Abstract
Both steatosis and inflammation are key pathological events in the progression of non-alcoholic fatty liver disease (NAFLD). Probiotics are beneficial for the prevention and treatment of NAFLD. Bifidobacterium animalis subsp. lactis V9 (V9) is a newly isolated strain with favorable probiotic properties. The study aims to evaluate the effects and mechanisms of V9 on the hepatic steatosis and inflammatory responses in a rat model of NAFLD induced by high-fat diets (HFD). Our results showed that administration of V9 significantly attenuated the HFD-induced increases in alanine transaminase (ALT) and aspartate aminotransferase (AST) levels, resulting in alleviated hepatic steatosis. V9 supplementation reduced the accumulation of hepatic triglyceride and free fatty acid,while increasing the levels of glycogen. Serum levels of glucose were also decreased in HFD rats administrated with V9. Meanwhile, the transcription of SREBP-1c and FAS was reduced, and the hepatic expression of phosphorylated-AMPK and PPAR-α was restored after V9 administration. V9 suppressed the production of inflammatory cytokines (e.g. IL-6, IL-1β, and TNF-α) in HFD-fed rats. The anti-inflammatory effects of V9 was found to be associated with the inhibition of hepatic expression of TLR4, TLR9, NLRP3, and ASC mRNA. Furthermore, the activation of ERK, JNK, AKT and NF-κB were suppressed by V9 treatment. These results indicate that Bifidobacterium lactis V9 improves NAFLD by regulating de novo lipid synthesis and suppressing inflammation through AMPK and TLR-NF-κB pathways, respectively.
Keywords: Probiotics, NAFLD, AMPK, TLR, NF-κB
Key points
V9 supplementation alleviates HFD-induced metabolic disorder.
V9 inhibits HFD-induced expression of FAS, SREBP1c and activates AMPK.
V9 suppresses the TLR-NF-κB signaling pathway in HFD-fed rats.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the chronic liver pathology that occurs when fat is deposited (steatosis) in the liver in the absence of a history of excessive alcohol consumption. NAFLD may progress to nonalcoholic steatohepatitis (NASH) with the help of fat deposition, oxidative stress and chronic inflammation in the liver. Some NAFLD patients may advance to develop cirrhosis and eventually hepatocellular carcinoma (Hashimoto and Tokushige 2011). The prevalence of NAFLD has been rapidly increasing due to lifestyle changes, such as increased consumption of high-fat diets (HFD) and the lack of exercise (Williams et al. 2011). NAFLD is now recognized as a worldwide health concern and it is closely associated with diabetes mellitus (type II) and cardiovascular diseases (Ratziu et al. 2010; Chacko and Reinus 2016). Besides lifestyle modifications (Malaguarnera et al. 2012) and bariatric procedures (Shouhed et al. 2017), there is no effective pharmacological treatment for NAFLD.
The pathogenesis of NAFLD is largely unknown. The widely accepted “two-hit” theory holds that the “first-hit” comprises of dysregulated lipid metabolism and insulin resistance followed by the hepatic “second-hit” including inflammatory cytokine production, oxidative stress-induced lipotoxicity and other mechanisms (Musso et al. 2010). Growing evidence indicates that the gut microbiota participates in NAFLD pathogenesis and lipotoxicity (Safari and Gérard 2019; Borrelli et al. 2018), through the metabolism of nutrients and a number of secretory factors that eventually target the liver (Szabo et al. 2011; Lu et al. 2016). Both experimental and clinical studies have shown that probiotics are beneficial in the prevention and treatment of NAFLD (Maddur and Neuschwander-Tetri 2014; Nobili et al. 2018). Administration of probiotic mixture of VSL#3 reduces HFD-induced inflammatory response in ApoE −/− mice (Chan et al. 2016). However, the molecular mechanisms of the beneficial effects of probiotics are not entirely understood.
Bifidobacterium animalis subsp. lactis V9 (V9) is a novel strain isolated from healthy Mongolian children, possessing favorable probiotic properties such as aciduricity and bile resistance (Sun et al. 2010). The strain has been used in the industrial fermentation of dairy starter cultures by Inner Mongolia Yili Industrial Group Co. Ltd, China. The results of a recent research have demonstrated that the fermentation of whole oat flour with V9 and Lactobacillus plantarum TK9 yield a synbiotic food rich in lactic acid bacteria and prebiotics (Wang et al. 2015). A study has shown that Bifidobacterium and dietary blueberry supplementation could attenuate hepatic lipid accumulation in NAFLD rats (Ren et al. 2014). Moreover, administration of Bifidobacterium longum and fructo-oligosaccharides reduces the production of pro-inflammatory cytokines, steatosis and NASH in patients (Malaguarnera et al. 2012). We are wondering if the novel probiotic V9 strain has beneficial effects for NAFLD patients. In this study, we established an HFD-induced rat model of NAFLD that reflects the path physiology of the disease in humans, including the development of fatty liver, hyperglycemia, as well as systemic inflammation. We then explored the potential effects and underlying mechanisms of V9 on the improvement of metabolic dysfunction and inflammation in HFD-fed rats by examining various physiological parameters, the changes of metabolism-related genes and the inflammatory signaling pathways. Our results indicate that Bifidobacterium lactis V9 is a potential candidate for the treatment of HFD-induced metabolic syndrome.
Materials and methods
Bacteria strain and growth conditions
The strain of Bifidobacterium animalis subsp. lactis V9 (V9) is a special probiotic starter identified and preserved by the Key Laboratory of Dairy Biotechnology and Bioengineering, Educational Ministry of China, and also submitted to CGMCC for preservation with a deposit number of No. 5470. The bacteria was isolated from healthy Mongolian children and identified by PCR-based 16 s rRNA amplication. The V9 strain shares 99% homology with Bifidobacterium animalis subsp. lactic Bb12 (Sun et al. 2010). The strain was cultured in a modified TPY agar medium under an anaerobic environment at 37 °C. The V9 cell pellets were harvested, washed twice with PBS and then lyophilized. The lyophilized powder of V9 was resuspended in physiological saline and adjusted to 1 × 109 CFU/ml for treatment of the rats.
