Abstract
MicroRNA (miRNA) dysregulation in cancer causes changes in gene expression programs regulating tumor progression and metastasis. Candidate metastasis suppressor miRNA are often identified by differential expression in primary tumors compared to metastases. Here, we performed comprehensive analysis of miRNA expression in The Cancer Genome Atlas (TCGA) skin cutaneous melanoma (SKCM) tumors (97 primary, 350 metastatic), and identified candidate metastasis-suppressor miRNAs. Differential expression analysis revealed miRNA significantly downregulated in metastatic tumors, including miR-205, miR-203, miR-200a-c, and miR-141. Furthermore, sequential feature selection and classification analysis identified miR-205 and miR-203 as the miRNA best able to discriminate between primary and metastatic tumors. However, cell-type enrichment analysis revealed that gene expression signatures for epithelial cells, including keratinocytes and sebocytes, were present in primary tumors and significantly correlated with expression of the candidate metastasis-suppressor miRNA. Examination of miRNA expression in cell lines revealed that candidate metastasis-suppressor miRNA identified in the SKCM tumors, were largely absent in melanoma cells or melanocytes, and highly restricted to keratinocytes and other epithelial cell types. Indeed, the differences in stromal cell composition between primary and metastatic tumor tissues is the main basis for identification of differential miRNA that were previously classified as metastasis-suppressor miRNAs. We conclude that future studies must consider tumor-intrinsic and stromal sources of miRNA in their workflow to identify bone fide metastasis-suppressor miRNA in cutaneous melanoma and other cancers.
Introduction
Cutaneous melanoma is the most aggressive form of skin cancer. Although accounting for only 5% to 10% of cases, more than 80% of skin cancer-related deaths are due to melanoma. Globally, close to 300,000 new cases of melanoma were diagnosed in 2018, and the incidence has been steadily increasing over the past several decades [1,2]. When diagnosed early, surgical resection of primary tumors can be curative. However, once the tumor becomes metastatic, 5-year survival decreases rapidly to less than 20% for patients with stage IV disease [1]. Staging and prognosis of melanoma tumors are based on various measurements of tumor invasion, such as the tumor thickness, and depth (in mm) to which it has penetrated (formerly measured as the Breslow’s depth), as well as the presence of regional lymph node metastases [3]. Recent advancements in the development and clinical use of targeted therapies like BRAF and immune checkpoint inhibitors have had success in extending patient survival. However, inherent or developed resistance still occurs in a large proportion of patients with metastatic disease, leaving many with limited treatment options [4,5]. For this reason, there is a critical need to enhance understanding of the mechanisms that drive metastatic melanoma to inform the development of new targeted therapies or better apply existing therapies to the patients most likely to benefit from them.
MicroRNA (miRNA) are a class of small non-coding RNA that regulate gene expression at a post-transcriptional level. Expression of miRNAs is frequently altered in cancer, leading to broad changes in target mRNAs, and key signaling pathways. These changes in gene expression can promote phenotypes that contribute to cancer progression, metastasis, and resistance to therapy [6,7]. Due to the relative abundance and often restricted expression patterns, many miRNAs can serve as biomarkers of cell-type and disease [7,8]. Identifying miRNA that are downregulated in metastasis, termed metastasis-suppressor miRNA, has been an area of considerable interest [[9], [10], [11], [12]]. Defining miRNA associated with metastasis in melanoma could provide insights into the genes and pathways that drive metastatic progression, and potentially lead to the discovery of new therapeutic targets or biomarkers. However, many studies aiming to characterize metastasis-suppressor miRNAs have suffered major caveats, including sub-optimal detection methods, and failure to consider tissue- and cell-type-specific expression patterns. As a result, extensive research efforts have pursued miRNAs that are unlikely to have direct biological relevance in tumor types where they have been characterized.
In the current study, miRNA expression profiles from primary and metastatic melanoma tumors in The Cancer Genome Atlas (TCGA) Skin Cutaneous Melanoma (SKCM) project [13] were analyzed using traditional and emerging methodologies to identify candidate metastasis-suppressor miRNAs. We show that primary and metastatic melanoma tumors have complex and heterogeneous miRNA expression profiles, identifying a previously unreported subgroup of patients with expression of the chromosome 19 miRNA cluster (C19MC). We also demonstrate that conventional differential expression, and supervised machine learning methods are both vulnerable to spurious identification of metastasis-suppressor miRNA due to cell type-specific expression patterns in tissue-level profiles. These findings support greater consideration of stromal cell populations when seeking to identify and functionally characterize putative metastasis-associated miRNAs using tumor tissue samples.
Methods
Datasets
Accession numbers and sample information for the datasets used in this study are provided in Table 1.
Table 1.
