Abstract
Background
The burden of COVID-19 was extremely severe in Northern Italy, an area characterized by high concentrations of particulate matter (PM), which is known to negatively affect human health. Consistently with evidence already available for other viruses, we initially hypothesized the possibility of SARS-CoV-2 presence on PM, and we performed a first experiment specifically aimed at confirming or excluding this research hyphotesys.
Methods
We have collected 34 PM10 samples in Bergamo area (the epicenter of the Italian COVID-19 epidemic) by using two air samplers over a continuous 3-weeks period. Filters were properly stored and underwent RNA extraction and amplification according to WHO protocols in two parallel blind analyses performed by two different authorized laboratories. Up to three highly specific molecular marker genes (E, N, and RdRP) were used to test the presence of SARS-CoV-2 RNA on particulate matter.
Results
The first test showed positive results for gene E in 15 out of 16 samples, simultaneously displaying positivity also for RdRP gene in 4 samples. The second blind test got 5 additional positive results for at least one of the three marker genes. Overall, we tested 34 RNA extractions for the E, N and RdRP genes, reporting 20 positive results for at least one of the three marker genes, with positivity separately confirmed for all the three markers. Control tests to exclude false positivities were successfully accomplished.
Conclusion
This is the first evidence that SARS-CoV-2 RNA can be present on PM, thus suggesting a possible use as indicator of epidemic recurrence.
Keywords: COVID-19, SARS-CoV-2, Particulate matter, Air pollution
Highlights
-
•
COVID-19 burden seems more severe in areas with high concentrations of PM.
-
•
Particulate matter is already known to have negative effects on human health.
-
•
This is the first evidence that SARS-CoV-2RNA can be found on particulate matter.
1. Introduction
Severe acute respiratory syndrome known as COVID-19 disease - due to SARS-CoV-2 virus - is recognized to spread via respiratory droplets and close contacts (World Health Organisation, 2020). The burden of COVID-19 was extremely severe in Lombardy and Po Valley (Northern Italy), (Italian Ministry of Health, 2020) an area characterized by high concentrations of particulate matter, which are already known to have negative effects on human health (European Environmental Agency, 2019). Regional figures are available for Italy at the date of May 1stshow that about 30% of currently positive people still live in Lombardy (about 40% if considering the overall cases confirmed from the beginning of the epidemic), followed by Emilia Romagna (13.5% of currently positive people), Piedmont (10.5%), and Veneto (10%). (Italian Ministry of Health, 2020) These four regions of the Po Valley account for 80% of total deaths recorded in Italy and 65% of Intensive Care Units admissions. (Italian Ministry of Health, 2020) A research carried out by the Harvard School of Public Health seems to confirm an association between increases in PM concentrations and mortality rates due to COVID-19. (Wu et al., 2020).
In previous communications, we have hypothesized the possibility that SARS-CoV-2 virus could be present on particulate matter (PM) during the spreading of the infection (Italian Society of Enviro, 2020; Setti et al., 2020), consistently with evidence already available for other viruses (Sedlmaier et al., 2009; Zhao et al., 2019a, 2019b; Ma et al., 2017a, 2017b; Sorensen et al., 2000; Glostera and Alexandersen, 2004; Reche et al., 2018; Qin et al., 2020). However, the issue of airborne PM-associated microbiome, especially in urban environments, remains largely under-investigated (Jiang et al., 2015), and – at the present – nobody has still carried out experimental studies specifically aimed at confirming or excluding the presence of the SARS-CoV-2 on PM. Here, we present the first results of the analyses that we have performed on 34 PM10 samples of outdoor/airborne PM10 from an industrial site of Bergamo Province, the epicenter of the Italian COVID-19 epidemic from 02/21/2020 to 03/11/2020.
2. Methods
Following the methodology described by Pan et al., in 2019 for the detection of airborne viruses (Pan et al., 2019), particulate matter has been collected in industrial area of Bergamo (Italy) – over a continuous 3-weeks period, from February 21st to March 13th,2020 – on quartz fiber filters by using a low-volume gravimetric air sampler (38.3 l/min for 24 h), compliant with the reference method EN12341:2014 for PM10 monitoring. This sampling procedure allows collection of aerosol and bioaerosol, by filtering 55 m3 per day, in a wide dimensional range, by using an approach considered adequate for screening/sentinel purposes.
