Abstract
In 2011 the Spanish Society of Medical Oncology (SEOM) and the Spanish Society of Pathology (SEAP) started a joint project to establish guidelines on biomarker testing in patients with advanced non-small-cell lung cancer (NSCLC) based on current evidence. As this field is constantly evolving, these guidelines have been updated, previously in 2012 and 2015 and now in 2019. Current evidence suggests that the mandatory tests to conduct in all patients with advanced NSCLC are for EGFR and BRAF mutations, ALK and ROS1 rearrangements and PD-L1 expression. The growing need to study other emerging biomarkers has promoted the routine use of massive sequencing (next-generation sequencing, NGS). The coordination of every professional involved and the prioritisation of the most suitable tests and technologies for each case remains a challenge.
Keywords: ALK, Biomarkers, Non-small-cell lung cancer, EGFR, BRAF, PD-L1, ROS1
Introduction
Non-small-cell lung cancer (NSCLC) is the solid tumour with the widest variety of potential therapeutic targets. It represents both a significant therapeutic opportunity and a challenge in predictive biomarkers determination. This third consensus statement update guidelines published in 2012 and 2015 focused on predictive biomarker testing in patients with advanced NSCLC [1, 2]. The current document is, supported by the Spanish Society of Pathology (SEAP) and the Spanish Society of Medical Oncology (SEOM).
Requirements for testing an optimal biological specimen
Obtaining enough and optimal quality specimen for biomarkers in a particular patient should be a responsibility shared by the entire tumour board. In order to do this, it is important that the professionals involved have sufficient knowledge of the advantages and disadvantages of each technology. It would be very helpful to establish automated and routine channels that could provide a solution when one or all tests fail, always taking into account adequate response times. When conducting molecular and immunohistochemical (IHC) tests, it is important to consider the tumour percentage and the amount of tumour cells in the specimen, and also the pre-analytical variables [3]. Most of the samples obtained are small biopsy and/or cytology-type specimens (for example cell blocks, smears and liquid-based cytology). All of these sample types are suitable for IHC and molecular studies. The use of one or another will depend on the experience and capacity of each laboratory [4]. The first step for obtaining an adequate specimen is the time between sample removal out of patient and its early fixation. This is why having seamless communication between the specialists involved is essential, as well as the availability of optimised diagnostic techniques. The general requirements for a specimen to be optimal are conservation in 10% buffered formalin for 6–12 h for small biopsies and 24–48 h for surgical resections [5], and the presence of at least 50–100 viable cells for IHC studies or fluorescence in situ hybridisation (FISH). For real-time polymerase chain reaction (PCR) tests, a minimum 5% of tumour cells in NSCLCs are recommended [1, 6]. This percentage should be increased to 20–30% for direct next-generation sequencing (NGS) studies [7]. Direct smears that are air-dried or ethanol-based fixation and liquid-based cytology are also suitable for FISH and molecular testing, but it is compulsory to perform appropriate validation studies in each laboratory following previously described recommendations [8–10]. The use of cytology specimens has not yet been validated to determine the expression of programmed death ligand-1 (PD-L1), despite the good correlation observed between cytology smears and cell blocks with biopsies [11]. Tissue-sparing protocols are recommended [12, 13]. For liquid biopsies, the two key technical factors to maintain optimal preservation of circulating cell-free DNA (cfDNA) are the storage and shipping conditions of the sample, and the elapsed time between specimen extraction and processing [14].
Which biomarkers should be tested in NSCLC and in which patients?
Table 1 summarises the essential biomarkers to be performed on tissue- and/or cytology-type samples from advanced NSCLC patients, including the predictive alterations and their testing methods.
Table 1.
Essential biomarkers in NSCLC patients
| Gene/protein | Predictive alteration | Methodology (in tissue) |
|---|---|---|
| EGFR | Mutation | PCR: sanger, real-time PCR and NGS |
| ALK | Rearrangement | IHC, FISH and NGS |
| ROS1 | Rearrangement | IHC (screening), FISH and NGS |
| BRAF V600 | Mutation | PCR: sanger, real-time PCR and NGS |
| PD-L1 | Overexpression | IHC |
EGFR epidermal growth factor receptor, FISH fluorescence in situ hybridisation, H&E haematoxylin/eosin, IHC immunohistochemistry, NGS next-generation sequencing, NSCLC non-small-cell lung cancer, PCR polymerase chain reaction, PD-L1 programmed death ligand-1
EGFR
In Spain, epidermal growth factor receptor (EGFR) mutations are present in 8–11% of advanced NSCLCs, and in 16–18% of lung adenocarcinomas [15]. The most common mutations (85–90%) are tyrosine-kinase inhibitors (TKIs) sensitivity mutations such as deletions in exon 19 and point mutations in exon 21. Other uncommon mutations may be clinically relevant (i.e. exon 20 insertions are typically intrinsically resistant to EGFR-TKI inhibitors and exon 18 alterations may be more sensitive to a specific TKI) [16]. EGFR-TKI inhibitor drugs are currently available, and administration as first-line therapy is standard in the main clinical guidelines [17], since these improve progression-free survival (PFS) and quality of life when compared to the administration of platinum doublet chemotherapy [17]. Therefore, the recommendations from the last SEOM/SEAP consensus statements are still valid [1]:
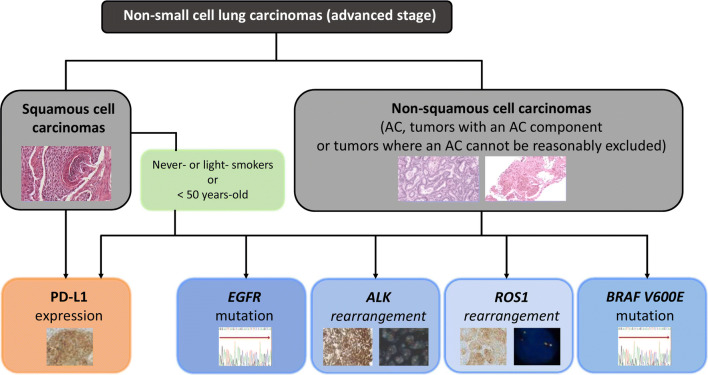
EGFR mutation tests in patients with advanced NSCLC should be conducted for all adenocarcinomas, non-squamous non-small-cell histologies and squamous cell carcinomas in patients younger than 50 years of age and/or with no or low tobacco use (i.e. < 15 pack-years) (Fig. 1);
The latest international consensus statements recommend that EGFR mutation tests should also be conducted on any small sample in which the tumour is poorly represented and in cases with an uncertain histological subtype;
Lastly, an upfront liquid biopsy is not recommended if tissue is available. This procedure could be selected for determining the T790M mutation at disease progression.
Fig. 1.
Diagnostic algorithm for biomarker testing in patients with advanced NSCLC. AC adenocarcinoma, EGFR epidermal growth factor receptor, PD-L1 programmed death ligand-1
Most patients with an EGFR mutation who receive first- or second-generation EGFR-TKIs will progress, and the most frequent molecular mechanism for acquired resistance is EGFR T790M mutation, that occurs in 50–60% of cases [18]. In patients who present with an EGFR T790M mutation after progression on first-line treatment with a first- or second-generation EGFR-TKI, osimertinib has shown a higher PFS than a platinum/pemetrexed regimen (10.1 months vs. 4.4 months, respectively; HR 0.30) [19]. Based on this data, osimertinib is considered the treatment of choice for these patients. Resistance mechanisms are less well known when osimertinib is used as first-line treatment [20, 21]. Determination of EGFR T790M in tumour tissue and in cfDNA are both valid alternatives. If EGFR T790M testing in plasma is negative, a new biopsy is recommended whenever possible.