Data sources and sample information
| Dataset | Method/platform | Samples | Accession | Reference |
|---|---|---|---|---|
| TCGA Level 3 SKCM miRNAseq | Illumina HiSeq 2000 | 97 primary tumors 350 metastatic tumors |
Broad Institute GDAC Firehose portal | [13] |
| TCGA Level 3 SKCM RNAseq | Illumina HiSeq 2000 | 103 primary tumors 367 metastatic tumors |
Broad Institute GDAC Firehose portal | [13] |
| TCGA SKCM tumor purity scores | ABSOLUTE | 439 primary and metastatic tumors | - | [14] |
| Andrews et al. LM-MEL panel small RNAseq | Illumina HiSeq 2000 | 57 patient-derived melanoma cell lines | GEO GSE89438 | [15] |
| Behren et al. LM-MEL panel mRNA array | Illumina HumanHT-12 V4.0 microarray | 56 patient-derived melanoma cell lines |
ArrayExpress E-MTAB-1496 |
[16] |
| Boyle et al. miRNA array | Agilent-016436 Human miRNA microarray 1.0 | 51 melanoma cell lines 2 pools of normal melanocytes |
GEO GSE25653 |
[17] |
| Caramuta et al. miRNA array | Aligent-016434 Human miRNA Microarray 1.0 | 21 patient-derived melanoma cell lines 3 normal melanocytes |
GEO GSE19387 |
[18] |
| microRNAome small RNAseq | Illumina and AB SOLiD Systems NGS platforms | 2 melanoma cell lines 5 normal melanocytes 5 keratinocytes 3 sebocytes 19 normal skin tissue |
NCBI BioProject database PRJNA358331 |
[19] |
| Mizrahi et al. small RNAseq | Illumina HiSeq 2500 | 3 pools of normal primary keratinocytes |
GEO GSE101192 |
[20] |
Data Preprocessing and Normalization
miRNA and RNA Sequencing
TCGA miRNAseq and RNAseq data were screened for batch effects and outliers, and normalized as previously described [13,21]. Filtering was performed to retain miRNA expressed above the 90th quantile of overall expression in at least one sample. The final dataset comprised of expression of 264 miRNA across 447 melanoma tumors (350 metastatic, 97 primary). The miRNA sequencing data from cell lines were processed and normalized in the same manner. All cell line datasets were analyzed separately due to potential differences between platforms and alignment pipelines.
miRNA and mRNA Microarrays
Raw Agilent miRNA microarray data were processed using the R/Bioconductor package AgiMicroRna [22]. Data were normalized by scaling log-ratios to have the same median-absolute-deviation across arrays. Probes with detection P-values above a 0.05 threshold in all samples were discarded. Raw mRNA expression measured by microarray for LM-MEL cell lines was processed and normalized using the R/Bioconductor package limma [23], as previously described [15].
Unsupervised Clustering
Hierarchical clustering was performed using normalized miRNA expression profiles after median-centering. Euclidean distance was used for rows (miRNAs) and Spearman correlation for columns (samples). Clustering of miRNA expression and xCell score correlations used Euclidean distance for both rows and columns. Average linkage was used in all cases.
Differential miRNA Expression Analysis
Differences in miRNA expression between primary and metastatic tumors were evaluated using the Mann-Whitney U test, corrected for multiple testing using the Benjamini-Hochberg adjustment. Effect sizes were calculated using the formula [24]. Z = Mann-Whitney U Z statistic, N = sample size.
Feature Selection and Classification Using Supervised Machine Learning Algorithms
Feature selection was performed using the MATLAB ‘sequentialfs’ function [25], along with a custom script incorporating five different classification algorithms (linear support vector machine (SVM), linear discriminant analysis, decision trees, k-nearest neighbor, and an ensemble model of boosted classification trees). Briefly, data were partitioned for 10-fold cross-validation. Training data were used to train the model, and the feature selection criterion was computed as the overall accuracy of the predictions on the test set. Feature selection was repeated ten times, resulting in the selection of 50 feature sets. The ability of the selected miRNA features to discriminate between primary and metastatic tumors was verified using the Classification Learner App in MATLAB, running a collection of 23 classification algorithms to classify the tumors with 10-fold cross-validation. The quality of each classification model was assessed using confusion matrices, and a receiver operating characteristic (ROC) curve.
Survival Analysis and Associations to Clinical Data
Kaplan-Meier survival analysis was performed using the observed survival interval (OSI), representing the time between tumor sample procurement and death or last follow-up. Patients were stratified into ‘high’ or ‘low’ groups based on median miRNA expression. Statistical significance between the survival curves was calculated using the log-rank test. Associations between miRNA expression and patient age and Breslow’s depth were assessed using Spearman’s correlation. Differences in expression of miRNA between groups with different clinical parameters were assessed using the Mann-Whitney U test for two groups, or the Kruskal-Wallis H test for three or more groups.
Cell-Type Enrichment Analysis
Tumor purity estimates produced using ABSOLUTE [26] were plotted for the SKCM tumors. RNAseq gene expression from tumors was used to assess the enrichment of 64 immune and stromal cell types using the xCell webtool (http://xCell.ucsf.edu/) [27]. Cell enrichment scores were computed and compared between primary and metastatic tumor samples. Significant differences were determined using the Mann-Whitney U test. The association between miRNA expression and cell-type enrichment scores was assessed using Spearman’s correlation. Associations between stromal-enriched miRNA and expression of cell-type specific markers (Keratinocytes: KRT1, KRT5, KRT14, CASP14, IVL, LOR; Melanocytes: DCT, GPNMB, TYRP1, TYR, MLANA, MITF; Immune cells: CD14, CD3E, CD3D, CD19, CD22) were assessed using Spearman’s correlation (Supplemental File 2), and visualized in clustered heatmaps.
