Abstract
Multidrug resistance (MDR) of bacteria is a major challenge due to the wide-spread use of antibiotics. While a range of strategies have been developed in recent years, suppression of bacterial activity and virulence via their network of extracellular amyloid has rarely been explored, especially with nanomaterials. Here we synthesized silver nanoparticles and nanoclusters (AgNPs and AgNCs) capped with cationic branched polyethylenimine (bPEI) polymer, and determined their antimicrobial potentials at concentrations safe to mammalian cells. Compared with the ultrasmall AgNCs, AgNPs entailed stronger binding to suppress the fibrillization of FapC, a major protein constituent of the extracellular amyloid matrix of Pseudomonas aeruginosa. Both types of nanoparticles exhibited concentration-dependent antibiofilm and antimicrobial properties against P. aeruginosa. At concentrations of 1 μM or below, both the bactericidal activity of AgNCs and the antibiofilm capacity of AgNPs were associated with their structure-mediated bio-nano interactions but not ion release. For AgNPs, specifically, their antibiofilm potency correlated with their capacity of FapC fibrillization inhibition, but not with their bactericidal activity. Our study demonstrated the antimicrobial potential of safe nanotechnology through the novel route of amyloidosis inhibition.
Keywords: antibiofilm, FapC, amyloid, silver nanoparticle, silver nanocluster
INTRODUCTION
Bacterial infections are a major source of human diseases and mortality, while the gut microbiota are a key constituent of the gut-brain axis implicated in the physio-pathogeneses of neurological disorders, type 2 diabetes, obesity, depression and cancer.[1–5] Current antibiotic strategies usually target bacterial cell wall as well as their translational and gene replication machineries,[6] but some bacteria can evade virtually all antibiotics by enzymatic degradation, modifications to the cell wall and cellular organelles, or elevated expression of efflux pumps.[7] As a result, multi-drug resistance (MDR) of bacteria has become a major public health threat, and alternative strategies involving novel mechanisms are urgently needed.
Amyloidosis refers to the aggregation of proteins and peptides into toxic oligomers, protofibrils and cross-beta amyloid fibrils, with the latter being a hallmark of Alzheimer’s disease, Parkinson’s disease and type 2 diabetes.[8] Functional amyloidosis, in contrast, is a relatively new concept, exemplified by melanin synthesis via Pmel-17 aggregation in the skin against UV exposure, as well as by the formation of bacterial amyloid network/biofilm.[9, 10] The bacterial amyloid provides support for polysaccharides deposition to a) reinforce the encased community,[11] b) enable cell adhesion, motility, cell-cell interaction and quorum sensing,[12] and c) hinder penetration of antimicrobial agents.[13] FapC, in particular, is a main protein component of the extracellular amyloid network of Pseudomonas aeruginosa, a Gram-negative opportunistic pathogen inhabiting plants and animals, including humans.[9, 14] P. aeruginosa has a particularly large armamentarium of resistance mechanisms and can become resistant against all currently available antibiotics (in monotherapy). Furthermore, P. aeruginosa strains commonly form biofilm, which makes infections by this pathogen even more challenging to treat. The aggregation of FapC into amyloid fibrils is similar to the assembly of CsgA of the curli system in the amyloid biogenesis of Escherichia coli.[14, 15] FapC fibrils strengthen the bacterial biofilm mechanically,[16] and small molecules which remodel or dissociate FapC fibrils weaken the biofilm against antibiotics[17] and reduce biofilm formation to an extent which correlates with their ability to inhibit FapC fibrillization in vitro.[18] Accordingly, controlling bacterial amyloids may entail new antimicrobial strategies.
Nanotechnology offers unique advantages in bactericidal drug development, as the versatile and tunable physicochemical properties of nanomaterials facilitate their adsorption and penetration into bacterial membranes, biocatalysis, drug delivery and ion release. Indeed, gold, silver and zinc oxide nanoparticles and nanoclusters are common antimicrobial agents utilizing their strong capacity in inducing membrane damage and toxic ion release.[7, 19–22] Recently, silver nanoclusters (AgNCs) packed with daptomycin have shown improved potency and synergy in compromising bacterial membrane integrity, eliciting DNA damage, and generating reactive oxygen species (ROS).[23] In another study, silver bromide nanoparticle-pyridinium polymeric composites adhered onto glass substrates and exhibited long-term biocidal properties against airborne and waterborne bacteria.[24] Furthermore, lanthanum hydroxide-graphene oxide nanocomposites (La@GO) conferred strong bactericidal effects on both wild-type and antibiotic-resistant E. coli, Lactobacillus crispatus, Staphylococcus aureus and P. aeruginosa strains.[25]
Within the context of amyloid inhibition, the catalytic or inhibitory effects of metal nanoparticles depend upon the surface functionalization, metal core and size of the particles.[26–28] For example, gold nanoparticles (AuNPs) capped with citrate or polyethylene glycol (PEG) accelerated amyloidosis, while AuNPs coated with milk proteins inhibited the fibrillization of amyloid proteins.[29–33] Larger metal nanoparticles elicited a stronger inhibitory effect on amyloidosis than smaller nanoparticles of similar surface functionalization.[27] Silver nanoparticles (AgNPs) capped with citrate, chitosan or natural products have also been reported to inhibit the aggregation of amyloid proteins.[34–36]
The surface charge and antimicrobial property of AgNPs often correlate. For example, cationic AgNPs are more toxic to the net negatively charged bacterial cells, as they promote membrane permeability and reduce bacterial drug efflux.[37] In comparison, ultra-small AgNCs (<3 nm) can target MDR bacteria by a membrane disruption mechanism and act as a broad spectrum antimicrobial agent.[38] Motivated by the tremendous health implications of the gut microbiota as well as the urgent need to develop safe and potent nanobactericides against a range of human diseases, here we examined the interactions between P. aeruginosa biofilms and AgNPs/AgNCs from the unique perspective of FapC amyloidosis. While AgNPs/AgNCs have been studied for their tremendous potential against wild type and drug-resistant bacteria,[39–41] the toxicity associated with AgNPs/AgNCs hinders their clinical implications. Here we exploited the anti-amyloid activities of AgNPs/AgNCs at sub-bactericidal doses to inhibit biofilm formation. Our working hypothesis is that we can compromise bacterial viability by weakening the bacterial biofilm through a reduction of functional amyloid formation. AgNPs/AgNCs were synthesized via the chemical reduction method by adjusting the ligand to reductant ratio,[38] and were functionalized with cationic branched polyethylenimine (bPEI) polymer. bPEI coating enhances the interaction of nanoparticles with amyloid proteins and negatively charged P. aeruginosa,[42, 43] and is known for its interaction with bacterial surface and its associated bactericidal activities.[44, 45]
The interactions between AgNPs/AgNCs and FapC were studied via a thioflavin T (ThT) assay (for amyloidosis and inhibition), transmission electron microscopy (TEM, for effect on aggregate morphology), high-angle annular dark-field (HAADF) imaging (for elemental imaging and analysis), energy-dispersive X-ray spectroscopy (EDAX, for elemental analysis), circular dichroism (CD) spectroscopy (for protein secondary structure) and cytotoxicity (for biocompatibility of bactericides), complemented by discrete molecular dynamics (DMD) simulations (for molecular details of FapC-AgNC binding). In addition, bacterial cell cultures of P. aeruginosa (PAO1) in the presence of AgNPs/AgNCs were examined with a biofilm assay (for viability), fluorescence microscopy (for biofilm architecture), TEM and helium ion microscopy (HIM) (for more detailed biofilm architecture). Our results demonstrated a safe and facile new antimicrobial strategy through amyloidosis inhibition with nanomaterials.