Recommendations:
All individual EGFR mutations with a frequency higher than 1% should be tested in tissue and/or cytology-type samples;
The pathologist should examine all available specimens and use the one with better cellularity and tumour proportion (biopsy or cytology) from the primary tumour or the metastases;
High-sensitivity detection methods should be used, especially for EGFR T790M mutation testing (5% detection limit) [22]. The most recent recommendations from the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) and the National Institute for Health and Care Excellence (NICE) suggest having two alternative methods to carry out a redundant molecular test, if necessary;
When the objective is to select patients to receive a therapy, IHC techniques (including EGFR mutation-specific antibodies) or copy number analysis should not be used;
If sufficient expertise is available, and if the extended biomarker panel is to be tested, it is preferable to determine the EGFR mutation with targeted NGS panels;
Cell blocks and other cytological preparations tested in laboratories with experience are also suitable specimens.
ALK
Anaplastic lymphoma kinase (ALK) rearrangements are present in 2–5% of advanced NSCLCs [17]. Due to the clinical benefit provided by targeted therapies in this disease, it is key of identify all patients with this molecularly driven type of lung cancer [23].
Recommendations:
The histological types eligible for ALK rearrangement tests include all adenocarcinomas, carcinomas with non-squamous histological evidence and squamous tumours in patients younger than 50 years of age and/or with low or no tobacco use (i.e. < 15 pack-years) (Fig. 1) [24]. In some neuroendocrine carcinomas, ALK expression is intense but rearrangement cannot be detected in the sequencing test [25, 26]. The key methods for detecting ALK gene rearrangement are IHC, FISH, PCR and NGS [22, 27]. Actually, IHC is an equivalent alternative to FISH. In this regard [17, 24] IHC is a quick and cost-effective method to determine low prevalence biomarkers. Cell integrity can be assessed, and the method can be applied to different biological specimens, such as biopsies or cytological samples. Its use in cytology smears is quite controversial, although recent studies have proven the suitability of the method [28]. The most commonly used antibodies are D5F3 (Ventana ALK [D5F3] CDx Assay, Tucson, Arizona, USA) and 5A4 (Novocastra, Leica Biosystems, Buffalo Grove, Illinois, USA), although the latter is not included in a diagnostic kit [29]. The role of FISH as the optimal standard methodology is currently under discussion. The technical rationale is being assessed, as well as its interpretation of complex molecular mechanisms [30], although there are automated reader algorithms approved by the food and drug administration (FDA) that greatly increase reliability [29]. When there is a positive IHC as manifested by strong granular cytoplasmic staining with either of the 5A4 or D5F3 antibodies, the current recommendation is that confirmation by a second technique is not mandatory [22]. However, it is advisable to do so in cases that are inconclusive. This diagnostic redundancy is also helpful if unusual FISH patterns are found [31]. The methods based on NGS and RNA-assays are highly specific and there are numerous studies that demonstrate their value for detecting fusions in patients who show negative results with other techniques [32, 33].
Lastly, variant testing for specific rearrangements in ALK, which has been reported as a crucial element in the clinical response to specific inhibitors, does not yet have sufficient data for recommendation, although it could be useful in the future [34].
ROS1
The c-ros oncogene 1 (ROS1) gene encodes a receptor with tyrosine kinase activity that appears to be translocated in approximately 1% of NSCLCs, especially in young, non-smoking patients. It is associated with adenocarcinoma histology, with the presence of a solid component and signet-ring cells. This histological profile is also typical of tumours harbouring an ALK translocation. In fact, both receptors have a 77% similarity in their ATP-binding domain.
Crizotinib is approved as a first- or second-line monotherapy in stage IV lung cancer patients with ROS1 rearrangement [35–37]. Other drugs, such as ceritinib, brigatinib, lorlatinib and entrectinib, are being studied but they are not approved for this indication yet.
Recommendations:
It is currently recommended to carry out ROS1 testing in patients with advanced stage (IIIB-IV) lung adenocarcinoma, regardless of its clinical characteristics [17] (Fig. 1). ROS1 testing is not recommended in squamous cell carcinoma, except in the context of patients with no or low tobacco exposure [17, 22].Essentially, there are three methodological approaches to detecting ROS1 rearrangements: (a) IHC; (b) cytogenetic techniques, particularly FISH; and (c) molecular techniques, such as reverse transcription PCR (RT-PCR) or NGS [30, 38]. To determine ROS1 translocation in clinical specimens, international guidelines recommend IHC as the screening method and confirmation of positive cases with another orthogonal method (cytogenetic or molecular) [22]. Currently there is no FDA-approved IHC assay for clinical routine, but there are two commercially available antibodies (D4D6, Cell Signalling Technology and SP384, Ventana Medical Systems) which show high sensitivity in most studies when compared to other techniques, in particular FISH or RT-PCR [22, 39]. However, according to the method and the criteria for positivity used, the specificity ranges from 70 to 100% [22, 39]. At present, there is no universally accepted system for how to score IHC results but it is recommended that the specimen includes at least 20 tumour cells and that each laboratory validates its own interpretation range [22, 24, 38, 40]. Moreover, it is important to consider that ROS1 expression without underlying rearrangement (false positives) has been described in nearly a third of tumours [41, 42]. The presence of other molecular abnormalities, such as EGFR, KRAS, BRAF or HER2 mutations and ALK rearrangements, has also been identified in some of these tumours [43].
Regarding FISH, usually considered as the gold-standard technique, the use of a dual-colour break-apart probes and a count of at least 50 tumour cells is recommended [22, 38–40]. A tumour should be considered positive when at least 50% of tumour cells have break-apart signals (separated by ≥ 1 signal diameter), and/or 3’ isolated signals (frequently marked with green fluorochrome) [38, 39]. False positives and false negatives have been described, attributable to both methodological and biological causes [38, 40]. In respect of this latter aspect, it is important to note that some commercial probes could not detect rearrangements due to their design, as is the case for the variant GOPC-ROS1 [38, 44].
Regarding RT-PCR and NGS (DNA or RNA-based), most published studies show high sensitivity and specificity data [33, 44, 45].
BRAF
BRAF mutations can be found in approximately 2% of lung carcinomas, both in smokers and non-smokers. Most of these are adenocarcinomas and tumours with papillary growth (Fig. 1) [46, 47]. Nearly all studies find a 50% frequency for BRAF V600E mutations [48], although in a European study the frequency is as high as 83% [49]. Additionally, BRAF V600E mutations are mostly mutually exclusive with most druggable abnormalities present in this tumour [46, 50]. It should be noted that certain BRAF mutations can co-exist with KRAS mutations [50]. Following robust results from clinical studies with BRAF inhibitors, whether or not associated with MEK inhibitors, both the EMA and FDA have approved dabrafenib and trametinib treatment for patients with the V600E mutation [51].
Recommendations:
It is currently recommended to study the BRAF V600 mutation in all patients with advanced non-squamous NSCLC (Fig. 1) [17, 22]
The BRAF test can be conducted with any PCR method, including NGS, but the methodology should always analyse exons 11 and 15 [46]. Along this lines, the FDA has included the panel Oncomine Dx Target Test® (ThermoFisher, Mass, USA) in its approval [52].
PD-L1
In randomised studies, immunotherapy with PD-1/PD-L1 (nivolumab, pembrolizumab, atezolizumab and durvalumab) and CTLA4 inhibitors (ipilimumab in combination with nivolumab) is shown to be effective in patients with advanced NSCLC [17]. PD-L1 is a type-1 transmembrane protein (B7-H1) that belongs to the B7 ligand family, which can be expressed both by haematopoietic cells (lymphocytes) and non-haematopoietic cells (tumour cells) [53]. In advanced NSCLC, overexpression of PD-L1 is predictive of clinical benefit with PD-1/PD-L1 inhibitor drugs. In metastatic disease, and as a first-line palliative therapy, it is clearly predictive of efficacy for monotherapy with pembrolizumab when PD-L1 ≥ 50% [54, 55]. In some studies, it is also predictive of efficacy for the combination of PD-1/PD-L1 inhibitors with chemotherapy [56, 57]. In pre-treated patients with advanced NSCLC, overexpression of PD-L1 is also predictive of efficacy with nivolumab, pembrolizumab and atezolizumab [17]. In general, there is a correlation between positive testing for the biomarker and efficacy, although this is a marker with a suboptimal negative predictive value [58].