Distinguishing Cutaneous Stromal-Enriched miRNA
Candidate metastasis-suppressor miRNA were defined as those that demonstrated a significant loss of expression between primary and metastatic tumors, identified in differential expression and feature selection analyses. Cutaneous stromal-enriched miRNAs were identified as candidate metastasis-suppressor miRNA that were significantly positively correlated (rho ≥ 0.3) with the enrichment scores for the cell types most overrepresented in primary tumors (Supplemental File 1). These were further refined to the miRNA expressed at >0.1% of total miRNA reads in keratinocytes and/or sebocytes, and expressed at <0.1% of total miRNA reads in both melanoma and normal melanocyte cell lines. Average expression across samples within the same dataset was considered (Table S1). Six miRNA met these criteria and were defined as cutaneous stromal-enriched (miR-205, miR-203, miR-200a-c, miR-141), and were filtered before repeating feature selection and classification analyses
Statistical Analysis Software
Data processing, visualization, and statistical analysis of gene expression datasets were performed using the Statistics and Machine Learning Toolbox and custom scripts in MATLAB R2016b (Mathworks Inc.), and R version 3.6.3 [28]. Statistical and survival analysis of clinical data was carried out using IBM SPSS Statistics, version 24 (IBM Corp).
Results
TCGA-SKCM Primary and Metastatic Melanoma Patient Characteristics
The TCGA SKCM project is the largest publicly available melanoma tumor cohort [13]. The inclusion of both primary and metastatic tumors provides a unique opportunity for comparison. Here, patients were grouped based on the primary or metastatic status of their tumors (N = 97 and N = 350, respectively). A significant difference of 7.7 years in mean age was detected between patients with primary and metastatic disease (Table 2; 95% CI, 4.2-11.2, P < .001), and both groups had more males than females (73% and 60%, respectively). A significant difference in Breslow’s depth was also observed (primary: 11.9 mm; metastatic: 3.4 mm, P < .001). This mean depth of local tumor invasion at the primary tumor site suggests that some tumors may have already been invasive at the time of resection.
Table 2.
Patient information for primary and metastatic groups within the SKCM cohort*
| Samples | N† | Age⁎ | Sex (male:female) | Breslow’s Depth⁎,# (mm) |
|---|---|---|---|---|
| Primary | 97 | 64 ± 13.9 | 56:41 | 11.9 ± 13.2 |
| Metastatic | 349 | 57 ± 15.7 | 218:131 | 3.4 ± 4.7 |
Median ± SD, # data missing from 25% of samples
n = 446 one patient had paired primary and metastatic tumors
Unsupervised Clustering Reveals Subsets of Patients with Unique miRNA Expression Profiles
Hierarchical clustering of tumor miRNA expression profiles resulted in the formation of complex nodes, and separation of primary and metastatic samples were highly heterogeneous (Figure 1A). However, two miRNA nodes with starkly variable expression patterns were observed (Figure 1A, nodes 1 and 2). Spearman correlation coefficients between all miRNA were ordered according to genomic location, revealing that the miRNA identified in nodes 1 and 2 were spatially related in the genome. A third large spatially-related group with more variable co-expression was also observed (Figure 1B). Indeed, all of the miRNA identified in node 1 mapped to the chromosome 19 miRNA cluster (C19MC) made up 46 genes (Figure 1C). Acquired expression of C19MC has been reported in other cancers [29,30], but this constitutes a novel finding in melanoma. The miRNAs in node 2 were made up of 14 genes in the miR-506/514 gene cluster, located on the X chromosome (Figure 1C). The miR-506/514 miRNA have been shown to promote a malignant phenotype in melanoma [31]. The third group of co-expressed miRNA in the correlation matrix corresponded to the chromosome 14 miRNA cluster (C14MC) but were mostly of low-abundance in the tumors. Despite identifying subgroups with unique miRNA expression profiles, hierarchical clustering was driven mainly by large, co-expressed miRNA gene clusters, and failed to resolve primary and metastatic tumors. To identify miRNA specifically associated with metastasis, additional methods were employed.
Figure 1.
Unsupervised clustering identifies subgroups of patients with unique miRNA expression patterns. (A) Hierarchical clustering of median-centered, normalized, and filtered miRNA expression. Columns represent patients, and rows represent miRNA. Metastatic samples are marked in pink, primary samples in blue. Two miRNA clusters of interest highlighted with black bars, 1 and 2. (B) Spearman rho correlation matrix for miRNA ordered by genomic location (top-bottom on the y-axis, left to right on the x-axis). Clusters 1 and 2 from (A) shown. (C) Schematics of the C19MC and mi-5045/514 miRNA clusters located on chromosome 19 and the X chromosome.
Differential Expression and Feature Selection Analyses Identify Metastasis-Associated miRNA in Melanoma Tumors
Differential expression analysis identified 90 miRNA significantly altered between primary and metastatic tumors (P < .05) (Table 3, Table S1). These included several frequently identified and highly studied melanoma- and metastasis-associated miRNAs that were highly downregulated in metastases (miR-205, miR-203, miR-200a-c, and miR-141) [[10], [11], [12],[32], [33], [34], [35], [36], [37], [38], [39]]. Although many differences were highly statistically significant, the effect sizes were relatively small (Table 3). Hierarchical clustering using the differentially expressed miRNA improved the separation of primary and metastatic samples compared to total miRNA profiles, resolving a subset of primary tumors largely defined by expression of miR-205, miR-203, and the miR-200 family. However, clustering of classes remained heterogeneous (Figure 2A). Differential expression of the miR-506/514 miRNA cluster was detected between primary and metastatic samples. However, expression of C19MC was not significantly altered between groups, which may contribute to the absence of reports in the literature.
Table 3.