Animal experiments
Male Wistar rats (120–140 g) were purchased from Vital River Laboratories Animal Co. Ltd. (Beijing, China) and housed under controlled conditions (20–22 °C, 55 ± 10% relative humidity, 12-h light/dark cycles). Experiments on rats began after 1 week of adaptation. During the adaption period, rats were fed a standard chow diet (10% energy from fat, Ke Ao Xie Li Diet Co. Ltd, Beijing, China) and received water ad libitum. All protocols for animal experiments were approved by the Animal Care and Use Committee at Inner Mongolia Agricultural University. Rats were randomly divided into five groups (n = 8 in each group): control, high-fat diet (HFD), HFD + Berberine, V9 treatment (HFD/V9) and V9 control (CON/V9). With the exception of the control and CON/V9 group, which were fed with a normal chow diet, rats in the remaining groups were given a high-fat diet (60% energy from fat, Ke Ao Xie Li Diet Co. Ltd, Beijing, China) to establish a model of NAFLD. After 6 weeks, all groups of rats were fed with a normal chow diet. Rats in the HFD/V9 and CON/V9 groups were gavaged with V9 (1 × 109 CFU) for 4 weeks. At the same time, the HFD + Berberine group of rats received oral administration of Berberine (300 mg/kg). At the end of the experiment, all the rats were fasted overnight and then anesthetized with isoflurane. Samples of blood and liver were collected. Parts of liver tissues were fixed in 10% formalin for histological analysis and other samples were stored in aseptic cryopreservation tubes. The tubes were immediately snap-frozen in liquid nitrogen followed by storage in a − 80 °C refrigerator.
Biochemical analysis
Serum samples were obtained by centrifuging the clotted blood samples at 3500×g for 10 min, which was then stored at − 80 °C for subsequent analysis. The levels of serum ALT, AST and glucose were measured using an automatic biochemical analyzer (Olympus 2700). Hepatic triglyceride (TG), glycogen, and free fatty acid (FFA) levels were measured by the corresponding kits according to the manufacturer’s instructions (Jiancheng Institute of Biotechnology, Nanjing, China). The lysate of liver samples (100–200 mg) was prepared by homogenization in modified RIPA buffer with protease inhibitor cocktail. The samples were processed mechanically by a dounce homogenizer with 15–20 strokes. Tissue and cell debris in the lysates was then removed by centrifugation. The protein concentration was quantified using the Sangon Biotech BCA protein kit according to the manufacturer’s instructions. The contents of TG, glycogen, and FFA were measured by spectrophotography and calculated based on the protein content of the liver homogenates.
Histological examination
The liver samples were fixed in 10% paraformaldehyde at room temperature for 24 h. After dehydration through a graded ethanol series, the samples were immersed in xylene, embedded in paraffin, and then cut into 5 μm sections for hematoxylin-eosin (H&E) staining. The images of HE-stained sections were taken using a light microscope and assessed blindly.
Enzyme-linked immunosorbent assay
Serum TNF-α, IL-1β, and IL-6 levels were detected using commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, USA) according to the manufacturer’s instructions.
Real-time PCR assay
Total mRNA from the liver tissues was extracted using a Trizol reagent (Invitrogen, Carlsbad, CA). RNA concentration and purity (OD260/280) were measured by a NanoDrop Lite spectrophotometer. The total mRNA (1 μg) was reversed into first strand cDNA using the RT-PCR kit (Takara, Dalian, China). Real-time PCR assay was performed using the specific primers as listed in Additional file 1: Table S1. GAPDH was used as an endogenous control to normalize the relative expression of target transcripts. Relative quantification of gene expression was performed by the 2−ΔΔCt method.
Western blot analysis
The liver proteins were extracted by using the tissue protein extraction solution or the nucleoprotein extraction kit (Sangon Biotech, Shanghai, China). The protein content was measured as previously described. The same amount of total protein from each sample was electrophoresed into SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were probed using rabbit polyclonal antibodies (Cell Signaling) against p-ERK1/2 (Thr202/Tyr204) (#9101), ERK1/2 (#9102), p-JNK(Thr183/Tyr185)(#9251), JNK (#9252), p-AKT (Ser473) (#9271), AKT(#4691), p-NF-κB (Ser468) (#3039), NF-κB (#3034) and p-AMPK (Thr172) (#2535), AMPK (#2532) overnight at 4 °C. After washing five times with TBST, the membranes were incubated with Goat anti-Rabbit IgG H&L (IRDye® 800CW) for 1 h. The images of specific proteins were detected and analyzed using the LI-COR Odyssey CLx.
Statistical analysis
The statistical analysis was performed using the SPSS 17 software package. All the data were expressed as mean ± standard deviation (SD) and analyzed using the one-way analysis of variance (ANOVA) with Duncan’s multiple-range test. P values of < 0.05 were regarded as statistically significant.
