1. Introduction
Skeletal muscle is a highly regenerative tissue that produces contractile forces required for respiration and locomotion and aids regulation of whole body energy homeostasis via post-prandial glucose uptake [1]. Impairment in skeletal muscle function and/or regenerative ability can occur due to cancer, aging, trauma and genetic muscular diseases, such as Duchenne Muscular Dystrophy (DMD). Currently, therapeutic options for these myopathies are limited due to inadequate understanding of the complex mechanisms of skeletal muscle repair, driving the need for novel cell, gene, and pharmaceutical therapies. Classically, drug discovery and validation for treatment of myopathies has been performed using two-dimensional (2D) cell culture platforms and animal models preceding clinical trials. However, this traditional drug development process has been highly inefficient with almost 90% of tested drugs failing to gain approval due to differential toxicity responses between animals and humans, and animal models not accurately replicating human disease progression, severity, or roles of the genetic and epigenetic diversity in patients [2]. In this editorial, we will describe the current state of three-dimensional (3D) human skeletal muscle tissue engineering and discuss its potential to fully replicate native skeletal muscle function, regeneration, maturation, and cellular complexity. We will further discuss how tissue-engineered human skeletal muscle models can be utilized as in vitro drug discovery platforms to complement preclinical animal studies and guide clinical trials to ultimately increase the successful translation of novel pharmacological therapies for muscle injury and disease.
2. In vitro skeletal muscle models
Mature skeletal muscle is comprised of post-mitotic multinucleated myofibers that provide a niche for resident muscle stem cells, called satellite cells (SCs). The SCs can be activated and differentiate to repair small muscle injuries and are the only robust source of myogenic progenitor cells (MPCs) in skeletal muscle. Human skeletal muscle has been modeled in vitro using primary, immortalized, and human induced pluripotent stem cell (hiPSC)-derived MPCs [1]. Primary MPCs are the current gold standard cell source but have limited proliferative ability, lose their myogenic and engraftment potential with serial passaging, and may be hard to obtain due to ethical reasons. Immortalized myoblast cell lines permit extensive expansion in culture, but have limited physiological relevance due to metabolic abnormalities and accumulated mitochondrial and genetic mutations. Recent advances in the derivation of muscle progenitor cells (iMPCs) and other muscle-resident cells starting from hiPSCs can permit the generation of more realistic heterocellular muscle models derived from a single human donor. Still, iMPCs generated with current reprogramming or directed differentiation protocols do not undergo complete myogenesis, yielding developmentally immature muscle fibers.
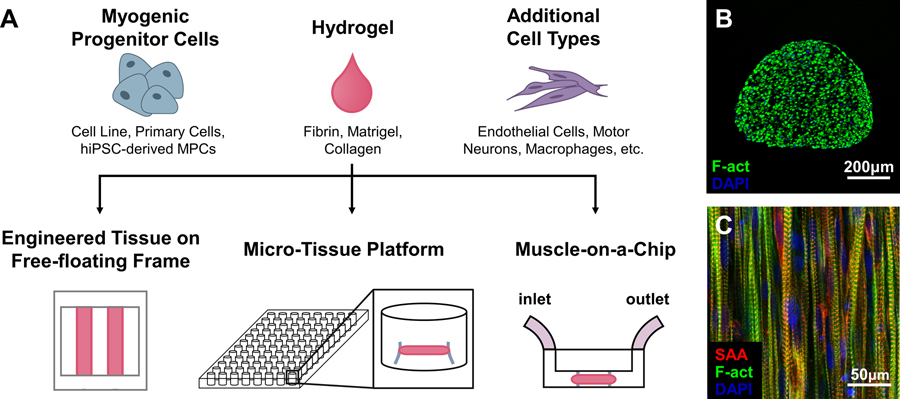
In native muscles, mature myofibers are aligned within a 3D laminin- and collagen IV-rich basal lamina matrix and surrounded by collagen I-rich connective tissue. In contrast, MPCs are traditionally cultured on 2D plates coated with collagen I, laminin, or Matrigel – a basement membrane extract rich in laminin and collagen IV. The 2D culture, in conjunction with various microfabrication techniques [3], enables the fusion of MPCs into aligned multinucleated myotubes; however, it does not permit long-term (>1wk) myotube culture and advanced muscle maturation. These limitations can be overcome by embedding MPCs in 3D hydrogel scaffolds (Figure 1A) that both enable cell alignment through passive tension and support spontaneous myotube contractions and long-term culture (>4wks), leading to higher expression of developmentally mature muscle proteins and improved ultrastructure and contractile function. Collagen I hydrogels were first utilized to mimic the 3D environment of native muscle, but their relatively high stiffness hampered long-term muscle culture, differentiation, and contractile force generation. More compliant fibrin hydrogels have overcome these limitations by providing a temporary matrix that cells can remodel and replace with endogenously generated extracellular matrix (ECM). Currently, use of composite fibrin-Matrigel hydrogels yields engineered skeletal muscle tissues with the highest force generation capacity. These tissues support both the formation of mature aligned muscle fibers (Figure 1B–C) and maintenance of a functional satellite-like cell pool, thereby allowing studies of muscle development, function, and regeneration in a single system [1].
