Summary
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) has caused a huge number of human deaths. Currently, there are no specific drugs or vaccines available for this virus (SARS-CoV-2). The viral polymerase is a promising antiviral target. Here, we describe the near-atomic-resolution structure of the SARS-CoV-2 polymerase complex consisting of the nsp12 catalytic subunit and nsp7-nsp8 cofactors. This structure highly resembles the counterpart of SARS-CoV with conserved motifs for all viral RNA-dependent RNA polymerases and suggests a mechanism of activation by cofactors. Biochemical studies reveal reduced activity of the core polymerase complex and lower thermostability of individual subunits of SARS-CoV-2 compared with SARS-CoV. These findings provide important insights into RNA synthesis by coronavirus polymerase and indicate adaptation of SARS-CoV-2 toward humans with a relatively lower body temperature than the natural bat hosts.
Keywords: SARS-CoV-2, non-structural proteins, polymerase, cofactors, RNA synthesis, cryo-EM
Graphical Abstract

Highlights
-
•
Cryo-EM structure of SARS-CoV-2 nsp12-nsp7-nsp8 core polymerase complex
-
•
The core complex of SARS-CoV-2 has lower enzymatic activity than SARS-CoV
-
•
SARS-CoV-2 nsp7-8-12 subunits are less thermostable than the SARS-CoV counterpart
Viral polymerase plays a central role in the virus life cycle and is an important antiviral drug target. Peng et al. report the cryo-EM structure of the SARS-CoV-2 core polymerase complex, finding that it has less efficient activity for RNA synthesis and lower thermostability of individual subunits compared with SARS-CoV.
Introduction
At the end of 2019, a novel coronavirus (2019-nCoV) caused an outbreak of pulmonary disease (Zhu et al., 2020) that was later officially named severe acute respiratory syndrome virus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). The pneumonia disease was named coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO). The outbreak has developed into a global pandemic affecting most countries in the world (Holshue et al., 2020, Kim et al., 2020, Shi, 2020). As of May 17, 2020, more than 4,700,000 human infections have been reported worldwide, including over 300,000 deaths (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The origin of this virus has not been identified, but multiple origins possibly exist, based on recent bioinformatics analyses of viral isolates from different countries (Andersen et al., 2020, Zhang and Holmes, 2020). So far, there are no specific drugs or vaccines available, which poses a great challenge for the treatment and control of related diseases.
SARS-CoV-2 belongs to the family of Coronaviridae, a group of positive-sense RNA viruses with a broad host spectrum (Vicenzi et al., 2004). Currently, seven human-infecting coronaviruses have been identified, among which SARS-CoV-2 displays the highest similarity in genome sequence to the SARS-CoV emerging in 2002–2003 (Zhong et al., 2003, Zhou et al., 2020). Both viruses utilize the same host receptor, angiotensin-converting enzyme 2 (ACE2), for cell entry and cause respiratory symptoms that may progress to severe pneumonia and lead to death (Lu et al., 2020, Zhou et al., 2020). However, compared with SARS-CoV, SARS-CoV-2 has a much higher transmission rate and lower mortality (Huang et al., 2020, Wang et al., 2020). Most infections result in mild symptoms, and a substantial number of asymptomatic infection cases have also been reported (Rothe et al., 2020). These properties allow SARS-CoV-2 to be transmitted among humans furtively, facilitating quantum leaps of pandemic expansion. Characterizing the infection and replication behavior of SARS-CoV-2 would provide critical information for understanding its unique pathogenesis and host adaption properties.
The replication of coronavirus is operated by a set of non-structural proteins (nsps) encoded by open reading frame 1a (ORF1a) and ORF1ab in its genome, which are initially translated as polyproteins, followed by proteolysis cleavage for maturation (Ziebuhr, 2005). These proteins assemble into a multi-subunit polymerase complex to mediate transcription and replication of the viral genome. Among them, nsp12 is the catalytic subunit with RNA-dependent RNA polymerase (RdRp) activity (Ahn et al., 2012). nsp12 itself is capable of conducting the polymerase reaction with extremely low efficiency, whereas the presence of nsp7 and nsp8 cofactors remarkably stimulates its polymerase activity (Subissi et al., 2014). The nsp12-nsp7-nsp8 subcomplex is thus defined as the minimal core component for mediating coronavirus RNA synthesis. To achieve complete transcription and replication of the viral genome, several other nsp subunits are required to assemble into a holoenzyme complex, including nsp10, nsp13, nsp14 and nsp16, for which the precise functions in RNA synthesis are not well understood (Adedeji et al., 2012, Lehmann et al., 2015, Sevajol et al., 2014, Ziebuhr, 2005). The viral polymerase has shown enormous promise as a highly potent antiviral drug target because of its higher evolutionary stability compared with the surface proteins, which are more prone to drift as a result of selection by host immunity (Shi et al., 2013). Therefore, understanding the structure and function of the SARS-CoV-2 polymerase complex is an essential prerequisite for developing novel therapeutic agents.
In this work, we determined the near-atomic-resolution structure of the SARS-CoV-2 nsp12-nsp7-nsp8 core polymerase complex by cryoelectron microscopy (cryo-EM) reconstruction and revealed its reduced polymerase activity and thermostability compared with the SARS-CoV counterpart. These findings improve our understanding of coronavirus replication and evolution which might indicate clues for the fitness of SARS-CoV-2 to human hosts.
