Abstract
Early detection of retroviruses including human T-cell lymphotropic virus and human immunodeficiency virus in the human body is indispensable to prevent retroviral infection propagation and improve clinical treatment. Until now, diverse techniques have been employed for the early detection of viruses. Traditional methods are time-consuming, resource-intensive, and laborious performing. Therefore, designing and constructing a selective and sensitive diagnosis system to detect serious diseases is highly demanded. Genetic detection with high sensitivity has striking significance for the early detection and remedy of disparate pathogenic diseases. The nucleic acid biosensors are based on the identification of specific DNA sequences in biological samples. Nanotechnology has an important impact on the development of sensitive biosensors. Different kinds of nanomaterials include nanoparticles, nanoclusters, quantum dots, carbon nanotubes, nanocomposites, etc., with different properties have been used to improve the performance of biosensors. Recently, DNA nanobiosensors are developed to provide simple, fast, selective, low-cost, and sensitive detection of infectious diseases. In this paper, the research progresses of nano genosensors for the detection of HIV-1 and HTLV-1 viruses, based on electrochemical, optical, and photoelectrochemical platforms are overviewed.
Keywords: Analytical chemistry, Infectious disease, Nanotechnology, Human immunodeficiency virus (HIV), Human T-cell lymphotropic virus (HTLV), Early detection, DNA nanobiosensors
Analytical chemistry; Infectious disease; Nanotechnology; Human immunodeficiency virus (HIV); Human T-cell lymphotropic virus (HTLV); Early detection; DNA nanobiosensors
1. Introduction
Viruses are nanoparticle infectious agents that are capable of causing various diseases [1]. Retrovirus is a RNA virus which its cDNA integrates into the chromosomal DNA of a host cell [2]. It is difficult to detect the provirus due to the rare proviral DNA expression in the infected host for some weeks, while the oncogenicity may be at high risk [3, 4]. Human Immunodeficiency Virus (HIV) and Human T-cell lymphotropic virus (HTLV) are two kinds of human retroviruses that cause diseases with a high mortality potential [3, 5, 6]. Despite the joint transmission routes between HIV-1 and HTLV-1, they lead to remarkable different diseases. The infection by HIV-1 results in the acquired immunodeficiency syndrome (AIDS) associated with CD4+ T cell depletion which results in many mortal diseases [7, 8]. HTLV-1 causes two main diseases including Adult T-cell leukaemia/lymphoma (ATLL) and HTLV-1 Associated Myelopathy/Tropical Spastic Paraparesis (HAM/TSP) and also some other disorders [9, 10, 11]. Moreover, the retroviral coinfection has been reported frequently in recent years [12, 13]. Approximately, HIV [14] and HTLV-1 [15] infected over about 36 and 20 million people, respectively. Therefore, they have been causing a significant concern worldwide and their early detection will help the control and prevention of spreading the viruses and ensure the appropriate treatment. Some serological tests are performed routinely for HIV-1 and HTLV-1 detection such as western blot and enzyme-linked immunosorbent assay (ELISA) assay which are very selective and sensitive, however, these methods suffer from some false positive and negative outcomes due to the reaction of samples with one or more of the antigens. Also, these methods need specialized laboratories and highly skilled personnel [16, 17, 18, 19, 20, 21, 22]. Due to a relatively long time from host infection to antibody production or insufficient antibody to be detected with immune assay-based during this period, new strategies would be developed to the detection of virus DNA sequences at the earliest possible time after human infection [23].
The development of nanoscience and nanotechnology has provided tremendous progress in the research activities. Toward this endeavor, some scientists have been interested in constructing biosensors to diagnosis virus DNA or RNA with high selectivity and sensitivity by applying nanomaterials with their unique and tunable electrochemical, optical, mechanical, catalytic, magnetic, surface, and biological properties [24]. These analytical diagnostic techniques can facilitate the early diagnosis of HIV-1 and HTLV-1 in the human body more quickly, accurately, sensitively, and affordable. In this way, these techniques will help to improve clinical therapy and prevention of virus propagation. Here we review the recently developed genosensors which are designed with applying nanomaterials for detection of HIV-1 and HTLV-1 in the recent 10 years, focused on electrochemical- and optical-based methods. Therefore, the comparison of the nanotechnology-based methods for early diagnosis of each virus will be provided and discussed.
2. Electrochemical detection
Electrochemical biosensors have many compelling advantages like inexpensive instrumentation, good specificity and sensitivity, underneath detection limit, and rapid detection [25]. The application of several electroactive nanomaterials to detect different analytes have been reported [26]. Up till now, some electrochemical gene sensors for HIV-1 and HTLV-1 gene detection have been developed using a variety of nanomaterials based on different detection methods.
2.1. Square wave voltammetry
The square wave voltammetry (SWV) is one of the substantial potentiostatic technique in which the current of working electrode is measured as a function of time and potential between the indicator and reference electrodes. The excitation signal comprises a base staircase potential superimposed by a symmetrical square-wave pulse [27, 28]. Adam and colleagues reported a method using paramagnetic microparticles which were covered by streptavidin. They modified particles by a specific biotin-labeled viral sequence to detect the human immunodeficiency virus [29]. The viral nucleic acids were detected employing carbon nanotubes-based screen-printed as the working electrodes. The potential step 5 mV and frequency 280 Hz were employed. The oxidation signal of adenine in the target nucleotide sequence was recorded at 1.15 ± 0.05 V. The detection limit was measured as 0.1 pg/μL that was 15-fold higher than that was performed using a hanging mercury drop electrode (HMDE).
2.2. Electrochemical impedance spectroscopy
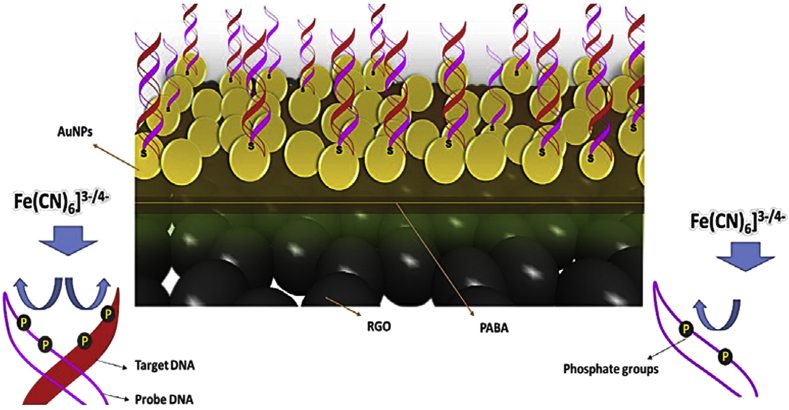
Electrochemical impedance Spectroscopy (EIS) is a sensitive and potent technique, in which a periodic tiny amplitude AC signal is applied to monitor the impedance over an appropriate frequency range [30]. EIS is an effective technique to understand the chemical transformation, interfacial reactions, and properties (attendance of adsorbed species on the conductive electrode surface) [31]. Hu et al. demonstrated a label-free HIV-1 gene diagnosis using functionalized graphene by EIS technique [32]. In this work, the carboxylated graphene sheets due to functionalized 10-perylene tetracarboxylic acid (PTCA) was employed. Afterward, the NH2-IL-stabilized gold nanoparticles (AuNPs) were monotonically dispersed onto the graphene surface. The resulting Au-IL/PTCA/graphene platform had vigorous electrostatic interaction to single-stranded DNA (ssDNA). After hybridizing ssDNA with its complementary target sequence, the formed double-stranded DNA (dsDNA) contained a more negative charge. Moreover, the enhancement of electrochemical impedance value was observed. The EIS hybridization was monitored by LOD of 3.4×10−14 M with the following EIS parameters: the voltage frequencies ranged from 0.1 Hz to 104 Hz and the AC voltage amplitude of 5 mV. To test the selectivity of the Au-IL/PTCA/graphene platform toward the DNA hybridization, the hybridization of 20-base oligonucleotides probes with the conserved sequence of the pol gene of HIV-1 as target was compared with disparate mismatched DNA oligonucleotides. The ΔRet values were to be 4.40%, 52.5%, and 26.4% of hybridization with cDNA for non-complementary, double-base, and four-base mismatched oligonucleotides, respectively. Gong et al. fabricated an impedimetric DNA sensor to determine the HIV-1 gene utilizing a graphene-Nafion composite-modified electrode [33]. The ssDNA probes were adsorbed on the graphene surface through π–π stacking. In the attendance of HIV gene oligonucleotides, probes hybridized with them and constituted ds DNA which led to exclusion from the graphene Nafion/glassy carbon electrode (GCE) surface. Conformational transition of DNA caused graphene-Nafion interfacial charge changes and these changes were monitored by EIS in the frequency ranged from 100 kHz to 0.1 Hz and the voltage amplitude of 5mV with the limit of detection of 2.3 ×10−14 M. Shamsipur and coworkers utilized a novel sandwich nanocomposite film to construct an impedimetric sensor which was capable of detecting sub-femtomolar concentrations of HIV-1 gene (3.4×10−17 M) by electrochemical impedance spectroscopy [34]. The designed sandwich array was prepared by electropolymerization of p-aminobenzoic acid (PABA) on the reduced graphene oxide layer. Then, AuNPs were electrodeposited resulting in the constitution of AuNPs/PABA/ERGO/GCE platform (Figure 1). A comparison between the obtained analytical parameters of the fabricated sensor based on probe-DNA immobilized on GCE modified with ERGO/PABA/AuNPs and another GCE modified with ERGO/AuNPs revealed a supreme performance for AuNPs/PABA/ERGO/GCE platform in comparison with that for AuNPs/ERGO/GCE. Moreover, increasing the mismatched bases causes less double helix formation on the ssDNA/AuNPs/PABA/ERGO modified electrode and lower signal intensity compared to the complementary sequences.
Figure 1.
Schematic diagram of the HIV gene detection using DNA immobilized on the ternary polymer-sandwiched composite as a sensor template. Reprinted with permission from [34].
2.3. Differential pulse voltammetry
Differential pulse voltammetry (DPV) is a kind of voltammetric technique that provides the accurate determination of analytes [35]. In this technique, the potential is scanned with the sequential regular pulses superimposed on the potential linear sweep or stairsteps. It can equilibrate the capacitive/background current which increases the signal to noise ratio [36, 37].
To detect the small quantities of DNA, a large amount of amplifying methods has been introduced such as rolling circle amplification (RCA) [38, 39], polymerase chain reaction (PCR) [40, 41], hybridization chain reaction (HCR) [42, 43, 44], and enzyme-assisted target recycling [45, 46].
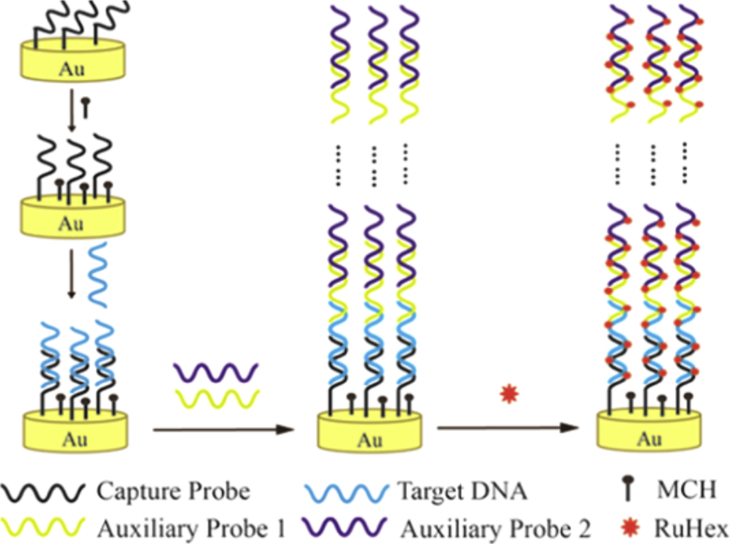
In 2011, Wang and colleagues proposed a voltammetric procedure for the diagnosis of HIV-1 oligonucleotide, in which a sandwich of multiwall carbon nanotubes modified by silver nanoparticles was used as the electrochemical signal amplification probe [47]. The thiolated ss-DNA self-assembled on the gold microelectrode was employed as the capture probes. In the attendance of HIV-1 oligonucleotide, capture probes hybridized with it and then with the electrochemical probe. The resulting sandwich conjugate structure provided a remarkable electrochemical oxidation signal that was linear in the range from1.0 to 100 pM of HIV-1 DNA with the detection limit of 0.5 pM. An ultrasensitive electrochemical-based DNA sensor for the diagnosis of HIV gene was constructed by Chen and coworkers [48]. They amplified the signal through carriers which were long-range self-assembled DNA structures. The designed sensor achieved a remarkable detection limit of 5 aM even in the cell lysate and human serum. They designed a three electrodes system comprising Ag/AgCl reference electrode, gold electrode assembled with DNA as a working electrode, and a platinum wire as counter electrode. Two auxiliary oligonucleotide probes named as AP1 and AP2 were designed. Many hybridizations between the two auxiliary probes occurred which led to 1D DNA nanostructures to the length of more than 20 μm. The Au–S bond was utilized to immobilization of capture probes on the surface of gold electrode [Figure 2, left panel]. DPV measurement was carried out using a potential range from 0.2 to -0.6 V (versus Ag/AgCl) in Ru(NH3)6]3, Hexaammineruthenium (III) chloride (RuHex) solution. After hybridization of capture probe with target, the higher number of RuHex cations was bound to them. Then, the surface of the electrode was exposed to AP1 and AP2 which led to the self-assembling of innumerable AP1 and AP2 onto the DNA nanostructures. Therefore, more RuHex bound to the DNA nanostructures which caused the amplification of the electrochemical signal. The sensor was able to discriminate the target from other sequences.