Significantly differentially expressed miRNA between primary and metastatic tumors
| miRNA | Log2 fold-change | FDR-adj P-value | abs(r)⁎ |
|---|---|---|---|
| mir-205 | -8.65 | 9.79E-30 | 0.56 |
| mir-203 | -4.98 | 5.21E-21 | 0.47 |
| mir-200b | -2.27 | 8.12E-19 | 0.44 |
| mir-200c | -2.65 | 5.82E-18 | 0.43 |
| mir-200a | -2.35 | 6.55E-18 | 0.43 |
| mir-141 | -2.64 | 1.75E-14 | 0.38 |
| mir-224 | -1.30 | 7.82E-10 | 0.32 |
| mir-675 | 1.54 | 4.05E-09 | 0.30 |
| mir-215 | 1.15 | 6.81E-07 | 0.26 |
| mir-625 | 0.64 | 4.29E-06 | 0.25 |
| mir-153-2 | 1.64 | 1.32E-05 | 0.24 |
| mir-1274b | 1.09 | 2.06E-05 | 0.23 |
| mir-206 | -2.43 | 5.35E-05 | 0.22 |
| mir-29c | 0.79 | 7.50E-05 | 0.22 |
| mir-452 | -0.76 | 7.50E-05 | 0.22 |
| mir-218-2 | 0.60 | 8.50E-05 | 0.21 |
| mir-508 | -1.85 | 8.50E-05 | 0.21 |
| mir-514-1 | -1.95 | 8.50E-05 | 0.21 |
| mir-514-2 | -1.90 | 8.50E-05 | 0.21 |
| mir-514-3 | -1.95 | 8.50E-05 | 0.21 |
Effect size (small 0.1, medium 0.3, large 0.5).
Figure 2.
Differential expression analysis and feature selection identify metastasis-associated miRNA in primary and metastatic melanoma tumors. (A) Unsupervised hierarchical clustering of all primary and metastatic samples using the 90 significantly differentially expressed miRNA. Columns (samples) were clusters using Spearman’s rank correlation, and rows (miRNA) were clustered using Euclidean distance, with average linkage. Metastatic tumors are shown in pink, and primary tumors, in blue. (B) Top 20% of miRNA features selected with the highest frequency. The y-axis shows the number of feature subsets a miRNA appeared in, out of a possible 50. (C) Hierarchical clustering of all primary and metastatic samples using only 24 miRNA identified in the top 20% of selected features. All parameters the same as previously stated. (D) Confusion matrices the Ensemble RUSBoosted decision tree classifier constructed using the miRNA identified in the top 20% of feature selection. (E) ROC curve for the Ensemble model with the current classifier indicating the prediction of primary tumors.
To further define miRNA that could discriminate primary and metastatic tumors, ten rounds of sequential feature selection were performed, and the top 20% of features with the highest selection frequency were considered (Figure 2B). Of these 24 miRNA, 14 were significantly differential between primary and metastatic tumors, as well as 10 additional miRNAs not identified in the previous analysis (Table 4). Of the differentially expressed miRNAs, miR-205 and miR-203 were the most stable discriminating features with the highest selection frequency. Interestingly, members of the miR-200 family that have been associated with melanoma metastasis were not identified [10,12,32,35,40]. Furthermore, two miRNA from the miR-506/514 cluster (miR-514-2 and miR-513c), and miR-517c from the C19MC cluster were selected as discriminating features. Hierarchical clustering using the 24 selected features improved the separation of primary and metastatic samples slightly compared to clustering using the differentially expressed miRNA (Figure 2C). However, the dimensionality was greatly reduced (24 miRNA compared to 90 for differential expression analysis).
Table 4.
The top 20% of miRNA identified during feature selection to classify primary and metastatic tumors
| Identified by differential expression and feature selection |
Identified by feature selection only |
||
|---|---|---|---|
| miRNA | Log2 fold-change | miRNA | Log2 fold-change |
| mir-205 | -8.65 | mir-155 | 0.33 |
| mir-203 | -4.98 | mir-34a | -0.29 |
| mir-215 | 1.15 | mir-517c | 1.27 |
| mir-130b | 0.26 | let-7c | -0.03 |
| mir-127 | -0.50 | mir-103-1 | -0.14 |
| mir-379 | -0.44 | mir-1323 | 1.01 |
| mir-514-2 | -1.90 | mir-23a | -0.03 |
| mir-1247 | 0.47 | mir-26b | 0.04 |
| mir-125b-2 | 0.29 | mir-28 | 0.11 |
| mir-218-2 | 0.60 | mir-381 | -0.36 |
| mir-29c | 0.79 | ||
| mir-3130-1 | 0.94 | ||
| mir-412 | -1.75 | ||
| mir-513c | -2.31 | ||
Next, classification analysis was used to validate the ability of the 24 miRNA identified by feature selection to discriminate primary and metastatic tumors. An ensemble decision tree model using the RUSBoost algorithm (appropriate for class imbalance) [41] performed the best at predicting tumor class, with an overall prediction accuracy of 86.6% and a specificity of 88% (true negatives). However, the sensitivity was only 80% (true positives), giving the model an overall precision of 66% (the proportion of results that are true positives; Figure 2D). Plotting the rate of true positives against the rate of false positives produced a ROC curve with an AUC = 0.93 (Figure 2E). Notwithstanding acceptable overall performance and classification of metastatic tumors, 20% of primary tumors were misclassified. Despite this, these analyses confirmed that the miRNA identified by feature selection could discriminate a large proportion of primary tumors from metastases.