Other bioaerosol/virus sampling methods – such as impactors, cyclones, liquid impingers, electrostatic precipitators, water-based condensation - are suitable for restricted size ranges (few minutes or hours monitoring) in virus viability studies. (Pan et al., 2019). The two LV PM10 samplers were positioned at 1150 m apart from each other, in the frame of air quality monitoring of an urban area close to an industrial complex. During the sampling period, average temperature, average relative humidity and irradiance have been respectively recorded as follows: 8.5 °C, 61% and 117.9 W/m2 for the period February 21st-27th; 6.8 °C, 69% and 80,9 W/m2 for the period February 28th to March 5th; 6.8 °C, 67% and 120.9 W/m2 for the period March 6th-11th. One of the two samplers had operational failure for 9 days out of 21. Average PM10 values for week 1 to week 3 have been 51.1 μg/m3 (n = 7), 27.8 μg/m3 (n = 7), 32.0 μg/m3 (n = 7) for sampling site A and 53.3 μg/m3 (n = 7), 26.8 μg/m3 (n = 4), 39.7 μg/m3 (n = 1) for sampling site B, respectively. Median values of PM10 for the overall 3 weeks were 52.1 μg/m3, 27.8 μg/m3, 38.0 μg/m3 for site A and 51.2 μg/m3, 25.1 μg/m3, 39.7 μg/m3 for sampling site B, respectively.
To assess eventual differences in mean or median values concerning the 34 PM10 samples collected by the two low volume air samplers in the whole period (from February 21st to March 11h), we performed inferential statistical analyses, which produced the following results: A) according to t-test, the two means (43.35 vs. 39.29, respectively) presented a point wise difference of 4.06 [90%IC: −15.40; 7.27; p = 0.55]; B) according to Wilcoxon rank sum test, the two medians (37.7 and 37.8, respectively) presented a point wise difference of 0.1 [90%IC: −6.6; 13.1; p = 0.70]. Being aware that missing values recorded for one of the samplers could affect the straight forward analysis, we repeated the calculation exploiting the permutation test approach (Pesarin and Salmaso, 2010), which is particularly suitable to manage small sample size; C) when performing a permutation test (with N = 300,000), the point wise difference of the means was 4.06 [90%IC: −12.31; 12.36; p = 0.30]; D) when performing a permutation test (with N = 300,000), the point wise difference of the medians was 0.1 [90%IC:-13.2; 13.1; p = 0.49].
Particulate matter was trapped on filters with 99.9% typical aerosol retention, properly stored and delivered to the laboratory of Applied and Comparative Genomics of Trieste University. Given the “environmental” nature of the sample, presumably rich in inhibitors of DNA polymerases, we proceeded with the extraction of RNA by using the Quick RNA fecal soil microbe kit adapted to the type of the filters (Zymoresearch Ldt, 2020). Half filter was rolled, with the top side facing inward, in a 5 ml polypropylene tube, together with the beads provided in the kit. From the initial 1 ml of lysis buffer, we were able to get about 400 μl of solution, which was then processed as defined by the standard protocols, resulting in a final eluate of 15 μl. Subsequently, 5 μl were used for the SARS-CoV-2 testing. Given the particular origin of the sample, the qScript XLT 1-Step RT-qPCRToughMix was used (Quantabio Ltd, 2020). The amplification systems were those of the protocol developed by Corman et al., published on the WHO website (Corman et al., 2020). The tests were explicitly aimed at confirming or excluding the presence of the SARS-CoV-2 RNA on particulate matter. Due to the scarce material available, we decided to test more than one marker gene on the collected PM10 samples. Therefore, up to three highly specific molecular marker genes (E, N, and RdRP) were used to test the presence of SARS-CoV-2 RNA on particulate matter.