Figure 2.
Schematic illustration of the enzyme- and label-free electrochemical diagnosis of HIV- DNA based on the self-assembled DNA nanostructures [48]. Copyright 2012, American Chemical Society.
A modified GCE by graphene stabilized gold nanocluster (GR/AuNCs) was developed by Wang et al. to diagnosis HIV-1- target using an exonuclease III (Exo III)-assisted target recycling amplification strategy [49]. GR/AuNCs were modified with aptamers (capture probes) labeled with cytosine (C)-rich base on 5′- end and methylene blue (MB) on 3′-end. Binding of aptamers on the GR/AuNCs platform led to the production of MB signal. The hybridization between target and aptamers formed a duplex DNA structure. Then, Exo III digested aptamers from its 3′-end leading to liberate MB molecules from the electrode. In this method, the signal alterations were followed by the DPV technique and identified as low as 30 aM of the HIV-1 target probe. The full matched target HIV DNA showed considerable DPV responses compared with the unmatched and mismatched DNAs. This platform demonstrated 99.8% of a recovery rate (10 fM of HIV-1 target probe) when used for the human blood samples.
A label-free bio-barcode sensor has been constructed by Zhang et al. for the quantitative determination of HTLV-1 and HTLV-2 target [50]. In this biosensor, the capture DNA functionalized with NH2 was firstly coupled with the magnetic beads (MBs) bearing the carboxyl functional group and then constituted a DNA sandwich through binding to the target self-assembled on AuNPs. Moreover, AuNPs were modified by the thiolated poly-A and poly-G. In order to detect two target DNAs, the integration of magnetic separation of MBs with the amplification feature of AuNPs and ploy A/G was performed. After magnetic separation, DDT was utilized to release the barcode sequences attached to the AuNP probes, dipurinization of the unleashed barcode DNA, aggregation at the modified electrode, and finally, DPV measurement due to the redox activity of free adenine (A) and guanine (G) nucleobases. The linearity was recorded in the range from 4.4×10−11 to 2.0×10−9 M. The detection limits were determined as1.71×10−12 M for HTLV 1-DNA and 1.55×10−12M for HTLV-2 DNA.
2.4. Hybrid methods
The hybrid methods are consisting of two or more electrochemical techniques for the detection of analytes. Dai and coworkers demonstrated the HIV DNA detection on Chitosan/Fe3O4/GEM platform using SWV and EIS techniques and MB as redox indicator [51]. The viral HIV sequence, 25-mer gene expression of modulator 91 abbreviated as GEM, was used as the immobilized DNA probe on CS/Fe3O4. The attendance of Fe3O4 superparamagnetic NPs in this system facilitated the electron transfer and subsequently enhanced the current response with a low detection limit (50 pM). The hybridizations with complementary (c-HIV), muted targets, and non-complementary were determined by SWV and also EIS employed to survey the impact of hybridization on the CS/Fe3O4 interfacial platform of screen-printed electrode (SPE) surface. The optimized SWV parameters were determined as potential ranges from -600 to 400 mV, the frequency 12.5 Hz, and amplitude 25 mV. The frequency ranging from 200 kHz to 10 mHz employing 5 mV alternating voltage were optimized as EIS parameters. The SWV response signal decreased with increasing concentration of HIV DNA target which caused by a higher affinity of MB for ssDNA rather than dsDNA as an intercalator. Gong et al. reported a sensing platform containing nanocomposite of polyaniline/graphene (PAN/GN) to diagnosis the target HIV-1 gene [52]. The presence of polyaniline greatly enhanced the stability and electrical conductivity at the modified GCE. The ssDNA probe was immobilized on the surface of modified electrode through π–π∗ stacking interactions. The [Fe(CN)6]3-/4- redox couple was used to determine the resistance against electron transfer of the electrode after any hybridization. Cyclic voltammetry (CV) and EIS were applied to find the electrochemical properties of this DNA biosensor. The ssDNA/PAN/GN/GCE hybridization with fully complementary, one-, two-, three-base mismatched DNAs, and noncomplementary sequences were tested to determine the hybridization specificity of the sensor. More mismatched bases led to decrease the formation of double helixes and also the production of the signal intensity compared with entirely complementary sequences. The sensor exhibited a LOD of 1.0×10−16 M.
Jia and colleagues produced a novel nanocomposites of CoO, NiCo2O4, and metallic Co/Ni oxide [53]. They developed the complex of metal oxides and mesoporous carbon derived from metal-organic frameworks (MOFs) as the platform of an electrochemical biosensor for the HIV-1 gene determination. The porous NiCo2O4/CoO/CNTs composite could simplify binding of the probe DNA strands through various interactions including π-π∗ interaction, hydrogen bonds, and electrostatic force. It elevated the sensing efficiency of the HIV-1 target. The techniques of CV and EIS were utilized to determine the electron transfer variations during the DNA assay in the detection of HIV-1 gene. The CV curves of the NiCo2O4/CoO/CNTs-based DNA assays were recorded over the potential range of −0.2–0.8 V. However, the EIS technique was also utilized to survey the efficiency of composite because of the insufficient detection sensitivity of the cyclic voltammetry technique. Electrochemical measurements disclosed that the developed strategy exhibited an excellent sensing performance with a low LOD of 16.7 fM to determine HIV-1 DNA. The selectivity of the NiCo2O4/CoO/CNTs-based assay for the complementary target HIV-1 DNA was assessed using noncomplementary and two-base mismatched DNA. Results revealed the high specificity of sensor for the fully complementary target.
3. Optical-based detection
Optical biosensors are known as the most promising type of biosensors that rely on the change of phase, polarization, amplitude, or frequency of the input light as a consequence of a biochemical reaction. Various optical techniques like colorimetry, fluorescence, luminescence, surface Plasmon resonance, etc. are employed in the optical biosensors [54].
3.1. Fluorescent-based assay
Fluorescence-based diagnosis is the most popular approach for biosensing, principally due to their high sensitivity, diversity, and ease of operation. Determining low quantities in the fluorescence methods can be difficult using organic dyes because of their limited quantum yields or extinction coefficients and the low dye-to reporter molecule labeling ratio. Nanomaterials as the new fluorophores with diverse optical properties could replace customary organic dyes [55]. The current review focuses on applying nanomaterials in the field of fluorescent-based sensors of HTLV-1 and HIV-1 DNA detection.
3.1.1. Quantum dots-based fluorescent assays
Semiconductor quantum dots (QDs) are nanocrystals that are used as the fluorescence labels in biosensing. Their compositions consist of hundreds to thousands of atoms of elements of II to VI (e.g., Cd, Zn, Se, Te) or III–V (e.g., In, P, As) groups. QD particles have numerous unique virtues like extensive UV-visible absorption spectra, slender emission bands, and tunable optical properties to any wavelength by merely changing their size. These features making QDs as the ideal fluorophore labels that are bright, non-photobleaching with narrow and symmetric emission spectra for high-throughput screening [56, 57, 58].
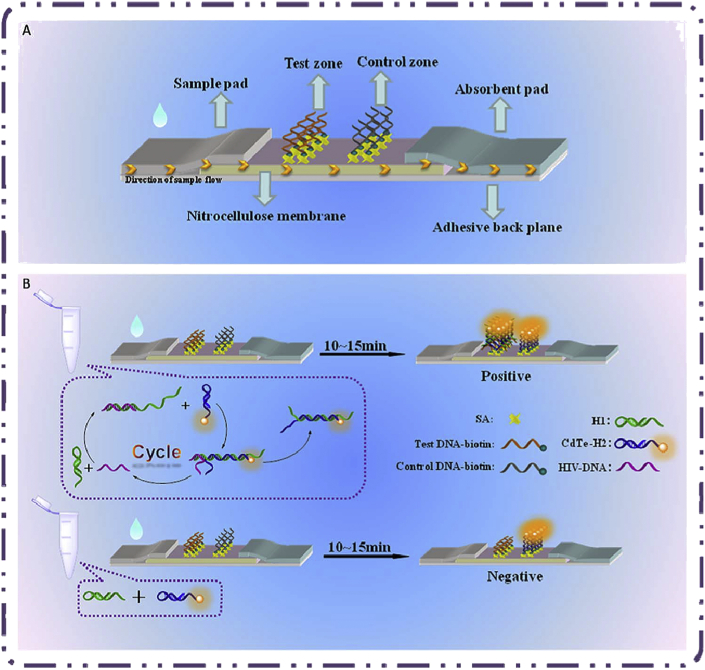
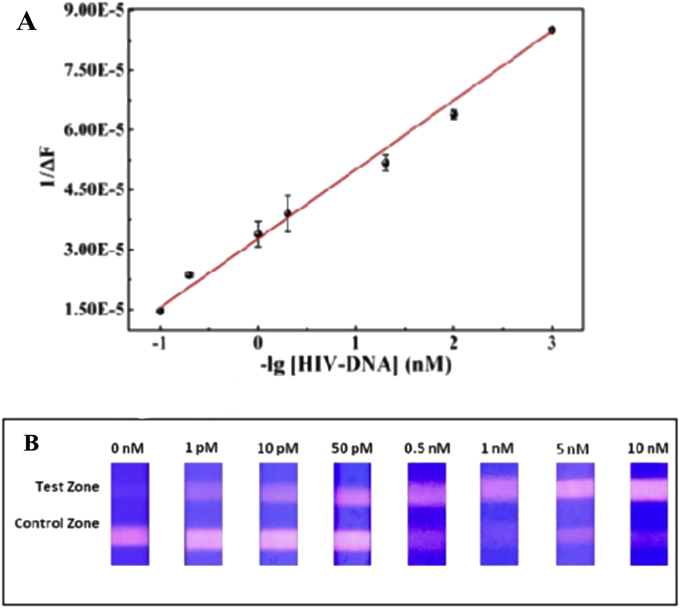
A cadmium tellurium quantum dots-carbon dots (CdTe-CDs) fluorescence biosensor was designed by Liang et al. for the diagnosis of HIV dsDNA. It comprises the water-soluble red-emitting 3-mercaptopropionic acid capped CdTe QDs and green-emitting carbon dots (CDs) [59]. In this system, Mitoxantrone (MTX), a synthetic anthraquinone drug containing an asymmetrical structure could affect the fluorescence intensity of the red-emitting CdTe QDs via electron transfer. The interaction of MTX with DNA led to a remarkable emission quenching in the MTX fluorescence spectra. In this work, the fluorescence of CdTe QDs was quenched by MTX by alteraion the red fluorescence to an “off” state by reduction of the emission peak at 599 nm. In the attendance of HIV DNA, MTX intercalated into the double helix DNA. The potent and specific attachment of dsDNA to MTX led to the recovery of QD fluorescence at wavelength of 599 nm. The linear response to the dsDNA concentration was observed in the range of 0–50 nM with a detection limit of 1.0 nM. Norouzi et al. constructed a HTLV-1 biosensor employing CdTe quantum dots [60]. It based on the biotin-labeled acceptor and NH2-reporter probes which were hybridized with target DNA and formed a sandwich complex on a well-containing streptavidin. The CdTe QDs were added to the sandwich complex and then they attached with the amine group of reporter probe through EDC/NHS. The presence of HTLV-1 DNA was detected by tracing the emission spectra of the quantum dots. The system showed a linear detection of HTLV from 10 pg/μl to 0.24 ng/μl with a detection limit as low as 19.5 pg/μl. Deng and coworkers utilized fluorescent lateral flow assay (LFA) strips to diagnosis HIV-DNA quantitatively and applied strand displacement amplification (SDA) technique and synthesized DNA-QDs [Fig. 3A, B] [61]. They chose strand displacement as the amplification technique, in which two hairpin oligonucleotide probes were included in the process without the need for enzyme catalysis or special reaction conditions. They applied QDs as a signifier for the detection experiments. Although a tiny amount of target contributed in the circular reaction, large QD-dsDNA nanostructures were generated which would be captivated by the test zone. Two kinds of hairpin DNAs (H1 and H2) were designed to have specific roles, H1 contained a trigger and complemented with target DNA and H2 was modified with CdTe QDs as a signal resource for diagnosing. After cultivating HIV DNA with two hairpin DNAs, it fully complemented with a sectional of H1. Moreover, it stimulated the despiralization of the H1 stem. Next, the rest of H1 would provoke the H2 stem to open and hybridize with it. Afterward, with strand displacement amplification, a tiny amount of HIV DNAs converted to a large number of H1–H2–CdTe structures, which elevated with increasing target and caught by t-DNA on the test zone [Fig. 4A, B]. The mismatched DNAs could not stimulate the despiralization of two hairpin DNAs, well. Thus, the strips have commendable selectivity for HIV-DNA. This strip exhibited a LOD as low as 0.76 pM.
Figure 3.
(A) The illustration of a LA assay strip. (B) The schematic of strand-displacement amplification and positive or negative expression on strips. Reprinted with permission from [61].
Figure 4.
(A) Linearity of HIV-DNA determination by the manufactured testing strips. (B) The outcomes of examination of various HIV-DNA concentrations of by the proposed LFA strips under the ultraviolet light. Reprinted with permission from [61].
Jimenez et al. extended a nanosystem for the identification of human papillomavirus (HPV) and HIV [62]. The construction was carried out using two components including magnetic glass particles (MGPs) loaded by target DNA oligonucleotides and CdTe QDs loaded with the complementary probe. Target DNAs were directly attached to the MGPs through the electrostatic interaction and CdTe QDs conjugated to the HIV probes by a robust covalent thiol binding. Afterward, MGPs helped to release the DNA from the sample solutions and eventually, the hybridization process of the bound DNA fragment with the corresponding probe (modified by QDs) was carried out. Unbound DNA fragments and CdTe QDs were removed by three times washing. The attendance of the target genes in the sample was clarified by single time fluorescence measurement.