Cutaneous Epithelial Cell-Types Are Enriched in Primary Melanoma Tumors
To further examine the candidate metastasis-suppressor miRNA identified by differential expression and feature selection (defined as those miRNAs downregulated between primary and metastatic tumors), Kaplan-Meier survival analysis was performed to assess patient survival in relation to miRNA expression. Interestingly, no significant differences in survival curves were identified for patients with high or low expression of miR-205, or miR-203 (miR-205 P = .805; miR-203 P = .265) (Figure S1, A and B), despite being the most differential miRNA between primary and metastatic tumors, and the most stable discriminating features. In addition, no differences in survival were detected for miR-200a, miR-200b, miR-200c, or miR-141 that were also highly differentially expressed in primary and metastatic tumors (data not shown). Visualizing miR-205 expression in primary and metastatic tumors revealed that a surprisingly large number had no expression of the miRNA (Figure 3A), raising questions as to the source of expression.
Figure 3.
Cutaneous epithelial cell-types are enriched in primary SKCM melanoma tumors. (A) Scatter plot visualizing expression of hsa-miR-205 in primary (blue), and metastatic (purple) SKCM tumors. (B) Violin plot of ABSOLUTE purity estimations for the SKCM tumors. (C) Average xCell cell enrichment scores for primary and metastatic SKCM tumors derived from RNA-seq gene expression quantification. Largest enrichments in primary tumors are: sebocytes, keratinocytes, and epithelial cells (P < .001, Mann Whitney U). (D) xCell enrichment scores for keratinocytes, sebocytes, epithelial cells, and melanocytes across all SKCM tumors.
To assess the possible sources of miRNA expression in the SKCM tumors, tumor purity was calculated using the ABSOLUTE method [26] across the cohort. The median tumor purity was 0.71 (71%), with a range of 0.1-1.0 (Figure 3B). To better understand the cellular composition of the tumors, xCell cell-type enrichment analysis was also performed using RNAseq data [27]. Cell-types enriched in primary tumors were considered as potential correlates with the apparent downregulation of miRNA upon metastasis. The largest differences identified were sebocytes, keratinocytes, and epithelial cell signatures (P < .001). Interestingly, a decrease in melanocyte-specific expression signatures was also observed between primary and metastatic tumors. This suggests that the metastatic tumors may have a greater proportion of infiltrating stromal cells, and supports the notion that metastasis is accompanied by a loss of melanocyte differentiation (Figure 3C) [40]. Visualizing the enrichment scores for the primary tumor-enriched cutaneous epithelial cell-types across all samples, it was noted that these signatures were not present in all primary tumors (Figure 3D). However, the proportion of primary tumors with enrichments was likely sufficient to influence differential and classification-based analyses of miRNA expression.
Candidate Metastasis-Suppressor miRNAs Are Associated with Enrichment of Stromal Cell-Type-Specific Gene Expression Signatures in SKCM Tumors
To examine the potential influence of stromal cell types on the identification of metastasis-suppressor miRNA in the SKCM tumors, Spearman correlations between expression of candidate metastasis-suppressor miRNA and cell-type enrichment scores were computed (File S1). Hierarchical clustering of rho values revealed subsets of miRNA correlated with enrichment of stromal cell and melanocyte signatures (Figure 4A). The miRNA most positively correlated with melanocyte signatures were miR-211 and members of the miR-506/514 miRNA cluster, which were significantly downregulated in metastatic tumors. However, miR-205, miR-203, and the miR-200 family were most positively correlated with the cutaneous epithelial cell-types enriched in the primary tumors (Figure 4A, Table 5).
Figure 4.
Expression of candidate metastasis-suppressor miRNA is associated with enrichment of stromal cell-type-specific gene expression signatures. (A) Hierarchical clustering of the Spearman’s rank correlation coefficient between miRNA significantly downregulated between primary and metastatic SKCM tumors, and xCell cell enrichment scores. Euclidean distance and average linkage used. Correlations between all miRNA and xCell scores provided in Supplemental File 1 (B) Heatmap of Spearman’s rank correlation coefficients between miRNA and mRNA expressed in the SKCM tumors. Purple: genes expressed in keratinocytes; grey: melanocytes; yellow: ubiquitous. (C) Heatmap of Spearman’s rank correlation coefficients between epithelial-enriched and melanocyte-enriched miRNA, MITF, and EMT transcription factors expressed in the SKCM tumors. Corresponding P-values for all correlations provided in Supplemental File 2.
Table 5.