Results
Overall Structure of the SARS-CoV-2 Core Polymerase Complex
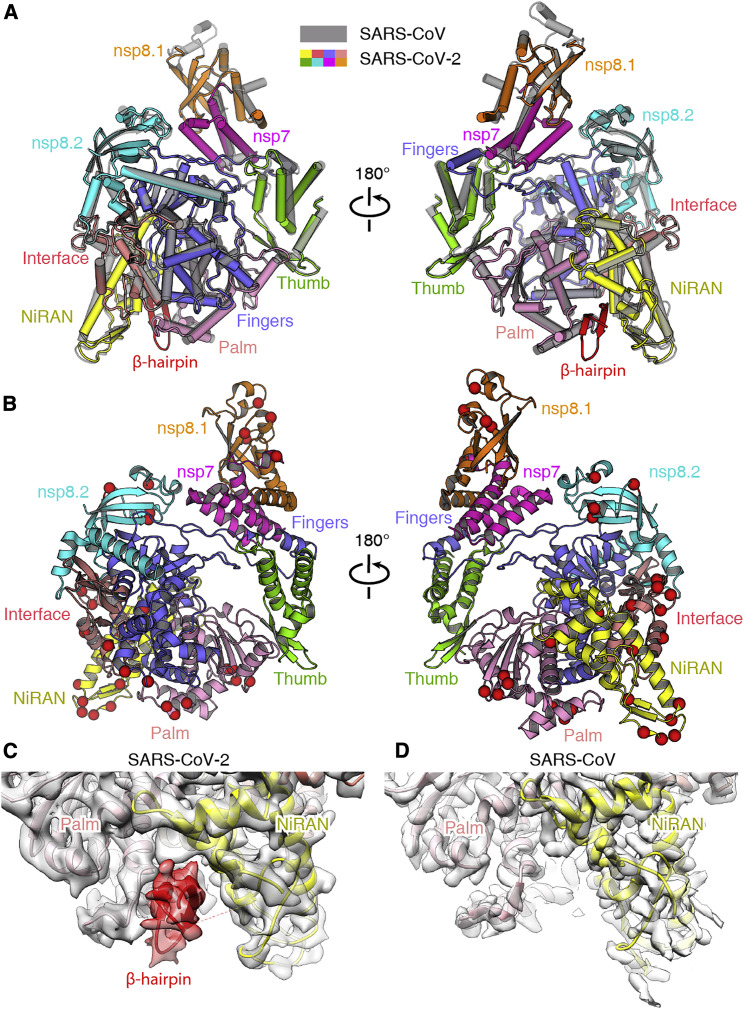
The SARS-CoV-2 nsp12 polymerase and nsp7-nsp8 cofactors were expressed using the baculovirus and Escherichia coli (E. coli) expression systems, respectively. The three protein subunits were mixed in vitro to constitute the core polymerase complex (Figure S1). The structure of the SARS-CoV-2 nsp12-nsp7-nsp8 complex was determined at 3.7-Å resolution, which clearly resolved the main-chain trace and the most bulky side chains of each subunit (Figures S2 and S3). Similar to the counterpart complex of SARS-CoV (Kirchdoerfer and Ward, 2019), the N-terminal ∼110 amino acids of nsp12 as well as the N-terminal ∼80 residues of nsp8 and a small portion of the nsp7 C-terminus could not be resolved in the density map (Figure 1 A). In total, approximately 80% of the 160-kDa complex was interpreted in the structure.
Figure 1.
Overall Structure of the SARS-CoV-2 Core Polymerase Complex
(A) Schematic diagram of the domain architecture for each subunit of the polymerase complex. Each domain is represented by a unique color. The unresolved region is colored in gray.
(B) Overall density map (left) and atomic model (right) of the SARS-CoV-2 nsp12-nsp7-nsp8 core complex in different views. The map and structural model are colored by domains with the same color code as in (A). The fingertip loop (one of the key catalytic motifs) is indicated by a red arrowhead, and the associated finger extension loops are indicated by a black arrow.
(C) The structure of the nsp12 polymerase subunit in different views, colored by domains with the same scheme as in (A).
See also Figures S1–S3 and Table S1.
The SARS-CoV-2 polymerase complex consists of a nsp12 core catalytic subunit bound with a nsp7-nsp8 heterodimer and an additional nsp8 subunit at a different binding site (Figure 1B). The N-terminal portion of the nsp12 polymerase subunit contains a nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain that is shared by all members of the Nidovirales order (Lehmann et al., 2015). This domain binds at the back side of the right-hand-configured C-terminal RdRp. Between them is an interface domain that links the NiRAN domain to the finger subdomain of RdRp (Figures 1B and 1C). The NiRAN and interface domains represent additional features of coronavirus RdRp compared with the polymerase subunit of flaviviruses, which is also a group of positive-sense RNA viruses (Duan et al., 2017, Godoy et al., 2017, Zhao et al., 2017). The C-terminal catalytic domain adopts a conserved architecture of all viral RdRps, composed of the finger, palm, and thumb subdomains (Figure 1C). A remarkable feature of the coronavirus RdRp is the long finger extension that intersects with the thumb subdomain to form a closed-ring structure (Figure 1C), in contrast to the smaller loop in segmented negative-sense RNA virus (sNSV) polymerases that results in a relatively open conformation, such as influenza virus, bunyavirus, and arenavirus polymerases (Figure S4; Gerlach et al., 2015, Peng et al., 2020, Pflug et al., 2014). Similar close contact between the fingers and thumb subdomains is also observed in the structures of poliovirus (PV) and Zika virus (ZIKV) polymerases (Figure S4; Godoy et al., 2017, Gong and Peersen, 2010), which might be a common feature of positive-sense RNA virus polymerases.
Structural Comparison of the SARS-CoV-2 and SARS-CoV nsp12-nsp7-nsp8 Complexes
Basically, the structure of the SARS-CoV-2 polymerase complex highly resembles that of SARS-CoV, with a global root-mean-square deviation (RMSD) of ∼1 Å for the α-carbon atoms (Figure 2 A). There are 1, 4, and 25 residue substitutions between the two viruses in the structurally visualized regions of the nsp7, nsp8, and nsp12 subunits, respectively (Figure 2B) (there are 1 and 7 additional substitutions in the unresolved regions of the nsp8 and nsp12 subunits, respectively). However, these variations did not result in obvious structural changes in the polymerase complex. During the review process of this manuscript, two other research groups also reported the high-resolution structure of the SARS-CoV-2 core polymerase complex, which revealed structural features similar to the SARS-CoV counterpart, consistent with our observations (Gao et al., 2020, Yin et al., 2020). All three structures reported by our group and others revealed similar structural features and allowed identification of a previously undefined β-hairpin motif in the N terminus of the nsp12 subunit that binds at the interface between the NiRAN domain and the palm subdomain of RdRp (Figure 2C). However, no extra density for this region was observed in the reconstruction of the SARS-CoV polymerase complex (Figure 2D), suggesting a different conformation or flexibility of this motif between the two viruses. Of note, in the structure determined by Gao et al. (2020), the N-terminal residues 51–117 of the nsp12 subunit are clearly resolved to constitute an almost complete NiRAN domain (Gao et al., 2020). In contrast, this region reveals poor density in the reconstructions by our group and Yin et al. (2020), suggesting moderate flexibility of this region, but the functional relevance remains elusive.