Results
V9 treatment attenuates HFD- induced liver injury
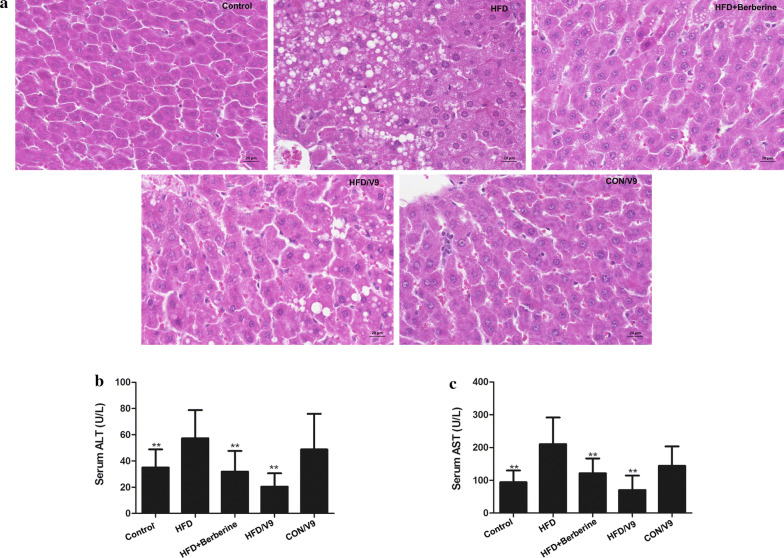
In order to explore the mitigating effects of Bifidobacterium hactis V9 on HFD-induced liver injury, the development of liver damage was examined histologically by H&E staining. Administration of the high-fat diet for 6 weeks caused evident damage in the liver as shown by the disturbed structure of lobules, hepatocyte hypertrophy and severe hepatic steatosis represented by the formation of numerous lipid droplets. In contrast, Bifidobacterium lactis V9 and Berberine treatment significantly improved the structure of liver lobules and alleviated hepatic steatosis (Fig. 1a). In addition, serum alanine transaminase (ALT) and aspartate aminotransferase (AST) levels were different across the various conditions. Compared with the HFD group, both V9 and Berberine treatment significantly decreased the serum ALT and AST levels (Fig. 1b and c). These results indicate that V9 treatment alleviates high fat diet-induced hepatic injury and steatosis.
Fig. 1.
Probiotic V9 improves hepatic steatosis and liver damage in HFD rats. a Paraffin-embedded liver tissues (n = 3 each group) were sectioned and stained with hematoxylin and eosin(H&E). Micrographs are representative pictures with magnification 200×. Serum ALT (b) and AST levels were measured (n = 8, each group). **P < 0.01 vs HFD group
V9 treatment improves HFD-induced disorder in lipid and glucose metabolism
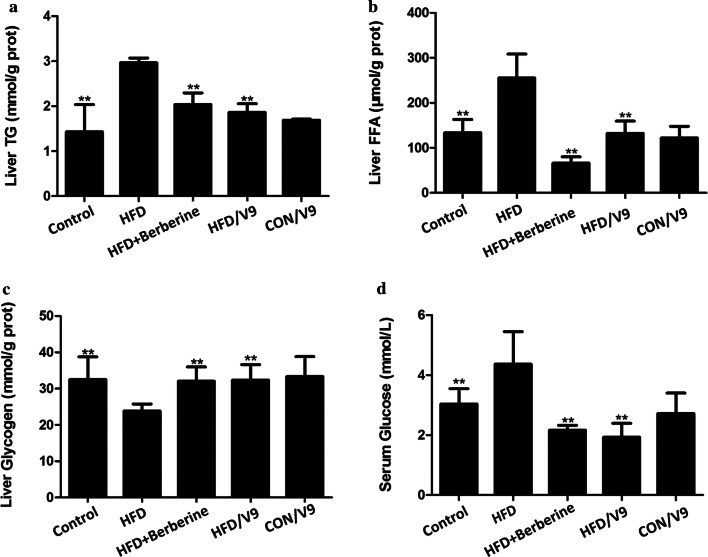
Next, we measured the parameters of lipid and glucose metabolism in the liver and the serum. The hepatic TG and FFA levels were increased in the HFD group, while these decreased in the V9 and Berberine treatment group (Fig. 2a and b). In addition, V9 and Berberine treatment restored the HFD-induced decrease in the hepatic glycogen content (Fig. 2c). Compared with the HFD group, V9 supplementation also induced a significant reduction in serum Total Cholesterol (TC) and fasting glucose levels (Fig. 2d). These results indicate that V9 supplementation improved the disorder in lipid and glucose metabolism.
Fig. 2.
V9 treatment improves HFD-induced metabolic disorder. a Hepatic levels of TG. b Hepatic levels of FFA. c Hepatic levels of Glycogen. d Fasting glucose levels in serum. Data are expressed as mean ± SD with n = 8, **P < 0.01 vs HFD group
V9 supplementation reverses the deregulation of genes involved in lipid and glucose metabolism
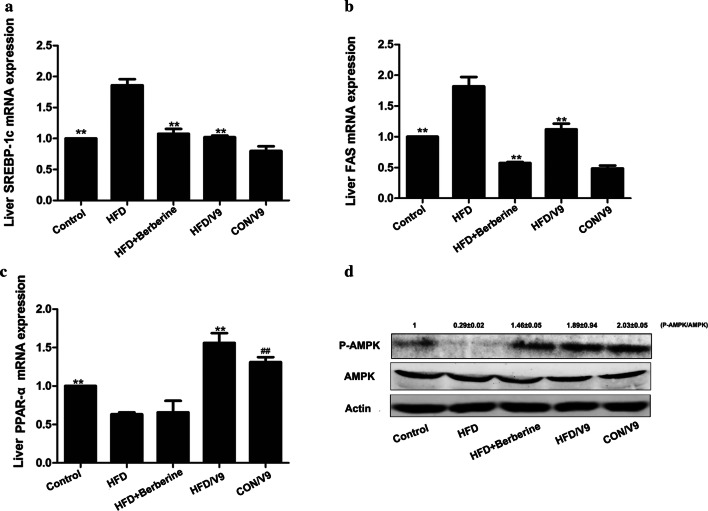
To understand the underlying mechanisms of the observed effects of V9 treatment, we evaluated the key signaling pathways involved in the modulation of lipid and glucose metabolism. The mRNA expression of sterol regulatory element-binding protein 1c (SREBP-1c) and the lipogenic enzyme fatty acid synthase (FAS) increased in HFD rats and was subsequently reduced by V9 and Berberine treatment (Fig. 3a and b). Hepatic PPAR-α mRNA expression was dramatically reduced by HFD challenge, which was restored by V9 treatment. On the other hand, there is no significant difference between the PPAR-α mRNA expression levels in the Berberine treatment group and the HFD group. Compared with the control group, an increase in the expression of PPAR-α mRNA was also observed in the CON/V9 group (Fig. 3c). Furthermore, western blotting analysis showed that treatment with V9 and Berberine restored the expression of phosphorylated-AMPK (Fig. 3d).