miRNA positively correlated with the cell-types most enriched in primary SKCM tumors
| Melanocytes | Sebocytes | Keratinocytes | Epithelial cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | rho | FDRpval | miRNA | rho | FDRpval | miRNA | rho | FDRpval | miRNA | rho | FDRpval |
| hsa-mir-211 | 0.81 | 1.91E-102 | hsa-mir-205 | 0.50 | 1.51E-26 | hsa-mir-205 | 0.49 | 1.82E-25 | hsa-mir-200c | 0.48 | 2.37E-24 |
| hsa-mir-514-3 | 0.51 | 1.18E-28 | hsa-mir-200c | 0.49 | 8.39E-26 | hsa-mir-200c | 0.48 | 1.80E-24 | hsa-mir-141 | 0.41 | 3.15E-17 |
| hsa-mir-514-2 | 0.51 | 1.18E-28 | hsa-mir-200a | 0.47 | 1.80E-23 | hsa-mir-200a | 0.48 | 1.80E-24 | hsa-mir-200a | 0.41 | 4.17E-17 |
| hsa-mir-514-1 | 0.51 | 1.18E-28 | hsa-mir-200b | 0.46 | 1.02E-22 | hsa-mir-200b | 0.47 | 1.63E-23 | hsa-mir-200b | 0.40 | 9.62E-17 |
| hsa-mir-508 | 0.51 | 1.63E-28 | hsa-mir-203 | 0.46 | 1.04E-22 | hsa-mir-203 | 0.44 | 9.61E-21 | hsa-mir-203 | 0.36 | 1.37E-12 |
| hsa-mir-509-3 | 0.49 | 3.15E-26 | hsa-mir-141 | 0.46 | 2.01E-22 | hsa-mir-141 | 0.43 | 4.43E-20 | hsa-mir-183 | 0.35 | 6.96E-12 |
| hsa-mir-509-2 | 0.48 | 8.01E-26 | hsa-mir-224 | 0.34 | 1.67E-11 | hsa-mir-224 | 0.35 | 1.50E-12 | hsa-mir-452 | 0.34 | 9.10E-12 |
| hsa-mir-509-1 | 0.48 | 8.80E-26 | hsa-mir-452 | 0.32 | 3.66E-10 | hsa-mir-452 | 0.33 | 1.02E-10 | hsa-mir-205 | 0.33 | 1.33E-10 |
| hsa-mir-506 | 0.45 | 5.14E-22 | hsa-mir-31 | 0.31 | 1.34E-09 | hsa-mir-224 | 0.31 | 1.17E-09 | |||
| hsa-mir-513c | 0.42 | 1.40E-18 | |||||||||
| hsa-mir-187 | 0.40 | 2.22E-17 | |||||||||
| hsa-mir-514b | 0.38 | 1.20E-15 | |||||||||
To further confirm these associations, we selected specific markers of keratinocytes (KRT1, KRT5, KRT14, IVL, CASP14, LOR [42]), and melanocytes (DCT, GPNMB, TYRP1, TYR, MLANA, and MITF [43]), and their expression validated in both the SKCM tumors and in the Ludwig Melbourne melanoma (LM-MEL) cell line panel comprised of 57 melanoma cell lines with a range of invasive phenotypes (Table S2, Figures S2-S4) [16]. Expression of the keratinocyte markers were undetectable in any melanoma cell lines, and all had significantly higher expression in primary SKCM tumors compared to metastatic (P < .001) (Figures S4). Next, we computed Spearman correlations between expression of the cell-specific markers and the cutaneous epithelial cell-enriched miRNA (miR-205, miR-203, and the miR-200 family [[44], [45], [46], [47], [48]]), melanocyte-enriched miR-211 [49], and ubiquitously expressed miR-21 in the SKCM tumors. The melanocyte markers were most positively correlated with expression of miR-211, while miR-205, miR-203, and the miR-200 family were most positively correlated with the keratinocyte markers, with no clear association to the melanocyte-specific genes. The ubiquitously expressed miR-21 was not appreciably correlated with markers for either cell type (Figure 4B).
The candidate metastasis-suppressor miRNAs miR-205, miR-203, and the miR-200 family are known to regulate epithelial-to-mesenchymal transition (EMT) and epithelial differentiation [[44], [45], [46], [47], [48]]. However, melanocytes are derived from the neural crest, and microphthalmia-associated transcription factor (MITF) is critical for driving lineage-specific transcriptional programs in melanocytes [50,51] that includes expression of miR-211 [49]. In normal melanocytes, EMT transcription factors SNAI2 and ZEB2 drive MITF-dependent melanocyte differentiation. Upon oncogenic transformation, a switch occurs whereby TWIST1 and ZEB1 are activated, resulting in a loss of E-cadherin and repression of differentiation [40]. In the SKCM tumors, expression of SNAI2 and CDH1 were elevated in primary tumors, while ZEB1 and CDH2 were elevated in metastatic tumors. Expression of TWIST1 and ZEB2 were more variable (Figure S5A-F). Expression of miR-211 and MITF were positively correlated with CDH1, ZEB2, and SNAI2, and negatively correlated with CDH2, TWIST1, and ZEB1 (Figure 4C). Furthermore, miR-205, miR-203, and the miR-200 family had weaker, and inconsistent associations with expression of the EMT transcription factors. However, all were negatively correlated with expression of ZEB2 [40]. Together, these findings suggest that expression of miRNAs that are enriched in populations of cutaneous epithelial cells, including keratinocytes and sebocytes, contribute to the identification of miRNA as putative metastasis-suppressors from tissue-level miRNA expression profiles. Therefore, any functional role of these miRNA in regulating EMT in melanoma may differ from what is understood for epithelial cells.