MATERIALS AND METHODS
Branched polyethylenimine (bPEI, MW: 800, 99% pure), silver nitrate (AgNO3), formaldehyde (35 wt%), 4-(2hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), dialyzing tube (molecular weight cut-off ~1 kDa), crystal violet and cation-adjusted Mueller-Hinton broth CAMHB (BD, Sparks, MD, USA) were obtained by Sigma Aldrich. All solutions were prepared in Milli-Q water (18.2 MQ) and the solvents used were of analytical grade. FapC was produced from P. aeruginosa (PAO1) according to reported literature and stored in 6 M GdmCl.[14]
Synthesis of bPEI-capped AgNCs and AgNPs
AgNCs were prepared by a modified silver mirror reaction.[46] Briefly, an aqueous solution of bPEI (0.1 M) was prepared and its pH was adjusted to 7 with 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES buffer). 2.5 mL of AgNO3 (0.01 M) was added drop-wise into 1 mL of the bPEI solution under vigorous stirring. The solution turned from colorless to light yellow, indicating the formation of bPEI-Ag complexes. Formaldehyde (100 μL, 30%) was then added dropwise to the mixture and incubated for 10 min at 70 °C and then at room temperature for 24 h to obtain bPEI-capped AgNCs. AgNCs were purified by dialysis against water in 1 kDa MWCO membranes for 24 h. UV-Vis absorption spectra exhibited peaks at 268 and 354 nm indicating the formation of AgNCs.[47]
For the synthesis of bPEI-capped AgNPs, an aqueous solution of bPEI (0.1 M, 5 mL) was heated up to 90 °C followed by a quick addition of AgNO3 (0.15 M, 5 mL). The reaction mixture was stirred continuously at 90 °C for 1 h, resulting in a change of color from colorless to yellow and then dark brown. The bPEI-capped AgNPs were cooled down to room temperature, purified by centrifugal washing (3×, 16,000 g for 30 min), and dispersed in water. The UV-Vis spectra of the resulted particles displayed a peak around 415 nm indicating the formation of AgNPs.[48] The nanoparticles were stored in dark at 4 °C for further use.
Sizes and zeta potentials of AgNCs and AgNPs
The hydrodynamic diameter (HDD) and zeta (ζ) potential of the AgNCs and AgNPs were acquired using a dynamic light scattering device (Zetasizer Nano-ZS, Malvern Instruments) at room temperature (25 °C) and with folded capillary zeta cells, and were recorded with Zeta software 7.03.
Thermogravimetric analysis
For thermogravimetric analysis (TGA) analysis to quantify bPEI grafted on the nanoparticle surfaces, 100 μL solutions of AgNCs or AgNPs were placed on a platinum pan. The samples were kept at 80 °C for 30 min to remove water and then the weight losses were measured from 80 to 700 °C at a rate of 10 °C/min under inert atmosphere of nitrogen. The measurements were conducted using a Shimadzu DTG-60AH analyzer.
Inductively coupled plasma optical emission spectroscopy
The concentrations of Ag in AgNCs and AgNPs were determined using inductively coupled plasma optical emission spectroscopy (ICP-OES) (iCAP 6000 Series, Thermo Scientific). The samples (1 mL) were digested by adding 9 mL of HNO3 (68%) and heating at 190, 150 and 100 °C (30 min at each step). The dried layer was reconstituted in 1 mL (4%) HNO3 and analyzed by ICP-OES.
ThT kinetic assay
A ThT kinetic assay was performed by preparing a 50 μL aqueous solution containing 75 μM ThT and 50 μM of FapC, in the presence or absence of AgNCs or AgNPs (1 μM), in a 96-well plate. The plates were incubated at 37 °C for 120 h without agitation and ThT fluorescence (excitation: 440 nm/emission: 485 nm) was recorded at different time points. ThT kinetic parameters of rate of fibrillization (k), time to reach half of the saturation point (T1/2) and lag time were calculated as described.[49]
Transmission electron microscopy and energy-dispersive X-ray spectroscopy
FapC was incubated with AgNCs or AgNPs at the same molar ratios as for the ThT assay. After 120 h of incubation, a 5 μL drop of each sample was placed on a glow-discharged copper grid and blotted after 1 min. The grid was negatively stained for 30 s with uranyl acetate (1%) and excess stain was blotted off. The prepared grid was air dried and imaged for TEM, HAADF and EDAX mapping spectra with an FEI Tecnai F20 microscope operated at 200 kV.
Circular dichroism spectroscopy
To determine the secondary structures of FapC monomers, fibrils and FapC fibrillated with or without AgNCs/AgNPs, 200 μL of each sample same as for the ThT assay was transferred into a CD cuvette. CD spectra were recorded from 190–240 nm, with a 1 nm step size. The data were deconvoluted and percentage secondary structures were obtained by Dichroweb.[50]
In vitro cytotoxicity
Human embryonic kidney 293 (HEK 293 from ATCC) cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) with 15% fetal bovine serum (FBS). For the viability assay, poly-L-lysine (70 μL) was coated on a costar black/clear bottom 96 well plate, incubated for 30 min at 37 °C and washed with DI water. 200 μL of the cells (density: 10,000 cells) were added to the wells. The cells were incubated at 37 °C for 24 h to attain 70–80% confluency. After replenishing the media, 1 μM PI dye, dissolved in DMEM, was added to the wells and incubated for 30 min. Samples of 25 μM FapC and 5 μM AgNCs or AgNPs (200 μL) were added into the wells. All samples were observed in triplicate and estimated in a live cell chamber (5% CO2, 37 °C) by Operetta (PerkinElmer, microscope objective: 20× PlanApo; numerical aperture: 0.7) after 14 h of treatment. The ratio of dead to total cell count was calculated by a built-in function, bright-field mapping of Harmony High-Content Imaging and Analysis software. The measurement was performed at 5 reads/well. Untreated cells were used as the control.
Bacterial viability and biofilm assay
Prior to the experiment, P. aeruginosa (wild-type reference strain PAO1) was freshly subcultured onto cation-adjusted Mueller-Hinton agar (CAMHA, containing 25 mg/L Ca2+ and 12.5 mg/L Mg2+) and incubated at 35 °C for 24 h. Random colonies (2–3) were selected and grown in 10 mL cation-adjusted Mueller-Hinton broth (CAMHB, containing 25 mg/L Ca2+ and 12.5 mg/L Mg2+) overnight, from which early-log-phase growth was obtained. The OD of the culture was adjusted to 0.5 MacFarland standard and 100 μL of the bacterial suspension was mixed with 100 μL of different concentrations of AgNPs or AgNCs (prepared by dilutions in CAMHB) and incubated for 24 h at 35 °C. A bacterial viability assay was performed by propidium iodide (PI) staining. Briefly, 1 μL of 2 mg/mL PI solution was added to each well and incubated for 20 min. Excess dye was removed by centrifugal washing at 5,000 g for 5 min and bacterial cells were resuspended in phosphate-buffered saline (PBS, 0.1 M, pH 7.4). Fluorescence corresponding to the PI-stained dead cells was recorded for excitation at 535 nm and emission at 617 nm. The percentage dead cells were recorded relative to the positive (0.1% Triton X-100) and negative (untreated) controls.[51]
Biofilm formation was assessed by crystal violet assay. Briefly 100 μL of bacterial suspension (OD adjusted to 0.5 MacFarland standard) was incubated at 35 °C with 100 μL of different concentrations of AgNPs or AgNCs. The medium was gently removed after 24 h and the wells were washed with PBS (0.1 M, pH 7.4) thrice. The microplates were air dried and then biofilms were stained with crystal violet (0.1%, 200 μL) for 15 min. Excess crystal violet dye was removed and the wells were washed again with PBS (0.1 M, pH 7.4) to remove traces of unbound dye. The biofilms stained with crystal violet were dissolved in acetic acid (200 μL, 33%) and transferred to new microplates. The absorbance was recorded at 595 nm, corresponding to the crystal violet-stained biofilms. Percentage biofilm formation was calculated relative to the positive and negative controls, similar to the viability assay.
Fluorescence microscopy and hyperspectral imaging
A fluorescent strain of P. aeruginosa (AH298-GFP) was employed to visualize the biofilm formation and viability of bacteria treated with AgNPs and AgNCs, respectively. After 24 h incubation of the bacteria with 1 μM of the nanoparticles (with respect to Ag content), 5 μL (100 μM) of ThT dye was added to the culture and incubated for another 10 min. Excess ThT dye was removed by centrifugal washing (5,000 g for 5 min, thrice) and bacterial cells were suspended in PBS (0.1 M, pH 7.4). 10 μL of this bacterial suspension was placed on poly-L-lysine-coated cover slips and imaged under the GFP channel of a fluorescence microscope (Nikon, Eclipse Ti).
Hyperspectral imaging (HSI) was performed to detect the surface plasmon resonance (SPR) spectra of AgNPs in AgNPs-treated bacteria. For this, bacterial cells were incubated with AgNPs for 24 h and then 10 μL of the culture was placed on a poly-L-lysine-coated glass slide, covered with a coverslip and imaged under an HSI dark field microscope (CytoViva) attached with a PixelFly CCD camera. Images were processed via ENVI 4.8 software. AgNPs alone were used as a control and a spectral library generated from AgNPs was averaged to single mean spectra and scanned against AgNPs-treated bacterial samples.[32] No scattering spectra were observed from untreated bacterial control.