The standard treatment of unresectable stage III NSCLC changed due to the positive results in terms of PFS and OS of the PACIFIC study [59]. This phase III double-blind, placebo-controlled trial randomized PD-L1 unselected patients with stage III, locally advanced, unresectable NSCLC who did not progressed after chemoradiotherapy in a 2:1 ratio to receive durvalumab or placebo every to 2 weeks for up to 12 months. PACIFIC allowed any level of PD-L1 expression and tumour tissue collection was not required. Nevertheless, the European Medicines Agency restricted the approval of durvalumab to treat patients with PD-L1 ≥ 1% tumour cell expression based on a post hoc exploratory analysis. Due to this, the determination of PDL1 status is now mandatory in unresectable stage III patients suitable to receive durvalumab once completed concurrent chemoradiotherapy in the absence of progressive disease.
Recommendations:
PD-L1 expression by IHC is currently accepted as the only validated biomarker for anti-PD-1/PD-L1 therapy in unresectable locally advanced (based on a controversial EMA decision) and advanced NSCLC [60]. Thus, in clinical practice, it should always be part of the diagnosis algorithm in order to select the best treatment option.
Evidence for the presence of the PD-L1 protein can be obtained in formalin-fixed paraffin-embedded tissue specimens. Regarding preanalytical conditions, the most critical step is an enough time of fixation (i.e. at least 6 h), but storage time could also be relevant (i.e. archival material fewer than 3 years is recommended) [61].
PD-L1 is expressed at the membrane level, while intracytoplasmic expression is less frequent (not considered a positive result) and it is observed in tumour and/or immune cells.
There are several PD-L1 clones available for IHC testing. The four most widely used in pathology labs are 22C3 and 28–8 by Agilent/Dako, which share the Autostainer LINK 48 diagnostic platform by Dako, SP263 by MedImmune/Ventana and SP142 by Spring/Bioscience/Ventana, which share the Ventana BenchMark diagnostic platform. For routine diagnostics, the most frequently used clones are any of the first three, since these have shown good expression correlation between them in several studies. With respect to the other clones, SP142 stains a lower proportion of tumour cells [62].
With these four clones, a positive result for PD-L1 is evaluated according to the percentage expression in tumour cells (partial or full membrane expression) at any intensity. With SP142, the proportion of the tumoral area occupied by immune cells is also evaluated [61].
In small biopsies, at least 50–100 viable cells should be tested in order to validate the test result.
At present, this can also be conducted with cytology [11, 63], but there is no study available to date that establishes a relationship with treatment response despite the good correlation observed between direct smears or cell blocks with biopsies [11].
Since indications change rapidly, it seems reasonable to recommend including all the quantifiable information (percentage of positive tumour cells and percentage of positive immune cells) in every report, and not only the qualitative value (positive vs. negative).
Which other biomarkers in NSCLC are currently of interest?
Table 2 summarises other biomarkers to be performed on tissue- and/or cytology-type samples from advanced NSCLC patients, including its predictive alteration and the method for testing.
Table 2.
Other biomarkers of interest in NSCLC patients
| Gene | Predictive alteration | Methodology (in tissue) |
|---|---|---|
| HER2 | Mutation | PCR: sanger, real-time PCR and NGS |
| Amplification | FISH, NGS, real-time PCR | |
| MET | Mutation | NGS |
| Amplification | FISH, NGS, real-time PCR | |
| RET | Rearrangement | FISH and NGS |
| NTRK | Rearrangement | IHC (screening) and NGS |
| TMB | Mutations* | NGS |
FISH fluorescence in situ hybridisation, IHC immunohistochemistry, NGS next-generation sequencing, NSCLC non-small-cell lung cancer, PCR polymerase chain reaction, TMB tumour mutation burden
*Measurement of somatic mutations present in tumour cells
HER2
The presence of HER2 abnormalities in advanced NSCLC patients can also be ancillary to targeted therapy, but the data to date is controversial, both from the clinical viewpoint and from the biomarker perspective. Two main deregulation mechanisms have been described that are mutually exclusive with other oncogenic abnormalities: (a) mutations of which 90% are in the kinase domain (exon 20), the most frequent being p.A775_G776insYVMA insertion, especially in adenocarcinomas, with an approximate frequency of 3% [64]; and (b) amplification/overexpression that occurs in a similar percentage of, and can overlap with mutations in 11% of cases [65]. HER2 mutations seem to be the best clinical benefit predictors [65]. It also should be noted that squamous cell lung carcinomas can present HER2 mutations, but outside the kinase domain, with certain clinical benefit data when treating with afatinib [66]. As a summary, the following points can be useful:
As an isolated biomarker, HER2 IHC may not be sufficient to select patients who can benefit from anti-HER2 therapies [67, 68];
HER2 mutations identified by NGS could give access to investigational targeted drugs in clinical trials [69];
HER2 amplification has been described as a resistance mechanism after therapy with EGFR-TKIs and also as a “de novo” alteration in pan-negative adenocarcinomas [16, 70].
MET
The MET gene encodes a tyrosine kinase receptor activated by its specific natural ligand: the hepatocyte growth factor receptor (HGFR). MET amplification (3–7%), as well as overexpression (25–75%), implies a worse prognosis for the patient, with the cut-off point with predictive value in dispute. Ten to twenty percent of patients with EGFR-mutated tumours acquire EGFR-TKI resistance through MET amplification, and the therapeutic implications of this are being explored [16]. Moreover, MET exon 14 (METex14) mutations are identified in approximately 3% of NSCLC cases. These are frequently concomitant with gene amplification, and present specific clinicopathological features (e.g. elderly patients, sarcomatoid histology or adenocarcinoma) [71, 72]. These mutations are predictive of benefit with specific MET-TKIs (crizotinib, tepotinib or capmatinib) [72, 73]. The preferred technique should be NGS. Sanger sequencing can detect METex14, but large deletions or low allelic frequency can hinder sensitivity. Quantitative RT-PCR (qRT-PCR), a method based on messenger RNA (mRNA), is sensitive and specific; therefore, it can be appropriate for selecting METex14 as a single gene test.
RET
Two main activation mechanisms have been described for the oncogenic kinase RET: point mutations and genetic rearrangements. Activating point mutations are most common in medullary thyroid cancer. RET fusions are observed in 10% of papillary thyroid cancers, 1–2% of NSCLC cases and other cancer subtypes, including colorectal, pancreatic and breast cancers [74]. In NSCLC, RET fusion presents mainly in adenocarcinomas of non-smoker patients, and the partner that is most frequently associated in this setting is KIF5B. In lung adenocarcinoma, the presence of calcifications in the form of psammoma bodies could be indicative of the possibility of finding this alteration [75]. Some multiple TKIs have shown activity in NSCLC with RET fusion, as well as in other cancer types. Recently, two molecules especially designed as strong and selective inhibitors, BLU-667 and LOXO 292, have shown promising activity in RET-positive NSCLCs, as well as in other tumours with RET mutations or rearrangements [74, 76]. NGS-based panels, including RET, may be more suitable than PCR-based diagnostic methods, as the former can detect abnormalities in multiple genes simultaneously. The FISH technique is also a valid alternative in this scenario [74, 77].
NTRK
The tropomyosin receptor kinase family is encoded by three genes (NTRK1, NTRK2 and NTRK3), and its activation by rearrangement is targetable. Several drugs are the subject of clinical trials, and at least two of them are approved or in the process of approval: larotrectinib (LOXO-101, a selective inhibitor) and entrectinib (also a ROS1 and ALK inhibitor) [78]. There is a very small proportion of lung carcinoma patients (especially with adenocarcinomas) that present rearrangements in NTRK1, NTRK2 or NTRK3 [79]. Although early studies showed higher percentages, recent publications suggest a prevalence of less than 1% [78]. It is worth stating that these three abnormalities are mutually exclusive and that they are not present together with the main targetable abnormalities in lung adenocarcinomas [70, 78]. Two strategies are recommended for detecting these abnormalities: (a) NGS with a panel that includes testing for the three genes and with mandatory RNA testing to avoid false negatives; (b) IHC screening, with subsequent confirmation of every positive result by FISH or NGS [80, 81]. The IHC assay should be used according to the recently released ESMO recommendations [81].