Tumor-Identified Metastasis-Suppressor miRNA Enriched in Cutaneous Epithelial Cells are Poorly Expressed in Melanoma Cell Lines
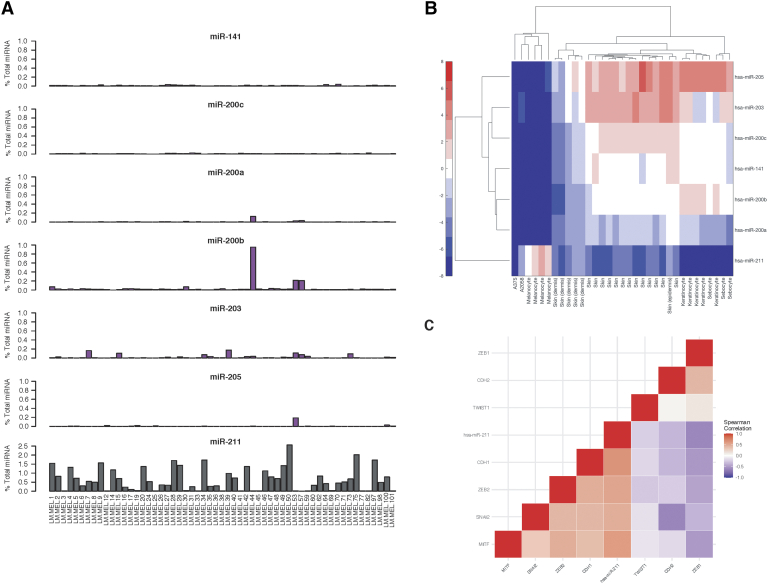
In light of the strong associations between a number of the candidate metastasis-suppressor miRNA and enrichments in stromal cell gene expression signatures in the SKCM tumors, further investigation into the sources of miRNA expression was warranted. To examine this, expression of the cutaneous epithelial cell-enriched miRNA (miR-205, miR-203, and the miR-200 family), along with melanocyte-associated miR-211 was surveyed in small RNAseq miRNA expression data from the LM-MEL cell line panel [15]. Only miR-200b exceeded 0.2% of total miRNA in three of 57 cell lines. Sparse expression of the other cutaneous epithelial cell-enriched miRNA, when detected, reached only 0.01-0.2% of the total miRNA reads. Conversely, miR-211 was expressed at level beyond 1% of total reads in many of the cell lines, consistent with having a functional role in melanoma (Figure 5A). To investigate these expression patterns, two additional melanoma cell line panels were examined. In a panel of 51 cell lines derived from both primary and metastatic tumors, expression of the miR-200 family was found to be very low or absent in all samples, while miR-205 and miR-203 did not meet minimal detection thresholds (Figure S6). An additional panel of 21 patient-derived melanoma cell lines followed the same pattern, with only miR-141/miR-200c exhibiting expression in one sample (Figure S7). Furthermore, none of the cutaneous epithelial cell-enriched miRNA were detected in the five normal melanocyte samples included in these datasets. Small RNAseq data harmonized by the microRNAome project [19] also revealed high levels of miR-205, miR-203, and the miR-200 family expressed in keratinocytes, sebocytes, and normal skin tissue samples, but absent in normal melanocytes and melanoma cell lines (A375, A2058) (Figure 5B). Interestingly, skin samples from the dermis had lower expression of the cutaneous epithelial cell-enriched miRNA as compared to the epidermis, which may account for reports of downregulation with increased thickness of primary tumors.
Figure 5.
Tumor-identified metastasis-suppressor miRNA enriched in keratinocytes are poorly expressed in melanoma cell lines. (A) Relative expression of the miR-200 family, miR-205, and miR-203, and melanocyte-enriched miR-211 in a panel of 57 patient-derived melanoma cell lines (LM-MEL, [15]). (B) Hierarchical clustering of hsa-miR-205, hsa-miR-203, and the hsa-miR-200 family expression in melanoma cell lines (A375 and A2058), normal melanocytes, keratinocytes, sebocytes, and normal skin tissue samples included in the microRNAome project [19]. (C) Heatmap of Spearman’s rank correlation coefficients between miRNA expressed in melanoma cell lines, MITF, and EMT transcription factors expressed in the LM-MEL cell line panel. Corresponding p-values for all correlations provided in Supplemental File 2.
Finally, expression of miR-211 in the LM-MEL cell line panel was found to be highly positively correlated with MITF, SNAI2, CDH1, and ZEB2, and negatively correlated with CDH2 and ZEB1 (Figure 5C, Figure S5G), consistent with its known functional role in maintaining melanocyte differentiation and suppressing EMT-like processes driving phenotypic plasticity [17,40,49]. Together, these findings suggest that the highly altered expression patterns of miR-205, miR-203, and the miR-200 family detected in the TCGA SKCM cohort are likely not derived from tumor cell-intrinsic changes in miRNA expression.
miRNA-Based Classification of Primary and Metastatic Melanoma Tumors Depends Largely on miRNA Highly Expressed in the Primary Tumor Stroma
In light of the expression patterns observed in cell lines, feature selection and classification analyses were repeated to examine the influence of the cutaneous epithelial cell-enriched miRNA on discrimination of primary and metastatic tumors. After filtering of cutaneous stromal-enriched miRNA (Table 6), 16 miRNA were among the top 20% with the highest selection frequency (Figure 6A). Nine were differentially expressed between primary and metastatic tumors, while an additional seven were not (Table 7). Only three miRNA (miR-215, miR-218-2, and miR-29c) overlapped with the first feature selection, suggesting that most others were identified due to interaction with one or more of the cutaneous stromal-enriched miRNAs removed. Classification analysis was repeated using the 16 features, and a Medium Gaussian Support Vector Machine (SVM) model predicted primary and metastatic classes with the highest overall accuracy of 85.7%. The specificity and sensitivity of this model were 98% and 42%, respectively. The overall precision was 84% (Figure 6B) and AUC = 0.83 (Figure 6C). However, 58% of primary tumors were misclassified, compared to 20% when the cutaneous stromal-enriched miRNAs were included in feature selection. Most apparent positive performance of the classifier was due to the skewing of the dataset towards metastatic tumors, such that a model that predicts all tumors as metastatic could still achieve more than 75% accuracy. As expected, based on the poor classification of primary tumors, hierarchical clustering using the 16 selected features resulted in no separation of primary and metastatic groups (Figure 6D). Further optimizing feature selection would improve classification; however, these findings demonstrate that the strongest discriminating features in the SKCM tumor miRNA expression profiles were those derived from epithelial stromal cells enriched at the cutaneous tissue site. Overall, this work highlights the challenges of interpreting tissue-level miRNA expression patterns between primary and metastatic tumors, where it is necessary to decode tumor-intrinsic changes in miRNA expression from those arising due to diversity in stromal cell populations (Figure 6E).