Transmission electron microscopy and helium ion microscopy of bacterial cells
TEM samples of treated and untreated bacterial cells were prepared by incubating the cells with AgNPs or AgNCs for 24 h, and washing them via repeated centrifugation. 5 μL of each sample was prepared on a grid as described for the nanoparticles in the previous section. Helium ion microscopy (HIM) was performed to image the surface morphology of fixed bacterial cells. Briefly, bacterial cells were treated with the nanoparticles for 24 h and then fixed by adding 200 μL of 2.5% paraformaldehyde. The bacterial cells were washed thrice via centrifugal washing and resuspended in PBS (0.1 M, pH 7.4). The bacterial cells were placed on poly-L-lysine-treated glass slides and incubated for 1 h. The slides were washed with excess DI water and then dehydrated in gradual concentrations of ethanol, i.e., 10, 20, 30, 50, 70, 90, 100 and 100% with 30 min incubation at each step. The slides were dried and imaged under HIM (Orion NanoFab, Zeiss, USA).
Reactive oxygen species generation
ROS generation by bacterial cells was measured by 2,7-dichlorofluorescein diacetate (DCFH-DA). The bacterial cell culture at OD 0.6 (100 μL) was incubated for 5 h with AgNPs or AgNCs (100 μL at concentrations similar to the viability assay). The culture was centrifuged at 5,000 g for 5 min and the supernatant was treated with DCFH-DA (100 μM) for 1 h. The fluorescence was recorded at excitation wavelength of 495 nm and emission wavelength at 529 nm. ROS generation was calculated relative to the positive control of 0.1% H2O2.
Discrete molecular dynamics simulations
We applied atomistic DMD simulations to simulate FapC aggregation. DMD is a rapid and predictive molecular dynamics algorithm which has been used to study protein aggregation and nano-bio interface,[32, 52] and detailed description of the DMD algorithm can be found elsewhere.[53–55] Based on our previous study,[56] we reconstructed an AgNC comprised of 38 silver atoms with a diameter of ~1 nm. We used the Ag (111) surface with five layers of atoms to approximate the relatively flat surface of an AgNP with a much larger radius. Each layer was comprised of 648 silver atoms with a dimension of ~7.0 × 6.8 nm2. To model the bPEI molecule, we started with a generation-3 PEI dendrimer[56] and randomly deleted terminal ethyleneimine groups iteratively until reaching the MW of ~ 800 Da as in the experiment. To construct the bPEI-AgNC complex, we performed initial binding simulations of one AgNC with nine bPEI molecules at 300K. Up to three bPEI molecules were found to bind the AgNC strongly. For the bPEI-AgNP complex, we performed an initial DMD simulation started with twelve bPEI molecules covering the surface, and found that only nine bPEI molecules were able to stay bound. Thus, an AgNC bound with three bPEI molecules and the nano-sized Ag (111) surface covered with nine bPEI molecules approximating an AgNP were used in our further simulations.
In addition to the N-terminal signaling sequence, FapC is comprised of three homologous repeat sequences (R1-R3) separated by two linker regions (L1-L2). We used the R1L1R2L2R3 region from the FapC sequence of the Pseudomonas strain UK4 (175 amino acids in total) in our computational modeling. For the full-length FapC (R1L1R2L2R3), equilibrated conformations were obtained with initial relaxation simulations at room temperature starting from a fully-stretched conformation. A cubic box with the periodic boundary condition and a dimension of 12 nm was used. To ensure sufficient sampling, ten independent simulations with different initial configurations were performed for a FapC monomer with and without the presence of a bPEI-capped AgNC. Each independent simulation lasted 350 ns at room temperature.
We used two fragments L1R2 and L2R3 separately to model the dimerization process in FapC aggregation and evaluated the impact of the bPEI-capped AgNC and bPEI-capped AgNP on FapC aggregation. For each dimer simulation in the presence and absence of AgNC or AgNP, 20 independent simulations each of 300 ns were performed. A simulation box with each dimension of 15 nm was used. The last 50 ns trajectories of all independent simulations, where the corresponding steady states were reached, were used in statistical analysis.
RESULTS AND DISCUSSION
Synthesis and characterizations of bPEI-capped AgNPs and AgNCs
Cationic bPEI[57] was employed to graft two types of Ag nanostructures (Fig. 1A,B) by chemical reduction.[38, 58] The size, morphology and zeta potential of the nanoparticles are summarized in Fig. 1C–H. The nanoparticles of ~2 nm in size and exhibiting UV-Vis absorption peaks at 268 nm and 354 nm were designated as bPEI-capped AgNCs, while the nanoparticles of ~10 nm in size and possessing a definite surface plasmon peak at 420 nm were labelled as bPEI-capped AgNPs (Fig. 1C,D). TEM micrographs revealed that both the AgNCs (2 nm) and AgNPs (10 nm) were well dispersed and near spherical (Fig. 1E,F). Dynamic light scattering (DLS) measurements indicated the corresponding hydrodynamic diameter and zeta potential for the bPEI-capped AgNCs and AgNPs were 6.2 nm and +33 mV, and 18.6 nm and +15 mV, respectively, and the nanoparticles were relatively monodispersed with a polydispersity index (PDI) of < 0.4 (Fig. 1G,H). The size distribution histograms of AgNCs and AgNPs, calculated from the TEM images, are presented in Fig. S1 (Supplementary Information). ICP-OES was used to quantify the concentrations of Ag in AgNCs and AgNPs. The samples were digested in aqua regia and the Ag contents were quantified to be 0.37 mM and 34 mM for AgNCs and AgNPs, respectively. The bPEI contents were quantified via TGA, at 0.95 mM for AgNCs and 13.2 mM for AgNPs, accordingly (Fig. S2).
Figure 1. Physical characteristics of bPEI-capped AgNCs and AgNPs.
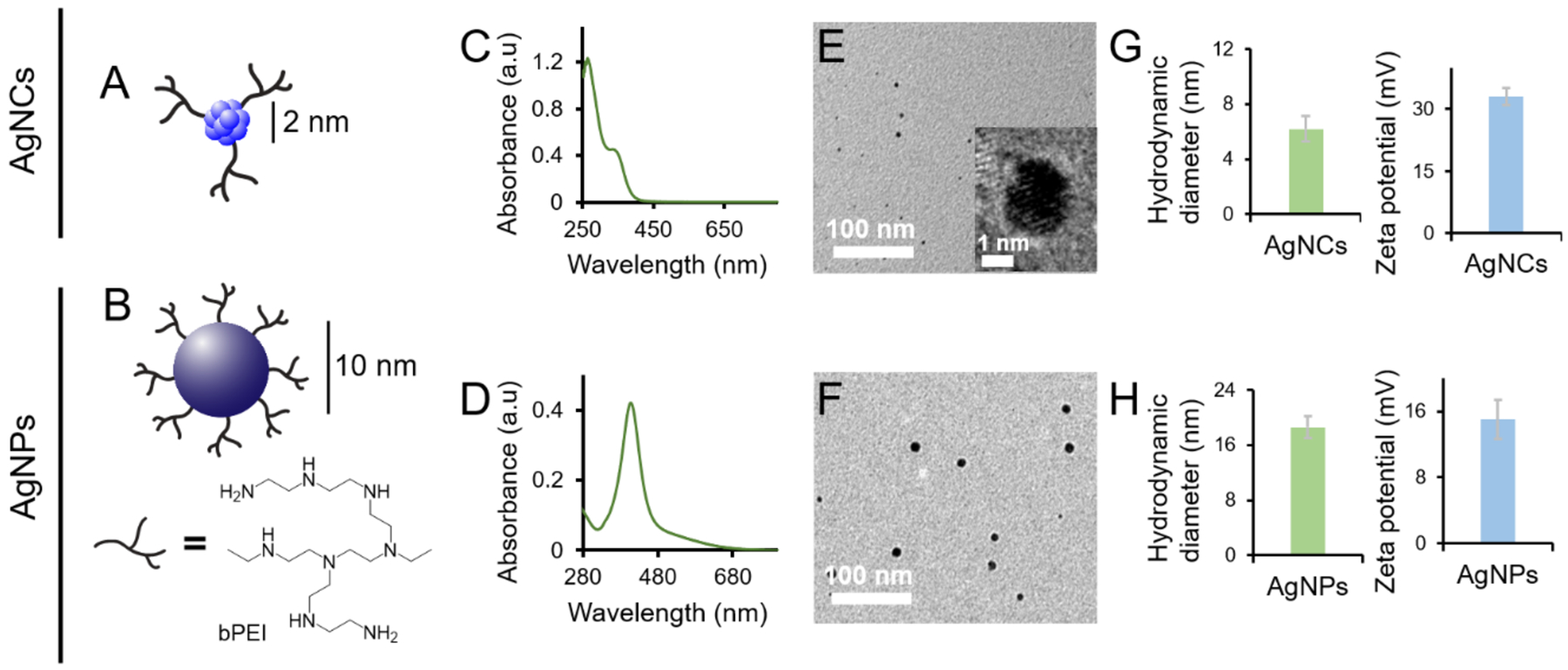
Graphical illustrations of bPEI-capped AgNCs (A) and AgNPs (B). UV-Vis absorption spectra of bPEI-capped AgNCs (C) and AgNPs (D). TEM micrographs of bPEI-capped AgNCs (E) and AgNPs (F). Zeta potentials and hydrodynamic diameters of bPEI-capped AgNCs (G) and AgNPs (H).