3.1.2. Fluorescence assays based on metallic nanoparticles, nanoclusters, and nanosheets
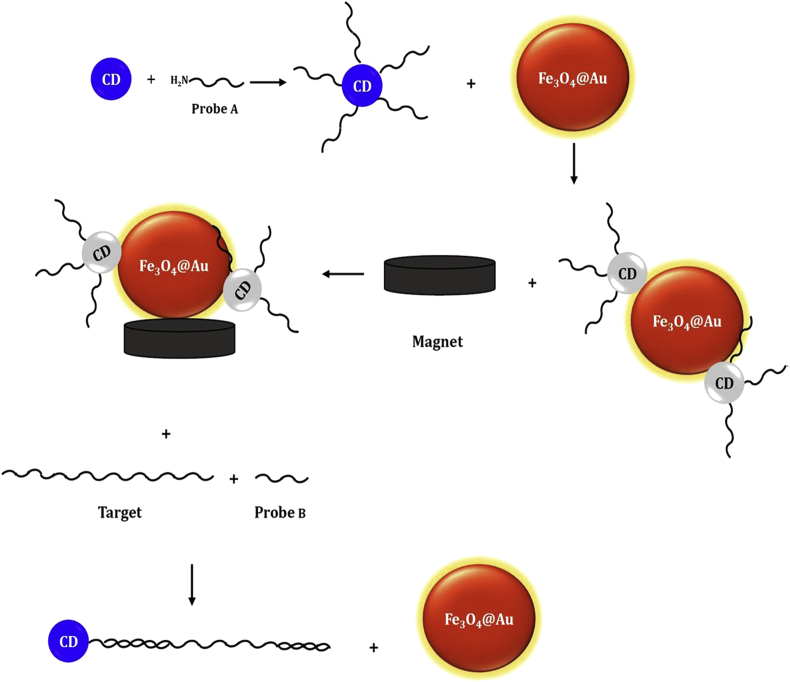
Metallic nanoparticles are nano-sized metals with dimensions within the size range of 1–100 nm. Their ideal electrical, optical, magnetic, and chemical properties might prove attractive in various bioanalytical applications. Metal nanoparticles possess large surface energy; hence they could be modified with diverse chemical functional groups and binding to antibodies, ligands, and drugs [63]. AuNPs have a significant role in the biological systems due to the extensive extinction coefficient, binding to the biomolecules, overlapping their absorption spectrum with the emission spectrum of joint energy donors, and their tunable attributes. AuNPs quench the fluorescence of a fluorophore and also carbon nanomaterials like carbon nanotube and graphene oxide through energy transfer [64, 65, 66, 67].
Hamd Qaddare and Salimi fabricated a homogeneous biosensor for the diagnosis of HIV DNA taking advantage of the fluorescence resonance energy transfer (FRET) strategy, between CDs as donors and AuNPs and AuNPs/GO as the fluorescence acceptors (quenchers) [68]. CDs with carboxyl groups covalently bound to functionalized 5-amino-labeled oligonucleotides as capture probes. The hybridization occurred between probe-CDs and oligonucleotides as the detection probes. In the absence of DNA target, CDs, AuNPs, and/or AuNPs/GO nanohybride were connected, which led to quenching the CD florescence. In the attendance of HIV DNA, CDs left the AuNPs surface. It led to the recovery of the CDs fluorescence because of the stronger hybridization between probe-CDs and detection probe than that of between AuNPs and probe-CDs. AuNPs/GO nanocomposite exhibited high quenching activities by significantly decreasing the CDs fluorescence compared with AuNPs, so, using AuNPs/GO as a quencher was preferred for improving the sensitivity of the approach. The sensor exhibited a quantitative behavior with a vast linear relationship between fluorescence intensity and concentration of HIV DNA target. The enhancement in the fluorescence response was found along with increasing the target DNA concentration and as a result, the increase of the dsDNAs. To assess the specificity of the suggested method, ssDNA-CDs was hybridized with the same concentrations of complementary, noncomplementary, and two-base mismatched DNAs. The complementary sequence showed a clear alteration in the signal compared with other sequences. The detection limit of 15 fM and 5 fM were acquired from an ssDNA-CD-AuNPs complex and ssDNA-CD-AuNPs/GO assembly, respectively. Zarei-Ghobadi et al. introduced a genosensor through quenching the CDs fluorescence in the attendance of Fe3O4-capped Au (Fe3O4@Au) for diagnosis of a special region of HTLV-1 genome [69]. Two dedicated probes were designed to diagnosis target DNA. They selected the 122-bases from the tax region of the HTLV-1 genome and complementary probes to diagnosis it. Also, the nucleotide BLAST tool was used to insure the specificity of probes toward the target. The functionalized CDs with one of the probes were adsorbed onto the surface of Fe3O4@Au, resulting in the quenching fluorescence emission of CDs. Upon attendance of target DNA and hybridization, no FRET signal was observed and the fluorescence emission of CDs was recovered due to lack of adsorption of dsDNA on the Fe3O4@Au. Eventually, a magnet was utilized to separate the unhybridized probes and targets adsorbed on the Fe3O4@Au [Figure 5]. The detection limit of this genosensor was determined as 10 nM with a linear response range from 10 to 320 nM. In another work, this group constructed a genosensor based on quenching the fluorescence of graphene oxide (GO) by AuNPs for long fragment diagnosis of the HTLV 1 genome [70]. Two thiolated 22 and 30 bases oligonucleotides were designed and functionalized with AuNPs. They were entirely complemented for the 122-base segment of the tax region of the HTLV-1 genome. The adsorbed DNA-AuNPs on the GO surface caused fluorescence energy transfer between AuNPs and GO. Therefore, the emission of GO fluorescence was quenched. Both probes were functionalized with AuNPs in order to place a greater number of AuNPs in the proximity to GO and consequently enhance the quenching efficiency. In the attendance of the target, the AuNPs-probes abandoned the surface of GO and hybridized with the target. Thus, the GO fluorescence emission was retrieved. The limit of detection was determined as low as 10 pg/mL. Lin et al. provided a new nanozyme-based fluorescent amplified assay for the diagnosis of HIV DNA based on the outstanding peroxidase-like activity of bimetallic nanoparticles, PtAu NPs, and separation by magnetic nanoparticles (MNPs) [71]. Nonfluorescent Amplex Red (AR) was chosen as the signal indicator which can be oxidized into fluorescent resorufin by peroxidase in the attendance of hydrogen peroxide (H2O2). The PtAu NPs catalyzed the non-fluorescent AR substrate into fluorescent resorufin, producing a stable and sensitive fluorescent signal. The complementary DNA of HIV (CHIV) was divided into two portions, CHIV1 and CHIV2. The mixture of CHIV1-PtAuNPs and CHIV2-MNPs was used as the sensing system. By adding HIV DNA, CHIV1-PtAuNPs and CHIV2-MNPs linked through hybridization of CHIV1/HIV and CHIV2/HIV to generate the CHIV1-PtAuNPs/HIV/CHIV2-MNPs sandwich-like complexes. Next, the unattached CHIV1-PtAuNPs formed complexes under the magnetic field. The developed strategy exhibited excellent specificity and could discern DNA target from the single-base mismatched mutant due to the formation of an impermanent sandwich-like complex. The fluorescence intensity changed linearly within a range from 10 pM to 500 pM of HIV DNA with the limit of detection of 5 pM.
Figure 5.
Schematic of the diagnosis stages of HTLV-1 DNA target based on the fluorescence quenching of carbon dots in the attendance of iron magnetic nanoparticle capped Au [69].
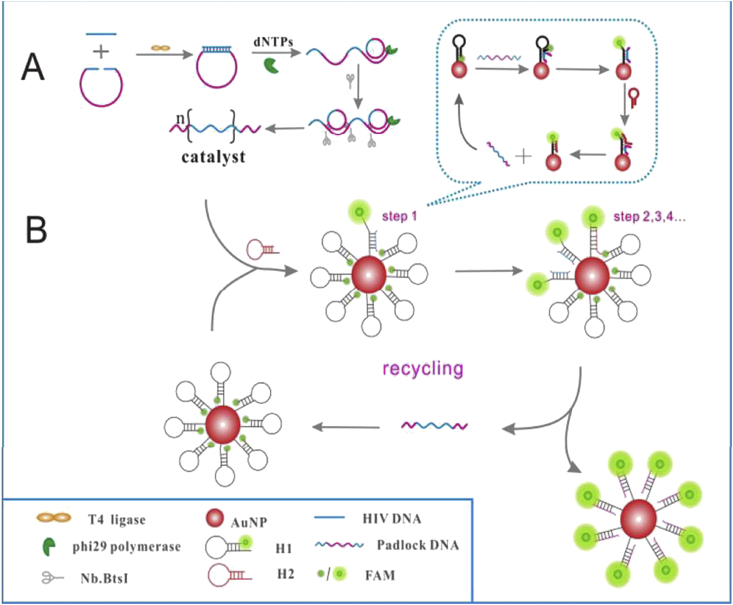
Zheng and colleagues introduced the convenient and simple DNA nanomachine based on AuNPs coupling rolling circle amplification (RCA) and DNA walker cascade amplification to an ultrasensitive determination of HIV DNA [72]. The proposed strategy included two steps: (I) RCA reaction and (II) DNA walker cascade amplification for hybridization onto the DNA-functionalized Au nanoparticle surfaces. In the attendance of target nucleic acid as the primer sequence, RCA reaction produced hundreds of repetitious catalytic strands triggering the DNA walker cascade amplification reaction and initiated conjugation of hairpin DNA (H1) labeled with FAM (5 carboxyfluorescein) on the AuNPs surface and following assembly of another hairpin DNA (H2) to generate numerous H1/H2 duplexes. Consequently, the FAM-labeled DNA payload was liberated and the fluorescence was turned on [Fig. 6A,B]. This strategy exhibited a linear relevance between the HIV DNA concentration and fluorescence intensity in the range from 5 fM to 1.67 pM with a detection limit of 1.46 fM. The specificity of the DNA nanomachine biosensor was studied by the same concentrations of target DNA, single- and two-base mismatched DNA, and noncomplementary sequence DNA. It was shown that only the target DNA could produce a strong signal due to highly hybridization efficiency.
Figure 6.
(A) Schematic image of the DNA nanomachine steps for the sensitive diagnosis of HIV DNA by DNA nanomachine. (B) The DNA walker cascade amplification process on AuNP surface. Reproduced from [72], with permission from the Royal Society of Chemistry.
Metal nanoclusters (NCs) are particles composed of few atoms with dimensions between metal atoms and nanoparticles and unique chemical and physical properties. Metal NCs exhibit the molecule-like properties such as excellent fluorescence emission and biocompatibility since their size is nearby the Fermi wavelength of electrons. Owing to their ultra-small size, powerful luminescence, enhanced photo-stability, great emission rates, and biocompatibility, they have been extensively applied to develop the optical biosensors. Fluorescent silver nanoclusters (Ag NCs) are promising volunteers in the biological usages because these nanoclusters can be easily synthesized with good biocompatibility. Some kind of different scaffolds is applied to synthesize Ag NCs, such as DNA, polymers, thiols, peptides, and proteins. Ag NCs which could be constituted within DNA strands containing abundant cytosine have been considered in the field of bioassay [73, 74]. Cao and coworkers reported the fluorescent molecular beacon based on the DNA protected silver nanoclusters (DNA-Ag NCs) probe to diagnosis HIV and HTLV genes [75]. The signal reporter probe contained a loop complement to the target DNA and labeled with DNA-Ag NCs at 3́ terminal and guanine-rich sequences (GRSs) at 5́ end. The hairpin-shape of the probe led to the fluorescence elevation of Ag NCs because the Ag NCs were in the vicinity of GRSs. At the attendance of target DNA, the hybridization made the hairpin loop structure opened and kept Ag NCs far from GRSs which resulted in the fluorescence reduction of Ag NCs. This biosensor detected HIV and HTLV-1 genes by the detection limits of 4.4 nM and 8.5 nM, respectively. The system showed that this approach could distinguish specific DNA from different mismatched DNA like single-base mismatched.
