 20: 567–581. [DOI] [PubMed] [Google Scholar]
- 40.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess S, Foley CN, Zuber V. Inferring Causal Relationships Between Risk Factors and Outcomes from Genome-Wide Association Study Data. Annu Rev Genomics Hum Genet 2018; 19: 303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 2013; 42: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013; 178: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.