Fig. 3.
Probiotic V9 regulate the expression of the genes in lipid metabolism. Relative SREAP-1c (a), FAS (b), and PPAR-α mRNA expression (c) are reported (n = 8 each group). #P < 0.05 vs control group, and **P < 0.01 vs HFD group. d Western blotting analysis of hepatic phospho-AMPK (p-AMPK) and total AMPK. The representative picture is one from three independent experiments. The densitometric analysis of bands from all samples was shown as the fold change relative to the control group
V9 supplementation reduces HFD-induced pro-inflammatory responses
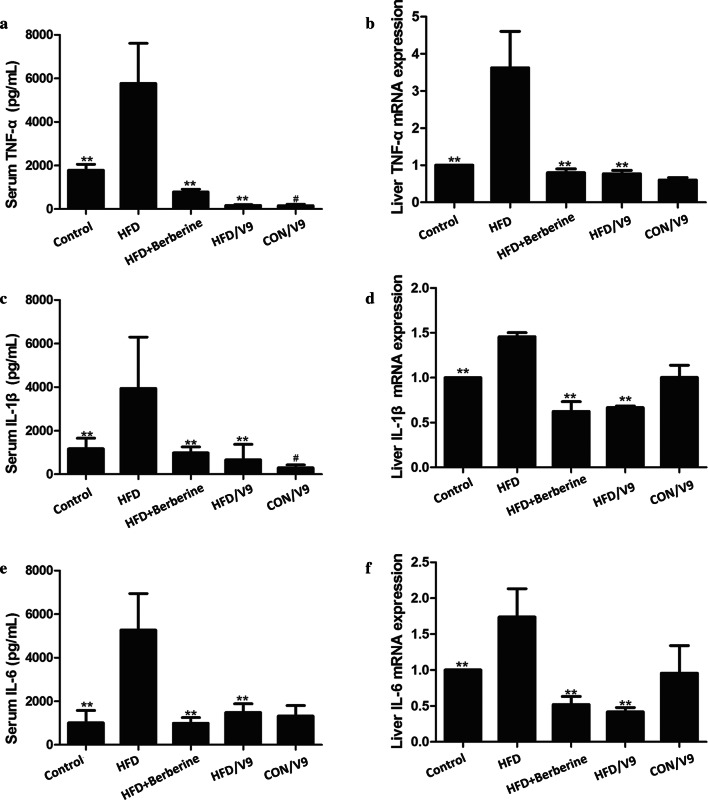
To investigate the anti-inflammatory effects of the probiotic V9, hepatic mRNA levels and serum concentrations of TNF-α, IL-1β, and IL-6 were measured. Compared with the control group, HFD challenge resulted in significantly increased levels of serum TNF-α, IL-1β and IL-6 which was also the case for the hepatic mRNA expression of these cytokines. V9 supplementation markedly inhibited HFD-induced expression of TNF-α, IL-1β, and IL-6 at both the mRNA and the protein level. A similar trend was found in the Berberine treatment group (Fig. 4a–f). These results indicate that both V9 and Berberine can reduce HFD-induced pro-inflammatory cytokine production.
Fig. 4.
V9 treatment reduces HFD-induced inflammatory responses. a Serum TNF-α production. b Hepatic TNF-α mRNA expression. c Serum IL-1β production. d Hepatic IL-1β mRNA expression. e Serum IL-6 production. f Hepatic IL-6 mRNA expression. Data are expressed as mean ± SD, with n = 8, #P < 0.05 vs control group, and **P < 0.01 vs HFD group
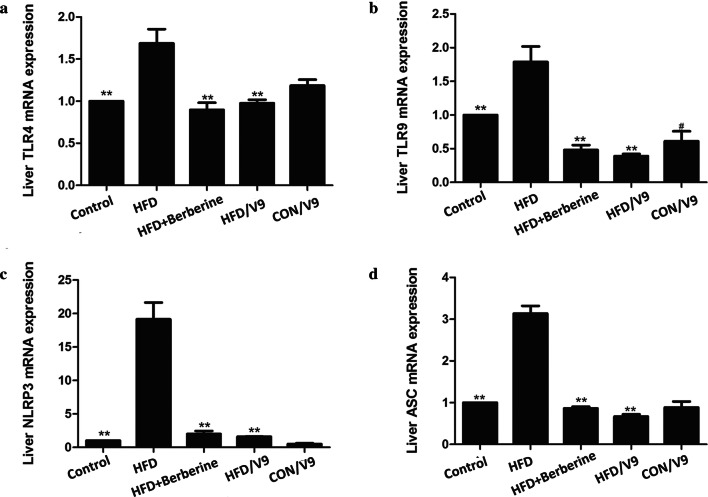
V9 suppresses the hepatic expression of TLRs and NLRP3
As shown in Fig. 5a and b, the hepatic TLR4 and TLR9 mRNA expression increased significantly in the HFD group in comparison with the control group. However, V9 and Berberine treatment significantly reduced the expression of TLR4 and TLR9 mRNA in HFD rats. We found a similar pattern in the TLR4 and TLR9 protein expression after the administration of V9 or Berberine (Additional file 1: Figure S1). A significant decline in NLRP3 and ASC mRNA expression was also found in the V9 and Berberine treatment groups (Fig. 5c and d).