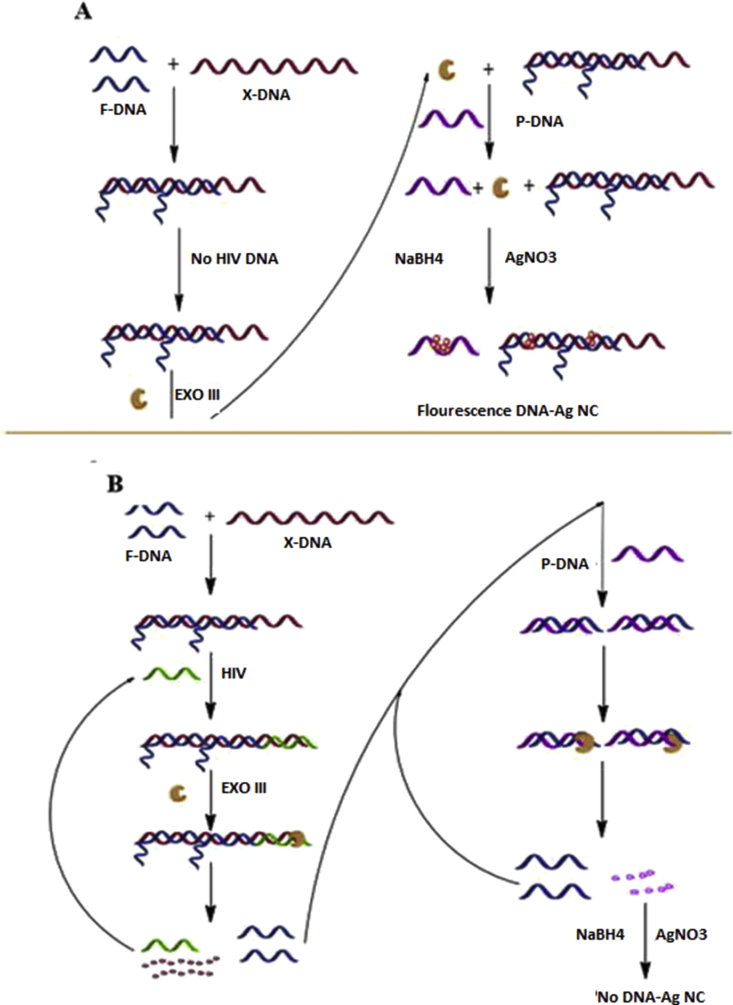
Ye et al. presented a sensing platform for assay of HIV-DNA based on the energy transfer from the DNA template Ag NCs as a donor to carbon nanoparticles oxide as acceptor [76]. They fabricated water-soluble CNPs oxide combined with dual Ag NCs probe with an excellent quenching ability which was employed as label-free fluorescent probes. This strategy was designed based on the significant discrepancy between the interaction of CNPs oxide with ssDNA and dsDNA. It was according to the quenching fluorescence of DNA-Ag NCs by CNPs oxide. In the attendance of target DNA, the hybridization led to the liberation of ssDNA template nanoclusters probes from CNPs oxide and the fluorescence recovery of Ag NCs. Applying dual Ag NCs probes, capturing the same DNA improved the detection. The fluorescence revering of Ag NCs with dsDNA was dependent on the HIV DNA concentration in the ranges of 1–50 nM, with the limit of detection of 0.4 nM. Enkin et al. reported two different sensing strategies based on FRET including Ag NCs as the fluorescents labels [77]. One sensing strategy involved the community of a three-component sensing platform containing a DNA Ag NCs and a BHQ (Black Hole Quencher)-modified oligonucleotides hybridized with a oligonucleotide scaffold which was complement with the target DNA. In the absence of target DNA, the fluorescence of the Ag NCs was quenched. By introducing target DNA to the sensing platform, the strand displacement of the scaffold occurred leading to the separation of the DNA Ag NCs and illuminating the NCs fluorescence. In the second sensing strategy, DNA Ag NCs and the BHQ-modified oligonucleotide were hybridized with a hairpin DNA scaffold. The fluorescence of the Ag NCs was dropped without target DNA. The attendance of the target DNA triggered the opening of the hairpin and switched on the fluorescence of the Ag NCs due to the resulting distance between Ag NCs and the quencher. To examine whether the sensing platform could sufficiently distinguish target DNA, one-, two-, and three-base mutations in HIV gene were analyzed. The intensity of the fluorescence alterations decreased with the increase in the number of mutations. Yang and coworkers developed a fluorescence multi-amplified biosensor for the label-free diagnosis of HIV-DNA based on the DNA/Ag NCs fluorescence and Exo III-assisted cyclic amplification strategy [78]. Three types of DNA were employed in this approach. One DNA called X-DNA contained three sectors: a sector was utilized to be complementary of HIV DNA and the other two were complementary to F-DNA. A X-DNA hybridized with one HIV-DNA and two F-DNA. The P-DNA which was employed as a platform to synthesize DNA/AG NCs was complementary to F-DNA. Firstly, X-DNA and F-DNA hybridized incompletely and the duplex structure with protruding 3-hydroxyl termini was manufactured. It hampered the dsDNA digestion by Exo III, so the F-DNA could not be released. After introduction of HIV DNA, it hybridized with dsDNA and then, a new double-stranded DNA (n dsDNA) was constituted. Afterward, the quadratic amplification system was actuated in association with the incision of P-DNA which played as the template of the DNA/Ag NCs, so the fluorescent of the solution was dropped [Fig. 7A,B]. The addition of HIV-DNA could result in the fluorescence alteration in the detection range of 50 pM to 5 nM with the detection limit of 35 pM. The evaluation of sequence specificity disclosed a substantial distinction between the relative fluorescence intensity of HIV-DNA and non-target DNA.
Figure 7.
Representation of the analysis of the HIV-DNA using DNA/AgNCs and autonomous exonuclease III (Exo III)-assisted recycling signal amplification, A) Fluorescence on; B) Fluorescence off. Reproduced from [78], with permission from the Royal Society of Chemistry.
An efficient and label-free fluorescent platform based on Ag NCs was developed by Zou et al. for the simultaneous detecting of two HIV related genes (HIV 1 and HIV-2) [79]. The sensing strategy was designed based on the fluorescence advancement of guanine (G)-rich and forming a nanoclusters dimer using a new double helix probe. The probe (green fluorescent Ag NCs/G) was used to detect HIV-1 based on the enhancement effect of G-rich sequence with the maximum emission peak of 565 nm. On the other hand, the orange fluorescent probe (AG NCs/AG NCs) was utilized for the detection of HIV-2 with the maximum emission peak of 630 nm. It produced fluorescence only in the form of nanoclusters dimer because of FRET. In the absence of target HIV DNAs, Ag NCs were lighted up at one end by G-rich sequence and were enhanced at the other end, depend on squeezing each other between two Ag NCs. In the attendance of the target HIV, the structure of the conjugate pairs was ruined through hybridized probe-HIV DNAs. The green fluorescence was dropped after adding the target HIV-1 because of the created distance between Ag NCs and the G-rich sequence. Moreover, the orange fluorescence was quenched after adding HIV-2 due to the segregation from its auxiliary. When both targets were added, the structure was ruined and both fluorescence signals were dropped. The nanoprobe showed a linear range of 0.2–700 nM, and the detection limit of HIV-1 and HIV-2 were both 12 pM. Another approach for the simultaneous diagnosis of several targets including HIV-1 gene based on DNA-Ag NCs was presented by Han and Wei [80]. This system was ascribed to the non-overlapping emission regions of Ag NCs. They designed the molecular beacon with three domains: the C-rich sequence for the Ag NCs formation, the complementary loop region of target DNA, and the blocking region. In the attendance of target DNA, the hairpin structure opened and formed an abiding duplex structure upon hybridization with the target. So, the C-rich DNA segment was liberated and DNA-Ag NCs were synthesized and illuminated the fluorescence signal. The molecular beacon can be utilized for the sensitive diagnosis of different targets by substituting the recognition sequence. A good linear relevance for HIV gene was observed in the range between 0–250 nM with the limit of detection of 0.12 nM. It was indicated that the Ag NCs-based molecular beacon had superior target selectivity and could discern even sequence with one mismatch. The layered transition-metal dichalcogenides (TMD) nanosheets (NS) are a family of two-dimensional materials (e.g., MoS2, TiS2, WS2, etc.) with interesting physical, electronic and chemical properties. Due to their simple synthesis and dispersion in aqueous, the TMD NS have many biomedical applications. Molybdenum disulfide nanosheets (MoS2 NS) shows the fluorescence quenching and various adsorption capabilities toward ssDNA against dsDNA [81]. A fluorescence sensing approach for the HIV DNA determination was developed by Wang and coworkers employing MoS2 nanosheet and Exo III-assisted amplification [81]. The study utilized the different affinity of MoS2 nanosheets for long and short ssDNA accompanied by exonuclease III-assisted target recycling signal amplification. Two principals have been proposed for developing this strategy: I) Exo III elects the stepwise exclusion of mononucleotides from blunt or recessed 3′-hydroxyl termini of dsDNA. However, it has no activity toward protruding 3′-termini of dsDNAs or ssDNA [82]. II) MoS2 NS has the great fluorescence quenching capability and different affinity for short oligonucleotide segment and ssDNA [83, 84]. In the absence of target DNA, Exo III was unable to remove the fluorescence-tagged ssDNA probes (FPs), so FPs absorbed on the MoS2 NS surface. Consequently, the fluorescence of the fluorophore was decreased by MoS2 NS. Upon adding target DNA, the FPs hybridized with target and constituted dsDNA. Then, the DNA probe in the duplex was hydrolyzed from the blunt 3 and mononucleotides by Exo III. Therefore, the stunted dye-tagged oligonucleotide segments were produced. Their low affinity toward the MoS2 NS caused remaining the fluorescence intensity. In this process, the HIV DNA target with a protruding 3′ terminus survived and hybridized with another FP. Afterward, many of FPs were hydrolyzed with the short oligonucleotide segments containing a low affinity to MoS2 NS. The limit of detection of this biosensor was 5.3 pM for HIV DNA.
The fluorescence intensity for the HIV DNA target was three-fold than that of single-base mismatched DNA and six-fold than that of three-base mismatched DNA and random DNA indicating the high specificity of this HIV DNA biosensor.
3.1.3. Fluorescence polarization assay
Fluorescence polarization (FP) or fluorescence anisotropy is a rapid, accurate, quantitative, and sensitive technique in the biochemical research in a broad range of experimental conditions [85, 86, 87, 88, 89]. The polarization value has reverberate relevance with the rotation rate of the fluorescently labeled molecule, which depends upon the molecular volume and molecular mass at a fixed solution viscosity and temperature [90]. The increase in the molecular mass or size of the fluorescent molecule slows down the rotation motion, thus causing a more fluorescence polarization value. Conversely, the fluorescence polarization value is smaller. To enhance the diagnosis sensitivity, numerous nanomaterials such as carbon nanotubes [91], AuNPs [92], and Ag nanoparticle [93], etc were utilized to amplify fluorescence polarization signals. Liang and coworkers constructed a fluorescence polarization assay system for diagnosis of HIV-DNA through AuNPs self-assembled by the functional dendritic macromolecules [94]. Firstly, AuNPs loaded by two DNA probes as reporters were prepared. In the attendance of HIV-DNA, the fluorescently labeled DNA probes and AuNPs-DNA dendritic macromolecules hybridized with the target DNA to constitute a sandwich DNA complex. The rotational speed of the probes was reduced because of the large size and molecular weight of AuNP-DNA dendritic macromolecules. It led to the substantial enhancement of the fluorescence polarization value. The values of fluorescence polarization decreased with the addition of the number of oligonucleotide mutations. The relative fluorescence polarization amounts for the non-complementary target DNA was almost the same as the background signal. The fluorescence polarization showed a linear trend in the concentration range of 150 pM to 6 nM HIV-DNA with the detection limit of 73 pM.
3.2. Chemiluminescence-based assays
Electrogenerated chemiluminescence known as electrochemiluminescence (ECL) is according to the generation of light at an electrode by producing the electronically excited-state intermediate via electron-transfer reactions at the electrode surfaces and then light-emitting. Possessing attractive benefits such as excellent selectivity, high sensitivity, fast response, trivial background signal, cheap instruments, and superior controllability, ECL biosensors are a powerful device for the diagnosis of the trivial amounts of biomolecules [95].
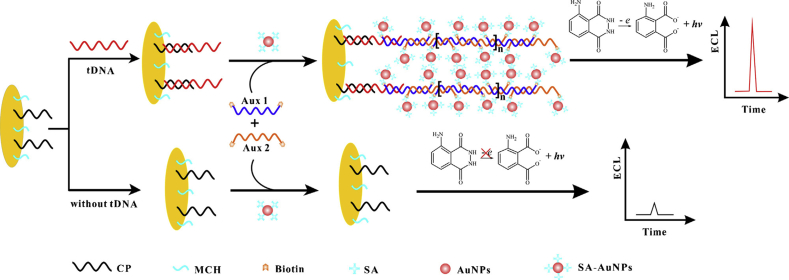
In 2015, Wang et al. designed a new strategy for the highly efficacious ECL diagnosis of HIV DNA taking advantage of HCR strategy and using AuNPs as a signal amplification component. As depicted in Figure 8, a capture probe (CP) immobilized on an electrode through an Au–S bond. In the attendance of tDNA, the lengthy concatamers containing plentiful intermittent biotinylated DNA probes (auxiliaries 1 and 2) could be formed automatically and were immobilized firmly on the gold electrode through tDNA and CP. Then, the streptavidin-coated AuNPs were attached to the biotinylated DNA and assembled on the DNA sensor surface. It catalyzed luminol to generate a substantial ECL signal. This ECL biosensor provided the sensitive diagnosis of HIV-1 with a detection limit of 5.0 fM. The noncomplementary DNA and the blank solution exhibited the same response. This approach revealed a notable proficiency to discern DNA target from the mismatched ones and high potential for single nucleotide polymorphism (SNP) analysis [96]. In another work, a new molecularly imprinted polymer (MIP) electrochemiluminescence (MIP-ECL) biosensor was used for the HIV-1 gene diagnosis in which Europium sulfide nanocrystals (EsNCs) was used to produce the luminescent signal [97]. Electropolymerization of the HIV aptamer and ophenylenediamine as the template and functional monomer, respectively, formed an ultrathin imprinted film on the ITO electrode surface. Incubation of MIP and the DNA target led to the constitution of the EuS NCs-DNA probe/HIV target DNA duplex. The EuS NCs-DNA probe could introduce to the electrode leading to the significant enhancement of the ECL signal. This biosensor exhibited the supreme specificity for the detection of HIV DNA in comparison to noncomplementary and sequence with two bases mismatch. The strong electrochemiluminescence emission of EuS NCs and good efficacy of molecular imprinting method caused the successful diagnosis of HIV DNA with a LOD of 0.3 fM and a linear range of 3.0 fM to 0.3 nM.
Figure 8.
Electrogenerated chemiluminescence (ECL) diagnosis of HIV- DNA with the DNA-based hybridization chain reaction. Reprinted with permission from [96].