Figure 2.
Comparison of the SARS-CoV-2 and SARS-CoV Core Polymerase Complex Structures
(A) Overlay of the nsp12-nsp7-nsp8 complexes of SARS-CoV-2 (colored by domains) and SARS-CoV (gray). The two structures could be well superimposed with high similarity.
(B) Residue substitutions between the SARS-CoV-2 and SARS-CoV core polymerase complexes. The structural model is shown as cartoons and colored by domains. The substitution sites are represented by red spheres to highlight their locations.
(C and D) Comparison of nsp12 N-terminal densities in the SARS-CoV-2 (C) and SARS-CoV (D) polymerase complexes. The density for the newly identified N-terminal β-hairpin of SARS-CoV-2 nsp12 subunit is highlighted in red. No corresponding density was observed in the reconstruction of the SARS-CoV polymerase complex.
See also Figure S4.
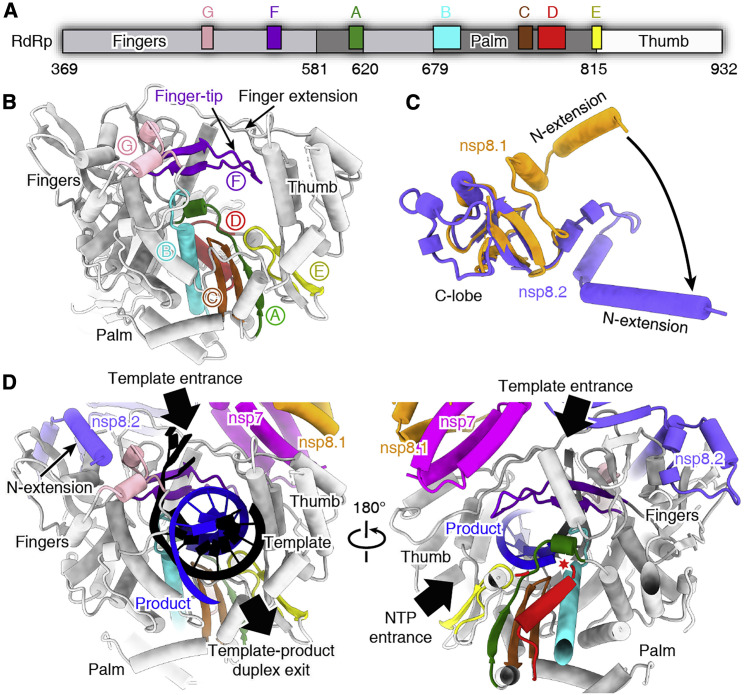
Conserved Catalytic Center of nsp12 and Interaction with Cofactors
The catalytic domain of the SARS-CoV-2 nsp12 subunit is arranged following the typical right-hand configuration shared by all viral RdRps, which includes seven critical catalytic motifs (A–G) (Figure 3 ). Among them, motifs A–F are highly conserved for all viral RdRps, and motif G is defined as a hallmark of primer-dependent RdRp in some positive-sense RNA viruses and interacts with the primer strand to initiate RNA synthesis (Figure S4). Motif C contains the critical 759-SDD-761 catalytic residues, which reside in a β-turn loop connecting two adjacent strands. Motif F forms a fingertip that protrudes into the catalytic chamber and interacts with the finger extension loops and the thumb subdomain (Figure 3B). It has been shown that some sNSV polymerases, e.g., influenza virus and bunyavirus polymerases, require binding of a conserved 5′ RNA hook to activate the activity for RNA synthesis by stabilizing the fingertip, which is otherwise highly flexible in the apo form (Gerlach et al., 2015, Hengrung et al., 2015, Peng et al., 2020, Pflug et al., 2014, Reich et al., 2014). In the structure of coronavirus polymerase, this fingertip loop is stabilized by the adjacent finger extension loops, which are secured by interactions with the nsp7-nsp8 heterodimer (Figures 1B and 1C). In the absence of this heterodimer, the finger extension loops displayed significant flexibility, as observed in the structure of the SARS-CoV nsp12-nsp8 subcomplex, which would destabilize the fingertip motif (Figures S4G and S4H; Kirchdoerfer and Ward, 2019). This evidence is consistent with the observation that nsp12 alone shows extremely weak activity for nucleotide polymerization, whereas this activity is remarkably stimulated upon binding of the nsp7-nsp8 cofactors (Ahn et al., 2012, Subissi et al., 2014).
Figure 3.
Catalytic Center of nsp12 RdRp and the Interactions with nsp7-nsp8 Cofactors
(A) Schematic diagram of the domain organization of the SARS-CoV-2 nsp12 RdRp region. The RdRp domain consists of fingers, palm, and thumb subdomains. The seven conserved catalytic motifs are indicated by different colors in corresponding locations.
(B) Atomic structure of nsp12, shown as cartoons, with the seven catalytic motifs colored differently as in (A).
(C) Structural comparison of the two nsp8 subunits in the complex. The C-terminal lobe could be well superimposed, whereas the N-terminal extension helix shows different conformations.
(D) Structural model of RNA synthesis by the coronavirus polymerase complex. The template (black) and product (blue) strands are modeled based on the elongation complex of PV polymerase (PDB: 3OL8). The template and NTP entrance and the template-product duplex exit tunnels are indicated by black arrows. The conserved RdRp catalytic motifs are colored with the same scheme as in (A). The catalytic site is highlighted with a red star.
See also Figures S4 and S5.