3. Results
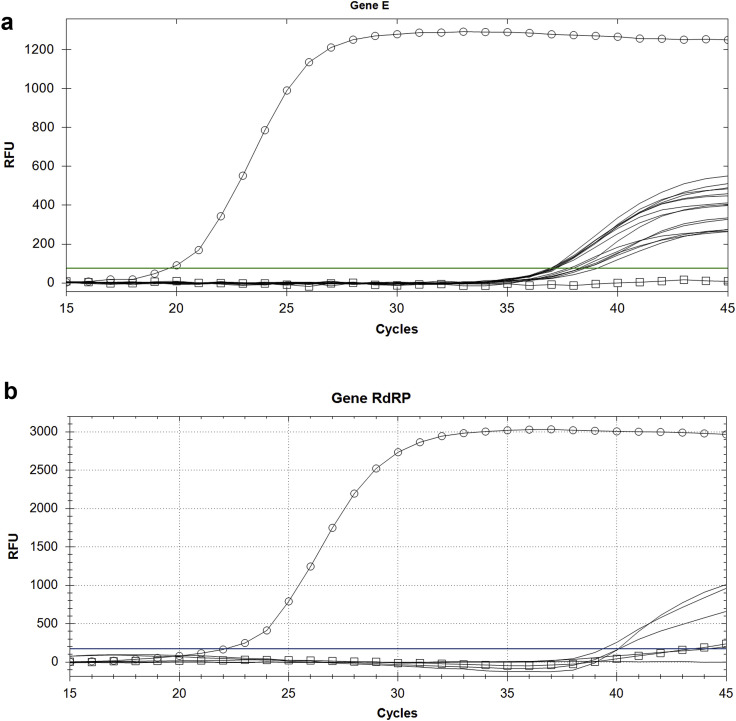
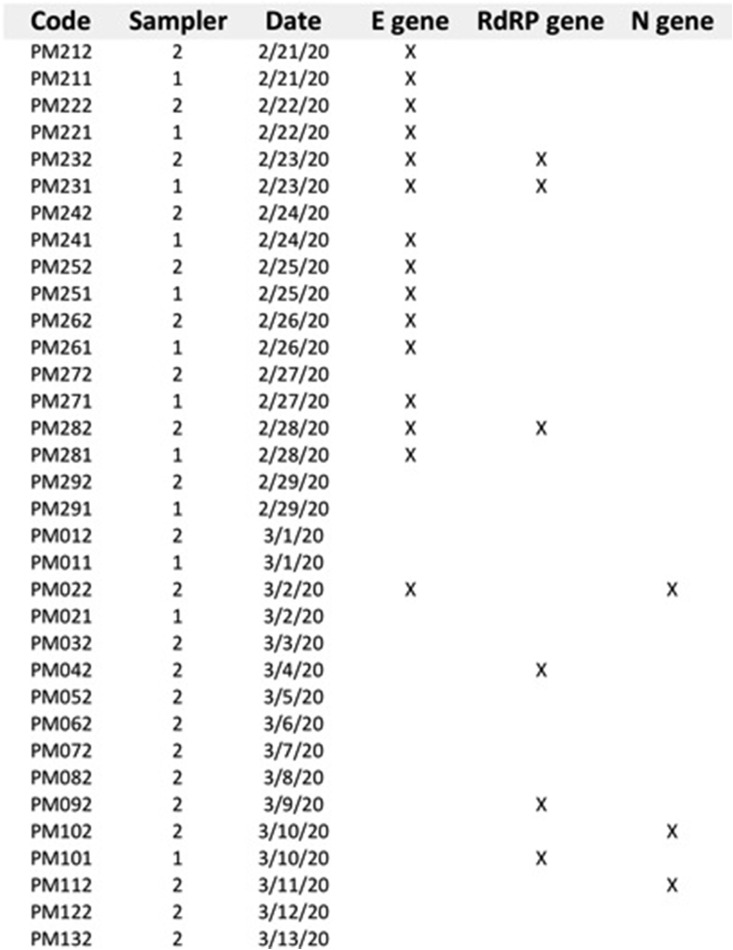
The first analysis used the “E gene”as a molecular marker and produced an impressive positive result on 15 out of 16 filters even if, as we could expect, the Ct was between 36 and 38 cycles. After that, we have replicated the analysis on 6 of the positive filters (already positive to “E gene”) by using the “RtDR gene” as a molecular marker – which is highly specific for SARS-CoV-2 – reaching 4 significant results of positivity. Control tests to exclude false positivities were also successfully performed (Fig. 1 ). To avoid the running out of the scarce sampling material available, the remaining extracted RNAs were delivered to the local University Hospital (one of the clinical centres authorized by the Italian Government for SARS-CoV-2 diagnostic tests), in order to perform a second parallel blind test. This second clinical laboratory resulted in 5 additional positive results for at least one of the three marker genes, including “N” gene. Overall, we tested 34 RNA extractions for the E, N and RdRP genes, reporting 20 positive results for at least one of the three marker genes, with positivity separately confirmed for all the three markers (Fig. 2 ).