3.3. Nano plasmonic assays
Surface plasmons (SPs) are surface electromagnetic waves resulting from surface plasmon resonance (SPRs) which propagate the metal/dielectric interface. Essentially, the waves trapped at the border of the conductor and the external medium. The resonant oscillations are highly dependent on the alteration of the interface like the molecule adsorption on the conducting surface. SPR is a substantial optical biosensing method due to its application for real-time, label-free, and noninvasive nature of method [98, 99].
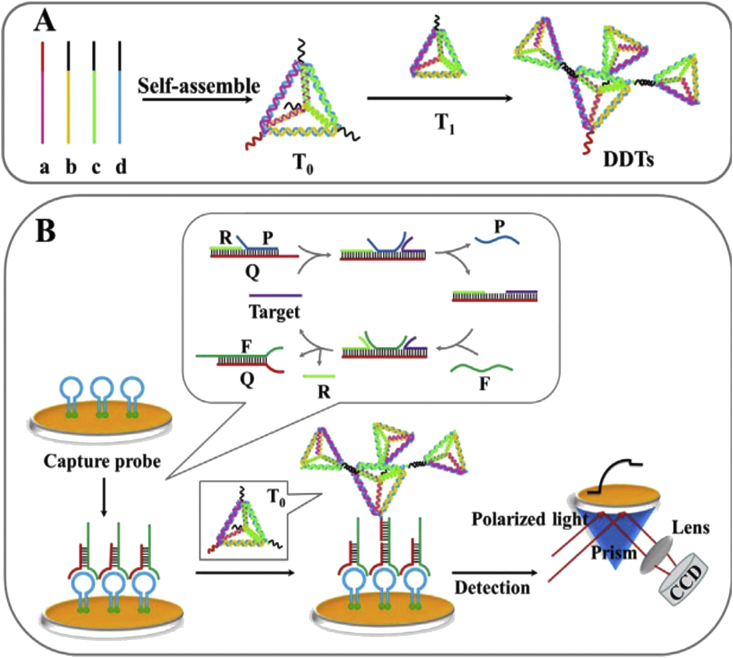
An SPR biosensing strategy was developed by Diao et al. for the HIV DNA diagnosis without enzyme or label based on double-layer DNA tetrahedrons (DDTs) and dynamic entropy-driven strand displacement reactions (ESDRs) [100]. The sensing chip surface was immobilized by the Hairpin capture probes via Au–S binding. When the stable three-stranded beacon complexes were formed, the DNA target could specifically attach to the toehold zone of three-stranded complexes and started toehold-mediated strand displacement reactions. It led to the employing DNA target and generation of the multiple double-stranded complexes, which specifically hybridized with the capture probes and exposed the binding sites of DDTs to the chip surface. Then, three-decker composites were generated on the SPR chip which dramatically improved signal [Figure 9A, B]. The response of DNA target was approximately 2.5-fold that of a single-base mismatch target and 6-fold that of double-base mismatch target and non-complementary sequence. The constructed SPR sensor could determine DNA target in a linear range from 1 pM to 150 nM with a LOD of 48 fM.
Figure 9.
A) The representation of the construction of double-layer DNA tetrahedrons (DDTs); (B) Schematic illustration of the SPR biosensing platform for the HIV-DNA detection based on the dynamic ESDRs and structural DDTs Nanodevices. Reprinted with permission from [100].
Surface-Enhanced Raman Scattering (SERS) spectroscopy is an indispensable technique that overcomes the traditional drawbacks of Raman scattering by greatly enhancing the weak Raman signal of metals as high as 1014-1015 which is sufficient to allow detection of even individual molecule [101, 102, 103, 104]. Two enhancement mechanisms were proposed for the large amplification of Raman signal namely electromagnetic enhancement and chemical enhancement. The electromagnetic theory is based on the oscillation of conducting electrons in the metal nanostructures due to the oscillation of the electric field. Chemical enhancement theory is according to the charge transfer of metals via holes or electrons to the adsorbed molecules [105, 106]. Raman modes are excited by Visible and Near-Infrared Radiation (NIR). Au NPs and Ag NPs are common nanoparticles used to enhance optical properties in SERS because of providing maximal SERS enhancement effect [107]. A novel SERS-based LF was proposed by Fu and colleagues for the quantitative diagnosis of HIV-1 DNA. It was designed based on the hybridization reaction of DNA conjugated AuNPs, capture DNA, and target DNA [108]. Raman reporter functionalized by Malachite green isothiocyanate (MGITC) AuNPs were applied as nanotags. Since MGITC exhibits a strong Raman enhancement effect, MGITC-AuNPs-DNA were used as SERS tags instead of using DNA-labeled AuNPs which were utilized in the conventional LF strips. The control and test lines were formed by immobilizing control and capture DNA probes on the surface of MGITC-AuNPs, respectively. The quantitative determination was performed through following the alteration in the Raman intensity of DNA conjugated AuNPs on the test line. Under the optimized condition, a linear relevance between the Raman intensity and concentration of HIV-1 was acquired with the LOD of 0.24 pg/ml in the range of 8 pg/ml to 64 ng/ml. In other work, Hu et al. developed an assay to diagnosis HIV-1 DNA based on the nanojunctions (NJs)-based biosensor using SERS. In this detection assay, the thiol-terminated capture probe complementary to a segment of HIV-1 DNA was stabilized on the surface of Au substrate and AuNPs were modified by two complementary Raman-labeled DNA sequences, separately and used as the detection probes. In the attendance of a target, at first, the detection probe I precipitated on the substrate with the capture probe and formed a sandwich structure resulting in the target amplification. Then, the distance between Au NPs and Raman tags was decreases after hybridization of two complementary diagnosis probes. Also, two conjugated Au NPs created the SERS ‘‘hot spots’’. Formation of the multi-metal-molecule-metal NJs led to reducing the distance between Au NPs and Raman labels, thus, the Raman signal of the tag-labeled diagnosis probes was substantially increased. To determine the selectivity and detect the target DNA, a series of comparative experiments using different mismatched sequences were surveyed. It was demonstrated that the proposed method had superior features for the diagnosis of target DNA due to the signal amplification. The SERS signal elevated linearly from 0 to 10−13 M and LOD as low as 10−19 M [109].
3.4. Detection with dynamic light scattering
Dynamic Light Scattering technique (DLS), also known as Photon Correlation Spectroscopy or Quasi-Elastic Light Scattering, is an optical technique to measure the size of different kinds of particles dispersed in a liquid medium, typically down to 1 nm. DLS measures the random changes in the intensity of laser light scattered by molecules in solution or suspension. These particles can be organic such as carbohydrates, polymers, proteins, and surfactants or inorganic like metals (silver, gold, and transition metal oxides). Also, the hybrid particles containing inorganic cores covered by organic molecules can be characterized [110, 111, 112].
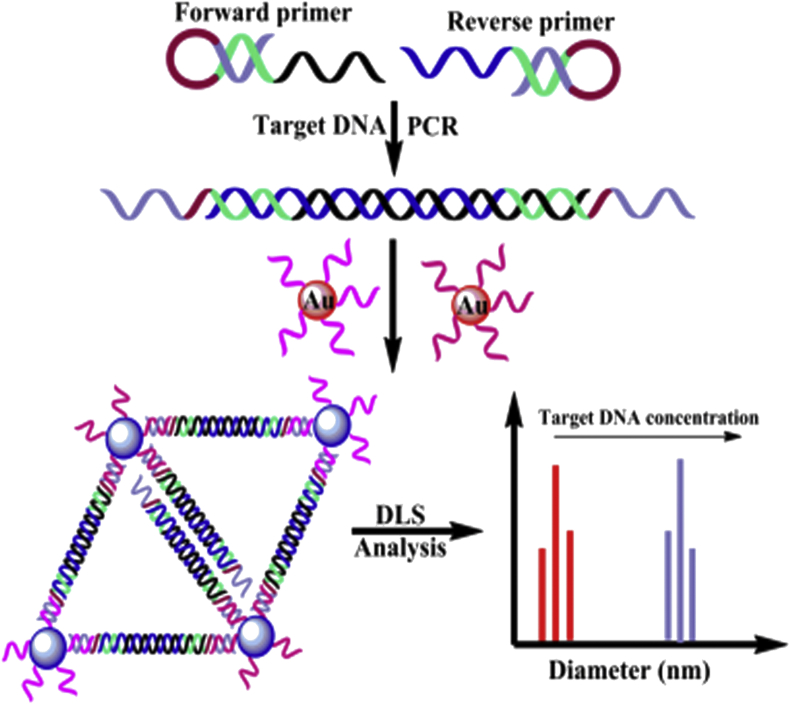
Zou et al. developed a novel PCR-DLS assay for the rapid diagnosis of HIV-1 gene considering the mean diameter alteration of AuNPs [113]. It is the first application of DLS technique as a signal read-out for the PCR assay. Before PCR amplification, the forward and reverse primers could not hybridize with AuNP probes, so, the AuNP probes dispersed well. The PCR process resulted in the unwinding hairpin and production of respective DNA templates, continuously. As illustrated in Figure 10, the PCR product contained liberate ssDNA tails at both ends could be produced by primers and hybridized with the DNA functionalized AuNP probes. It led to the aggregation of AuNPs and a remarkable increase of the particle size which measured using DLS with a LOD of 1.8 aM.
Figure 10.
PCR-DLS assay for the diagnosis of HIV- DNA based on the mean diameter change of AuNPs. Reprinted with permission from [113]. Copyright 2018, American Chemical Society.
4. Photoelectrochemical detection
The photoelectrochemical (PEC) process involves transforming photon-to-electricity due to the charge transfer after absorption of photons during light irradiation. PEC sensing is a novel strategy for bioanalysis via charge transfer reactions between a photoelectrochemically active material, analyte, and electrode upon light illumination. In the PEC detection, the excitation source and detection signal are light and photocurrent, respectively. PEC sensors have the advantages of both electrochemical and optical sensors because of the ability to couple the photoexcitation process with the electrochemical diagnosis. Furthermore, the resources of excitation and detection in the PEC process are separated which make PEC biosensors highly sensitive with low background signal [114].
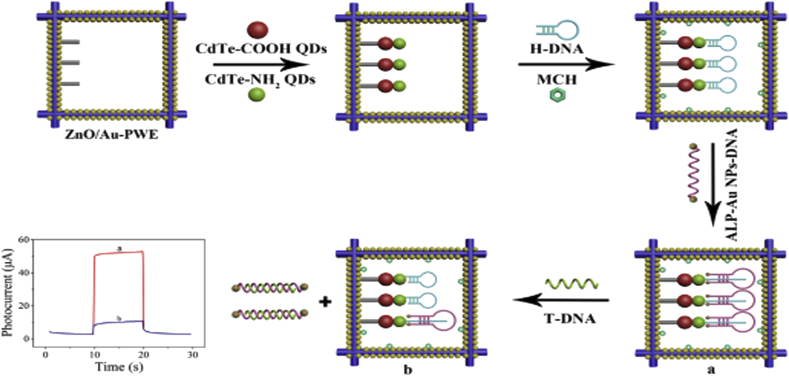
Shi et al. constructed a photoelectrochemical biosensor for the HTLV-1 DNA diagnosis by the association of λ-exonuclease (λ-Exo) aided target recycling, HCR, and enzyme catalysis for cascade signal amplification [115]. The ITO/RGO/CdS/ZnS PEC electrode was utilized to stabilize DNA sequences. They designed a hairpin structure (h1DNA) consisted of two zones comprising the recognition zone which was complementary to the target DNA (tDNA) at the 5′terminal and the output zone which was somewhat complementary to capture DNA (cDNA). In the lack of tDNA, the overhang phosphorylated terminal of h1 DNA could not be cleaved by λ-Exo. However, in the attendance of target DNA (tDNA) of HTLV-1, h1DNA could form a dsDNA and the blunt 5 phosphorylated terminal dsDNA 1. It also triggered the hybridization of oDNA with cDNA, which led to loading manifold biotin-labeled hairpin DNAs onto the photoelectrode through the HCR. Eventually, the electrode was functionalized with avidin-labeled alkaline phosphatase (avidin-ALP). The ALP catalyzed dephosphorylation of phospho-L-ascorbic acid trisodium salt (AAP) to produce ascorbic acid (AA) as an efficient electron donor and consequently extremely elevating the photocurrent signal. The intensity of the photocurrent signal was commensurate to the concentration of tDNA. To determine the specificity of the DNA sensor, the photocurrent signals response of various sequences including target DNA, one-base mismatch, and noncomplementary DNA sequences were surveyed. The higher photocurrent response of HTLV-1 DNA was observed than those of mismatch DNA, which confirmed that the constructed biosensor could efficiently discern various DNA sequences. The fabricated photoelectrochemical biosensor showed an ultrasensitive diagnosis of HTLV-1 DNA (11.3 aM). A PEC biosensor was fabricated by Wang et.al. to diagnosis HIV-1 DNA utilizing the triple-helix molecular switch and cascaded photoactive materials. The ZnO nanorods were attached to the gold-paper working electrode (Au-PWE) of a microfluidic paper-based analytical device (μ- PAD). The various sizes of CdTe quantum dots (CdTe–NH2 QDs and CdTe–COOH QDs) were employed to constitute a cascaded photoactive interface for amplifying the signal.
A hairpin structure DNA as capture probe (H-DNA) was immobilized on the photoactive surface and 6-mercaptohexanol (MCH) was utilized to cover non-specific regions. Afterward, a gold nanoparticle modified ssDNA, which was labeled by alkaline phosphatase (ALP-Au NPs-DNA) at each termini was added to be complementary of the H-DNA and form a triple-helix structure. In the absence of T-DNA, the triple-helix molecule conformation was unchanged and the ALP of ALP-Au NPs-DNA could catalyze AAP to generate AA as electron donors, resulting in a substantial photocurrent signal. When T-DNA was introduced to the system, it hybridized with ALP-Au NPs-DNA of the triple-helix molecule which disassembled the triple-helix structure. Thus, the basic structure of H-DNA was retrieved and compelled the ALP-Au NPs-DNA to release from the electrode surface. Subsequently, the photocurrent response was significantly reduced [Figure 11]. To assess the specificity of the PEC sensor, the impact of various DNA sequences on the photocurrent changes were surveyed. The increase in the number of mismatch bases led to the enhancement of the photocurrent responses. It confirmed the superior specificity of biosensor. This PEC biosensor had a broad detection range of 1fM-1nM and also a low LOD of 0.65 fM [116].