The nsp7-nsp8 heterodimer binds above the thumb subdomain of RdRp and sandwiches the finger extension loops in between to stabilize its conformation (Figure 1B). This interaction is mainly mediated by nsp7 within the heterodimer, whereas nsp8 (nsp8.1) contributes few contacts to the nsp12 polymerase subunit (Figures 1B and 3D). The other nsp8 (nsp8.2) subunit clamps the top region of the finger subdomain and forms additional interactions with the interface domain (Figures 1B and 3D). The two nsp8 subunits display significantly different conformations with substantial refolding of the N-terminal extension helix region that mutually preclude binding at the other molecular context (Figure 3C). The importance of both cofactor-binding sites has been validated by previous biochemical studies of SARS-CoV polymerase, which revealed their essential roles in stimulating activity of the nsp12 polymerase subunit (Subissi et al., 2014).
Based on the elongation complex of PV polymerase (Gong and Peersen, 2010), we modeled the RNA template and product strands into the catalytic chamber of the SARS-CoV-2 nsp12 subunit. This pseudo-elongation intermediate structure reveals that the template entrance is supported by the finger extension loops and the fingertip to guide the 3′ viral RNA (vRNA) to the catalytic chamber (Figure 3D). The nucleotide triphosphate (NTP) substrate enters through a channel at the back side of the palm subdomain to reach the active site. The template and product stands form a duplex to exit the polymerase chamber in the front (Figure 3D). Because the viral genome and sub-genomic mRNA products are functional in single-stranded form, it requires further steps assisted by other nsp subunits to separate the duplex for complete transcription and replication processes.
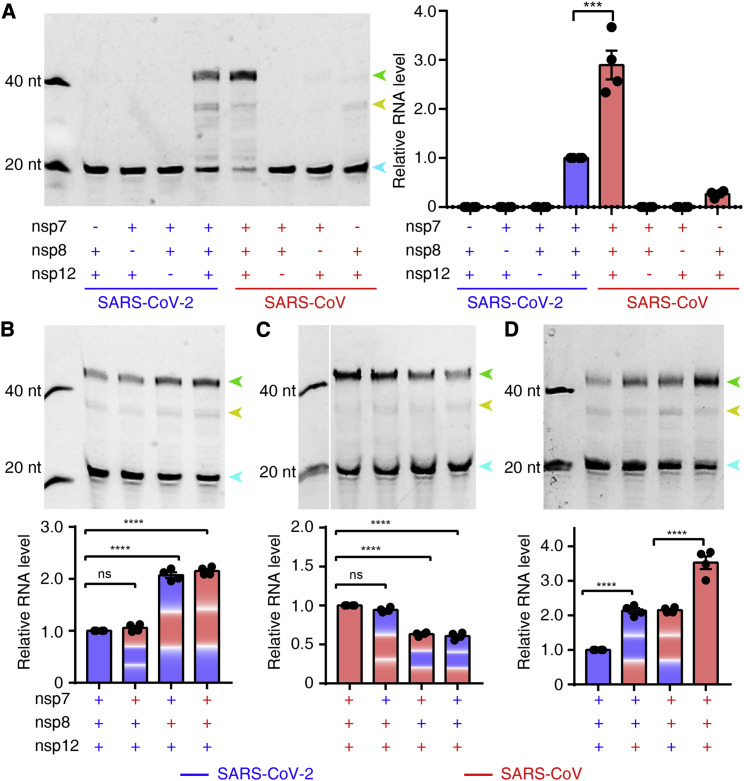
Reduced Activity of the SARS-CoV-2 Core Polymerase Complex
Given the residue substitutions between SARS-CoV-2 and SARS-CoV polymerase subunits but the high degree of overall sequence similarity, we compared the enzymatic behaviors of the viral polymerases, aiming to analyze their properties in terms of viral replication. Both sets of core polymerase complex could well mediate primer-dependent RNA elongation reactions templated by the 3′ vRNA. Intriguingly, the SARS-CoV-2 nsp12-nsp7-nsp8 complex displayed a much lower efficiency (∼35%) for RNA synthesis compared with the SARS-CoV counterpart (Figure 4 A). Because all three nsp subunits harbor some residue substitutions between the two viruses, we further conducted cross-combination analysis to evaluate the effects of each subunit on the efficiency of RNA production. In the context of the SARS-CoV-2 nsp12 polymerase subunit, replacement of the nsp7 cofactor subunit with that of SARS-CoV did not result in an obvious effect on polymerase activity, whereas introduction of the SARS-CoV nsp8 subunit greatly boosted the activity to ∼2.1 times of that of the homologous combination. Simultaneous replacement of the nsp7 and nsp8 cofactors further enhanced the efficiency of RNA synthesis to ∼2.2 times of that for the SARS-CoV-2 homologous complex (Figure 4B). Consistent with this observation, combination of the SARS-CoV-2 nsp7-nsp8 subunits with the SARS-CoV nsp12 polymerase subunit compromised its activity compared with the native cognate cofactors, among which the nsp8 subunit exhibited a more obvious effect than nsp7 (Figure 4C). This evidence suggested that the variations in the nsp8 subunit had a significant negative effect on the polymerase activity of SARS-CoV-2 nsp12. The non-significant effect of nsp7 on polymerase activity was quite conceivable because only one residue substitution occurred between the two viruses (Figure 2B). In addition, we also compared the polymerase activity of different nsp12 subunits in the same context of nsp7-nsp8 cofactors. Combined with either cofactor set, the SARS-CoV-2 nsp12 polymerase showed a lower efficiency (∼50%) for RNA synthesis compared with the SARS-CoV counterpart (Figure 4D). This observation demonstrated that the residue substitutions in nsp12 also contributed to the reduction of its polymerase activity, with a similar effect on the variations in the nsp8 cofactor.
Figure 4.
In Vitro Polymerase Activity of nsp12 and Regulatory Effects of Cofactors
(A) Comparison of RNA synthesis activities of the SARS-CoV-2 and SARS-CoV core polymerase complex. Efficient activity of nsp12 polymerase requires the presence of the nsp7 and nsp8 cofactors. Apart from the fully elongated product (green arrowhead), some aberrant termination products were also observed (yellow arrowhead). The excess primer band is indicated by a cyan arrowhead.
(B and C) Comparison of the regulatory effects of the nsp7 and nsp8 cofactors in the context of the SARS-CoV-2 (B) and SARS-CoV (C) nsp12 polymerase, respectively.