Fig. 1.
Amplification curves of E (A) and RdRP genes (B): green lines represent tested filters; cross line represents reference filter extractions; red lines represent the amplification of the positive samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Positive results (marked with X) for E, N and RdRP genes obtained for all the 34 PM10 filters tested and confirmed by the two parallel analyses performed.
4. Discussion
As a first-choice approach for monitoring the presence of airborne viruses on environmental samples, standard low volume PM10 samplers were selected because it is an easy and widely diffuse operational methodology, promptly available on the field. The selected monitoring approach is well established among environmental protection agencies, researchers and professionals and it is able to determine inactivation of viruses due to dehydration or extraction from filters (Pan et al., 2019), an issue that can be regarded as a positive safety feature for operators. Both liquid impingers and water-based condensation devices maintain viability of viruses - a drawback for operators involved in standard environmental surveys – while impactors, cyclones, electrostatic precipitators have low collection efficiency for submicrometre virus particles, that can be of interest in infection spreading. The proposed sampling option can represent a good starting point for detecting airborne RNA traces of the virus, highlighting the sussistence of potential viral hazard that can be followed by focused short term aerosol collection on liquids (e.g. by impingers or liquid cyclone (Brisebois et al., 2018)) for virus viability assessment on environmental samples. Because of the nature of the sample, and considering that the sampling has not been carried out for clinical diagnostic purposes but for environmental pollution tests (taking also into account that filters were stored for at least four weeks before undergoing molecular genetic analyses, as a consequence of the Italian shutdown), we can confirm to have reasonably demonstrated the presence of SARS-CoV-2 viral RNA by detecting highly specific “RtDR gene” on 8 filters. However, due to the lack of additional materials from the filters, we were not able to repeat enough number of tests to show positivity for all the 3 molecular markers simultaneously.
Concerning the approach chosen for the isolation of viral RNA and its molecular detection, it should be highlighted that the use of protocols developed for genetic analysis on environmental matrices may have been the crucial element for the success of the perfomed analyses. As stated, the available material was limited and would not have allowed to perform as many as replications by using different methodologies, but thanks to the methodology applied – testing more than a single molecular marker gene – we have evidence that amplifications with a DNA polymerase developed, despite the scarce materials available and impure templates, resulted in a higher percentage of positives tests compared to what we could have obtained by using the enzymes cocktail generally used in the commercially available diagnostic kits.
This is the first evidence that SARS-CoV-2 RNA can be present on outdoor particulate matter, thus suggesting that, in conditions of atmospheric stability and high concentrations of PM,SARS-CoV-2 could createclusters with outdoor PM10 and – by reducing their diffusion coefficient – enhance the persistence of the virus in the atmosphere. Further confirmations of this preliminary evidence are ongoing in Milan and Naples (Italy), Madrid and Barcelona (Spain), Bruxelles (Belgium), and New York – under the RESCOP (Research group on COVID-19 and Particulate Matter) International Research Initiative, promoted by the Italian/International Society of Environmental Medicine (SIMA/ISEM) with the aim of using the presence of SARS-COV-2 on PM10 as early indicator of epidemic recurrence – and should include real-time assessment about the vitality of the SARS-CoV-2 as well as its virulence when adsorbed on particulate matter where possible (Setti et al., 2020). At the present, no assumptions can be made concerning the presence of the virus on PM and COVID-19 outbreak diffusion among the population as we still do not know if the virus remains vital on PM and – in this case – how long. Other issues to be specifically addressed are the average concentrations of PM10 eventually required for a potential “boost effect” of the contagion in the areas experiencing the most dramatic burden of COVID-19, or even the theoretic possibility of immunization consequent to minimal dose exposures at lower thresholds of PM10.