Fig. 5.
The effect of Probiotic V9 on hepatic TLRs, NLRP3 and ASC. a Hepatic mRNA levels of TLR4. b Hepatic mRNA levels of TLR9. c Hepatic mRNA levels of NLRP3. d Hepatic mRNA levels of ASC. Data are expressed as mean ± SD with n = 8, #indicates P < 0.05 vs Control group; **P < 0.01 vs HFD group
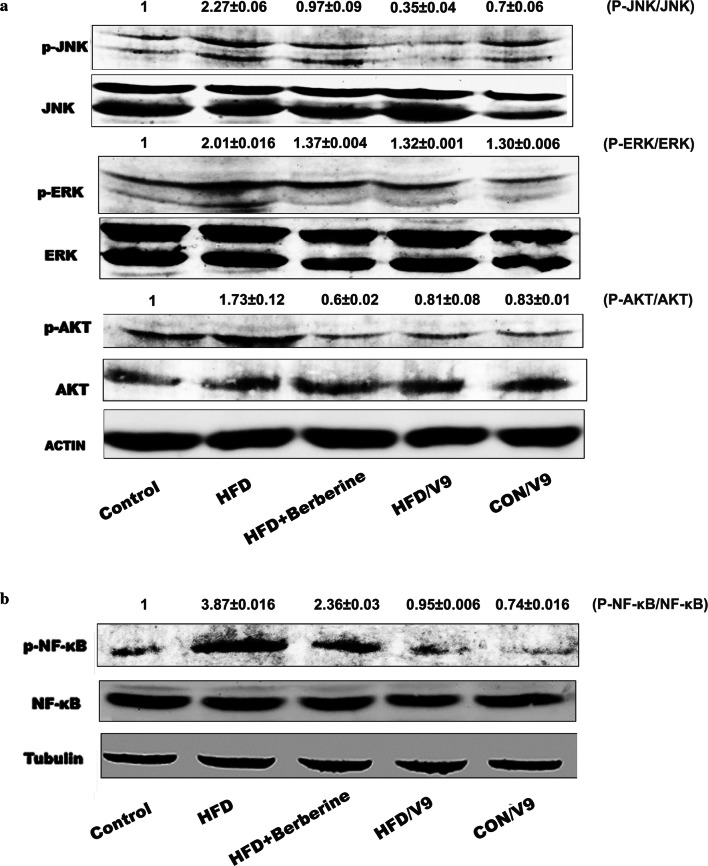
V9 inhibits the activation of MAPK, AKT and NF-κB
In order to further study the mechanisms of the protective effects of V9 against HFD-induced NAFLD, we went on to evaluate the changes of the TLR-mediated downstream signaling pathways. As shown in Fig. 6a, HFD challenge resulted in elevated phosphorylation of JNK, ERK and AKT. However, V9 and Berberine supplementation evidently reduced the activation of JNK, ERK and AKT. V9 treatment also inhibited HFD-induced phosphorylation of NF-κB (Fig. 6b). These results indicate that modulation of the activation of MAPK, AKT and NF-κB contribute to the anti-inflammatory effects of V9.
Fig. 6.
The effects of probiotic V9 on hepatic MAPK, AKT and NF-κB activation in HFD-challenged rats. a Western blotting analysis of phospho –JNK (p-JNK), total JNK, phospho-ERK (p-ERK), total ERK, phospho-AKT (p-AKT) and total AKT. b Western blotting analysis of nuclear phospho-NF-κB (p-NF-κB) and total NF-κB. The Representative pictures in a and b are shown from three independent experiments. The densitometric analysis of bands from all samples was shown as the fold change relative to the control group
Discussion
The chronic liver disease NAFLD has emerged as a worldwide health burden, without effective treatments. Probiotics confer health benefits on the host with NAFLD by regulating glycolipid metabolism and steatosis-induced inflammatory responses (Kim et al. 2017; Ritze et al. 2014), which are dependent on the strains and the dosages (Xu et al. 2012; Ferolla et al. 2015). In this study, our results reveal that the novel strain of probiotic V9 exhibits beneficial effects in the HFD-induced NAFLD setting through its hypolipidemic and anti-inflammatory potential.
In our study, a rat model of NAFLD was successfully established, as evidenced by elevated serum glucose levels, compounded by the accumulation of hepatic TG and FFA. In addition, histopathological observations revealed severe steatosis in the NAFLD rat model. Studies have demonstrated that the levels of ALT and AST are increased in patients of NAFLD with insulin resistance (Sheng et al. 2018). Similarly, our study also shows that ALT and AST levels are significantly increased in HFD-challenged rats. All these results indicate that the HFD-challenged rats exhibit metabolic dysfunction, hepatic steatosis, and hepatotoxicity, which are in concordance with signs presented by NAFLD patients.
Evidence from a randomized, double-blind, placebo-controlled study demonstrated that administration of a multispecies probiotic mixture reduces the intrahepatic fat and body weight in obese NAFLD patients (Ahn et al. 2019). Animal studies have also shown that co-administration with cholesterol-lowering probiotics (two strains) and anthraquinone from Cassia obtusifolia L. has potential therapeutic effects for NAFLD (Mei et al. 2015). A similar study also indicates that supplementation with Bifidobacterium longum and Lactobacillus acidophilus attenuate liver fat accumulation in NAFLD rats, with a superior observed from treatment with Bifidobacterium longum (Xu et al. 2012). Our results are consistent with previously published studies, showing that V9 treatment improved hepatic steatosis and HFD-induced lipid and glucose disorder. Serum levels of ALT, AST, TG and glucose were markedly reduced, accompanied by reduced hepatic lipid accumulation (TG and FFA) and increased hepatic glycogen content in the V9 treatment group. These results suggest the single Bifidobacterium strain V9 has beneficial effects,and attenuates the progression of NAFLD. Whether the V9 strain is superior in terms its ability to alleviate liver fat accumulation than other identified Lactobacillus strains needs to be clarified in the future.