(D) Comparison of the activity of the nsp12 polymerase of different viruses in the same context of cofactors. Polymerase activity was quantified by integrating the intensity of the fully elongated product bands. The results, in histograms, are presented as the means ± SD, where error bars represent SDs. The significance of difference was tested by one-way ANOVA based on the results of four independent experiments (n = 4) using different protein preparations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S4.
Effects of Amino Acid Substitutions on the Core Polymerase Subunits
Despite the amino acid substitutions in all three subunits of the core polymerase complex between SARS-CoV-2 and SARS-CoV, none of these residues is located at the polymerase active site or the contacting interfaces between adjacent subunits (Figure 2B), suggesting that these substitutions do not affect the inter-subunit interactions for assembly of the polymerase complex. To test this hypothesis, we measured the binding kinetics between different subunits of the two viruses by surface plasmon resonance (SPR) assays. Each interaction pair exhibited similar kinetic features for the two viruses, all with sub-micromolar-range affinities (Figures 5A and 5B). We also tested cross-binding between subunits of the two viruses, which revealed similar affinities for heterologous pairs compared with the native homologous interactions (Figures 5C and 5D).
Figure 5.
SPR Binding Kinetics of Protein Pairs in the Polymerase Complex of SARS-CoV-2 and SARS-CoV
(A and B) The binding profiles of the homologous protein subunits of SARS-CoV-2 (A) and SARS-CoV (B), respectively.
(C and D) The cross-binding kinetics between protein subunits of SARS-CoV-2 and SARS-CoV. All analytes were measured with serially diluted concentrations as shown in (A).
The title is presented as "analyte/immobilized ligand" to facilitate comparison. The binding between nsp12 and the nsp7L8 fusion protein was fitted with the heterogeneous binding mode because the nsp7-nsp8 heterodimer exhibited non-uniform conformations in solution (Zhai et al., 2005). It can occupy either cofactor binding site as a stable nsp7-nsp8 complex or free nsp8 when nsp7 detaches from the heterodimer. Both equilibrium binding constant values (KD1 and KD2) were calculated in this mode. The data shown are a representative result of three independent experiments using different protein preparations, all of which produced similar results. See also Figure S4.
We then compared the thermostability of each component in the polymerase complex of the two viruses (Figure 6 ). Consistent with the almost identical sequences, nsp7 of both viruses displayed comparable melting behaviors in the circular dichroism (CD) profiles, demonstrating similar thermostabilities of the two proteins (Figures 6A and 6D). In contrast, both the nsp8 and nsp12 subunits of SARS-CoV-2 showed lower melting temperature (Tm) values compared with the corresponding subunits of SARS-CoV, suggesting poorer thermostability of SARS-CoV-2 proteins (Figure 6B, 6C, 6E, and 6F).
Figure 6.
CD Profiles of Individual Subunits in the Core Polymerase Complex
(A–F) The multi-wavelength (215–260 nm) CD spectra of the SARS-CoV-2 (A–C) and SARS-CoV (D–F) polymerase components at different temperatures. The Tm values were calculated to evaluate the thermostability of each protein subunit. The data shown are representative results of two independent experiments using different protein preparations. See also Figure S4.
Taken together, the residue substitutions in the SARS-CoV-2 nsp12 polymerase subunit and nsp7-nsp8 cofactors compromise the efficiency of RNA synthesis by the core polymerase complex and reduce the thermostability of individual protein subunits compared with the counterparts of SARS-CoV. These changes may indicate some clues regarding the adaptive evolution of SARS-CoV-2 in favor of human hosts. It has been shown that humans have a relatively lower body temperature than bats, which are potentially the natural host of a panel of zoonotic viruses, including SARS-CoV and SARS-CoV-2 (O’Shea et al., 2014, Zhou et al., 2020).
Discussion
The structural information of coronavirus polymerase interaction with cofactors suggests a common theme of viral RdRp activation despite being executed by different structural components. The coronavirus polymerase subunit requires multiple cofactors or subunits for complete transcription and replication functions, similar to the related flaviviruses, which also harbor a positive-sense RNA genome (Aktepe and Mackenzie, 2018, Sevajol et al., 2014, Ziebuhr, 2005). In contrast, sNSVs utilize fewer multi-subunit or multi-domain protein components to accomplish similar processes, which could be activated by RNA segments instead of proteins (Gerlach et al., 2015, Peng et al., 2020, Pflug et al., 2014). As revealed by the coronavirus core polymerase structures, it lacks the essential component for unwinding the template-product hybrid, which is required to release the single-stranded mRNA and viral genome for protein expression and virion assembly. In the structure of sNSV polymerases, a lid domain is present at the intersection region of template and product exit tunnels to force duplex deformation before leaving the polymerase chamber (Gerlach et al., 2015, Peng et al., 2020, Reich et al., 2014). The nsp13 subunit has been shown to have RNA helicase activity, suggesting its involvement in RNA synthesis at the post-catalytic stage (Adedeji et al., 2012). Further investigations are required to understand how this process takes place.
Of note, we demonstrate that the amino acid substitutions in the polymerase and cofactors of SARS-CoV-2 lead to obviously reduced activity for RNA synthesis compared with the SARS-CoV core polymerase complex. Indeed, these observations are based on partial components of the multiple-subunit holoenzyme for coronavirus replication, which also involves proofreading and capping by other nsp subunits; e.g., the nsp10-nsp14 exonuclease subcomplex, nsp13 RNA 5′-triphosphatase, and the nsp14 and nsp16 methyltransferases (Sevajol et al., 2014, Ziebuhr, 2005). Among them, nsp14 may directly interact with nsp12 to assemble a processive elongating complex and ensure the fidelity of RNA synthesis (Subissi et al., 2014). Moreover, nsp16 may also be involved in formation of a stable holoenzyme, which, together with nsp10 and nsp14, constitutes the capping modules in transcription (Chen et al., 2011, Sevajol et al., 2014). These steps would also render important determinants for the efficiency and accuracy of RNA synthesis by the coronavirus replication machinery. Thus, the collective behavior of SARS-CoV-2 polymerase complex in the context of an authentic viral replication cycle still remains an open question to be explored further. On the other hand, the lower thermostability of SARS-CoV-2 polymerase subunits might indicate its adaptation to humans, who have a relatively lower body temperature compared with bats, the potential natural hosts of SARS-CoV-2 (O’Shea et al., 2014, Zhou et al., 2020). Interestingly, we also found that the closely related bat coronavirus RaTG13 showed an extremely high sequence identity of core polymerase subunits to SARS-CoV-2, in which the nsp7 and nsp8 cofactors are strictly identical and the nsp12 catalytic subunit harbors only four residue replacements between the two viruses (Figure S5), suggesting similar enzymatic properties and thermostabilities of their polymerase components. This observation indicates that the RaTG13 coronavirus may have been well adapted to human hosts in terms of viral replication machinery and would further support the probable bat origin of SARS-CoV-2 (Zhou et al., 2020).