Figure 1.
In vitro tissue-engineered models of human skeletal muscle. (a) Components and platforms for engineering human skeletal muscle tissues for drug testing. Free-floating, dynamic culture improves muscle maturation and enables measurement of force–length relationships; micro-tissue platforms allow for high-throughput drug screening in individual muscle tissues; muscle-on-a-chip platforms have microfluidic feeds for interfacing with other organ-on-a-chip systems. MPC, myogenic progenitor cell. (b-c) Transverse (b) and longitudinal (c) sections of cylindrically shaped engineered human skeletal muscles showing viable myotubes throughout the entirety of the tissue; myotubes are aligned and cross-striated as characteristic of native skeletal muscle. SAA, sarcomeric-actinin; F-actin, filamentous actin; DAPI, nuclei. Tissues in b and c are cultured within free-floating frames on a rocker.
In traditional 2D cultures, functional outcomes of experimental interventions are inferred indirectly from histological, transcriptomic, or protein changes. The most important benefit of 3D culture is the ability to directly quantify and study muscle function (i.e. force generation and fatigue resistance), the key clinical measure of therapeutic success. Contractile function can be measured in absolute terms using a force transducer, or estimated (with a higher throughput) through deformation of micro-posts attached to engineered muscle. While engineered muscle tissues generate forces an order of magnitude lower than native muscle, they replicate native muscle force-length and force-frequency relationships [4, 5]. Additional assessments of calcium transient amplitude, biomarker release, and multi-omic analyses can be multiplexed with functional tests to permit detailed experimental characterization [4–6].
3. Expert Opinion
Ideally, the widespread utilization of engineered muscle tissues for drug discovery would require the ability to: 1) replicate known responses to well-characterized pharmaceuticals, 2) accurately predict drug toxicity, 3) faithfully model human disease and predict drug efficacy, and 4) offer relatively low-cost and high-throughput testing capabilities. Engineered human skeletal muscle tissues have been shown to accurately model functional responses to both positive (e.g. IGF-1 and β2 agonists) and negative (e.g. statins, glucocorticoids, and mitochondrial toxins) modulators of muscle function [5]. However, successfully modeling patient-specific myotoxicity or predicting clinical toxicity of novel compounds are likely to require incorporation of liver and intestine tissues to model first-pass drug metabolism as well as additional organs (e.g. fat, endothelium, blood-brain barrier) to predict drug biodistribution, unexpected adverse effects, and organ-organ interactions [7]. The first such “human-on-a-chip” models have already shown success in identifying unanticipated toxicities, including cardiotoxicity driven by bleomycin-induced lung inflammatory factors [8]. Furthermore, the Crabtree Effect, where in vitro cultured cells utilize glycolysis but not oxidative phosphorylation to generate ATP, prevents accurate predication of mitochondrial toxicity, the most common cause of preclinical failure of drug candidates identified from in vitro screens. Shifts to greater oxidative metabolism in engineered muscle tissues could be achieved by decreasing media glucose concentration and adding galactose and/or relevant fatty acids.
Another anticipated application of engineered muscle platforms is modeling of patient-specific myopathic mutations, disease modifiers, and pharmacogenomic responses. Among different muscle pathologies, this is of particular importance for DMD (which can be caused by >4000 mutations in the dystrophin gene), where disease progression is determined by various modifiers and is poorly modeled in traditional mouse models. 2D cell cultures utilizing primary or hiPSC-derived DMD MPCs can model some disease symptoms including increased biomarker release, impaired myoblast fusion, and impaired calcium-handling. However, mature membrane-bound dystrophin is not seen in 2D cultures limiting their physiological relevance. In contrast, mature dystrophin localization is present in both human primary and hiPSC-derived 3D muscle tissues indicating greater maturation and a more physiologically relevant system for modeling human disease [4, 6]. While 3D muscle tissues derived from immortalized dystrophic mouse myoblasts model some expected pharmacological responses [9], more advanced clinical DMD features such as myofiber branching, fibrosis, lipid accumulation, and increased membrane permeability have not been demonstrated. Moreover, discovering patient-specific genotype-phenotype relationships and drug responses requires use of human- rather than mouse-derived platforms.
Despite recent advances, engineered muscle tissues still lack the maturity and complexity of native skeletal muscle, which limits their pathophysiological relevance and suitability for predictive drug testing. Development of high-fidelity in vitro engineered muscle platforms for drug discovery will require identification of culture conditions to either maintain identity and function of primary SCs or more efficiently derive MPCs from hiPSCs. This includes the ability to expand large numbers of aged SCs and/or identify conditions to faithfully age primary SCs or iMPCs for accurate modeling of sarcopenia. Optimized regimes of biophysical stimulation and combinatorial uses of growth factors, small molecules, and metabolic fuel sources will be needed to accelerate engineered muscle maturation in weeks rather than years [1, 6]. Biomimetic incorporation of other muscle-resident cells such as fibroblasts, motor neurons [10], and immune [11] and vascular [12, 13] cells will allow more accurate modeling of native muscle composition, function, regeneration, and complex disease phenotypes. Incorporating macrophages, in particular, was critical to successful in vitro and in vivo regeneration of adult rat engineered muscle, highlighting the increased physiological relevance of multicellular models [11]. Miniaturization of engineered human muscle size [14] without the loss of maturation, structure, or functional phenotype will be required for increased experimental throughput.