TMB
The tumour mutation burden (TMB), also known as mutation load, is an independent biomarker for immunotherapy in many types of tumours including lung cancer [82, 83]. TMB refers to the number of somatic mutations present in the tumour, after eliminating polymorphisms and germline mutations from all variants expressed per megabase (MB) in the studied exome. The mutations acquired by tumour cells can be reflected as an abnormal protein structure, and consequently, in the expression of neoantigens that can be related to the immunotherapy response. With regard to testing, targeted NGS is considered to be a good alternative to more complex massive sequencing [84]. Although this biomarker is not yet validated for clinical practice, it may be helpful in selecting patients for immunotheraphy as NSCLCs with a high mutation burden are more sensitive to these treatments [83]. Furthermore, there are implementation difficulties due to the tissue requirements, the definition of TMB, the need for validating interconnectivity between different NGS studies, with the heterogeneity of the numbers of included genes, horizontal coverage, the required optimal depth, and the chemical sequencing type, etc. and also, and more importantly, because the algorithms are continuously developing [84]. If eventually drugs are approved based on TMB cut-offs, the harmonization efforts underway could be very useful [84].
Other biomarkers
The KRAS gene appears to be mutated in about 20% of all cases of NSCLC, especially in adenocarcinomas and smokers. Although its prognostic value has not been clearly demonstrated, it is the most common oncogenic mutation in lung cancer. In fact, many treatment strategies (such as the farnesyl transferase inhibitors, MEK and CDK4/6) have failed in this context [85]. For this reason, KRAS testing is currently not indicated as an individual test but it is appropriate that the study of the KRAS gene is included in extended panels [21, 22].
With regards to other potential biomarkers predictive of an immune response, the microsatellite instability and the immune microenvironment study should be highlighted, from the viewpoint of RNA expression and tissue determination of multiple immune cells [17, 58, 86, 87].
How to prioritise the use of biological specimens for an accurate diagnosis
Most previous recommendations regarding sample prioritisation and its preservation for multiple biomarkers testing in advanced NSCLC patients are still valid [1, 13]. However, there are several new aspects that require an update on sample-sparing procedures [22, 24, 40].
Regarding histological diagnosis, it is still advisable to use the smallest amount of tissue for tumour typing, with a reasonable use of classificatory IHC [88]. This means using no more than two markers (i.e. TTF1 and p40) in cases without any clear morphological differentiation. It is worth noting that a different degree of TTF1 positivity has been described for adenocarcinomas, depending on the clone used (for example, the 8G7G3/1 antibody shows higher specificity and lower sensitivity than other clones) [89].
Regarding the testing of both molecular and immune biomarkers (see previous sections), it is still important to remember two principles: (a) the fewer times paraffin-embedded material (tissue or cytological as cell blocks) is placed in a microtome, the more will be spared; and (b) the order of biomarker prioritisation is important, as the tissue can be depleted [13]. To meet at least the first principle, testing should be always planned in advance for every NSCLC patient.
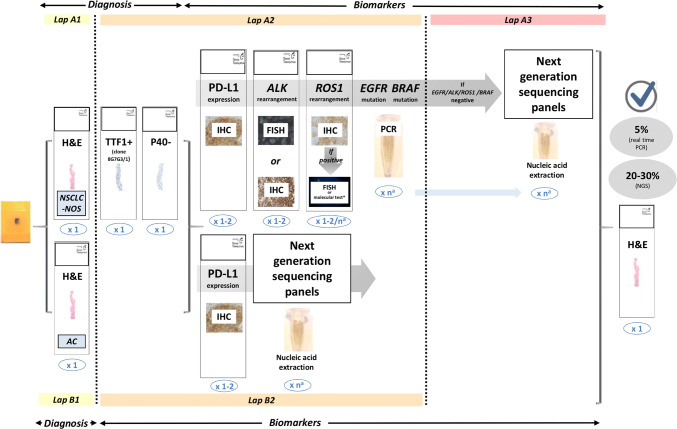
Regarding the molecular biomarkers to be analysed, apart from EGFR mutations and ALK translocations, it is currently mandatory to include testing for ROS1 rearrangements and BRAF mutations [17, 24]. The study of biomarkers such as MET, RET, HER2, NTRK and KRAS as individual tests is currently not indicated, but instead it is advised to include these biomarkers in extended panels performed either initially in all advanced NSCLCs or when previous EGFR/ALK/ROS1/BRAF testing is negative [17, 24]. The recommended protocol to follow is shown in Fig. 2, which includes the new biomarkers, detailing the recommended techniques and showing the two alternative pathways. Both routes are equally valid but upfront NGS could be a more cost/effective approach [90]. An aspect that undoubtedly improves overall quality is to use validated tests and to take part in quality control programs (see below).
Fig. 2.
Protocol for multiple biomarker testing on samples from patients with advanced NSCLC. The number of sections for each test is shown in blue. aThe requirements for nucleic acid extraction for individual molecular testing or for extended genetic panels (NGS) are variable. AC adenocarcinoma, EGFR epidermal growth factor receptor, FISH fluorescence in situ hybridisation, H&E haematoxylin and eosin, IHC immunohistochemistry, NGS next-generation sequencing, NSCLC-NOS non-small-cell lung carcinoma – not otherwise specified, PCR polymerase chain reaction, PD-L1 programmed death ligand-1
(Adapted protocol from international guidelines ASCO/CAP, ESMO and NCCN [17, 22, 24, 40]. Figure modified from Conde et al. (confidential, submitted))
One issue under discussion and without reference in international guidelines is how to incorporate immune biomarkers. The proposed sequence of previous steps includes one slice (or two) for PD-L1, together with the slides needed for ALK and ROS1, or before nucleic acids extraction [61].
Other issue being debated is whether the biomarkers should be reflex tested (by the pathologist at diagnosis) or in response to the clinical request. Ideally, this should be done as reflex testing so the pathologist can prioritise the sample, avoid the need to review preparations when the molecular tests are requested, and minimise response time, although this is not always possible [91, 92].
The whole concept of sample prioritization and multidisciplinary coordination should be organized through a molecular tumour board [93].
The role of NGS in NSCLC
Following the discovery of new low-frequency abnormalities, there is an increased need for multigene testing, as opposed to single approaches. Testing should include RET, HER2, NTRK, KRAS and MET for cases in which the usual oncogenic drivers (EGFR, ALK, ROS1 and BRAF) give a negative result and whenever an adequate technique is available [24]. The advantage of ultrasequencing and transcriptome analysis is the possibility of conducting mass screening without any loss of sensitivity and specificity, reducing the use of minimal biological specimens.
The development of this technique results in three categories for biomarker classification: (a) key biomarkers that should be tested to identify patients who are to be treated with approved therapies; (b) additional biomarkers that are desirable for identifying those patients who can benefit from clinical trials; and (c) other biomarkers that, at present, are only used in research and are not used in clinical practice (exome, genome, transcriptome) [22].
Biomarkers can be tested simultaneously with NGS. This technique is capable of detecting not only point mutations or insertions/deletions (indels) but also rearrangements and copy number variations, as well as a wide range of structural variants [30]. As mentioned previously, NGS appears to be a optimal technique for TMB testing [84].
The fact that most available specimens in routine healthcare are fixed in formalin and embedded in paraffin, and that double testing for DNA and RNA is necessary, is decisive when considering whether a technique is ideal for clinical use. Additionally, the specimens most commonly available for lung cancer have a low tumour cell content. In fact, most recommended techniques are those that are capable of detecting molecular abnormalities in samples with at least 20–30% of cancer cells [24].