In summary, our structural and biochemical analyses of the SARS-CoV-2 core polymerase complex improve our understanding of the mechanisms of RNA synthesis by different viral RdRps and highlight a common theme for polymerase activation by stabilizing critical catalytic motifs via diverse means. In addition, the different biochemical properties of the polymerase components of SARS-CoV-2 and SARS-CoV suggest clues for adaptive evolution of coronaviruses in favor of human hosts.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli: BL21 (DE3) competent cells | Thermo Fisher | Cat.# EC0114 |
| E. coli: DH10Bac competent cells | Invitrogen | Cat.# 10361-012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Insect-XPRESS™ Protein-free Insect Cell Medium | Lonza | Cat.# 12-730Q |
| FuGENE 6 Transfection Reagent | Promega | Cat.# E2691 |
| TCEP | Sigma | Cat.# C4706 |
| ATP | Takara | Cat.# 4041 |
| CTP | Takara | Cat.# 4043 |
| UTP | Takara | Cat.# 4044 |
| GTP | Takara | Cat.# 4042 |
| IPTG | Sigma | Cat.# 6758 |
| nsp7 subunit from SARS-CoV-2 | This study | N/A |
| nsp8 subunit from SARS-CoV-2 | This study | N/A |
| nsp12 subunit from SARS-CoV-2 | This study | N/A |
| nsp7L8 from SARS-CoV-2 | This study | N/A |
| nsp7 subunit from SARS-CoV | This study | N/A |
| nsp8 subunit from SARS-CoV | This study | N/A |
| nsp12 subunit from SARS-CoV | This study | N/A |
| nsp7L8 from SARS-CoV | This study | N/A |
| Deposited Data | ||
| Cryo-EM map and model of nsp12 bound to nsp7 and nsp8 from SARS-CoV-2 | This study | 7BW4 (PDB); 30226(EMDB) |
| Experimental Models: Cell Lines | ||
| High Five cells | Invitrogen | Cat.# B85502 |
| SF9 cells | Invitrogen | Cat.# 11496015 |
| Oligonucleotides | ||
| 40-nt Template RNA strand CUAUCCCCAUGUGAUUUUAAUAGCUUCUUAGG AGAAUGAC |
Takara | N/A |
| 20-nt Primer RNA strand . FAM-GUCAUUCUCCUAAGAAGCUA | Takara | N/A |
| Recombinant DNA | ||
| Expression plasmid for nsp7 of SARS-CoV-2 | This study | N/A |
| Expression plasmid for nsp8 of SARS-CoV-2 | This study | N/A |
| Expression plasmid for nsp7L8 of SARS-CoV-2 | This study | N/A |
| Expression plasmid for nsp12 of SARS-CoV-2 | This study | N/A |
| Expression plasmid for nsp7 of SARS-CoV | This study | N/A |
| Expression plasmid for nsp8 of SARS-CoV | This study | N/A |
| Expression plasmid for nsp7L8 of SARS-CoV | This study | N/A |
| Expression plasmid for nsp12 of SARS-CoV | This study | N/A |
| Software and Algorithms | ||
| Relion 3.0 | Zivanov et al., 2018 | https://github.com/3dem/relion |
| MotionCor 2.0 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-motioncor2 |
| CTFFIND 4.1 | Rohou and Grigorieff, 2015 | https://grigoriefflab.umassmed.edu/ctffind4 |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu/ |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net/ |
| PyMOL | schrodinger | https://pymol.org/2/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
| IBM SPSS Statistics | IBM | https://www.ibm.com/cn-zh/analytics/spss-statistics-software |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Biacore 8K 481 Evaluation Software (version 1.1.1.7442) | GE Healthcare | https://www.biacore.com/lifesciences/service/downloads/software_and_associated_documents/index.html |
| Global 3 Thermal Global Analysis Software | Applied Photophysics | https://www.photophysics.com/support-and-service/documents-and-software/global-3-thermal-global-analysis-software |
| Other | ||
| HisTrap 5 ml column | GE Healthcare | Cat.# 17524802 |
| StrepTrap 5 ml column | GE Healthcare | Cat.# 28907548 |
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cat.# 28990944 |
| CM5 chips | GE Healthcare | Cat.# 29149603 |
| Amicon Ultra-15 (30 kDa cut-off) | Millipore | Cat.# UFC903008 |
| Amicon Ultra-15 (15 kDa cut-off) | Millipore | Cat.# UFC901008 |
| Quantifoil R1.2/1.3 Cu 300 | Quantifoil | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yi Shi (shiyi@im.ac.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The cryo-EM density map and atomic coordinates have been deposited to the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with the accession codes EMD-30226 and 7BW4, respectively. All other data are available from the authors on reasonable request.
Experimental Model and Subject Details
Escherichia coli BL21 (DE3) strain (EC0114) and MAX Efficiency DH10Bac Competent cells (10361-012) were incubated in LB medium with corresponding antibiotics at 37°C in a shaking incubator (180 rpm). Sf9 (11496015) and High Five (B85502) cells were cultured at 27°C in Insect-XPRESS (12-730Q) with continuous shaking (120-130 rpm).