In order to elucidate the underlying mechanisms of the hypolipidemic effect of V9, the expression of lipogenic sterol regulatory element binding protein 1c (SREBP1c) and its target gene fatty acid synthase (FAS) was analyzed. The decreased expression of SREBP1c and FAS in the HFD/V9 group indicates that de novo lipid synthesis was suppressed by V9 treatment. Peroxisome proliferative-activated receptors (PPARs) are transcription factors belonging to the nuclear receptor superfamily, which are involved in lipid metabolism, energy homeostasis and inflammation (Derosa et al. 2017). PPARα is highly expressed in the liver, and is the master regulator of hepatic lipid flux by modulating fatty acid transport and β-oxidation. In addition, PPARα activation counteracts the expression of the inflammatory genes induced by NF-κB (Tanaka et al. 2017; Alves et al. 2017; Staels et al. 1998). Experimental evidence shows that PPARα deficiency leads to susceptibility to NAFLD, NASH and hepatic inflammatory responses (Ip et al. 2003). Thus, PPARα is an potential therapeutic target for hyperlipidemia. Research has shown that selective activation of PPARa by compound K-877 improves lipid metabolism in mice (Takei et al. 2017). The results of our study showed that HFD-induced decreased expression of hepatic PPAR-α mRNA was restored in the V9 treatment group but not in the Berberine group, suggesting different underlying mechanisms of action of V9 and Berberine.
AMP-activated kinase (AMPK), a key regulator in energy metabolic homeostasis, has been identified as a crucial target for drugs (Hardie et al. 2012). Activated AMPK phosphorylates SREBP and attenuates hepatic steatosis and atherosclerosis in mice with insulin resistance (Li et al. 2011). Treatment with V9 and Berberine induced the phosphorylation of AMPK, indicating that both of them alleviated hepatic steatosis through promoting the activation of AMPK. A recent study demonstrated that Lactobacillus plantarum NA136 could activate the AMPK signaling pathway to suppress the SREBP-1/FAS signaling, which in turn inhibits de novo lipogenesis in mice with NAFLD (Zhao et al. 2019). Our study is consistent with the study of Lactobacillus plantarum NA136. And thus, we conclude that V9 supplementation alleviates HFD-induced metabolic disorder through upregulating the AMPK and PPARα signaling pathways.
It has been demonstrated that TLR4 plays a key role in the progression from hepatic steatosis to NASH in mice with obesity-induced NAFLD (Ye et al. 2012). In addition, upregulation of TLR9 is increased in NASH models and activation of TLR9 signaling lead to inflammatory cell recruitment (Mridha et al. 2017a, b). Our results concur with previous reports by showing that the expression of TLR4 and TLR9 was increased in the NAFLD model group. However, V9 supplementation downregulated the expression of TLR4 and TLR9 (Fig. 5), which correlated with reduced production of pro-inflammatory cytokines in V9 treatment group. This suggests that the beneficial effects of V9 on NAFLD are associated with the downregulation of TLR4 and TLR9-induced inflammatory responses.
The role of NLRP3 in the progression from steatosis to NASH has also been documented recently (Rossato et al. 2020). Blocking NLRP3 by a small-molecule inhibitor MCC950 mitigates hepatic inflammation and fibrosis, as shown in two mice models of NASH (Mridha et al. 2017a, b). In addition, the study of Xiao et al. (2016) showed that bee honey attenuates the progression of NASH partly through suppressing the thioredoxin-interacting protein (TXNIP)-NLRP3 pathway. In our study, increased transcription of NLRP3 and its adaptor protein ASC was present in the NAFLD model group. Treatment with V9 and Berberine significantly decreased the expression of NLRP3 and ASC, suggesting that regulation on the expression of NLRP3 inflammasome is one of the underlying mechanisms of the anti-inflammatory effect of V9.
To further support the anti-inflammatory effect of V9 on HFD-induced inflammatory responses, we studied the TLR4 downstream signaling changes. Our results showed that V9 and Berberine treatment reduced the phosphorylation of JNK, ERK, AKT, and NF-κB (Fig. 6). As activation of NF-κB leads to the priming of NLRP3 inflammasome, it is rational to deduce that reduced activation of NF-κB contributes to the reduced expression of NLRP3, which was verified in our study. In summary, the anti-inflammatory effect of V9 against NAFLD is associated with the down-regulation on TLR-NF-κB-NLRP3 signaling.
In summary, our results demonstrate that the administration of probiotic V9 alleviates HFD-induced liver damage, mediated by a reduction in hepatic fat accumulation (through increasing AMPK-mediated fatty acid β-oxidation and reduced de novo lipid synthesis) and its anti-inflammatory activity (through downregulation of TLR-NF-κB signaling pathways). Therefore, we demonstrate that the probiotic V9 has the potential to be used as an treatment for NAFLD.
Supplementary information
Additional file 1: Table S1. The specific primer sequences of genes of interest. Figure S1. Western blot analysis of hepatic TLR4 and TLR9. The representative picture is one from three independent experiments.
Acknowledgements
We show our thanks to Haijie Wu, Fei Zhang, Yihong Wu, Huan Tian for their help in animal experiment. We thank Dr. Yexin Xie for proofreading the revised manuscript.