There are studies that show very good concordance data between different technical solutions based on paraffin-embedded tissue. All these procedures urgently require quality control on the pre-test fixation parameters, as well as control of tumour cellularity and quality control for the nucleic acids [94]. Although there are advantages and limitations associated with amplicon-based and hybrid capture solutions, the most important thing to keep in mind is the need to use RNA when looking for druggable fusions to avoid a significant risk of false negatives [95].
The role of liquid biopsy in NSCLC
Tumour biopsies are often insufficient for molecular study or are impossible to obtain. This is why liquid biopsies have been proposed as an alternative. It has many advantages, as it is a minimally invasive technique that can be used at diagnosis or during follow-up. [96, 97]. Circulating tumour cells (CTCs), circulating tumour DNA (ctDNA), circulating exosomes, platelet RNA, and circulating tumour RNA (ctRNA) are included in the definition of liquid biopsy. ctDNA represents the whole genomic picture of the tumour and is used in current clinical practice for liquid biopsies to test for genetic and epigenetic abnormalities specific to the tumour [14]. It can be detected in blood and also in urine, pleural fluids and saliva, among others [98].
Test methods for ctDNA can have a high specificity [14]. Therefore, when a mutation is detected in a clinical setting, it can be used to determine a targeted therapy. Since levels of ctDNA vary significantly and can be as low as 0.01% of all cfDNA, detection techniques must have a high sensitivity in order to detect the DNA from tumour cells, from 15 to 0.01% being the most widely used [98]. These techniques include ARMS (amplification refractory mutation system) PCR, qPCR, digital PCR (dPCR), BEAMing (beads, emulsions, amplification and magnetics) and the recommended NGS technique when ALK and ROS1 fusions are to be tested.
From a clinical perspective, these methods offer alternative diagnostic techniques when a tissue biopsy is insufficient or not viable to determine EGFR T790M resistance mutation in NSCLC patients who harbour EGFR mutations, and also when the disease progresses. Nevertheless, a negative result with liquid biopsy requires testing with conventional techniques, such as tumour biopsy. Although validation and clinical usefulness are not sufficiently determined as yet, this is a promising technique for diagnosing other molecular abnormalities and their resistance mechanisms. It offers different possible applications, such as response monitoring, tumour recurrence detection, determination of residual disease after full tumour resection, early detection of lung cancer and for immuno-oncology [14].
Main requirements for implementing optimal quality control
Quality test control is important, necessary and should be incorporated into the quality plan of the laboratory or service conducting the tests. In Spain, it is recommended that the lab has an ISO 9001 certification, and also that the different tests be accredited by the UNE-EN ISO15189 standard that has started to be enforced by pathology and molecular laboratories and evaluated by the Spanish National Certification Entity (ENAC).
The roadmap of processes and quality indices should include (a) the staff involved (technicians, biologists, pathologists etc.) and their training, experience and standard operating procedures; (b) instrumentation, with CE certifications for use and maintenance; and (c) reagents. For more information, the SEAP [99], CAP [22], and Association of Directors of Anatomic and Surgical Pathology (ADASP) recommendations can be reviewed [100].
As a summary outline, the controls for every test can be: (a) internal, such as positive and negative controls associated with each test; (b) external, such as quality control schemes (SEAP, EMQN, UK-NEQAS) (Table 3); and (c) results control, to verify that the percentage of mutations found corresponds to the frequency described depending on the type of tested samples. To facilitate this last control, it is advised to take part in case registration programs set up in collaboration with SEAP (Lungpath or ALKanza) in order to compare results with those obtained in similar hospitals.
Table 3.
Examples of european quality assurance shemes
| Supplier | Name | Starting material | Aim | Format |
|---|---|---|---|---|
| EMQN | Molecular testing of cfDNA in plasma for EGFR gene mutations (pilot) | Plasma containing cfDNA | Mutations in the EGFR gene | Five mock clinical cases with matching samples |
| Molecular testing in lung cancer | Mix of real tissue and artificial FFPE materials | Mutations in the EGFR, PIK3CA, KRAS and BRAF genes | Ten mock clinical cases with matching samples | |
| DNA Sequencing–NGS (vSomatic) | DNA sample derived from FFPE material | Any NGS strategy can be used | One mock clinical case with matching samples | |
| Oncogene panel testing | Rolled sections of FFPE materials | Mutations in the EGFR, PIK3CA, KRAS, HRAS, NRAS, KIT, TP53 and BRAF genes | Three mock clinical cases with matching samples | |
| ESP | ALK FISH | Slides | ALK rearrangements | Five resections, five digital cases |
| ALK IHC | Slides | ALK rearrangements | Five resections | |
| EGFR, KRAS (optional), BRAF (optional) | Slides/rolled sections | Mutations | Ten resection specimens, possible cell-line | |
| ROS1 fish | Slides | ROS1 rearrangements | Five resections or possibly cell-lines, five digital cases | |
| ROS1 IHC | Slides | ROS1 rearrangements | Five resections or possibly cell-lines | |
| PD-L1 | Slides | PD-L1 overexpression | Eight resections (TMAs) and four digital cases | |
| MET EQA scheme (ex 14 skipping) for DNA and RNA | Slides/rolled sections | MET exon 14 mutations | Five resections | |
| NordiQC | Companion PD-L1 | Slides | PD-L1 overexpression | One preparation with multiple cases and one in-house case |
| SEAP | ALKanza MODULE | Slides | ALK rearrangements | One slide with four cases + one in house |
| EGFR | Slides/rolled sections | EGFR mutations | Four consecutive slides | |
| UKNQEQAS | NSLCC ALK IHC | Slides | ALK and ROS1 rearrangements | One slide with several cases + one in house |
| NSLCC ALK/ROS1 FISH (pilot) | Slides | ALK and ROS1 rearrangements | One slide with several cases + one in house | |
| NSLCC PD-L1 IHC (pilot) | Slides | PD-L1 overexpression | One slide with several cases + one in house | |
| Gen QA | Lung cancer | Slides/rolled sections | EGFR, ALK (optional), KRAS (optional), BRAF (optional) | 5–4 cases |
| Circulating tumour DNA (pilot) | Plasma | EGFR mutations | Five cases | |
| Additional lung cancer biomarkers | Slides/rolled sections | ROS1, RET and MET (amplification) | Four cases |
cfDNA cell-free DNA, EGFR epidermal growth factor receptor, FFPE formalin-fixed paraffin-embedded, FISH fluorescence in situ hybridisation, IHC immunohistochemistry, NGS next-generation sequencing, PD-L1 programmed death ligand-1, TMA tissue microarrays
The most frequent quality indices are: (a) response time, with around 7–10 working days recommended, on the understanding that this refers to having all biomarker results available within this time frame, both from individual tests and targeted NGS; (b) results from previously described quality controls; and (c) discrepancy/error analysis. Therefore, the creation of multidisciplinary committees for analysing the molecular diagnoses will facilitate the establishment of these indices. Table 4 specifies the information that should be contained in the results report for any biomarker test.
Table 4.
Proposed pathology results report
| Identification of the patient and the doctor who ordered the test (or, failing that, the authorised person) |
| Pathological diagnosis |
| Type of specimen submitted: |
|
Previous treatment (yes/no) Time of biopsy (initial/relapse/progression) Date on which the specimen was collected |
| The external code in the case of referral centres |
| The medium in which the specimen was received (fresh, frozen, paraffin-embedded, etc.) |
| The anatomical origin of the specimen |
| The order date, the specimen receipt date and the date on which the results were issued |
| The biomarker test method used, specifying detectable mutations and/or other abnormalities. In the case of commercial kits, the commercial name, the batch number and whether they are an approved ‘in vitro diagnostics’ product should be stated |
| The quality of the sample, specifying the percentage of cancer cells and whether the sample was enriched by micro- or macrodissection, as well as DNA concentration and purity |
| Comments about the adequate or inadequate nature of the sample |
| The test result, defining the type of molecular abnormality detected or the absence of molecular abnormalities |
| Identification of the professional responsible for the test (all phases) |
| Identification of the laboratory supervisor (optional) |
| Any additional information or comments of interest to the doctor who ordered the test |
| Accreditation or participation in quality programs |
Conclusions
The mandatory tests for every patient with advanced NSCLC are EGFR and BRAF mutations, ALK and ROS1 rearrangements [17, 24, 40], and PD-L1 expression [17, 40]. However, the growing need to study emerging biomarkers (HER2, MET, RET, NTRK and TMB) warrants the establishment of a routine and more comprehensive molecular assessment with targeted NGS. The coordination between all professionals and prioritisation of the proper tests and technologies for each case remains a challenge. Thus, adequate multidisciplinary communication is essential in order to provide the information within the required time frame, with the required quality and at a reasonable cost.