In vitro interactions between FapC and bPEI-capped AgNPs and AgNCs
Amyloidosis inhibition with nanomaterials is an emerging frontier against amyloid diseases,[28, 31, 59, 60] but has rarely been attempted before for antimicrobial purposes.[61] Here the effects of bPEI- capped AgNPs or AgNCs on FapC fibrillization were systematically investigated to inhibit biofilm formation by P. aeruginosa. FapC monomers (50 μM) were incubated alone or with bPEI-capped AgNPs or AgNCs (equivalent to 1 μM Ag) for 5 days at 37 °C. In the absence of agitation, FapC fibrillization (monitored by the fluorescence of the amyloid-binding ThT) was slow and showed a lag time of 50 h, after which it reached a plateau within the next ~25 h (Fig. 2A). When FapC was incubated with bPEI-capped AgNPs, FapC fibrillization was completely inhibited. In contrast, AgNCs shortened the lag phase to ~25 h but also reduced the end-level ThT fluorescence by 4-fold (Fig. 2A, key kinetic parameters summarized in Table S1). CD spectroscopy revealed changes in the secondary structure of FapC before and after fibrillization, in the presence and absence of AgNPs or AgNCs (Fig. 2B). According to the deconvolution program DichroWeb, the α-helical content of FapC was decreased from 28.4% (monomers) to 5.9% (fibrils), while the β-sheet content was increased from 15.6% (monomers) to 47.7% (fibrils). When incubated with AgNPs, the secondary structure of the AgNPs + FapC complexes was similar to that of FapC monomers, at 25% for α helices and 18% for β sheets. This suggests that the AgNPs inhibited FapC fibrillization by sequestering monomeric FapC (Fig. 2C).[32] The secondary structure contents of the AgNCs + FapC complexes appeared to be between that for FapC monomers and fibrils, indicating limited formation of cross-β amyloid fibrils (Fig. 2B,C). Similarly, the zeta potential of AgNCs + FapC complexes was −25 mV, compared to a zeta potential of −36 mV for FapC amyloid fibrils and +30 mV for AgNCs, indicating the presence of fibrils in the AgNCs + FapC complexes (Fig. 2D). Also, the zeta potential of AgNPs dropped from +16 mV to +10 mV, after binding with FapC, confirming sequestration of FapC by AgNPs. The influence of AgNPs/AgNCs on FapC amyloidosis was further studied by TEM. No FapC amyloids were observed in the FapC + AgNPs sample, indicating full inhibition of FapC amyloidosis by AgNPs (Fig. 2E), while AgNCs clustered together with FapC monomers and supported the formation of short FapC fibrils (Fig. 2F). The white signals from the HAADF images (Fig. 2G,H) indicated the presence of Ag metal (EDAX mapping spectra in Fig. 2I,J). The Cu signal was from the Cu grid. The presence of AgNCs in the core of clustered AgNCs + FapC complexes was further confirmed by HAADF imaging (Fig. 2H). Fig. 2K,L presents graphical illustrations of the binding between FapC and AgNPs or AgNCs. bPEI and AgNO3 did not elicit any effect on FapC fibrillization, at a concentration equivalent to 1 μM (with respect to Ag) of AgNPs/NCs (Fig. S3). No toxicity of FapC was observed against HEK 293 cells, while AgNPs and AgNCs were biocompatible at 1 μM Ag equivalent concentration (Fig. S4).
Figure 2. Binding between FapC and bPEI-capped AgNPs or AgNCs.
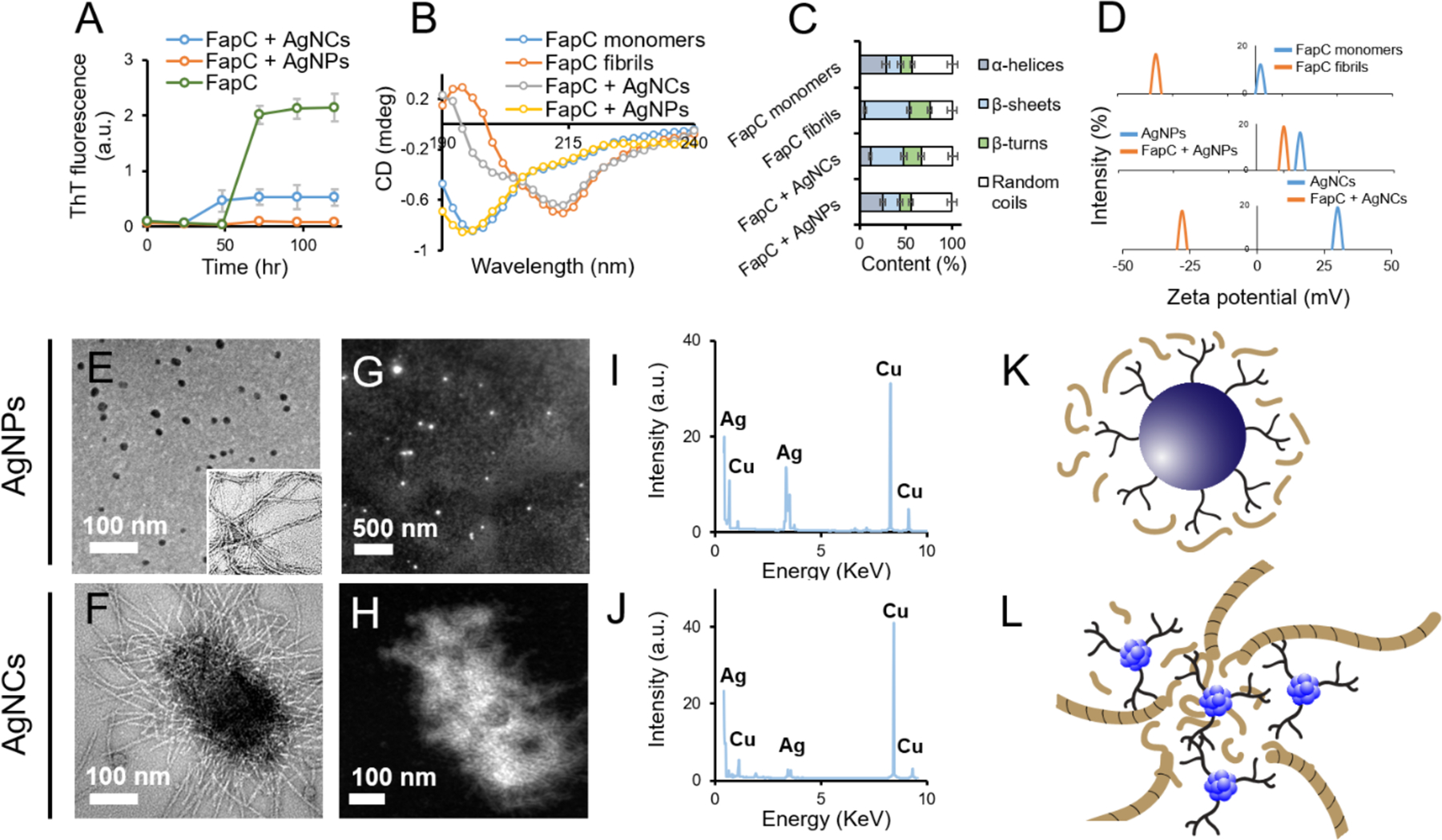
ThT kinetic assay of FapC mixed with AgNPs or AgNCs (A). CD spectra (B) and percentage secondary structure (C) of FapC monomers, fibrils and FapC monomers after incubation with bPEI-capped AgNPs or AgNCs, calculated using DichroWeb. Changes in the zeta potential of FapC monomers before and after incubation with bPEI-capped AgNPs and AgNCs (D). TEM micrographs of FapC + AgNPs (E) and FapC + AgNCs (F). The inset shows control FapC fibers. Panels G,H and I,J present the respective HAADF images and EDAX spectra of FapC + AgNPs and FapC + AgNCs. Panels K and L represent graphical illustrations of bPEI-capped AgNPs and AgNCs interacting with FapC.