5. Conclusion
This is the first evidence that SARS-CoV-2 RNA can be present on outdoor particulate matter in defined conditions of atmospheric stability and high concentrations of PM10, thus suggesting a possible use of this test as indicator of epidemic recurrence.
Credit author statement
Leonardo Setti, Fabrizio Passarini, Gianluigi De Gennaro, Pierluigi Barbieri, Maria Grazia Perrone, Massimo Borelli, Jolanda Palmisani, Alessia Di Gilio, Valentina Torboli, Francesco Fontana, Libera Clemente, Alberto Pallavicini, Maurizio Ruscio, Prisco Piscitelli, Alessandro Miani conceived, wrote, approved and recise the manuscript. Maurizio Ruscio, Valentina Torboli, Francesco Fontana, Libera Clemente, Alberto Pallavicini performed the genetic and molecular analyses; Leonardo Setti, Fabrizio Passarini, Gianluigi De Gennaro, Pierluigi Barbieri, Maria Grazia Perrone, Massimo Borelli, Jolanda Palmisani, Alessia Di Gilio set the methodology for air sampling;
Declaration of competing interest
On behalf of all the co-authors we declare no conflict of interests.
Contributor Information
Leonardo Setti, Email: leonardo.setti@unibo.it.
Fabrizio Passarini, Email: fabrizio.passarini@unibo.it.
Gianluigi De Gennaro, Email: gianluigi.degennaro@uniba.it.
Pierluigi Barbieri, Email: barbierp@units.it.
Maria Grazia Perrone, Email: mariagrazia.perrone@tcrtecora.com.
Massimo Borelli, Email: borelli@units.it.
Jolanda Palmisani, Email: jolanda.palmisano@uniba.it.
Alessia Di Gilio, Email: alessia.digilio@uniba.it.
Valentina Torboli, Email: torboli@units.it.
Francesco Fontana, Email: francesco.fontana@asugi.sanita.fvg.it.
Libera Clemente, Email: libera.clemente@asugi.sanita.fvg.it.
Alberto Pallavicini, Email: pallavic@units.it.
Maurizio Ruscio, Email: maurizio.ruscio@asugi.sanita.fvg.it.
Prisco Piscitelli, Email: priscofreedom@hotmail.com.
Alessandro Miani, Email: alessandro.miani@unimi.it.
References
- Brisebois E., Veillette M., Dion-Dupont V., Lavoie J., Corbeil J., Culley A., Duchaine C. Human viral pathogens are pervasive in wastewater treatment center aerosols. J. Environ. Sci. 2018;67:45–53. doi: 10.1016/j.jes.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Mulders D.G. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269/ available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glostera J., Alexandersen S. New directions: airborne transmission of foot-and-mouth disease. Virus Atmos. Environ. 2004;38(3):503–505. [Google Scholar]
- Italian Ministry of Health, 2020. Daily bulletin Covid-19 outbreak in Italy, available at: http://www.salute.gov.it/imgs/C_17_notizie_4451_0_file.pdf.
- European Environmental Agency, 2019. Air Quality in Europe 2019 Report; No 10/2019; European Environment Agency: Copenhagen, Denmark, availbale at: https://www.eea.europa.eu/publications/air-quality-in-europe-2019.
- Italian Society of Environmental Medicine (SIMA), Position Paper Particulate Matter and COVID-19, available at: http://www.simaonlus.it/wpsima/wp-content/uploads/2020/03/COVID_19_position-paper_ENG.pdf.