Method Details
Protein expression and purification
The codon-optimized sequences of nsp7 and nsp8 were synthesized with an N-terminal 6 × histidine tag and inserted into pET21a vector for expression in E. coli (Synbio Tec, Suzhou, China). For the nsp7L8 fusion protein, the sequence was also codon-optimized for E. coli expression system and a 6 × histidine linker was introduced between the nsp7 and nsp8 subunits (Genewiz Tec, Suzhou, China). Protein production was induced with 1 mM isopropylthio-galactoside (IPTG) and incubated for 14-16 hours at 16°C. Bacterial cells were harvested by centrifugations (12,000 rpm, 10 min), resuspended in buffer A (20 mM HEPES, 500 mM NaCl, 2 mM Tris (2-carboxyethyl) phosphine (TCEP), pH 7.5, and lysed by sonication. Cell debris were removed via centrifugation (12,000 rpm, 1 h) and filtration with a 0.22 μm cut-off filter. The supernatant was loaded onto a HisTrap column (GE Healthcare) for initial affinity purification. The target proteins were eluted using buffer A supplemented with 300 mM imidazole. Fractions were pooled and subjected to size-exclusion chromatography (SEC) with a Superdex 200 10/300 increase column (GE Healthcare). The final product was concentrated and stored at −80°C.
For nsp12 proteins, the genes were codon-optimized for Spodoptera frugiperda and incorporated into the pFastBac-1 plasmid with a C-terminal thrombin proteolysis site, a 6 × histidine and two tandem Strep tags. Proteins were expressed with High Five cells at 27°C for 48 h post infection. Cells were collected by centrifugation (3,000 rpm, 10 min) and resuspended in buffer B (25 mM HEPES, 300 mM NaCl, 1 mM MgCl2, and 2mM TCEP, pH 7.4. The cell suspension was lysed by sonication and the lysate was clarified using ultracentrifugation (30,000 rpm, 2 h) and filtered with 0.22 μm cut-off membranes. The resulting supernatant was applied to a StrepTrap column (GE Healthcare) to capture the target proteins. The bound proteins were eluted with buffer B supplemented with 2.5 mM desthiobiotin. Target fractions were pooled and subjected to further purification by SEC using a Superdex 200 10/300 increase column (GE Healthcare). The final product was concentrated and stored at −80°C before use.
Cryo-EM sample preparation and imaging
An aliquot of 3 μL protein solution (0.6 mg/mL) was applied to a glow-discharged Quantifiol 1.2/1.3 holey carbon grid and blotted for 2.5 s in a humidity of 100% before plunge-freezing with an FEI Vitrobot Mark IV. Cryo-samples were screened using an FEI Tecnai TF20 electron microscope and transferred to an FEI Talos Arctica operated at 200 kV for data collection. The microscope was equipped with a post-column Bioquantum energy filter (Gatan) which was used with a slit width of 20 eV. The data were automatically collected using SerialEM software (https://bio3d.colorado.edu/SerialEM/). Images were recorded with a Gatan K2-summit camera in super-resolution counting mode with a calibrated pixel size of 0.8 Å at the specimen level. Each exposure was performed with a dose rate of 10 e-/pixel/s (approximately 15.6 e-/Å2/s) and lasted for 3.9 s, resulting in an accumulative dose of ∼60 e-/Å2 which was fractionated into 30 movie-frames. The final defocus range of the dataset was approximately −1.4 to −3.4 μm.
Image processing
The image drift and anisotropic magnification was corrected using MotionCor2 (Zheng et al., 2017). Initial contrast transfer function (CTF) values were estimated with CTFFIND4.1 (Rohou and Grigorieff, 2015) at the micrograph level. Images with an estimated resolution limit worse than 5 Å were discarded. Particles were automatically picked with RELION-3.0 (Zivanov et al., 2018) following the standard protocol. In total, approximately 1,860,000 particles were picked from ∼4,200 micrographs. After 3 rounds of extensive 2D classification, ∼924,000 particles were selected for 3D classification with the density map of SARS-CoV nsp12-nsp7-nsp8 complex (EMDB-0520) as the reference which was low-pass filtered to 60 Å resolution. After two rounds of 3D classification, a clean subset of ∼101,000 particles was identified, which displayed clear features of secondary structural elements. These particles were subjected to 3D refinement supplemented with per-particle CTF refinement and dose-weighting, which led to a reconstruction of 3.65 Å resolution estimated by the gold-standard Fourier shell correlation (FSC) 0.143 cut-off value. The local resolution distribution of the final density map was calculated with ResMap (Kucukelbir et al., 2014).
Model building and refinement
The structure of SARS-CoV nsp12-nsp7-nsp8 complex (PDB: 6NUR) was rigidly docked into the density map using CHIMERA (Pettersen et al., 2004). The model was manually corrected for local fit in COOT (Emsley et al., 2010) and the sequence register was corrected based on alignment. The initial model was refined in real space using PHENIX (Adams et al., 2010) with the secondary structural restraints and Ramachandran restrains applied. The model was further adjusted and refined iteratively for several rounds aided by the stereochemical quality assessment using MolProbity (Chen et al., 2010). The representative density and atomic models are shown in Figure S3. The statistics for image processing and model refinement are summarized in Table S1. Structural figures were rendered by either CHIMERA (Pettersen et al., 2004) or PyMOL (https://pymol.org/2/).
In vitro polymerase activity assay
The activity of SARS-CoV-2 polymerase complex was tested as previously described for SARS-CoV nsp12 with slight modifications. Briefly, a 40-nt template RNA (5′-CUAUCCCCAUGUGAUUUUAAUAGCUUCUUAGGAGAAUGAC-3′, Takara) corresponding to the 3′ end of the SARS-CoV2 genome was annealed to a complementary 20-nt primer containing a 5′-fluorescein label (5′FAM- GUCAUUCUCCUAAGAAGCUA-3′, Takara). To perform the primer extension assay, 1 μM nsp12, nsp7 and nsp8 were incubated for 30 min at 30°C with 1 μM annealed RNA and 0.5 mM NTP in a reaction buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM KCl, 1 mM beta-mercaptoethanol and 2 mM MgCl2 (freshly added prior usage). The products were denatured by boiling (100°C, 10 min) in the presence of formamide and separated by 20% PAGE containing 9 M urea run with 0.5 × TBE buffer. Images were taken using a Vilber Fusion system and quantified with the ImageJ software.