The differences in the size and curvature of AgNPs and AgNCs can explain their differential behavior against FapC fibrillization. The large size of AgNPs (10 nm) matched the cross sections of single amyloid fibrils (~5–10 nm) and also provided a surface area large enough to physically interface with FapC monomers,[8] thereby enabling surface-assisted binding with the protein to prevent FapC from aggregation.[29, 30] In the case of AgNCs, their small size (3 nm) resulted in a shortened nucleation phase (<24 h), likely due to their large surface area for FapC adsorption. However, due to the heterogeneity of the AgNCs + FapC aggregates, elongation of FapC protofibrils into fibrils could be entropically unfavorable, leading to a reduced yield of fibrils compared with FapC alone (Fig. 2A).
In silico interactions of bPEI-capped AgNC with FapC monomer
Given the much larger size (and, hence, greater computational cost) of AgNPs compared to AgNCs, we first focused on the interactions of a bPEI-capped AgNC on FapC monomer on the molecular level (Methods). From control simulations of a full-length FapC (R1L1R2L2R3, 175 residues excluding the N-terminal signaling sequence) monomer alone, the isolated protein mainly adopted a random coil conformation as also observed experimentally (Fig. 3A & Fig. 2C). In the presence of a bPEI-capped AgNC, the two linker regions – L1 and L2 – corresponded to the major binding sites for the positively charged AgNCs. Residues with binding probabilities higher than 0.7 for the AgNC included three negatively charged residues (49D, 119D, 125E), two polar residues (53T, 117S, 121H), and two hydrophobic residues (118L, 122F). There were also two other binding peaks around 29D and 103D, together confirming that the binding was mainly driven by electrostatic interactions between the positively charged bPEI and the negatively charged residues of FapC, in addition to H-bonding and hydrophobic interactions. The binding of bPEI-AgNC resulted in local perturbations to β sheets and α helices near the binding sites (Fig. 3A&3B, & Fig. S5), although the overall content changes of β sheets and α helices were small (e.g., the representative snapshots of FapC monomer with and without AgNC binding in Fig. 3C).
Figure 3. DMD simulations of a bPEI-capped AgNC interacting with a FapC monomer and a FapC fragment dimer.
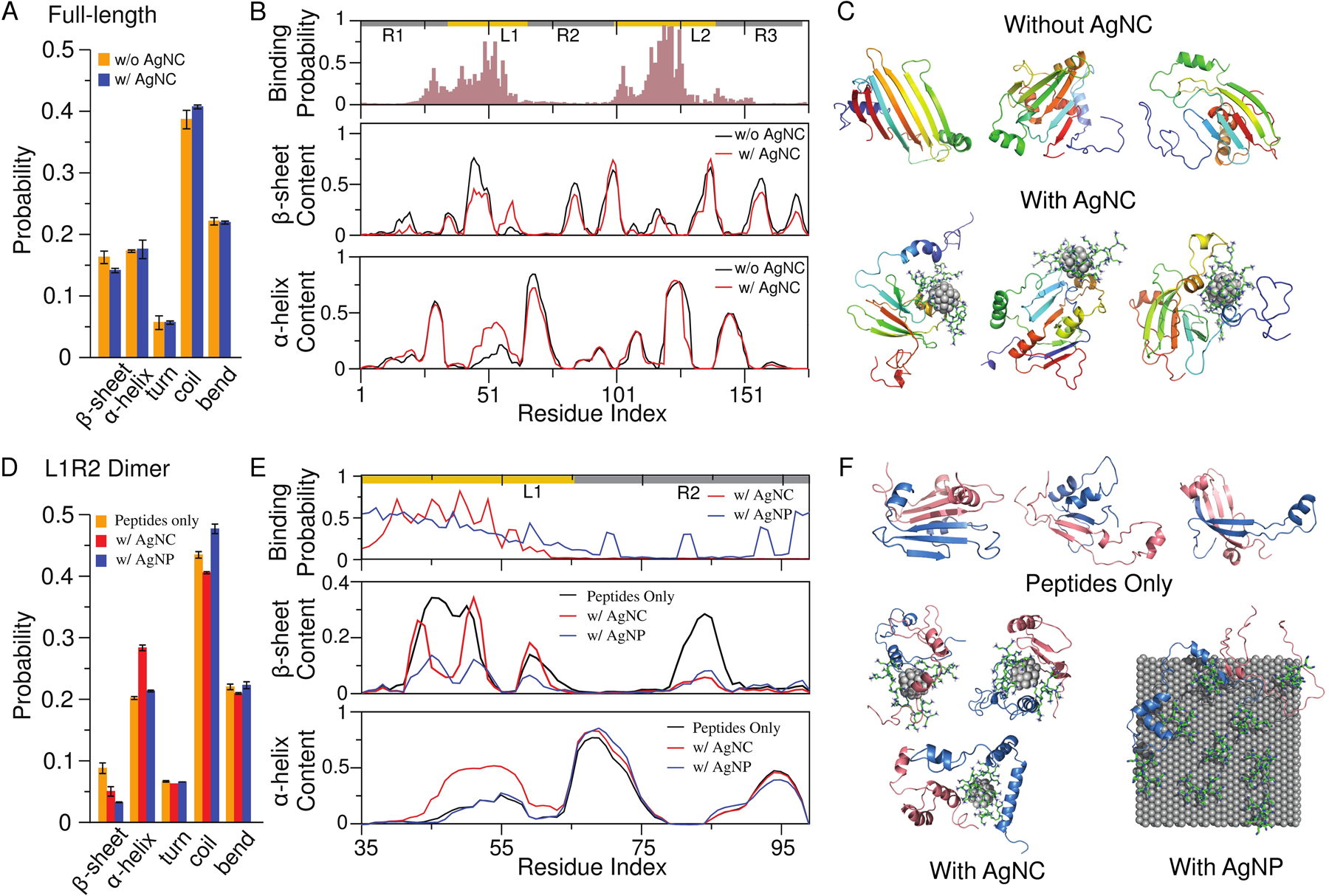
(A-C) Full-length FapC monomer (R1L1R2L2R3) with and without the presence of the bPEI-capped AgNC. (A) Overall secondary structure contents of the full-length FapC monomer. (B) The bPEI-capped AgNC-binding probability, and the β-sheet and α-helix propensities of each residue. (C) Three representative structures of FapC in the absence (upper) and presence (lower) of the AgNC. The peptide shown as cartoon is colored rainbow from blue (N-terminus) to red (C-terminus). The core of the AgNC is shown as spheres and bPEIs as sticks. (D-E) A L1R2 dimer with and without the presence of the bPEI-capped AgNC. (D) Overall secondary structure contents of the dimer. (E) The binding probability with bPEI-capped AgNC or AgNP, and the β-sheet and α-helix propensities of each residue. (F) Representative structures of the L1R2 dimer in the absence (upper) and presence of the bPEI-capped AgNC (lower left) and AgNP (lower right). The two peptides are colored differently.
The large size of FapC (175 residues) made the aggregation simulation computationally expensive. On the other hand, analysis of intra-chain contact frequency for FapC monomer simulations indicated that the R1 region had the least interactions among itself and with the rest of the protein (Fig. S5A), suggesting that R1 likely played a weaker role in driving the self-assembly of FapC. In addition, because the bPEI-capped AgNC preferred to bind the two linker regions (L1 and L2), we adopted the divide-and-conquer approach and used two smaller FapC fragments – L1R2 and L2R3 – to study their aggregation. Next, we performed DMD simulations to evaluate the effects of the bPEI-capped AgNC and AgNP on the dimer formation of the two fragments as an initial step towards FapC aggregation.
In silico interactions of bPEI-capped AgNC and AgNP on FapC fragment dimer
As in an early study,[56] we modeled the effect of large AgNPs by a nano-sized (111) silver surface with five atom layers, where each layer was comprised of 648 atoms occupying a dimension of ~7.0 × 6.8 nm2. In contrast to the 38-atom AgNC coated with three bPEIs, only nine bPEIs with strong electrostatic repulsion with each other were able to bind the nano AgNP surface due to differences in radii and thus surface curvatures of the two types of nanostructures (Methods). Compared to AgNC, the lower bPEI coating density of the simulated AgNP was consistent with its smaller zeta potential observed experimentally (Fig. 1).