- Jiang W., Laing P., Wang B., Fang J., Lang J., Tian G., Jiang J., Zhu T.F. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 2015;10:768–779. doi: 10.1038/nprot.2015.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhou J., Yang S., Zhao Y., Zheng X. Assessment for the impact of dust events on measles incidence in western China. Atmos. Environ. 2017;157:1–9. [Google Scholar]
- Ma Y., Zhou J., Yang S., Zhao Y., Zheng X. Assessment for the impact of dust events on measles incidence in western China. Atmos. Environ. 2017;157:1–9. [Google Scholar]
- Pan M., Lednicky J.A., Wu C.-Y. Collection, particle sizing and detection of airborne viruses. J. Appl. Microbiol. 2019;127:1596–1611. doi: 10.1111/jam.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesarin F., Salmaso L. John Wiley&Sons, Ltd; 2010. Permutation Tests for Complex Data: Theory, Applications and Software.Wiley Series in Probability and Statistics. 9780470689516. [Google Scholar]
- Qin N., Liang P., Wu C., Wang G., Xu Q., Xiong X., Wang T., Zolfo M., Segata N., Qin H., Knight R., Gilbert J.A., Zhu T.F. Longitudinal survey of microbiome associated with particulate matter in a megacity. Genome Biol. 2020;21:55. doi: 10.1186/s13059-020-01964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche I., D'Orta G., Mladenov N., Winget D.M., Suttle C.A. Deposition rates of viruses and bacteria above the atmosperic boundary layer. ISME J. 2018;12:1154–1162. doi: 10.1038/s41396-017-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlmaier N., Hoppenheidt K., Krist H., Lehmann S., Lang H., Buttner M. Generation of avian influenza virus (AIV) contaminated fecal fine particulate matter (PM2.5): genome and infectivity detection and calculation of immission. Vet. Microbiol. 2009;139:156–164. doi: 10.1016/j.vetmic.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Barbieri P., Pallavicini A., Ruscio M., Piscitelli P., Colao A., Miani A. Searching for SARS-COV-2 on particulate matter: a possible early indicator of COVID-19 epidemic recurrence. Int. J. Environ. Res. Publ. Health. 2020;17:2986. doi: 10.3390/ijerph17092986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Barbieri P., Perrone M.G., Borelli M., Palmisani J., Di Gilio A., Piscitelli P., Miani A. Int. J. Environ. Res. Public Health; 17: 2020. Airborne Transmission Route of COVID-19: Why 2 Meters/6 Feet of Inter-Personal Distance Could Not Be Enough; p. 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen J.H., Mackay D.K.J., Jensen C.Ø., Donaldson A.I. An integrated model to predict the atmospheric spread of foot-and-mouth disease virus Epidemiol. Infection. 2000;124:577–590. doi: 10.1017/s095026889900401x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantabio Ltd, 2020. Descriprion of the product, available at: https://www.quantabio.com/qscript-xlt-1-step-rt-qpcr-toughmix.
- World Health Organisation, Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations, Scientific brief; available at:https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations (29 March 2020).
- Zhao Y., Richardson B., Takle E., Chai L., Schmitt D., Win H. Airborne transmission may have played a role in the spread of 2015 highly pathogenic avian influenza outbreaks in the United States. Sci. Rep. 2019;9:11755. doi: 10.1038/s41598-019-47788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Richardson B., Takle E., Chai L., Schmitt D., Win H. Airborne transmission may have played a role in the spread of 2015 highly pathogenic avian influenza outbreaks in the United States. Sci. Rep. 2019;9:11755. doi: 10.1038/s41598-019-47788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Wu, Rachel C. Nethery, M. Benjamin Sabath, Danielle Braun, Francesca Dominici, 2020. Exposure to air pollution and COVID-19 mortality in the United States, available at: https://www.medrxiv.org/content/10.1101/2020.04.05.20054502v2.
- Zymoresearch Ldt, 2020. product description, available at: https://www.zymoresearch.com/products/quick-rna-fecal-soil-microbe-microprep-kit.