SPR assay
The affinities between nsp12, nsp7 and nsp8 or nsp7L8 proteins were measured at room temperature (r.t.) using a Biacore 8K system with CM5 chips (GE Healthcare). The nsp12 protein was immobilized on the chip with a concentration of 100 μg/mL (diluted by 0.1 mM NaAc, PH 4.0), and the nsp7 protein was immobilized with a concentration of 50 μg/mL (diluted by 0.1 mM NaAc, PH 4.5). For all measurements, the same running buffer was used which consists of 20 mM HEPES, pH 7.5, 150 mM NaCl and 0.005% tween-20. Proteins were pre-exchanged into the running buffer by SEC prior to loading to the system. A blank channel of the chip was used as the negative control. Serially diluted protein solutions were then flowed through the chip surface. The Multi-cycle binding kinetics was analyzed with the Biacore 8K Evaluation Software (version1.1.1.7442) and fitted with a two-state reaction binding model (for ligand nsp8) or heterogeneous ligand binding model (for ligand nsp7L8).
Circular dichroism measurement
The thermostability of nsp12, nsp7 and nsp8 were tested by measuring the CD spectra of each protein at different temperatures. The multi-wavelength (215-260 nm) CD spectra of each protein were recorded with a Chirascan spectrometer (Applied Photophysics) using a thermostatically controlled cuvette at rising temperatures from 25 to 99°C with 0.5°C intervals and an elevating rate of 1°C/min. The data were analyzed using Global3 software and the Tm values were calculated for each sample.
Quantification and Statistical Analysis
Data are presented as the means ± SD and were analyzed with GraphPad Prism and SPSS programs. The one-way analysis of variance (ANOVA) was used to compare the significance of differences between different groups. The p values less than 0.05 were considered significant. Statistical differences are indicated as follows: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. Biological replicates are indicated in the figure legends.
Acknowledgments
We thank all staff members at the Center of Biological Imaging (CBI), Institute of Biophysics (IBP), Chinese Academy of Sciences (CAS) for assistance with data collection. We are grateful to the Core Facility of the Institute of Microbiology, CAS for assistance with SPR experiments. This study was supported by the Strategic Priority Research Program of the CAS (XDB29010000), the National Science and Technology Major Project (2018ZX10101004), the National Key Research and Development Program of China (2020YFC0845900), the National Natural Science Foundation of China (NSFC) (82041016, 81871658, and 81802010), and a grant from the Bill and Melinda Gates Foundation. M.W. is supported by the National Science and Technology Major Project (2018ZX09711003) and National Natural Science Foundation of China (NSFC) (81802007). R.P. is supported by the Young Elite Scientist Sponsorship Program (YESS) of the China Association for Science and Technology (CAST) (2018QNRC001). Y.S. is also supported by the Excellent Young Scientist Program from the NSFC (81622031) and the Youth Innovation Promotion Association of CAS (2015078).
Author Contributions
Conceptualization, Y. Shi, Q.P., and R.P.; Methodology, Q.P., R.P., and M.W.; Investigation, Q.P., R.P., B.Y., J.Z., M.W., X.W., Q.W., Y. Sun, Z.F., and J.Q.; Data Curation, Q.P. and R.P.; Writing – Original Draft, Q.P., R.P., M.W., and Y. Shi; Writing – Review & Editing, all authors; Visualization, Q.P., R.P., and M.W.; Supervision, Y. Shi; Project Administration, Q.P. and R.P.; Funding Acquisition, Y. Shi., R.P., and M.W.
Declaration of Interests
The authors declare no competing interests.
Published: May 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107774.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktepe T.E., Mackenzie J.M. Shaping the flavivirus replication complex: It is curvaceous! Cell. Microbiol. 2018;20:e12884. doi: 10.1111/cmi.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., Song H., Wang H., Chai Y., Su C., Qi J., Shi Y., Gao G.F. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 2017;36:919–933. doi: 10.15252/embj.201696241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach P., Malet H., Cusack S., Reguera J. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015;161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy A.S., Lima G.M., Oliveira K.I., Torres N.U., Maluf F.V., Guido R.V., Oliva G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017;8:14764. doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengrung N., El Omari K., Serna Martin I., Vreede F.T., Cusack S., Rambo R.P., Vonrhein C., Bricogne G., Stuart D.I., Grimes J.M., Fodor E. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature. 2015;527:114–117. doi: 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Choe P.G., Oh Y., Oh K.J., Kim J., Park S.J., Park J.H., Na H.K., Oh M.D. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J. Korean Med. Sci. 2020;35:e61. doi: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea T.J., Cryan P.M., Cunningham A.A., Fooks A.R., Hayman D.T.S., Luis A.D., Peel A.J., Plowright R.K., Wood J.L.N. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., Xu X., Jing J., Wang M., Peng Q., Liu S., Wu Y., Bao X., Wang P., Qi J. Structural insight into arenavirus replication machinery. Nature. 2020;579:615–619. doi: 10.1038/s41586-020-2114-2. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pflug A., Guilligay D., Reich S., Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crépin T., Hart D., Lunardi T., Nanao M., Ruigrok R.W.H., Cusack S. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. New virus, new challenge. The Innovation. 2020;1:100005. doi: 10.1016/j.xinn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Xie Y., Shi L., Xu W. Viral RNA polymerase: a promising antiviral target for influenza A virus. Curr. Med. Chem. 2013;20:3923–3934. doi: 10.2174/09298673113209990208. [DOI] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicenzi E., Canducci F., Pinna D., Mancini N., Carletti S., Lazzarin A., Bordignon C., Poli G., Clementi M. Coronaviridae and SARS-associated coronavirus strain HSR1. Emerg. Infect. Dis. 2004;10:413–418. doi: 10.3201/eid1003.030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. Published online May 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Yi G., Du F., Chuang Y.C., Vaughan R.C., Sankaran B., Kao C.C., Li P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017;8:14762. doi: 10.1038/ncomms14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map and atomic coordinates have been deposited to the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with the accession codes EMD-30226 and 7BW4, respectively. All other data are available from the authors on reasonable request.