Acknowledgements
The authors are grateful for the editorial assistance of Beatriz Gil-Alberdi of HealthCo (Madrid, Spain) in the production of this manuscript. F. López-Ríos thanks T. Crean for his support.
Funding
SEAP and SEOM have received financial support for this project in the form of unrestricted grants from AstraZeneca, Novartis, Pfizer, Roche, Sysmex and Takeda. F. López-Rios, E. Conde and L. Paz-Ares thank the support of iLUNG-CM (B2017/BMD-3884).
Compliance with ethical standards
Conflict of interest
The authors declare that, when writing and revising the text, they did not know the names of the pharmaceutical companies that provided financial support for this project, so this support has not influenced the content of this article.
Ethical approval
The study has been performed in accordance with the ethical standards of the Declaration of Helsinki and its later amendments. This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study formal consent is not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
P. Garrido, Email: pilargarridol@gmail.com
E. Conde, Email: econde@hmhospitales.com
J. de Castro, Email: javier.decastro@salud.madrid.org
J. J. Gómez-Román, Email: josejavier.gomez@scsalud.es
E. Felip, Email: efelip@vhebron.net
L. Pijuan, Email: Lpijuan@parcdesalutmar.cat
D. Isla, Email: lola.isla@gmail.com
J. Sanz, Email: jsanzo.hcsc@salud.madrid.org
L. Paz-Ares, Email: lpazaresr@seom.org
F. López-Ríos, Email: flopezrios@hmhospitales.com
References
- 1.Felip E, Concha A, de Castro J, Gomez-Roman J, Garrido P, Ramirez J, et al. Biomarker testing in advanced non-small-cell lung cancer: a National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin Transl Oncol. 2015;17:103–112. doi: 10.1007/s12094-014-1248-9. [DOI] [PubMed] [Google Scholar]
- 2.Garrido P, de Castro J, Concha A, Felip E, Isla D, Lopez-Rios F, et al. Guidelines for biomarker testing in advanced non-small-cell lung cancer. A National Consensus of the Spanish Society of Medical Oncology (SEOM) and the Spanish Society of Pathology (SEAP) Clin Transl Oncol. 2012;14:338–349. doi: 10.1007/s12094-012-0806-2. [DOI] [PubMed] [Google Scholar]
- 3.Eberhard DA, Giaccone G, Johnson BE, Non-Small-Cell Lung Cancer Working G Biomarkers of response to epidermal growth factor receptor inhibitors in non-small-cell lung cancer working group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 4.Malapelle U, Mayo-de-Las-Casas C, Molina-Vila MA, Rosell R, Savic S, Bihl M, et al. Consistency and reproducibility of next-generation sequencing and other multigene mutational assays: a worldwide ring trial study on quantitative cytological molecular reference specimens. Cancer Cytopathol. 2017;125:615–626. doi: 10.1002/cncy.21868. [DOI] [PubMed] [Google Scholar]
- 5.Dietel M, Bubendorf L, Dingemans AM, Dooms C, Elmberger G, Garcia RC, et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): recommendations of the European expert group. Thorax. 2016;71:177–184. doi: 10.1136/thoraxjnl-2014-206677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angulo B, Conde E, Suarez-Gauthier A, Plaza C, Martinez R, Redondo P, et al. A comparison of EGFR mutation testing methods in lung carcinoma: direct sequencing, real-time PCR and immunohistochemistry. PLoS One. 2012;7:e43842. doi: 10.1371/journal.pone.0043842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doxtader EE, Cheng YW, Zhang Y. Molecular testing of non-small cell lung carcinoma diagnosed by endobronchial ultrasound-guided transbronchial fine-needle aspiration. Arch Pathol Lab Med. 2018;143:670–676. doi: 10.5858/arpa.2017-0184-RA. [DOI] [PubMed] [Google Scholar]
- 9.Jain D, Roy-Chowdhuri S. Molecular pathology of lung cancer cytology specimens: a concise review. Arch Pathol Lab Med. 2018;142:1127–1133. doi: 10.5858/arpa.2017-0444-RA. [DOI] [PubMed] [Google Scholar]
- 10.Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing—the MD Anderson experience. Arch Pathol Lab Med. 2016;140:1191–1199. doi: 10.5858/arpa.2016-0117-RA. [DOI] [PubMed] [Google Scholar]
- 11.Noll B, Wang WL, Gong Y, Zhao J, Kalhor N, Prieto V, et al. Programmed death ligand 1 testing in non-small cell lung carcinoma cytology cell block and aspirate smear preparations. Cancer Cytopathol. 2018;126:342–352. doi: 10.1002/cncy.21987. [DOI] [PubMed] [Google Scholar]
- 12.Auger M, Brimo F, Kanber Y, Fiset PO, Camilleri-Broet S. A practical guide for ancillary studies in pulmonary cytologic specimens. Cancer Cytopathol. 2018;126(Suppl 8):599–614. doi: 10.1002/cncy.22028. [DOI] [PubMed] [Google Scholar]
- 13.Conde E, Angulo B, Izquierdo E, Paz-Ares L, Belda-Iniesta C, Hidalgo M, et al. Lung adenocarcinoma in the era of targeted therapies: histological classification, sample prioritization, and predictive biomarkers. Clin Transl Oncol. 2013;15:503–508. doi: 10.1007/s12094-012-0983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 16.Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24:3108–3118. doi: 10.1158/1078-0432.CCR-17-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: esmo clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;129:iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 18.Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018;29:i10–i19. doi: 10.1093/annonc/mdx703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321–346. doi: 10.5858/arpa.2017-0388-CP. [DOI] [PubMed] [Google Scholar]
- 23.Solomon BJ, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, et al. Final overall survival analysis from a study comparing first-line crizotinib versus chemotherapy in ALK-mutation-positive non-small-cell lung cancer. J Clin Oncol. 2018;36:2251–2258. doi: 10.1200/JCO.2017.77.4794. [DOI] [PubMed] [Google Scholar]
- 24.Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura H, Tsuta K, Yoshida A, Shibata T, Wakai S, Asamura H, et al. Aberrant anaplastic lymphoma kinase expression in high-grade pulmonary neuroendocrine carcinoma. J Clin Pathol. 2013;66:705–707. doi: 10.1136/jclinpath-2012-201329. [DOI] [PubMed] [Google Scholar]
- 26.Conde E, Hernandez S, Prieto M, Martinez R, Lopez-Rios F. Profile of Ventana ALK (D5F3) companion diagnostic assay for non-small-cell lung carcinomas. Expert Rev Mol Diagn. 2016;16:707–713. doi: 10.1586/14737159.2016.1172963. [DOI] [PubMed] [Google Scholar]
- 27.Thunnissen E, Allen TC, Adam J, Aisner DL, Beasley MB, Borczuk AC, et al. Immunohistochemistry of pulmonary biomarkers: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med. 2018;142:408–419. doi: 10.5858/arpa.2017-0106-SA. [DOI] [PubMed] [Google Scholar]
- 28.Lozano MD, Echeveste JI, Abengozar M, Mejias LD, Idoate MA, Calvo A, et al. Cytology smears in the era of molecular biomarkers in non-small cell lung cancer: doing more with less. Arch Pathol Lab Med. 2018;142:291–298. doi: 10.5858/arpa.2017-0208-RA. [DOI] [PubMed] [Google Scholar]