Figure 11.
The fabrication of PEC biosensor for the diagnosis of HIV-1 photoactive materials and triple-helix molecular switch. Reprinted with permission from [116].
5. Conclusions and outlook
Traditional strategies for recognition of viruses and viral components are laborious, time-consuming, and expensive, thus, simple, rapid, and inexpensive viral biosensors are urgently demanded the early diagnosis of viral infections. Nucleic acids are one of the most significant biomarkers which have a significant role in the basic studies and clinical diagnosis specially at the ultralow concentration. Researchers have developed DNA nanobiosensors that offer a great prospect for the detection of viruses' genome using nanotechnology integrated methods and DNA structures. Utilizing these kinds of sensors that have been discussed here, highlight the sequence-specific hybridization detection of HIV-1 and HTLV-1 DNA and also their important parameters such as detection limit/range of designed bioassays. While all the reviewed DNA nanobiosensors demonstrate the remarkable capabilities to detect HIV-1 and HTLV-1 gene in the controlled lab environment. It is necessary to consider the characteristics required for the real application throughout the entire development process. Tables 1 and 2 summarize the characterized parameters of published reports on the HIV-1 and HTLV-1 genome detection, respectively. The tables indicate that the electrochemical based DNA nanobiosensors have the characteristics of large linear range, lower LOD, and superior sensitivity, however, their developments for the professional settings may take a long time and be very costly. Applying nanomaterials and nanostructured interfaces improved the performance of the developed DNA nanobiosensors, especially their sensitivity, ease of use, and a trace amount of sample. On the other hand, the limitation of current methods is using multiple steps and requiring amplification techniques in some of the designed methods that may lead to an increase in the time of detection. In addition, it should be considered that the DNA probe structures used for sensing have to resist in various solution conditions and remain stable during the detection. So, to overcome these defections further improvement in designing the protocols and trying to provide a clever and highly precise design concept assays is needed. DNA nanostructures offer many advantages over the previous approaches and enable studying multiplexed detection systems and networks [117, 118, 119]. Future DNA nanobiosensores will need to be improved in order to have superior transduction, simpler processing, more faithful signal conversion in the form of developed DNA bio strip and biochips with no need for amplification.
Table 1.
Comparison of analytical performance (linear range and LOD) of the different nano biosensing systems for detection of HIV-1.
| Detection Method | Strategy | Limit of Detection | Linear Range | Ref. | |
|---|---|---|---|---|---|
| Electrochemical | Square Wave Voltammetry (SWV) | CNT screen printed working electrode | 0.1 pg/μl | 0.2–25 μg/ml | [29] |
| PTCA/graphene sheets functionalized with AnNPs | 34 f M | 10 pM - 1μM | [32] | ||
| Electrochemical Impedance Spectroscopy (EIS) | graphene/nafion nanofilm modified GC electrode | 0.23 f M | 10 pM–10 nM | [33] | |
| GC electrode modified with AuNPs/PABA/ERGO nanocomposite film | 340 f M | 10 fM – 10 μM | [34] | ||
| Signal amplification of multiwall carbon nanotubes loaded AgNPs | 0.5 pM | 1–100 pM | [47] | ||
| Differential Pulse Voltammetry (DPV) | Long range self-assembled DNA nanostructures on gold electrode | 5 aM | 2 aM–10 pM | [48] | |
| Graphene stabilized AuNCs modified GC electrode | 10 f M | 0.1 fM – 100 nM | [49] | ||
| Hybride Methods | Chitosan/Fe3O4 immobilized DNA probe | 50 pM | 50–100 pM | [51] | |
| ssDNA/polyaniline/graphene nanocomposites platform | 50 fM | 50 fM -10 nM | [52] | ||
| Binding of DNA probe to porous NiCo2O4/CoO/CNTs composite | 16.7 fM | 0.1 pM-2nM | [53] | ||
| Optical | Fluorescence | FRET between CdTe QDs and CDs | 1 nM | 0–50 nM | [59] |
| Integration of Lateral Flow assay strips and QDs | 0.76 pM | 1 pM–10 nM | [61] | ||
| CdTe QDs tagged complementary DNA probe | 50 pM | 5μM–2000 μM | [62] | ||
| FRET between AuNPs/graphene oxide and CDs | 5 fM | 5μM–0.4 nM | [68] | ||
| Peroxidase like activity of Pt/Au NPs and separation of magnetic NPs | 5 pM | 10pM–500pM | [71] | ||
| DNA nanomachine based on Au NPs and RCA | 1.46 fM | 5 fM-1.67 pM | [72] | ||
| Hairpin shaped probe tagged with AG NCs | 4.4 nM | 10–200 nM | [75] | ||
| FRET between CNPs oxide and Ag NCs | 0.4 nM | 1–50 nM | [76] | ||
| FRET between DNA Ag NCs and BHQ | 35 pM | 50 pM–5 nM | [77] | ||
| DNA/AG NCs probe and Exo III-assisted cyclic amplification | 35 pM | 50 pM- 5 nM | [78] | ||
| Fluorescence enhancement of Ag NCs by dimer formation and auxiliary G rich sequence probe | 12 pM | 0.2–700 nM | [79] | ||
| Non overlapping emission of different molecular beacon template Ag NCs | 0.12 nM | 0–250 nM | [80] | ||
| Different interaction of ss DNA and ds DNA with MoS2 sheet | 5.3 pM | 0.01–0.25 nM | [81] | ||
| Fluorescence Polarization | Self-assembly of Au NPs to form the dendric macromolecules | 73 pM | 150 pM- 6 nM | [94] | |
| Chemiluminescence | Hybridization chain reaction and AuNPs | 5 fM | 0.02–1 pM | [96] | |
| Molecularly Imprinted Polymer and Europium sulfide nano crystals | 0.3 fM | 0.3–3 fM | [97] | ||
| Nano plasmonic | SPR biosensing strategy based on dynamic entropy-driven strand displacement reactions and double-layer DNA tetrahedrons | 48 fM | 1 pM–150 nM | [100] | |
| SERS biosensing strategy based on hybridization of DNA conjugated on MGTIC/AuNPs | 0.24 pg/ml | 8 pg/ml – 64 ng/ml | [108] | ||
| SERS biosensing strategy based on formation of multi metal molecule DNA nanoconjugation | 100 AM | 0–10 pM | [109] | ||
| Dinamic Light Scattering (DLS) | Combination of polymerase chain reaction and DLS | 1.8 aM | 10 aM–1.9 pM | [113] | |
| Photoelectrochemical | HCR and enzyme catalysis for cascade signal amplification | 11.3 aM | 50 aM–100 pM | [115] | |
Table 2.
Comparison of analytical performance (linear range and LOD) of the different nano biosensing systems for detection of HTLV-1.
| Detection Method | Strategy | Limit of Detection | Linear Range | Ref. | |
|---|---|---|---|---|---|
| Electrochemical | Differential Pulse Voltammetry (DPV) | Self-assembly of DNA on Au NPs and magnetic separation | 1.7 pM | 44 pM–2 nM | [50] |
| Optical | Fluorescence | CdTe modified DNA probes | 1.95 pg/μl | 10 pg/μl – 0.24 ng/μl | [60] |
| FRET between CDs and Fe3O4@Au NPs | 10 nM | 10–320 nM | [69] | ||
| CdTe QDs tagged complementary DNA probe | 10 pg/ml | 10 pg/ml – 21 ng/ml | [70] | ||
| Hairpin shaped probe tagged with AG NCs | 8.5 nM | 10–200 nM | [75] | ||
| Photoelectrochemical | Cascade photoactive materials and triple helix molecular switch | 0.65 fM | 1 fM – 1 nM | [115] | |
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Abrescia N.G., Bamford D.H., Grimes J.M., Stuart D.I. Structure unifies the viral universe. Annu. Rev. Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 2.Boeke J.D., Chapman K.B. Retrotransposition mechanisms. Curr. Opin. Cell Biol. 1991;3(3):502–507. doi: 10.1016/0955-0674(91)90079-e. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y., Chen J., Chen G. A label-free electrochemical biosensor for detection of HIV related gene based on interaction between DNA and protein. Sensor. Actuator. B Chem. 2013;184:113–117. [Google Scholar]
- 4.Tagaya Y., Gallo R.C. The exceptional oncogenicity of HTLV-1. Front. Microbiol. 2017;8:1425. doi: 10.3389/fmicb.2017.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fani M., Rezayi M., Meshkat Z., Rezaee S.A., Makvandi M., Abouzari-Lotf E. Current approaches for detection of human T-lymphotropic virus Type 1: a systematic review. J. Cell. Physiol. 2019;234(8):12433–12441. doi: 10.1002/jcp.28087. [DOI] [PubMed] [Google Scholar]
- 6.Abedi F., Mozhgani S.-H., Rahimzadegan M., Gudarzi H., Valizadeh N., Rezaee S.A. Prevalence and phylogenic study of human T-lymphotropic virus 1 in patients with thalassemia in the northeast of Iran. Future Virol. 2017;12(5):253–258. [Google Scholar]
- 7.Nadal D., Boni J., Kind C., Varnier O.E., Steiner F., Tomasik Z. Prospective evaluation of amplification-boosted ELISA for heat-denatured p24 antigen for diagnosis and monitoring of pediatric human immunodeficiency virus type 1 infection. J. Infect. Dis. 1999;180(4):1089–1095. doi: 10.1086/315012. [DOI] [PubMed] [Google Scholar]
- 8.Mozhgani S.-H., Ebrahimian S.A., Gudarzi H., Jazayeri S.M., Jahanbakhsh F., Mohraz M. CRF35-AD as the main circulating genotype of human immunodeficiency virus type 1 infection in Iran: a phylogenetic and demographic-based study. Intervirology. 2017;60(4):144–148. doi: 10.1159/000484691. [DOI] [PubMed] [Google Scholar]
- 9.Cavrois M., Gessain A., Gout O., Wain-Hobson S., Wattel E. Common human T cell leukemia virus type 1 (HTLV-1) integration sites in cerebrospinal fluid and blood lymphocytes of patients with HTLV-1-associated myelopathy/tropical spastic paraparesis indicate that HTLV-1 crosses the blood-brain barrier via clonal HTLV-1-infected cells. J. Infect. Dis. 2000;182(4):1044–1050. doi: 10.1086/315844. [DOI] [PubMed] [Google Scholar]
- 10.Mozhgani S.H., Jaberi N., Rezaee S.A., Bustani R., Jazayeri S.M., Akbarin M.M. Evaluation of HTLV-1 HBZ and proviral load, together with host IFN λ3, in pathogenesis of HAM/TSP. J. Med. Virol. 2017;89(6):1102–1107. doi: 10.1002/jmv.24721. [DOI] [PubMed] [Google Scholar]
- 11.Mozhgani S.-H., Jahantigh H.R., Rafatpanah H., Valizadeh N., Mohammadi A., Basharkhah S. Interferon lambda family along with HTLV-1 proviral load, tax, and HBZ implicated in the pathogenesis of myelopathy/tropical spastic paraparesis. Neurodegener. Dis. 2018;18:150–155. doi: 10.1159/000490058. [DOI] [PubMed] [Google Scholar]
- 12.Pilotti E., Bianchi M.V., De Maria A., Bozzano F., Romanelli M.G., Bertazzoni U. HTLV-1/-2 and HIV-1 co-infections: retroviral interference on host immune status. Front. Microbiol. 2013;4:372. doi: 10.3389/fmicb.2013.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isache C., Sands M., Guzman N., Figueroa D. HTLV-1 and HIV-1 co-infection: a case report and review of the literature. IDCases. 2016;4:53–55. doi: 10.1016/j.idcr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green D., Kharono B., Tordoff D.M., Akullian A., Bershteyn A., Morrison M. Demographic and risk group heterogeneity across the UNAIDS 90-90-90 targets: a systematic review and meta-analysis protocol. Syst. Rev. 2019;8(1):110. doi: 10.1186/s13643-019-1024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Saleem J., Dirksen W.P., Martinez M.P., Shkriabai N., Kvaratskhelia M., Ratner L. HTLV-1 Tax-1 interacts with SNX27 to regulate cellular localization of the HTLV-1 receptor molecule, GLUT1. PloS One. 2019;14(3) doi: 10.1371/journal.pone.0214059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber B., Gurtler L., Thorstensson R., Michl U., Muhlbacher A., Burgisser P. Multicenter evaluation of a new automated fourth-generation human immunodeficiency virus screening assay with a sensitive antigen detection module and high specificity. J. Clin. Microbiol. 2002;40(6):1938–1946. doi: 10.1128/JCM.40.6.1938-1946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proffitt M.R. Acquired immunodeficiency syndrome. Anal. Chem. 1993;65(12):396–400. doi: 10.1021/ac00060a605. [DOI] [PubMed] [Google Scholar]