Current engineered human tissue platforms are beginning to provide assays for additional validation of existing drug discovery studies [15]. Combining multicellular tissue engineering with high-throughput, non-destructive assessment of drug responses will improve the robustness and cost effectiveness of engineered muscle platforms, while further organ multiplexing will increase their clinical relevance. The ultimate success of these platforms will be contingent upon continued progress in muscle stem cell biology, biomimetic tissue engineering, device microfabrication, robotics, and non-invasive imaging, combined with the exhaustive validation of the predicted drug responses in animal and clinical studies. Though likely decades away, we anticipate that the traditional drug development pipelines for skeletal muscle diseases will transition away from dependence on preclinical animal models and shift toward increased utilization of more predictive, patient-specific, “clinical trials in a dish”.
Acknowledgments
Funding:
T Broer and N Bursac are both funded by National Institutes of Health (NIH) grants R01AR070543 while T Broer is also funded by NIH grant T32GM00855. N Bursac and A Khodabukus are also supported by NIH grants R01AR065873 and UG3TR002142 as well as by a grant from the Jain Foundation. N Bursac also has received grant support from the NIH with grant U01EB028901.
Footnotes
Declaration of Interest:
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- [1].Wang J, Khodabukus A, Rao L, Vandusen K, Abutaleb N, Bursac N, Engineered skeletal muscles for disease modeling and drug discovery, Biomaterials, 221 (2019) 119416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J, Clinical development success rates for investigational drugs, Nat Biotechnol, 32 (2014) 40–51. [DOI] [PubMed] [Google Scholar]
- [3].Lam MT, Sim S, Zhu X, Takayama S, The effect of continuous wavy micropatterns on silicone substrates on the alignment of skeletal muscle myoblasts and myotubes, Biomaterials, 27 (2006) 4340–4347. [DOI] [PubMed] [Google Scholar]
- [4].Rao L, Qian Y, Khodabukus A, Ribar T, Bursac N, Engineering human pluripotent stem cells into a functional skeletal muscle tissue, Nat Commun, 9 (2018) 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N, Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs, eLife, 4 (2015) e04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Khodabukus A, Madden L, Prabhu NK, Koves TR, Jackman CP, Muoio DM, Bursac N, Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle, Biomaterials, 198 (2019) 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prantil-Baun R, Novak R, Das D, Somayaji MR, Przekwas A, Ingber DE, Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips, Annu Rev Pharmacol Toxicol, 58 (2018) 37–64. [DOI] [PubMed] [Google Scholar]
- [8].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ, Shrike Zhang Y, Shin SR, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Jin Lee S, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A, Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform, Scientific reports, 7 (2017) 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vandenburgh H, Shansky J, Benesch-Lee F, Skelly K, Spinazzola JM, Saponjian Y, Tseng BS, Automated drug screening with contractile muscle tissue engineered from dystrophic myoblasts, FASEB J, 23 (2009) 3325–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Osaki T, Uzel SGM, Kamm RD, Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons, Sci Adv, 4 (2018) eaat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Juhas M, Abutaleb N, Wang JT, Ye J, Shaikh Z, Sriworarat C, Qian Y, Bursac N, Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration, Nat Biomed Eng, 2 (2018) 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Perry L, Landau S, Flugelman MY, Levenberg S, Genetically engineered human muscle transplant enhances murine host neovascularization and myogenesis, Commun Biol, 1 (2018) 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bersini S, Gilardi M, Ugolini GS, Sansoni V, Talo G, Perego S, Zanotti S, Ostano P, Mora M, Soncini M, Vanoni M, Lombardi G, Moretti M, Engineering an Environment for the Study of Fibrosis: A 3D Human Muscle Model with Endothelium Specificity and Endomysium, Cell reports, 25 (2018) 3858–3868 e3854. [DOI] [PubMed] [Google Scholar]
- [14].Mills RJ, Parker BL, Monnot P, Needham EJ, Vivien CJ, Ferguson C, Parton RG, James DE, Porrello ER, Hudson JE, Development of a human skeletal micro muscle platform with pacing capabilities, Biomaterials, 198 (2019) 217–227. [DOI] [PubMed] [Google Scholar]
- [15].Stillitano F, Hansen J, Kong CW, Karakikes I, Funck-Brentano C, Geng L, Scott S, Reynier S, Wu M, Valogne Y, Desseaux C, Salem JE, Jeziorowska D, Zahr N, Li R, Iyengar R, Hajjar RJ, Hulot JS, Modeling susceptibility to drug-induced long QT with a panel of subject-specific induced pluripotent stem cells, eLife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]