Abbreviations
- NAFLD
Non-alcoholic fatty liver disease
- V9
Bifidobacterium animalis subsp.lactis V9
- HFD
High-fat diets
- ALT
Alanine transaminase
- AST
Aspartate aminotransferase
- TG
Triglyceride
- FFA
Free fatty acid
- NASH
Nonalcoholic steatohepatitis
- CGMCC
China General Microbiological Culture Collection Center
- HE
Hematoxylin–eosin
- ELISA
Enzyme-linked immunosorbent assay
- SREBP1c
Sterol regulatory element binding protein 1c
- FAS
Fatty acid synthase
- PPARs
Peroxisome proliferative-activated receptors
- AMPK
AMP-activated kinase
Authors' contributions
YW and GZ conceived the study, analyzed the data, and revised the manuscript; YY and CL executed most of the experiments, collected the data and prepared the manuscript; SZ and XW performed the Real-time PCR and ELISA assay; JW did the HE staining and analyzed the photos; HZ help to prepare with the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Grants from the program for Yong Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (No. NJYT-19-A14), the National Natural Science Foundation of China (Nos. 81560677, 31560443), the Natural Science Foundation of Inner Mongolia (2019MS03019). The authors thank the China Scholarship Council for financial support to YW (No. 201808155035).
Ethics approval and consent to participate
This article does not contain any studies with human participants performed by any of the authors. All experiments were approved by the Animal Care and Use Committee of Inner Mongolia Agricultural University (China) according to the Chinese Council on Animal Care guidelines.
Consent for publication
All authors listed have read the complete manuscript and have approved submission of the paper.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Yan and Chunyan Liu contributed equally to the work
Contributor Information
Yuzhen Wang, Email: wangyuzhen817@126.com.
Guofen Zhao, Email: guofenzhao@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-01038-y.
References
- Ahn SB, Jun DW, Kang BK, Lim JH, Lim S, Chung MJ. Randomized, double-blind, placebo-controlled study of a multispecies probiotic mixture in nonalcoholic fatty liver disease. Sci Rep. 2019;9:5688. doi: 10.1038/s41598-019-42059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CC, Waitzberg DL, de Andrade LS, Dos Santos Aguiar L, Reis MB, Guanabara CC, Júnior OA, Ribeiro DA, Sala P. Prebiotic and synbiotic modifications of beta oxidation and lipogenic gene expression after experimental hypercholesterolemia in rat liver. Front Microbiol. 2017;8:2010. doi: 10.3389/fmicb.2017.02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli A, Bonelli P, Tuccillo FM, Goldfine ID, Evans JL, Buonaguro FM, Mancini A. Role of gut microbiota and oxidative stress in the progression of non-alcoholic fatty liver disease to hepatocarcinoma: current and innovative therapeutic approaches. Redox Biol. 2018;15:467–479. doi: 10.1016/j.redox.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko KR, Reinus J. Extrahepatic complications of nonalcoholic fatty liver disease. Clin Liver Dis. 2016;20:387–401. doi: 10.1016/j.cld.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Chan YK, El-Nezami H, Chen Y, Kinnunen K, Kirjavainen PV. Probiotic mixture VSL#3 reduce high fat diet induced vascular inflammation and atherosclerosis in ApoE(−/−) mice. AMB Express. 2016;61:61. doi: 10.1186/s13568-016-0229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa G, Sahebkar A, Maffioli P. The role of various peroxisome proliferator-activated receptors and their ligands in clinical practice. J Cell Physiol. 2017;233:153–161. doi: 10.1002/jcp.25804. [DOI] [PubMed] [Google Scholar]
- Ferolla SM, Armiliato GN, Couto CA, Ferrari TC. Probiotics as a complementary therapeutic approach in nonalcoholic fatty liver disease. World J Hepatol. 2015;7:559–565. doi: 10.4254/wjh.v7.i3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol. 2012;19:1222–1236. doi: 10.1016/j.chembiol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto E, Tokushige K. Prevalence, gender, ethnic variations, and prognosis of NASH. J Gastroenterol. 2011;46(Suppl 1):63–69. doi: 10.1007/s00535-010-0311-8. [DOI] [PubMed] [Google Scholar]
- Ip E, Farrell GC, Robertson G, Hall P, Kirsch R, Leclercq I. Central role of PPAR alpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology. 2003;38:123–132. doi: 10.1053/jhep.2003.50307. [DOI] [PubMed] [Google Scholar]
- Kim DH, Jeong D, Kang IB, Kim H, Song KY, Seo KH. Dual function of Lactobacillus kefiri DH5 in preventing high-fat-diet-induced obesity: direct reduction of cholesterol and upregulation of PPAR-α in adipose tissue. Mol Nutr Food Res. 2017;61:1700252. doi: 10.1002/mnfr.201700252. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Hou X, Jiang B, Luo Z, Cohen RA. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Fan C, Li P, Lu Y, Chang X, Qi K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci Rep. 2016;6:37589. doi: 10.1038/srep37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddur H, Neuschwander-Tetri BA. More evidence that probiotics may have a role in treating fatty liver disease. Am J Clin Nutr. 2014;99:425–426. doi: 10.3945/ajcn.113.082636. [DOI] [PubMed] [Google Scholar]
- Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, Mastrojeni S, Malaguarnera G, Mistretta A, Li Volti G, Galvano F. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012;57:545–553. doi: 10.1007/s10620-011-1887-4. [DOI] [PubMed] [Google Scholar]