- 18.Gallo D., Hoffman M.N., Cossen C.K., Diggs J.L., Hurst J.W., Penning L.M. Comparison of immunofluorescence, enzyme immunoassay, and Western blot (immunoblot) methods for detection of antibody to human T-cell leukemia virus type I. J. Clin. Microbiol. 1988;26(8):1487–1491. doi: 10.1128/jcm.26.8.1487-1491.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabino E.C., Zrein M., Taborda C.P., Otani M.M., Ribeiro-Dos-Santos G., Saez-Alquezar A. Evaluation of the INNO-LIA HTLV I/II assay for confirmation of human T-cell leukemia virus-reactive sera in blood bank donations. J. Clin. Microbiol. 1999;37(5):1324–1328. doi: 10.1128/jcm.37.5.1324-1328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiktor S.Z., Pate E.J., Weiss S.H., Gohd R.S., Correa P., Fontham E.T. Sensitivity of HTLV-I antibody assays for HTLV-II. Lancet. 1991;338(8765):512–513. doi: 10.1016/0140-6736(91)90585-d. [DOI] [PubMed] [Google Scholar]
- 21.Thorstensson R., Albert J., Andersson S. Strategies for diagnosis of HTLV-I and -II. Transfusion. 2002;42(6):780–791. doi: 10.1046/j.1537-2995.2002.00114.x. [DOI] [PubMed] [Google Scholar]
- 22.Mahieux R., Horal P., Mauclere P., Mercereau-Puijalon O., Guillotte M., Meertens L. Human T-cell lymphotropic virus type 1 gag indeterminate western blot patterns in Central Africa: relationship to Plasmodium falciparum infection. J. Clin. Microbiol. 2000;38(11):4049–4057. doi: 10.1128/jcm.38.11.4049-4057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W.-W., Han Y.-M., Zhu Y.-C., Zhang N., Xu J.-J., Chen H.-Y. DNA labeling generates a unique amplification probe for sensitive photoelectrochemical immunoassay of HIV-1 p24 antigen. Anal. Chem. 2015;87(11):5496–5499. doi: 10.1021/acs.analchem.5b01360. [DOI] [PubMed] [Google Scholar]
- 24.Koduru J.R., Kailasa S.K., Bhamore J.R., Kim K.-H., Dutta T., Vellingiri K. Phytochemical-assisted synthetic approaches for silver nanoparticles antimicrobial applications: a review. Adv. Colloid Interface Sci. 2018;256:326–339. doi: 10.1016/j.cis.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Rackus D.G., Shamsi M.H., Wheeler A.R. Electrochemistry, biosensors and microfluidics: a convergence of fields. Chem. Soc. Rev. 2015;44(15):5320–5340. doi: 10.1039/c4cs00369a. [DOI] [PubMed] [Google Scholar]
- 26.Goud K.Y., Kailasa S.K., Kumar V., Tsang Y.F., Lee S.E., Gobi K.V. Progress on nanostructured electrochemical sensors and their recognition elements for detection of mycotoxins: a review. Biosens. Bioelectron. 2018;121:205–222. doi: 10.1016/j.bios.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Osteryoung J.G., Osteryoung R.A. Square wave voltammetry. Anal. Chem. 1985;57(1):101–110. [Google Scholar]
- 28.Ramaley L., Krause M.S. Theory of square wave voltammetry. Anal. Chem. 1969;41(11):1362–1365. [Google Scholar]
- 29.Adam V., Huska D., Hubalek J., Kizek R. Easy to use and rapid isolation and detection of a viral nucleic acid by using paramagnetic microparticles and carbon nanotubes-based screen-printed electrodes. Microfluid. Nanofluidics. 2010;8(3):329–339. [Google Scholar]
- 30.Hassen W.M., Chaix C., Abdelghani A., Bessueille F., Leonard D., Jaffrezic-Renault N. An impedimetric DNA sensor based on functionalized magnetic nanoparticles for HIV and HBV detection. Sensor. Actuator. B Chem. 2008;134(2):755–760. [Google Scholar]
- 31.Rezaei B., Majidi N., Rahmani H., Khayamian T. Electrochemical impedimetric immunosensor for insulin like growth factor-1 using specific monoclonal antibody-nanogold modified electrode. Biosens. Bioelectron. 2011;26(5):2130–2134. doi: 10.1016/j.bios.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y., Hua S., Li F., Jiang Y., Bai X., Li D. Green-synthesized gold nanoparticles decorated graphene sheets for label-free electrochemical impedance DNA hybridization biosensing. Biosens. Bioelectron. 2011;26(11):4355–4361. doi: 10.1016/j.bios.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 33.Gong Q., Wang Y., Yang H. A sensitive impedimetric DNA biosensor for the determination of the HIV gene based on graphene-Nafion composite film. Biosens. Bioelectron. 2017;89(Pt 1):565–569. doi: 10.1016/j.bios.2016.02.045. [DOI] [PubMed] [Google Scholar]
- 34.Shamsipur M., Samandari L., Taherpour A.A., Pashabadi A. Sub-femtomolar detection of HIV-1 gene using DNA immobilized on composite platform reinforced by a conductive polymer sandwiched between two nanostructured layers: a solid signal-amplification strategy. Anal. Chim. Acta. 2019;1055:7–16. doi: 10.1016/j.aca.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Wang J. 3 ed. Wiley; 2006. Analytical Electrochemistry. [Google Scholar]
- 36.Scholz F. Springer Berlin Heidelberg; 2009. Electroanalytical Methods: Guide to Experiments and Applications. [Google Scholar]
- 37.Yuan L., Giovanni M., Xie J., Fan C., Leong D.T. Ultrasensitive IgG quantification using DNA nano-pyramids. NPG Asia Mater. 2014;6(7):e112–e. [Google Scholar]
- 38.Wang Q., Yang C., Xiang Y., Yuan R., Chai Y. Dual amplified and ultrasensitive electrochemical detection of mutant DNA Biomarkers based on nuclease-assisted target recycling and rolling circle amplifications. Biosens. Bioelectron. 2014;55:266–271. doi: 10.1016/j.bios.2013.12.034. [DOI] [PubMed] [Google Scholar]
- 39.Song W., Zhang Q., Sun W. Ultrasensitive detection of nucleic acids by template enhanced hybridization followed by rolling circle amplification and catalytic hairpin assembly. Chem. Commun. (Camb.) 2015;51(12):2392–2395. doi: 10.1039/c4cc09453k. [DOI] [PubMed] [Google Scholar]
- 40.Cheglakov Z., Weizmann Y., Beissenhirtz M.K., Willner I. Ultrasensitive detection of DNA by the PCR-Induced generation of DNAzymes: the DNAzyme primer approach. Chem. Commun. 2006;(30):3205–3207. doi: 10.1039/b605205c. [DOI] [PubMed] [Google Scholar]
- 41.Du Y.Q., Gao P.F., Wang W., Wang T.T., Chang Y., Wang J. A simple rapid detection method of DNA based on ligation-mediated real-time fluorescence PCR. Analyst. 2013;138(19):5745–5750. doi: 10.1039/c3an00763d. [DOI] [PubMed] [Google Scholar]
- 42.Choi H.M.T., Chang J.Y., Trinh L.A., Padilla J.E., Fraser S.E., Pierce N.A. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Lin Y.H., Li J., Lin L.S., Chen G.N., Yang H.H. A simple and ultrasensitive electrochemical DNA biosensor based on DNA concatamers. Chem. Commun. (Camb.) 2011;47(44):12116–12118. doi: 10.1039/c1cc15695k. [DOI] [PubMed] [Google Scholar]
- 44.Yang C., Shi K., Dou B., Xiang Y., Chai Y., Yuan R. In situ DNA-templated synthesis of silver nanoclusters for ultrasensitive and label-free electrochemical detection of MicroRNA. ACS Appl. Mater. Interfaces. 2015;7(2):1188–1193. doi: 10.1021/am506933r. [DOI] [PubMed] [Google Scholar]
- 45.Tao C., Yan Y., Xiang H., Zhu D., Cheng W., Ju H. A new mode for highly sensitive and specific detection of DNA based on exonuclease III-assisted target recycling amplification and mismatched catalytic hairpin assembly. Chem. Commun. 2015;51(20):4220–4222. doi: 10.1039/c5cc00385g. [DOI] [PubMed] [Google Scholar]
- 46.Zuo X., Xia F., Xiao Y., Plaxco K.W. Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J. Am. Chem. Soc. 2010;132(6):1816–1818. doi: 10.1021/ja909551b. [DOI] [PubMed] [Google Scholar]
- 47.Wang R., Xue C., Gao M., Qi H., Zhang C. Ultratrace voltammetric method for the detection of DNA sequence related to human immunodeficiency virus type 1. Microchimica Acta. 2011;172(3):291–297. [Google Scholar]
- 48.Chen X., Hong C.-Y., Lin Y.-H., Chen J.-H., Chen G.-N., Yang H.-H. Enzyme-free and label-free ultrasensitive electrochemical detection of human immunodeficiency virus DNA in biological samples based on long-range self-assembled DNA nanostructures. Anal. Chem. 2012;84(19):8277–8283. doi: 10.1021/ac3017828. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Bai X., Wen W., Zhang X., Wang S. Ultrasensitive electrochemical biosensor for HIV gene detection based on graphene stabilized gold nanoclusters with exonuclease amplification. ACS Appl. Mater. Interfaces. 2015;7(33):18872–18879. doi: 10.1021/acsami.5b05857. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X., Su H., Bi S., Li S., Zhang S. DNA-based amplified electrical bio-barcode assay for one-pot detection of two target DNAs. Biosens. Bioelectron. 2009;24(8):2730–2734. doi: 10.1016/j.bios.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 51.Tran L.D., Nguyen B.H., Van Hieu N., Tran H.V., Nguyen H.L., Nguyen P.X. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen printed electrodes. Mater. Sci. Eng. C. 2011;31(2):477–485. [Google Scholar]
- 52.Gong Q., Han H., Yang H., Zhang M., Sun X., Liang Y. Sensitive electrochemical DNA sensor for the detection of HIV based on a polyaniline/graphene nanocomposite. J. Materiomics. 2019;5(2):313–319. [Google Scholar]
- 53.Jia Z., Ma Y., Yang L., Guo C., Zhou N., Wang M. NiCo2O4 spinel embedded with carbon nanotubes derived from bimetallic NiCo metal-organic framework for the ultrasensitive detection of human immune deficiency virus-1 gene. Biosens. Bioelectron. 2019;133:55–63. doi: 10.1016/j.bios.2019.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Qureshi A., Gurbuz Y., Niazi J.H. Biosensors for cardiac biomarkers detection: a review. Sensor. Actuator. B Chem. 2012;171–172:62–76. [Google Scholar]
- 55.Zhong W. Nanomaterials in fluorescence-based biosensing. Anal. Bioanal. Chem. 2009;394(1):47–59. doi: 10.1007/s00216-009-2643-x. [DOI] [PubMed] [Google Scholar]
- 56.Ruan G., Agrawal A., Smith A.M., Gao X., Nie S. Quantum dots as fluorescent labels for molecular and cellular imaging. In: Geddes C.D., Lakowicz J.R., editors. Reviews in Fluorescence 2006. Springer US; Boston, MA: 2006. pp. 181–193. [Google Scholar]
- 57.Smith A.M., Nie S. Chemical analysis and cellular imaging with quantum dots. Analyst. 2004;129(8):672–677. doi: 10.1039/b404498n. [DOI] [PubMed] [Google Scholar]
- 58.Klostranec J.M., Chan W.C.W. Quantum dots in biological and biomedical research: recent progress and present challenges. Adv. Mater. 2006;18(15):1953–1964. [Google Scholar]
- 59.Liang S.-S., Qi L., Zhang R.-L., Jin M., Zhang Z.-Q. Ratiometric fluorescence biosensor based on CdTe quantum and carbon dots for double strand DNA detection. Sensor. Actuator. B Chem. 2017;244:585–590. [Google Scholar]
- 60.Norouzi M., Zarei Ghobadi M., Golmimi M., Mozhgani S.-H., Ghourchian H., Rezaee S.A. Quantum dot-based biosensor for the detection of human T-lymphotropic virus-1. Anal. Lett. 2017;50(15):2402–2411. [Google Scholar]
- 61.Deng X., Wang C., Gao Y., Li J., Wen W., Zhang X. Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection. Biosens. Bioelectron. 2018;105:211–217. doi: 10.1016/j.bios.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 62.Jimenez Jimenez A.M., Moulick A., Richtera L., Krejcova L., Kalina L., Datta R. Dual-color quantum dots-based simultaneous detection of HPV-HIV co-infection. Sensor. Actuator. B Chem. 2018;258:295–303. [Google Scholar]
- 63.Venkatesh N. 2018. Metallic Nanoparticle: A Review. [Google Scholar]
- 64.Jung J.H., Cheon D.S., Liu F., Lee K.B., Seo T.S. A graphene oxide based immuno-biosensor for pathogen detection. Angew Chem. Int. Ed. Engl. 2010;49(33):5708–5711. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]
- 65.Pustovit V.N., Shahbazyan T.V. Fluorescence quenching near small metal nanoparticles. J. Chem. Phys. 2012;136(20):204701. doi: 10.1063/1.4721388. [DOI] [PubMed] [Google Scholar]
- 66.Anger P., Bharadwaj P., Novotny L. Enhancement and quenching of single-molecule fluorescence. Phys. Rev. Lett. 2006;96(11):113002. doi: 10.1103/PhysRevLett.96.113002. [DOI] [PubMed] [Google Scholar]
- 67.Wang J., Wang L., Liu X., Liang Z., Song S., Li W. A gold nanoparticle-based aptamer target binding readout for ATP assay. Adv. Mater. 2007;19(22):3943–3946. [Google Scholar]