In the dimer simulations of both L1R2 (Fig. 3D–F) and L2R3 (Fig. S6), the bPEI-capped AgNC preferred to bind the linker regions as also observed in the monomer simulations. Compared to the full-length FapC (Fig. 3A), fragments had a lower overall β-sheet content (Figs. S6&S7). Nevertheless, with the addition of a bPEI-capped AgNC, significant inhibition of β sheets and promotion of α helices were observed for fragment L1R2 (Fig. 3D,E,F). Similarly, the bPEI-capped AgNC also significantly inhibited the formation of β sheets by the L2R3 dimer (Fig. S6). In the presence of bPEI-capped AgNP, stronger inhibition of β sheets and promotion of coil were observed for both L1R2 (Fig. 3D,E,F) and L2R3 (Fig. S6). Compared to bPEI-capped AgNC, the low coating density of bPEI molecules in AgNP resulted in more surface silver atoms exposed to interact with the peptides. As a result, bPEI-capped AgNP could bind more FapC regions than bPEI-capped AgNC (Fig. 3E&S6), leading to the observed stronger inhibition of β sheets and promotion of coils. Overall, the dimer simulation results were consistent with the CD measurement showing increased α helices and decreased β sheets upon addition of bPEI-capped AgNCs or AgNPs (Fig. 2B,C), and also consistent with the experimental observation that bPEI-capped AgNPs rendered stronger inhibition against the formation of β-rich aggregates. The analysis of inter-peptide contact frequencies (Fig. S7) indicated that the linker regions (especially L1) played an important role in dimerization by forming inter-peptide β sheets in the absence of the AgNC or AgNP. By binding to the linker regions multi-valently (e.g., typical snapshots in Figs. 3F&S6), the bPEI-capped AgNC disrupted β-sheet formation with a long-range order to mitigate FapC fibrillization. In the case of bPEI-capped AgNP, the binding with additional regions than the linkers in FapC resulted in stronger disruption of inter-molecular β-sheet formation and subsequent FapC fibrillization.
Effect of bPEI-capped AgNPs and AgNCs on biofilm formation
Following the in vitro characterizations of FapC interactions with AgNPs or AgNCs, the effect of such interactions on P. aeruginosa biofilm formation was studied by incubating the nanoparticles with the bacteria in microtiter plate wells for 24 h. First, the effects of AgNPs and AgNCs on biofilm formation and planktonic bacterial viability were studied. AgNCs completely suppressed biofilm formation ≥ 500 nM, but below 500 nM AgNCs, the bactericidal effect diminished. At all concentrations, biofilm formation correlated with bacterial viability (Fig. 4A). In contrast, 250 nM to 1 μM AgNPs suppressed biofilm formation while displaying an insignificant bactericidal effect (Fig. 4B). Below 250 nM, the antibiofilm activity of AgNPs became less prominent while the bacteria remained intact. At concentrations higher than 1 μM, the bactericidal activity of AgNPs eliminated the availability of viable bacteria from establishing biofilm.[38] The in-vitro anti-FapC activities of AgNPs/AgNCs were correlated with their antibiofilm activity. The relative bactericidal and anti-FapC activity of AgNCs and AgNPs can be explained based on their sizes and surface areas per particle. The larger AgNPs (10 nm) have more contact area per particle, which enabled them to sequester more FapC molecules per AgNP to mitigate FapC fibrillization. This is consistent with the literature where larger nanoparticles were more efficient amyloid inhibitors than their smaller counterparts.[62, 63] However, in terms of antibacterial efficacy, smaller AgNPs have been shown to be more efficient,[64] as they possess a greater affinity for bacterial cell wall and can be efficiently internalized by bacteria.[65]
Figure 4. Effects of bPEI-capped AgNPs and AgNCs on the biofilm formation of P. aeruginosa.
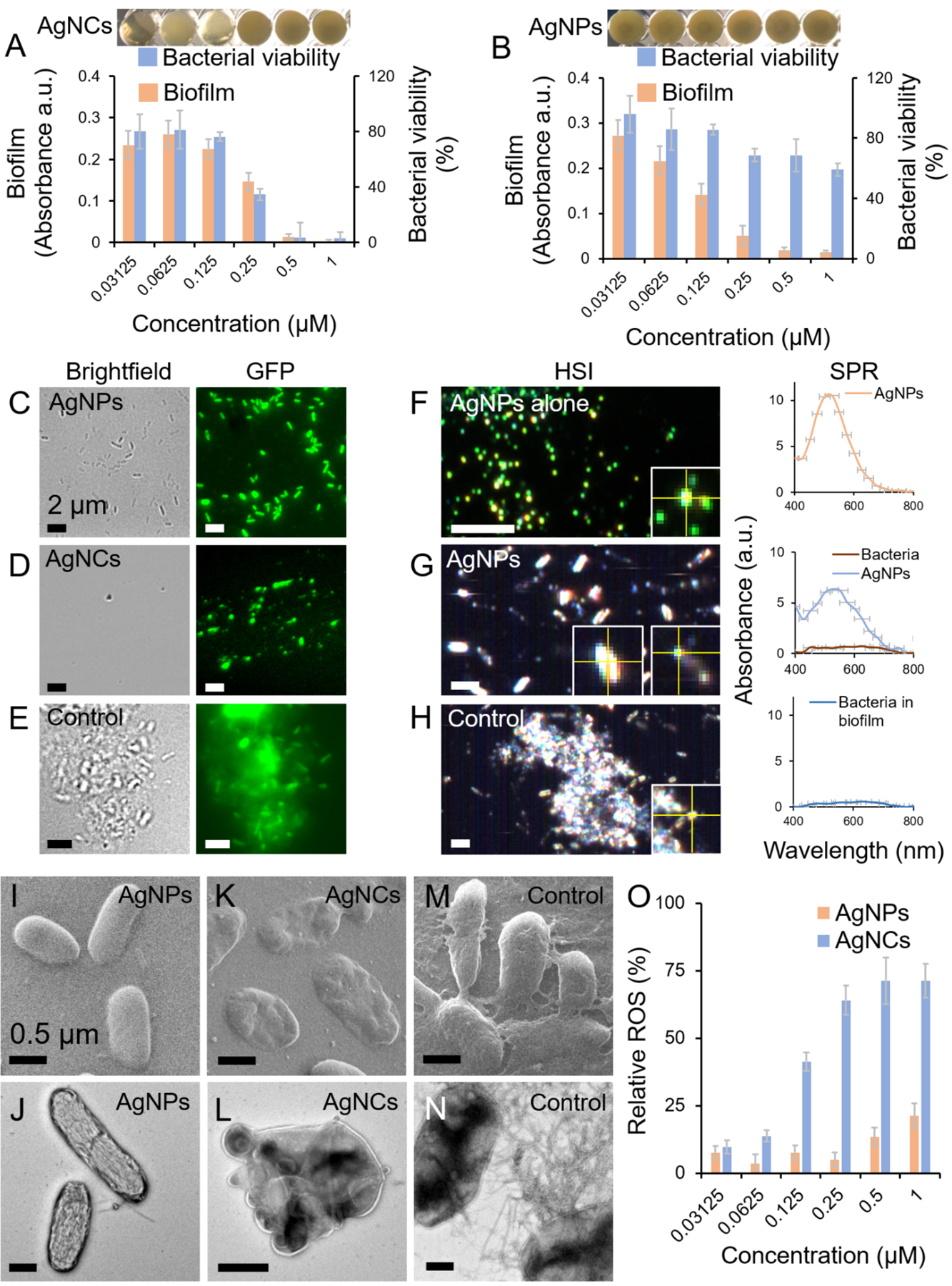
Relative biofilm formation and bacterial viability of P. aeruginosa PAO1 in the presence of different concentrations of bPEI-capped AgNCs (A) and AgNPs (B). Fluorescence microscopy images of P. aeruginosa AH298-GFP incubated with 1 μM of AgNPs (C) and AgNCs (D) and untreated control that formed biofilms (E). A fluorescent strain of P. aeruginosa was used for fluorescence microscopy and ThT dye was used to stain the biofilms. Panels F, G and H represent the hyperspectral images (HSI) along with the SPR spectra of AgNPs, P. aeruginosa incubated with 1 μM AgNPs and P. aeruginosa control, respectively. Helium ion microscopy (HIM) and TEM micrographs of P. aeruginosa incubated with AgNPs (HIM: panel I, TEM: panel J), P. aeruginosa incubated with AgNCs (HIM: panel K, TEM: panel L) and P. aeruginosa control (HIM: panel M, TEM: panel N). ROS generation by P. aeruginosa, relative to positive control (H2O2), upon incubation with AgNPs and AgNCs (panel O).