- Mei L, Tang Y, Li M, Yang P, Liu Z, Yuan J, Zheng P. Co-Administration of Cholesterol-Lowering Probiotics and Anthraquinone from Cassia obtusifolia L. Ameliorate Non-Alcoholic Fatty Liver. PLoS ONE. 2015;10:e0138078. doi: 10.1371/journal.pone.0138078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mridha AR, Haczeyni F, Yeh MM, Haigh WG, Ioannou GN, Barn V, Ajamieh H, Adams L, Hamdorf JM, Teoh NC, Farrell GC. TLR9 is upregulated in human and murine NASH: pivotal role for inflammatory recruitment and cell survival. Clin Sci. 2017;131:2145–2159. doi: 10.1042/CS20160838. [DOI] [PubMed] [Google Scholar]
- Mridha AR, Van Rooyen DM, Haczeyni F, Teoh NC. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037–1046. doi: 10.1016/j.jhep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M. Non-alcoholic fatty liver disease from pathogenesis to management: an update. Obes Rev. 2010;11:430–445. doi: 10.1111/j.1467-789X.2009.00657.x. [DOI] [PubMed] [Google Scholar]
- Nobili V, Putignani L, Mosca A, Chierico FD, Vernocchi P, Alisi A, Stronati L, Cucchiara S, Toscano M, Drago L. Bifidobacteria and lactobacilli in the gut microbiome of children with non-alcoholic fatty liver disease: which strains act as health players? Arch Med Sci. 2018;14:81–87. doi: 10.5114/aoms.2016.62150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratziu V, Bellentani S, Cortez-Pinto H, Day C, Marchesini G. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol. 2010;53:372–384. doi: 10.1016/j.jhep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Ren T, Huang C, Cheng M. Dietary blueberry and Bifidobacteria attenuate nonalcoholic fatty liver disease in rats by affecting SIRT1-mediated signaling pathway. Oxid Med Cell Longev. 2014;2014:469059. doi: 10.1155/2014/469059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritze Y, Bárdos G, Claus A, Ehrmann V, Bergheim I, Schwiertz A, Bischoff SC. Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS ONE. 2014;9:e80169. doi: 10.1371/journal.pone.0080169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossato M, Di Vincenzo A, Pagano C, El Hadi H, Vettor R. The P2X7 receptor and NLRP3 axis in non-alcoholic fatty liver disease: a brief review. Cells. 2020 doi: 10.3390/cells9041047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell Mol Life Sci. 2019;76:1541–1558. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X, Che H, Ji Q, Yang F, Lv J, Wang Y, Xian H, Wang L. The relationship between liver enzymes and insulin resistance in type 2 diabetes patients with nonalcoholic fatty liver disease. Horm Metab Res. 2018;50:397–402. doi: 10.1055/a-0603-7899. [DOI] [PubMed] [Google Scholar]
- Shouhed D, Steggerda J, Burch M, Noureddin M. The role of bariatric surgery in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol. 2017;11:797–811. doi: 10.1080/17474124.2017.1355731. [DOI] [PubMed] [Google Scholar]
- Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, Delerive P, Fadel A, Chinetti G, Fruchart JC, Najib J, Maclouf J, Tedgui A. Activation of human aortic smooth-muscle cells is inhibited by PPARα but not by PPARγ activators. Nature. 1998;393:790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- Sun Z, Chen X, Wang J, Gao P, Zhou Z, Ren Y, Sun T, Wang L, Meng H, Chen W, Zhang H. Complete genome sequence of probiotic Bifidobacterium animalis subsp. lactis strain V9. J Bacteriol. 2010;192:4080–4081. doi: 10.1128/JB.00369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Bala S, Petrasek J, Gattu A. Gut-liver axis and sensing microbes. Dig Dis. 2011;28:737–744. doi: 10.1159/000324281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Han SI, Murayama Y, Satoh A, Oikawa F, Ohno H, Osaki Y, Matsuzaka T, Sekiya M, Iwasaki H, Yatoh S. Selective peroxisome proliferator-activated receptor-α modulator K-877 efficiently activates the peroxisome proliferator-activated receptor-α pathway and improves lipid metabolism in mice. J Diabetes Investig. 2017;8:446–452. doi: 10.1111/jdi.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Aoyama T, Kimura S, Gonzalez FJ. Targeting nuclear receptors for the treatment of fatty liver disease. Pharmacol Ther. 2017;179:142–157. doi: 10.1016/j.pharmthera.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HK, Ng YK, Koh E, Yao L, Chien AS, Lin HX, Lee YK. RNA-Seq reveals transcriptomic interactions of Bacillus subtilis natto and Bifidobacterium animalis subsp. lactis in whole soybean solid-state co-fermentation. Food Microbiol. 2015;51:25–32. doi: 10.1016/j.fm.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Xiao J, Liu Y, Xing F, Leung TM, Liong EC, Tipoe GL. Bee’s honey attenuates non-alcoholic steatohepatitis-induced hepatic injury through the regulation of thioredoxin-interacting protein-NLRP3 inflammasome pathway. Eur J Nutr. 2016;55:1465–1477. doi: 10.1007/s00394-015-0964-4. [DOI] [PubMed] [Google Scholar]
- Xu RY, Wan YP, Fang QY, Lu W, Cai W. Supplementation with probiotics modifies gut flora and attenuates liver fat accumulation in rat nonalcoholic fatty liver disease model. J Clin Biochem Nutr. 2012;50:72–77. doi: 10.3164/jcbn.11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Li FY, Lam KS, Li H, Jia W, Wang Y, Man K, Lo CM, Li X, Xu A. Toll-like receptor-4 mediates obesity-induced non-alcoholic steatohepatitis through activation of X-box binding protein-1 in mice. Gut. 2012;61:1058. doi: 10.1136/gutjnl-2011-300269. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wang C, Zhang L, Zhao Y, Duan C, Zhang X, Gao L, Li S. Lactobacillus plantarum NA136 improves the non-alcoholic fatty liver disease by modulating the AMPK/Nrf2 pathway. Appl Microbiol Biotechnol. 2019 doi: 10.1007/s00253-019-09703-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The specific primer sequences of genes of interest. Figure S1. Western blot analysis of hepatic TLR4 and TLR9. The representative picture is one from three independent experiments.