- 29.Conde E, Suarez-Gauthier A, Benito A, Garrido P, Garcia-Campelo R, Biscuola M, et al. Accurate identification of ALK positive lung carcinoma patients: novel FDA-cleared automated fluorescence in situ hybridization scanning system and ultrasensitive immunohistochemistry. PLoS One. 2014;9:e107200. doi: 10.1371/journal.pone.0107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wistuba I, Sholl N, Lindeman N. ALK and ROS1 testing with NGS. In: Tsao MS, Hirsch FR, Yatabe Y, editors. IASLC atlas of ALK and ROS1 testing in lung cáncer. 2. FL: Editorial Rx Press; 2016. [Google Scholar]
- 31.Kerr KM, Lopez-Rios F. Precision medicine in NSCLC and pathology: how does ALK fit in the pathway? Ann Oncol. 2016;27(Suppl 3):iii16–iii24. doi: 10.1093/annonc/mdw302. [DOI] [PubMed] [Google Scholar]
- 32.Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, et al. Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016;21:762–770. doi: 10.1634/theoncologist.2015-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reguart N, Teixido C, Gimenez-Capitan A, Pare L, Galvan P, Viteri S, et al. Identification of ALK, ROS1, and RET fusions by a multiplexed mRNA-based assay in formalin-fixed, paraffin-embedded samples from advanced non-small-cell lung cancer patients. Clin Chem. 2017;63:751–760. doi: 10.1373/clinchem.2016.265314. [DOI] [PubMed] [Google Scholar]
- 34.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7:137–155. doi: 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazieres J, Zalcman G, Crino L, Biondani P, Barlesi F, Filleron T, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol. 2015;33:992–999. doi: 10.1200/JCO.2014.58.3302. [DOI] [PubMed] [Google Scholar]
- 36.Moro-Sibilot D, Faivre L, Zalcman G, Pérol M, Barlesi F, Otto J, et al. Crizotinib in patients with advanced ROS1-rearranged non-small cell lung cancer (NSCLC). Preliminary results of the ACSé phase II trial. J Clin Oncol. 2015;33:8065. [Google Scholar]
- 37.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bubendorf L, Buttner R, Al-Dayel F, Dietel M, Elmberger G, Kerr K, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469:489–503. doi: 10.1007/s00428-016-2000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conde E, Hernandez S, Martinez R, Angulo B, De Castro J, Collazo-Lorduy A, et al. Assessment of a new ROS1 immunohistochemistry clone (SP384) for the identification of ROS1 rearrangements in non-small cell lung carcinoma patients: the ROSING study. J Thorac Oncol. 2019 doi: 10.1016/j.jtho.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 40.NCCN. Non-small cell lung cancer. In: NCCN clinical practice guidelines in oncology. 2018. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed 14 Dec 2018.
- 41.Sholl LM, Sun H, Butaney M, Zhang C, Lee C, Janne PA, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol. 2013;37:1441–1449. doi: 10.1097/PAS.0b013e3182960fa7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida A, Tsuta K, Wakai S, Arai Y, Asamura H, Shibata T, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol. 2014;27:711–720. doi: 10.1038/modpathol.2013.192. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J, Chen X, Zheng J, Kong M, Wang B, Ding W. A genomic and clinicopathological study of non-small-cell lung cancers with discordant ROS1 gene status by fluorescence in situ hybridisation and immunohistochemical analysis. Histopathology. 2018;73:19–28. doi: 10.1111/his.13492. [DOI] [PubMed] [Google Scholar]
- 44.Davies KD, Le AT, Sheren J, Nijmeh H, Gowan K, Jones KL, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13:1474–1482. doi: 10.1016/j.jtho.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu YL, Yang JC, Kim DW, Lu S, Zhou J, Seto T, et al. Phase II study of crizotinib in east asian patients with ROS1-positive advanced non-small-cell lung cancer. J Clin Oncol. 2018;36:1405–1411. doi: 10.1200/JCO.2017.75.5587. [DOI] [PubMed] [Google Scholar]
- 46.Planchard D, Johnson BE. BRAF adds an additional piece of the puzzle to precision oncology-based treatment strategies in lung cancer. Arch Pathol Lab Med. 2018;142:796–797. doi: 10.5858/arpa.2018-0088-ED. [DOI] [PubMed] [Google Scholar]
- 47.Yousem SA, Nikiforova M, Nikiforov Y. The histopathology of BRAF-V600E-mutated lung adenocarcinoma. Am J Surg Pathol. 2008;32:1317–1321. doi: 10.1097/PAS.0b013e31816597ca. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen-Ngoc T, Bouchaab H, Adjei AA, Peters S. BRAF alterations as therapeutic targets in non-small-cell lung cancer. J Thorac Oncol. 2015;10:1396–1403. doi: 10.1097/JTO.0000000000000644. [DOI] [PubMed] [Google Scholar]
- 49.Gautschi O, Milia J, Cabarrou B, Bluthgen MV, Besse B, Smit EF, et al. Targeted therapy for patients with BRAF-mutant lung cancer: results from the European EURAF cohort. J Thorac Oncol. 2015;10:1451–1457. doi: 10.1097/JTO.0000000000000625. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110:2812–2820. doi: 10.1038/bjc.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leonetti A, Facchinetti F, Rossi G, Minari R, Conti A, Friboulet L, et al. BRAF in non-small cell lung cancer (NSCLC): pickaxing another brick in the wall. Cancer Treat Rev. 2018;66:82–94. doi: 10.1016/j.ctrv.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Odogwu L, Mathieu L, Blumenthal G, Larkins E, Goldberg KB, Griffin N, et al. FDA approval summary: dabrafenib and trametinib for the treatment of metastatic non-small cell lung cancers harboring BRAF V600E mutations. Oncologist. 2018;23:740–745. doi: 10.1634/theoncologist.2017-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Boyle TA, Zhou C, Rimm DL, Hirsch FR. PD-L1 expression in lung cancer. J Thorac Oncol. 2016;11:964–975. doi: 10.1016/j.jtho.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopes G, Wu YL, Kudaba I, Kowalski D, Cho BC, Castro G, et al. Pembrolizumab (pembro) versus platinum-based chemotherapy (chemo) as first-line therapy for advanced/metastatic NSCLC with a PD-L1 tumor proportion score (TPS) ≥ 1%: open-label, phase 3 KEYNOTE-042 study. J Clin Oncol. 2018;36:LBA4. [Google Scholar]
- 55.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 56.Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 57.Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378:2288–2301. doi: 10.1056/NEJMoa1716948. [DOI] [PubMed] [Google Scholar]
- 58.Mehnert JM, Monjazeb AM, Beerthuijzen JMT, Collyar D, Rubinstein L, Harris LN. The challenge for development of valuable immuno-oncology biomarkers. Clin Cancer Res. 2017;23:4970–4979. doi: 10.1158/1078-0432.CCR-16-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 60.Peters S, Dafni U, Boyer M, De Ruysscher D, Faivre-Finn C, Felip E, et al. Position of a panel of international lung cancer experts on the approval decision for use of durvalumab in stage III non-small-cell lung cancer (NSCLC) by the committee for medicinal products for human use (CHMP) Ann Oncol. 2019;30:161–165. doi: 10.1093/annonc/mdy553. [DOI] [PubMed] [Google Scholar]
- 61.IASLC. IASLC atlas of PD-L1 testing in lung cancer. 2017. https://www.iaslc.org/publications/iaslc-atlas-pd-l1-testing-lung-cancer. Accessed 1 Feb 2019.