To image the interactions of the nanoparticles with P. aeruginosa, a fluorescent strain of P. aeruginosa (AH298-GFP) was incubated with AgNPs or AgNCs while ThT dye was used to stain the biofilm. Consistent with Fig. 4A,B, individually dispersed bacterial cells and no biofilm mass were observed with AgNPs (Fig. 4C), while debris of dead bacteria were evident in the AgNC-associated sample (Fig. 4D). Intact bacteria embedded in biofilm mass were present in the untreated control (Fig. 4E). The HSI spectra derived from the SPR of AgNPs, bacterial cells + AgNPs and untreated control are presented in Fig. 4F–H. The SPR spectra of AgNPs were observed in the sample of bacterial cells + AgNPs with no traces of biofilms. The morphologies of control bacterial cells as well as cells exposed to AgNPs or AgNCs were also imaged via HIM and TEM. Intact bacterial cells with no biofilm or fibrillary aggregates were observed with AgNPs (Fig. 4I,J), while dead bacterial cells displaying flattened, blebbing, dents and protrusions in the outer membranes were found for the AgNCs-treated sample (Fig. 4K,L). In untreated control, fibrillar aggregates of FapC biofilms were evident between the cells (Fig. 4M,N). Furthermore, to determine the oxidative effect conferred by AgNPs or AgNCs to bacterial cells, ROS production in the cells was quantified. AgNPs and AgNCs induced ROS generation in a concentration dependent manner (Fig. 4O) but to very different extents, consistent with their respective bactericidal activities (Fig. 4A,B). This further indicates that biofilm inhibition by AgNPs was based on the inhibition of FapC fibrillization and not by their bactericidal activity as was the case with AgNCs. AgNO3 and bPEI alone were used as controls at concentrations equivalent to their concentration in AgNCs/NPs (Figs. S8&S9). At 1 μM concentration, specifically, soluble Ag+ provided by AgNO3 did not induce any bactericidal or antibiofilm effect. This is quite as expected, as the minimum inhibition concentration (MIC) of AgNO3 reported against P. aeruginosa is 17.6 μM.[66] This indicates that the bactericidal effect of AgNCs and the antibiofilm effect of AgNPs were associated with their nano-sized architectures and not with ion release at this given silver concentration. The enhanced bactericidal activity of AgNCs can be attributed to their ultrastructural disruption of the outer membranes followed by ROS generation,[23, 67] a mechanism that is shared with Ag+ released from AgNPs.[68]
CONCLUSION
We have examined the antibiofilm and antimicrobial potentials of bPEI-capped AgNPs and AgNCs, at concentrations below their toxicity thresholds for mammalian HEK 293 cells. As revealed by the ThT kinetic assay, both types of nanoparticles inhibited the amyloid aggregation of FapC, the protein constituent of the extracellular amyloid network of P. aeruginosa, through electrostatic interaction as well as H-bonding and hydrophobic interaction between the bPEI polymer and the protein. AgNPs sequestered FapC monomers while the much smaller AgNCs clustered with FapC monomers to render short amyloid fibrils, as revealed by TEM, HAADF and EDAX experiments. These biophysical phenomena entailed contrasting consequences in the antibiofilm application of the nanoparticles. Specifically, with increased nanoparticle concentrations, AgNPs inhibited P. aeruginosa biofilm formation without killing the bacteria through suppression of FapC amyloidosis and their associated extracellular amyloid network. In comparison, AgNCs impaired biofilm formation and were rendered bactericidal through structural-based disruption of cell membranes as well as elevated ROS production. Previously, AgNPs of biogenic origin were supposed to inhibit the bacterial biofilm formation by interfering with exopolysaccharides synthesis.[69] Here we demonstrated that AgNPs at low concentrations directly sequestered FapC to inhibit the bacterial biofilm activity. In agreement with previous work targeting functional amyloid to compromise biofilm,[17, 18] this study demonstrated the potential of exploiting functional amyloidosis inhibition with nanomaterials as a safe and facile antimicrobial strategy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ARC Project No. CE140100036 (Davis), NSF CAREER CBET-1553945 (Ding), NIH MIRA R35GM119691 (Ding). TEM imaging was performed at Bio21 Advanced Microscopy Facility, the University of Melbourne. HIM imaging was performed at Materials Characterization and Fabrication Platform (MCFP) at University of Melbourne. ZH and IH acknowledge the financial support by Pakistan Higher Education Commission for ZH PhD studentship and IRSIP fellowship. D.E.O. is supported by the Independent Research Foundation Denmark | Technical Sciences (grant no. 6111-00241B).
Footnotes
Competing financial interests: The authors declare no conflicting financial interests.
Supplementary Information
Kinetic parameters of FapC fibrillization in the presence of AgNCs and AgNPs (Table S1). Size distributions of the silver nanoparticles (Fig. S1). TGA surface ligand qualification (Fig. S2). ThT kinetic assay for bPEI and AgNO3 controls (Fig. S3). Cell viability of the silver nanoparticles with and without FapC (Fig. S4). Contact frequency maps of a FapC monomer with and without a bPEI-capped AgNC (Fig. S5). A FapC L2R3 dimer binding with a bPEI-capped AgNC or AgNP (Fig. S6). Contact frequency maps of a FapC dimer with and without a bPEI-capped AgNC or AgNP (Fig. S7). Antibiofilm and antibacterial activities of bPEI and AgNO3 controls (Fig. S8). Confocal fluorescence microscopy of P. aeruginosa (PAO1, AH298-GFP) with bPEI and AgNO3 as controls (Fig. S9).
References
- 1.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, Brennan CA, Chun E, Ngo L, Samson LD, Science 2019, 363, eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Nat. Microbiol 2019, 4, 623. [DOI] [PubMed] [Google Scholar]
- 3.Lebovitz Y. Modulation of Neurodevelopmental Outcomes using Lactobacillus in a Model of Maternal Microbiome Dysbiosis. Virginia Tech, 2019. [Google Scholar]
- 4.Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H, Endocr. Rev 2017, 39, 133–153. [DOI] [PubMed] [Google Scholar]
- 5.Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, Cell 2018, 175, 947–961. e17. [DOI] [PubMed] [Google Scholar]
- 6.Taglialegna A, Lasa I, Valle J, J. Bacteriol 2016, 198, 2579–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Hu C, Shao L, Int. J. Nanomed 2017, 12, 1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke PC, Sani M-A, Ding F, Kakinen A, Javed I, Separovic F, Davis TP, Mezzenga R, Chem. Soc. Rev 2017, 46, 6492–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dueholm MS, Petersen SV, Sønderkær M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Mo. Microbiol 2010, 77, 1009–1020. [DOI] [PubMed] [Google Scholar]
- 10.Fowler DM, Koulov AV, Balch WE, Kelly JW, Trends Biochem. Sci 2007, 32, 217–224. [DOI] [PubMed] [Google Scholar]
- 11.Flemming H-C, Wingender J, Nat. Rev. Microbiol 2010, 8, 623. [DOI] [PubMed] [Google Scholar]
- 12.Seviour T, Hansen SH, Yang L, Yau YH, Wang VB, Stenvang MR, Christiansen G, Marsili E, Givskov M, Chen Y, J. Biol. Chem 2015, 290, 6457–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AW, Adv. Drug Deliv. Rev 2005, 57, 1539–1550. [DOI] [PubMed] [Google Scholar]