- 68.Qaddare S.H., Salimi A. Amplified fluorescent sensing of DNA using luminescent carbon dots and AuNPs/GO as a sensing platform: a novel coupling of FRET and DNA hybridization for homogeneous HIV-1 gene detection at femtomolar level. Biosens. Bioelectron. 2017;89(Pt 2):773–780. doi: 10.1016/j.bios.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 69.Zarei-Ghobadi M., Mozhgani S.-H., Dashtestani F., Yadegari A., Hakimian F., Norouzi M. A genosensor for detection of HTLV-I based on photoluminescence quenching of fluorescent carbon dots in presence of iron magnetic nanoparticle-capped Au. Sci. Rep. 2018;8(1):15593. doi: 10.1038/s41598-018-32756-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghobadi M.Z., Mozhgani S.-H., Hakimian F., Norouzi M., Rezaee S.A., Ghourchian H. Long segment detection of HTLV-1 genome based on the fluorescence quenching technique. Heliyon. 2018;4(12) doi: 10.1016/j.heliyon.2018.e00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin X., Liu Y., Tao Z., Gao J., Deng J., Yin J. Nanozyme-based bio-barcode assay for high sensitive and logic-controlled specific detection of multiple DNAs. Biosens. Bioelectron. 2017;94:471–477. doi: 10.1016/j.bios.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 72.Zheng J., Ji X., Du M., Tian S., He Z. Rational construction of a DNA nanomachine for HIV nucleic acid ultrasensitive sensing. Nanoscale. 2018;10(36):17206–17211. doi: 10.1039/c8nr05206a. [DOI] [PubMed] [Google Scholar]
- 73.Kermani H.A., Hosseini M., Dadmehr M., Ganjali M.R. Rapid restriction enzyme free detection of DNA methyltransferase activity based on DNA-templated silver nanoclusters. Anal. Bioanal. Chem. 2016;408(16):4311–4318. doi: 10.1007/s00216-016-9522-z. [DOI] [PubMed] [Google Scholar]
- 74.Kermani H.A., Hosseini M., Dadmehr M. DNA-templated silver nanoclusters for DNA methylation detection. In: Zuccheri G., editor. DNA Nanotechnology: Methods and Protocols. Springer New York; New York, NY: 2018. pp. 173–182. [DOI] [PubMed] [Google Scholar]
- 75.Cao Q., Teng Y., Yang X., Wang J., Wang E. A label-free fluorescent molecular beacon based on DNA-Ag nanoclusters for the construction of versatile Biosensors. Biosens. Bioelectron. 2015;74:318–321. doi: 10.1016/j.bios.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 76.Ye Y.D., Xia L., Xu D.D., Xing X.J., Pang D.W., Tang H.W. DNA-stabilized silver nanoclusters and carbon nanoparticles oxide: a sensitive platform for label-free fluorescence turn-on detection of HIV-DNA sequences. Biosens. Bioelectron. 2016;85:837–843. doi: 10.1016/j.bios.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 77.Enkin N., Wang F., Sharon E., Albada H.B., Willner I. Multiplexed analysis of genes using nucleic acid-stabilized silver-nanocluster quantum dots. ACS Nano. 2014;8(11):11666–11673. doi: 10.1021/nn504983j. [DOI] [PubMed] [Google Scholar]
- 78.Yang W., Tian J., Wang L., Fu S., Huang H., Zhao Y. A new label-free fluorescent sensor for human immunodeficiency virus detection based on exonuclease III-assisted quadratic recycling amplification and DNA-scaffolded silver nanoclusters. Analyst. 2016;141(10):2998–3003. doi: 10.1039/c6an00184j. [DOI] [PubMed] [Google Scholar]
- 79.Zou R., Zhang F., Chen C., Cai C. DNA-programming multicolor silver nanoclusters for sensitively simultaneous detection of two HIV DNAs. Sensor. Actuator. B Chem. 2019;296:126608. doi: 10.1016/j.snb.2019.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han D., Wei C. A molecular beacon based on DNA-templated silver nanoclusters for the highly sensitive and selective multiplexed detection of virulence genes. Talanta. 2018;181:24–31. doi: 10.1016/j.talanta.2017.12.049. [DOI] [PubMed] [Google Scholar]
- 81.Wang L., Dong L., Liu G., Shen X., Wang J., Zhu C. Fluorometric determination of HIV DNA using molybdenum disulfide nanosheets and exonuclease III-assisted amplification. Microchim. Acta. 2019;186(5):286. doi: 10.1007/s00604-019-3368-y. [DOI] [PubMed] [Google Scholar]
- 82.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 83.Zhu C., Zeng Z., Li H., Li F., Fan C., Zhang H. Single-layer MoS2-based nanoprobes for homogeneous detection of biomolecules. J. Am. Chem. Soc. 2013;135(16):5998–6001. doi: 10.1021/ja4019572. [DOI] [PubMed] [Google Scholar]
- 84.Xiao M., Man T., Zhu C., Pei H., Shi J., Li L. MoS2 nanoprobe for MicroRNA quantification based on duplex-specific nuclease signal amplification. ACS Appl. Mater. Interfaces. 2018;10(9):7852–7858. doi: 10.1021/acsami.7b18984. [DOI] [PubMed] [Google Scholar]
- 85.Cui L., Zou Y., Lin N., Zhu Z., Jenkins G., Yang C.J. Mass amplifying probe for sensitive fluorescence anisotropy detection of small molecules in complex biological samples. Anal. Chem. 2012;84(13):5535–5541. doi: 10.1021/ac300182w. [DOI] [PubMed] [Google Scholar]
- 86.Jia T., Fu C., Huang C., Yang H., Jia N. Highly sensitive naphthalimide-based fluorescence polarization probe for detecting cancer cells. ACS Appl. Mater. Interfaces. 2015;7(18):10013–10021. doi: 10.1021/acsami.5b02429. [DOI] [PubMed] [Google Scholar]
- 87.Varriale A., Pennacchio A., Pinto G., Oliviero G., D'Errico S., Majoli A. A fluorescence polarization assay to detect steroid hormone traces in milk. J. Agric. Food Chem. 2015;63(41):9159–9164. doi: 10.1021/acs.jafc.5b03689. [DOI] [PubMed] [Google Scholar]
- 88.Kermani H.A., Hosseini M., Dadmehr M., Hosseinkhani S., Ganjali M.R. DNA methyltransferase activity detection based on graphene quantum dots using fluorescence and fluorescence anisotropy. Sensor. Actuator. B Chem. 2017;241:217–223. [Google Scholar]
- 89.Samokhvalov A.V., Safenkova I.V., Eremin S.A., Zherdev A.V., Dzantiev B.B. Use of anchor protein modules in fluorescence polarisation aptamer assay for ochratoxin A determination. Anal. Chim. Acta. 2017;962:80–87. doi: 10.1016/j.aca.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 90.Jameson D.M., Ross J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110(5):2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang Y., Shi M., Zhao L., Zhao S., Hu K., Chen Z.-F. Carbon nanotube signal amplification for ultrasensitive fluorescence polarization detection of DNA methyltransferase activity and inhibition. Biosens. Bioelectron. 2014;54:285–291. doi: 10.1016/j.bios.2013.10.065. [DOI] [PubMed] [Google Scholar]
- 92.Gao Y., Xu J., Li B., Jin Y. Nanoparticle-aided amplification of fluorescence polarization for ultrasensitively monitoring activity of telomerase. ACS Appl. Mater. Interfaces. 2016;8(22):13707–13713. doi: 10.1021/acsami.6b02271. [DOI] [PubMed] [Google Scholar]
- 93.Chen Z., Li H., Jia W., Liu X., Li Z., Wen F. Bivalent aptasensor based on silver-enhanced fluorescence polarization for rapid detection of lactoferrin in milk. Anal. Chem. 2017;89(11):5900–5908. doi: 10.1021/acs.analchem.7b00261. [DOI] [PubMed] [Google Scholar]
- 94.Liang S., He G., Tian J., Zhao Y., Zhao S. Fluorescence polarization gene assay for HIV-DNA based on the use of dendrite-modified gold nanoparticles acting as signal amplifiers. Microchim. Acta. 2018;185(2):119. doi: 10.1007/s00604-018-2673-1. [DOI] [PubMed] [Google Scholar]
- 95.Nezami A., Dehghani S., Nosrati R., Eskandari N., Taghdisi S.M., Karimi G. Nanomaterial-based biosensors and immunosensors for quantitative determination of cardiac troponins. J. Pharmaceut. Biomed. Anal. 2018;159:425–436. doi: 10.1016/j.jpba.2018.07.031. [DOI] [PubMed] [Google Scholar]
- 96.Wang X., Ge L., Yu Y., Dong S., Li F. Highly sensitive electrogenerated chemiluminescence biosensor based on hybridization chain reaction and amplification of gold nanoparticles for DNA detection. Sensor. Actuator. B Chem. 2015;220:942–948. [Google Scholar]
- 97.Babamiri B., Salimi A., Hallaj R. A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 2018;117:332–339. doi: 10.1016/j.bios.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 98.Barnes W.L., Dereux A., Ebbesen T.W. Surface plasmon subwavelength optics. Nature. 2003;424(6950):824–830. doi: 10.1038/nature01937. [DOI] [PubMed] [Google Scholar]
- 99.Oates T.W.H., Wormeester H., Arwin H. Characterization of plasmonic effects in thin films and metamaterials using spectroscopic ellipsometry. Prog. Surf. Sci. 2011;86(11):328–376. [Google Scholar]
- 100.Diao W., Tang M., Ding S., Li X., Cheng W., Mo F. Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices. Biosens. Bioelectron. 2018;100:228–234. doi: 10.1016/j.bios.2017.08.042. [DOI] [PubMed] [Google Scholar]
- 101.Nie S., Emory S.R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275(5303):1102. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 102.Wang Y., Tang L.-J., Jiang J.-H. Surface-enhanced Raman spectroscopy-based, homogeneous, multiplexed immunoassay with antibody-fragments-decorated gold nanoparticles. Anal. Chem. 2013;85(19):9213–9220. doi: 10.1021/ac4019439. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y., Yan B., Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem. Rev. 2013;113(3):1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 104.Yoon J., Choi N., Ko J., Kim K., Lee S., Choo J. Highly sensitive detection of thrombin using SERS-based magnetic aptasensors. Biosens. Bioelectron. 2013;47:62–67. doi: 10.1016/j.bios.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 105.Albrecht M.G., Creighton J.A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977;99(15):5215–5217. [Google Scholar]
- 106.Jeanmaire D.L., Van Duyne R.P. Surface Raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977;84(1):1–20. [Google Scholar]
- 107.Bartlett P.N., Baumberg J.J., Coyle S., Abdelsalam M.E. Optical properties of nanostructured metal films. Faraday Discuss. 2004;125:117–132. doi: 10.1039/b304116f. [DOI] [PubMed] [Google Scholar]
- 108.Fu X., Cheng Z., Yu J., Choo P., Chen L., Choo J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016;78:530–537. doi: 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- 109.Hu J., Zheng P.C., Jiang J.H., Shen G.L., Yu R.Q., Liu G.K. Sub-attomolar HIV-1 DNA detection using surface-enhanced Raman spectroscopy. Analyst. 2010;135(5):1084–1089. doi: 10.1039/b920358c. [DOI] [PubMed] [Google Scholar]
- 110.Chu B. Elsevier Science; 2012. Laser Light Scattering: Basic Principles and Practice. [Google Scholar]
- 111.Li Z., Wang Y., Shen J., Liu W., Sun X. The measurement system of nanoparticle size distribution from dynamic light scattering data. Optic Laser. Eng. 2014;56:94–98. [Google Scholar]
- 112.Ramos A.P. 4 - dynamic light scattering applied to nanoparticle characterization. In: Da Róz A.L., Ferreira M., de Lima Leite F., Oliveira O.N., editors. Nanocharacterization Techniques. William Andrew Publishing; 2017. pp. 99–110. [Google Scholar]
- 113.Zou L., Ling L. Ultrasensitive detection of HIV DNA with polymerase chain reaction–dynamic light scattering. Anal. Chem. 2018;90(22):13373–13377. doi: 10.1021/acs.analchem.8b03052. [DOI] [PubMed] [Google Scholar]
- 114.Devadoss A., Sudhagar P., Terashima C., Nakata K., Fujishima A. Photoelectrochemical biosensors: new insights into promising photoelectrodes and signal amplification strategies. J. Photochem. Photobiol. C Photochem. Rev. 2015;24:43–63. [Google Scholar]
- 115.Shi X.M., Fan G.C., Tang X., Shen Q., Zhu J.J. Ultrasensitive photoelectrochemical biosensor for the detection of HTLV-I DNA: a cascade signal amplification strategy integrating lambda-exonuclease aided target recycling with hybridization chain reaction and enzyme catalysis. Biosens. Bioelectron. 2018;109:190–196. doi: 10.1016/j.bios.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 116.Wang S., Zhao J., Zhang Y., Yan M., Zhang L., Ge S. Photoelectrochemical biosensor of HIV-1 based on cascaded photoactive materials and triple-helix molecular switch. Biosens. Bioelectron. 2019;139:111325. doi: 10.1016/j.bios.2019.111325. [DOI] [PubMed] [Google Scholar]
- 117.Giovanni M., Setyawati M.I., Tay C.Y., Qian H., Kuan W.S., Leong D.T. Electrochemical quantification of Escherichia coli with DNA nanostructure. Adv. Funct. Mater. 2015;25(25):3840–3846. [Google Scholar]
- 118.Tay C.Y., Yuan L., Leong D.T. Nature-inspired DNA nanosensor for real-time in situ detection of mRNA in living cells. ACS Nano. 2015;9(5):5609–5617. doi: 10.1021/acsnano.5b01954. [DOI] [PubMed] [Google Scholar]
- 119.Lee D.S., Qian H., Tay C.Y., Leong D.T. Cellular processing and destinies of artificial DNA nanostructures. Chem. Soc. Rev. 2016;45(15):4199–4225. doi: 10.1039/c5cs00700c. [DOI] [PubMed] [Google Scholar]