- 62.Buttner R, Gosney JR, Skov BG, Adam J, Motoi N, Bloom KJ, et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. J Clin Oncol. 2017;35:3867–3876. doi: 10.1200/JCO.2017.74.7642. [DOI] [PubMed] [Google Scholar]
- 63.Heymann JJ, Bulman WA, Swinarski D, Pagan CA, Crapanzano JP, Haghighi M, et al. PD-L1 expression in non-small cell lung carcinoma: comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol. 2017;125:896–907. doi: 10.1002/cncy.21937. [DOI] [PubMed] [Google Scholar]
- 64.Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med. 2018;24:638–646. doi: 10.1038/s41591-018-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters S, Curioni-Fontecedro A, Nechushtan H, Shih JY, Liao WY, Gautschi O, et al. Activity of afatinib in heavily pretreated patients with ERBB2 mutation-positive advanced NSCLC: findings from a global named patient use program. J Thorac Oncol. 2018;13:1897–1905. doi: 10.1016/j.jtho.2018.07.093. [DOI] [PubMed] [Google Scholar]
- 66.Goss GD, Felip E, Cobo M, Lu S, Syrigos K, Lee KH, et al. Association of ERBB mutations with clinical outcomes of afatinib- or erlotinib-treated patients with lung squamous cell carcinoma: secondary analysis of the LUX-lung 8 randomized clinical trial. JAMA Oncol. 2018;4:1189–1197. doi: 10.1001/jamaoncol.2018.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li BT, Shen R, Buonocore D, Olah ZT, Ni A, Ginsberg MS, et al. Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: results from a phase II basket trial. J Clin Oncol. 2018;36:2532–2537. doi: 10.1200/JCO.2018.77.9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peters S, Stahel R, Bubendorf L, Bonomi P, Villegas A, Kowalski DM, et al. Trastuzumab emtansine (T-DM1) in patients with previously treated HER2-overexpressing metastatic non-small cell lung cancer: efficacy, safety, and biomarkers. Clin Cancer Res. 2019;25:64–72. doi: 10.1158/1078-0432.CCR-18-1590. [DOI] [PubMed] [Google Scholar]
- 69.Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, et al. Actionable activating oncogenic ERBB2/HER2 transmembrane and juxtamembrane domain mutations. Cancer Cell. 2018;34(792–806):e5. doi: 10.1016/j.ccell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J Clin Oncol. 2016;34:721–730. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 72.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–849. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Felip E, Sakai H, Patel J, Horn L, Veillon R, Griesinger F, et al. OA12.01 phase II data for the MET inhibitor tepotinib in patients with advanced NSCLC and MET exon 14-skipping mutations. J Thorac Oncol. 2018;13:S347. [Google Scholar]
- 74.Drilon A, Hu ZI, Lai GGY, Tan DSW. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol. 2018;15:150. doi: 10.1038/nrclinonc.2017.188. [DOI] [PubMed] [Google Scholar]
- 75.Mukhopadhyay S, Pennell NA, Ali SM, Ross JS, Ma PC, Velcheti V. RET-rearranged lung adenocarcinomas with lymphangitic spread, psammoma bodies, and clinical responses to cabozantinib. J Thorac Oncol. 2014;9:1714–1719. doi: 10.1097/JTO.0000000000000323. [DOI] [PubMed] [Google Scholar]
- 76.Subbiah V, Velcheti V, Tuch BB, Ebata K, Busaidy NL, Cabanillas ME, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol. 2018;29:1869–1876. doi: 10.1093/annonc/mdy137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drilon A, Wang L, Arcila ME, Balasubramanian S, Greenbowe JR, Ross JS, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32:147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 79.Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, et al. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol. 2018;2:1–12. doi: 10.1200/PO.18.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41:1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchio C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019;30(9):1417–1427. doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 82.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Büttner R, Longshore JW, López-Ríos F, Merkelbach-Bruse S, Normanno N, Rouleau E, et al. Implementing TMB measurement in clinical practice: considerations on assayrequirements. ESMO Open. 2019;4:e000442. doi: 10.1136/esmoopen-2018-000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferrer I, Zugazagoitia J, Herbertz S, John W, Paz-Ares L, Schmid-Bindert G. KRAS-mutant non-small cell lung cancer: from biology to therapy. Lung Cancer. 2018;124:53–64. doi: 10.1016/j.lungcan.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 86.Conde E, Caminoa A, Dominguez C, Calles A, Walter S, Angulo B, et al. Aligning digital CD8(+) scoring and targeted next-generation sequencing with programmed death ligand 1 expression: a pragmatic approach in early-stage squamous cell lung carcinoma. Histopathology. 2018;72:270–284. doi: 10.1111/his.13346. [DOI] [PubMed] [Google Scholar]
- 87.Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25:3753–3758. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- 88.Yatabe Y, Dacic S, Borczuk AC, Warth A, Russell PA, Lantuejoul S, et al. Best practices recommendations for diagnostic immunohistochemistry in lung cancer. J Thorac Oncol. 2018;14:377–407. doi: 10.1016/j.jtho.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woo JS, Reddy OL, Koo M, Xiong Y, Li F, Xu H. Application of immunohistochemistry in the diagnosis of pulmonary and pleural neoplasms. Arch Pathol Lab Med. 2017;141:1195–1213. doi: 10.5858/arpa.2016-0550-RA. [DOI] [PubMed] [Google Scholar]
- 90.Layfield LJ, Hammer RD, White SK, Furtado LV, Schmidt RL. Molecular testing strategies for pulmonary adenocarcinoma: an optimal approach with cost analysis. Arch Pathol Lab Med. 2019;143:628–633. doi: 10.5858/arpa.2018-0218-OA. [DOI] [PubMed] [Google Scholar]
- 91.Levy BP, Chioda MD, Herndon D, Longshore JW, Mohamed M, Ou SH, et al. Molecular testing for treatment of metastatic non-small cell lung cancer: how to implement evidence-based recommendations. Oncologist. 2015;20:1175–1181. doi: 10.1634/theoncologist.2015-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lim C, Sekhon HS, Cutz JC, Hwang DM, Kamel-Reid S, Carter RF, et al. Improving molecular testing and personalized medicine in non-small-cell lung cancer in Ontario. Curr Oncol. 2017;24:103–110. doi: 10.3747/co.24.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van de Haar J, Hoes L, Voest E. Advancing molecular tumour boards: highly needed to maximise the impact of precision medicine. ESMO Open. 2019;4:e000516. doi: 10.1136/esmoopen-2019-000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rogers TM, Arnau GM, Ryland GL, Huang S, Lira ME, Emmanuel Y, et al. Multiplexed transcriptome analysis to detect ALK, ROS1 and RET rearrangements in lung cancer. Sci Rep. 2017;7:42259. doi: 10.1038/srep42259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santarpia M, Liguori A, D’Aveni A, Karachaliou N, Gonzalez-Cao M, Daffina MG, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis. 2018;10:S882–S897. doi: 10.21037/jtd.2018.03.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Taus A, Camacho L, Rocha P, Hardy-Werbin M, Pijuan L, Piquer G, et al. Dynamics of EGFR mutation load in plasma for prediction of treatment response and disease progression in patients with EGFR-mutant lung adenocarcinoma. Clin Lung Cancer. 2018;19(387–94):e2. doi: 10.1016/j.cllc.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 98.Sholl LM, Aisner DL, Allen TC, Beasley MB, Cagle PT, Capelozzi VL, et al. Liquid biopsy in lung cancer: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med. 2016;140:825–829. doi: 10.5858/arpa.2016-0163-SA. [DOI] [PubMed] [Google Scholar]
- 99.SEAP. “Reglas y Consejos” sobre buenas prácticas profesionales en anatomía patológica. 2013. https://www.seap.es/c/document_library/get_file?uuid=c08d2461-7bac-46e1-8eee-f9db7ec5e95a&groupId=10157. Accessed 14 Dec 2018.
- 100.Nakhleh RE, Nose V, Colasacco C, Fatheree LA, Lillemoe TJ, McCrory DC, et al. Interpretive diagnostic error reduction in surgical pathology and cytology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center and the Association of Directors of Anatomic and Surgical Pathology. Arch Pathol Lab Med. 2016;140:29–40. doi: 10.5858/arpa.2014-0511-SA. [DOI] [PubMed] [Google Scholar]