- 14.Dueholm MS, Søndergaard MT, Nilsson M, Christiansen G, Stensballe A, Overgaard MT, Givskov M, Tolker‐Nielsen T, Otzen DE, Nielsen PH, Microbiologyopen 2013, 2, 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans ML, Chorell E, Taylor JD, Åden J, Götheson A, Li F, Koch M, Sefer L, Matthews SJ, Wittung-Stafshede P, Mol. Cell 2015, 57, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng G, Vad BS, Dueholm MS, Christiansen G, Nilsson M, Tolker-Nielsen T, Nielsen PH, Meyer RL, Otzen DE, Front. Microbiol 2015, 6, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenvang M, Dueholm MS, Vad BS, Seviour T, Zeng G, Geifman-Shochat S, Søndergaard MT, Christiansen G, Meyer RL, Kjelleberg S, Nielsen PH, Otzen DE, J. Biol. Chem 2016, 291, 26540–26553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najarzadeh Z, Mohammad-Beigi H, Pedersen JN, Christiansen G, Sønderby TV, Shojaosadati SA, Morshedi D, Strømgaard K, Meisl G, Sutherland D, Pedersen JS, Otzen DE, Biomolecules 2019, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y, Liu Y, Yang J, Liu Y, Hu F, Zhu K, Jiang X, Angew. Chem. Int. Ed. Engl 2018, 57, 3958–3962. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, He B, Liu L, Qu G, Shi J, Hu L, Jiang G, Metallomics 2018, 10, 557–564. [DOI] [PubMed] [Google Scholar]
- 21.Sirelkhatim A, Mahmud S, Seeni A, Kaus N, Ann L, Bakhori S, Hasan H, Mohamad D, Kinetics model Linear Equation Parameters Values at 0.15 mg mL-1 DTT Values at 0.35 mg mL-1 DTT 189, 110. [Google Scholar]
- 22.Kim JS, Kuk E, Yu KN, Kim J-H, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang C-Y, Nanomedicine: NBM 2007, 3, 95–101. [DOI] [PubMed] [Google Scholar]
- 23.Zheng K, Setyawati MI, Lim T-P, Leong DT, Xie J, ACS nano 2016, 10, 7934–7942. [DOI] [PubMed] [Google Scholar]
- 24.Sambhy V, MacBride MM, Peterson BR, Sen A, J. Am. Chem. Soc 2006, 128, 9798–9808. [DOI] [PubMed] [Google Scholar]
- 25.Zheng H, Ji Z, Roy KR, Gao M, Pan Y, Cai X, Wang L, Li W, Chang CH, Kaweeteerawat C, Chen C, Xia T, Zhao Y, Li R, ACS Nano 2019, 13, 11488–11499. [DOI] [PubMed] [Google Scholar]
- 26.John T, Gladytz A, Kubeil C, Martin LL, Risselada HJ, Abel B, Nanoscale 2018, 10, 20894–20913. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y, Park J-H, Lee H, Nam J-M, Sci. Rep 2016, 6, 19548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke PC, Pilkington EH, Sun Y, Javed I, Kakinen A, Peng G, Ding F, Davis TP, Adv. Mat 2019, 0, 1901690 DOI 10.1002/adma.201901690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javed I, He J, Kakinen A, Faridi A, Yang W, Davis TP, Ke PC, Chen P, ACS Appl. Mater. Interfaces 2019, 11, 10462–10471. [DOI] [PubMed] [Google Scholar]
- 30.Gladytz A, Abel B, Risselada HJ, Angew. Chem. Int. Ed. Engl 2016, 55, 11242–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javed I, Sun Y, Adamcik J, Wang B, Kakinen A, Pilkington EH, Ding F, Mezzenga R, Davis TP, Ke PC, Biomacromolecules 2017, 18, 4316–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Javed I, Peng G, Xing Y, Yu T, Zhao M, Kakinen A, Faridi A, Parish CL, Ding F, Davis TP, Ke PC, Lin S, Nat. Commun 2019, 10, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao YH, Chang YJ, Yoshiike Y, Chang YC, Chen YR, Small 2012, 8, 3631–3639. [DOI] [PubMed] [Google Scholar]
- 34.Anand BG, Dubey K, Shekhawat DS, Kar K, Biochemistry 2016, 55, 3345–3348. [DOI] [PubMed] [Google Scholar]
- 35.Sen S, Konar S, Das B, Pathak A, Dhara S, Dasgupta S, DasGupta S, RSC Adv. 2016, 6, 43104–43115. [Google Scholar]
- 36.Wang M, Kakinen A, Pilkington EH, Davis TP, Ke PC, Biomater. Sci 2017, 5, 485–493. [DOI] [PubMed] [Google Scholar]
- 37.Rizzello L, Pompa PP, Chem. Soc. Rev 2014, 43, 1501–1518. [DOI] [PubMed] [Google Scholar]
- 38.Huma Z.-e., Gupta A, Javed I, Das R, Hussain SZ, Mumtaz S, Hussain I, Rotello VM, ACS Omega 2018, 3, 16721–16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng G, Zil-e-Huma MU, Hussain I, Javed I, Metal Nanoparticles for Drug Delivery and Diagnostic Applications 2019, 119. [Google Scholar]
- 40.Durán N, Durán M, De Jesus MB, Seabra AB, Fávaro WJ, Nakazato G, Nanomedicine: NBM 2016, 12, 789–799. [DOI] [PubMed] [Google Scholar]
- 41.Deshmukh S, Patil S, Mullani S, Delekar S, Mater. Sci. Eng. C 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmal S, Jana NR, Jana NR, J. Phys. Chem. C 2014, 118, 21630–21638. [Google Scholar]
- 43.Wagner SC, Roskamp M, Pallerla M, Araghi RR, Schlecht S, Koksch B, Small 2010, 6, 1321–1328. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Ma B, Pan T, Chen S, Sun J, Adv. Funct. Mater 2016, 26, 569–576. [Google Scholar]
- 45.Park J, Kim J, Singha K, Han D-K, Park H, Kim WJ, Biomaterials 2013, 34, 8766–8775. [DOI] [PubMed] [Google Scholar]
- 46.Manoth M, Manzoor K, Patra M, Pandey P, Vadera S, Kumar N, Mater. Res. Bull 2009, 44, 714–717. [Google Scholar]
- 47.Dong JX, Qu F, Li NB, Luo HQ, RSC Advances 2015, 5, 6043–6050. [Google Scholar]
- 48.Desai R, Mankad V, Gupta SK, Jha PK, Nanosci. Nanotechnol. Lett 2012, 4, 30–34. [Google Scholar]
- 49.Cabaleiro-Lago C, Quinlan-Pluck F, Lynch I, Lindman S, Minogue AM, Thulin E, Walsh DM, Dawson KA, Linse S, J. Am. Chem. Soc 2008, 130, 15437–15443. [DOI] [PubMed] [Google Scholar]
- 50.Whitmore L, Wallace BA, Biopolymers: Original Research on Biomolecules 2008, 89, 392–400. [DOI] [PubMed] [Google Scholar]
- 51.Alakomi H-L, Skyttä E, Saarela M, Mattila-Sandholm T, Latva-Kala K, Helander I, Appl. Environ. Microbiol 2000, 66, 2001–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Kakinen A, Xing Y, Faridi P, Nandakumar A, Purcell AW, Davis TP, Ke PC, Ding F, Small 2019, 15, 1805166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shirvanyants D, Ding F, Tsao D, Ramachandran S, Dokholyan NV, J Phys. Chem. B 2012, 116, 8375–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Proctor EA, Dokholyan NV, Curr. Opin. Str. Biol 2016, 37, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding F, Dokholyan NV, Discrete molecular dynamics simulation of biomolecules In Computational Modeling of Biological Systems, Springer: 2012; pp 55–73. [Google Scholar]
- 56.Wang B, Seabrook SA, Nedumpully-Govindan P, Chen P, Yin H, Waddington L, Epa VC, Winkler DA, Kirby JK, Ding F, Phys. Chem. Chem. Phys 2015, 17, 1728–1739. [DOI] [PubMed] [Google Scholar]
- 57.Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, Zink JI, Nel AE, ACS Nano 2009, 3, 3273–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Díez I, Ras RH, Nanoscale 2011, 3, 1963–1970. [DOI] [PubMed] [Google Scholar]
- 59.Javed I, Yu T, Peng G, Sánchez-Ferrer A, Faridi A, Kakinen A, Zhao M, Mezzenga R, Davis TP, Lin S, Ke PC, Nano Lett. 2018, 18, 5797–5804. DOI 10.1021/acs.nanolett.8b02446. [DOI] [PubMed] [Google Scholar]
- 60.Wang M, Sun Y, Cao X, Peng G, Javed I, Kakinen A, Davis TP, Lin S, Liu J, Ding F, Ke PC, Nanoscale 2018, 10, 19995–20006. DOI 10.1039/C8NR07180B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Kadiyala U, Qu Z, Elvati P, Altheim C, Kotov NA, Violi A, VanEpps JS, ACS nano 2019, 13, 4278–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Mao X, Yu Y, Wang CX, Yang YL, Wang C, Adv. Mater 2013, 25, 3780–3801. [DOI] [PubMed] [Google Scholar]
- 63.Vertegel AA, Siegel RW, Dordick JS, Langmuir 2004, 20, 6800–6807. [DOI] [PubMed] [Google Scholar]
- 64.Tang S, Zheng J, Adv. Healthc. Mater 2018, 7, 1701503. [DOI] [PubMed] [Google Scholar]
- 65.Jin J-C, Wu X-J, Xu J, Wang B-B, Jiang F-L, Liu Y, Biomater. Sci 2017, 5, 247–257. [DOI] [PubMed] [Google Scholar]
- 66.Richards R, Microbios 1981, 31, 83–91. [PubMed] [Google Scholar]
- 67.Jin J-C, Wu X-J, Xu J, Wang B-B, Jiang F-L, Liu Y, Biomater. Sci 2017, 5, 247–257. [DOI] [PubMed] [Google Scholar]
- 68.Gurunathan S, Han JW, Kwon D-N, Kim J-H, Nanoscale Res. Lett 2014, 9, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalishwaralal K, BarathManiKanth S, Pandian SRK, Deepak V, Gurunathan S, Colloids Surf. B: Biointerfaces 2010, 79, 340–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