Table 6.
Expression of candidate metastasis-suppressor miRNA in melanoma and cutaneous stromal cell-types (% total miRNA reads)
| miRNA | Keratinocytesa (n=3) | Keratinocytesb (n=5) | Sebocytesb (n=3) | Melanoma cellsc (n=57) | Melanoma cellsb (n=2) | Melanocytesb (n=5) |
|---|---|---|---|---|---|---|
| mir-141 | 2.79 ± 1.75 | 0.52 ± 0.04 | 0.31 ± 0.13 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.00 ± 0.00 |
| mir-200a | 0.21 ± 0.20 | 0.08 ± 0.08 | 0.04 ± 0.02 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| mir-200b | 1.45 ± 0.20 | 0.93 ± 0.19 | 0.36 ± 0.06 | 0.04 ± 0.13 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| mir-200c | 3.27 ± 0.35 | 0.43 ± 0.10 | 0.29 ± 0.12 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| mir-203 | 3.64 ± 1.42 | 0.43 ± 0.44 | 0.76 ± 0.41 | 0.02 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| mir-205 | 0.78 ± 0.53 | 8.97 ± 1.82 | 7.29 ± 4.00 | 0.01 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| miR-211 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.61 ± 0.63 | 0.00 ± 0.00 | 1.55 ± 0.88 |
Figure 6.
miRNA-based classification of primary and metastatic melanoma tumors depends largely on miRNA highly expressed in the primary tumor stroma. (A) Top 20% of miRNA features selected with the highest frequency after removal of putative keratinocyte-derived miRNA. The y-axis shows the number of feature subsets a miRNA appeared in, out of a possible 50. (B) Confusion matrices for the Medium Gaussian SVM classifier constructed using the miRNA identified in the top 20% of feature selection. (C) ROC curve for the same SVM classification model. (D) Hierarchical clustering of all primary and metastatic samples using the 16 miRNA identified in the top 20% of selected features. All parameters the same as previously stated. (E) Schematic showing how differences in stromal cell composition between primary and metastatic tissue sites are reflected in tissue-level miRNA expression. Differences in miRNA expression between primary and metastatic tumors will reflect both tumor cell-intrinsic changes, as well as changes in cell-type enriched miRNA that are dependent on stromal cell populations. Primary cutaneous melanoma tumors share the property of all being situated in the skin, while metastatic tumors can have a diverse set of stromal contexts. For this reason, candidate “metastasis-suppressor” miRNA with large apparent negative fold changes between primary and metastatic sites should be investigated for cell-type enriched expression patterns.
Table 7.
The top 20% of miRNA identified during feature selection to classify primary and metastatic tumors after filtering of cutaneous stromal-enriched miRNA
| Identified by differential expression and feature selection |
Identified by feature selection |
||
|---|---|---|---|
| miRNA | Log2 fold-change | miRNA | Log2 fold-change |
| mir-202 | -1.34 | mir-520h | 0.80 |
| mir-218-2 | 0.60 | miR-525 | 0.46 |
| mir-29c | 0.79 | let-7a-1 | 0.07 |
| mir-452 | -0.76 | mir-197 | 0.27 |
| mir-215 | 1.15 | mir-498 | 0.58 |
| mir-223 | -0.69 | mir-146b | 0.13 |
| mir-505 | 0.43 | mir-518a-1 | 0.84 |
| mir-378c | 0.59 | ||
| mir-675 | 1.53 | ||
Discussion
Despite recent advancements in targeted therapies to treat cutaneous melanoma, resistance is common, and survival is remains poor for patients with metastatic disease [4,5]. In the current study, differential expression analysis and sequential feature selection using classification algorithms identified candidate metastasis-suppressor miRNA in the TCGA-SKCM cohort. However, expression of the most highly downregulated miRNA between primary and metastatic tumors (miR-205, miR-203, and the miR-200 family) were found to be specifically associated with enrichments of cutaneous epithelial cell signatures, including keratinocytes and sebocytes, in the primary tumors. Subsequent comparisons in cell lines revealed that the presence of cutaneous epithelial cell-types likely accounted for the large apparent differences in a number of candidate metastasis-suppressor miRNA downregulated between primary and metastatic tumor tissues. Filtering of cutaneous stromal-enriched miRNA greatly reduced the performance of classification models, suggesting a major role for stromal-derived, rather than tumor cell-intrinsic miRNA expression in discriminating primary from metastatic melanoma tumors. This study demonstrates the importance of considering tissue source and composition when identifying putative metastasis-suppressor miRNA in tissue samples, and supports validation of cell-type-specific miRNA expression patterns to ensure that physiologically relevant models of melanoma progression will be created and studied in the future.
Some of the most highly studied metastasis-suppressor miRNA in melanoma include miR-205, miR-203, and the miR-200 family (made up of miR-200a/miR-200b and miR-200c/miR-141), with numerous reports of downregulation in metastatic tumors, and tumor and metastasis-suppressive functions [10,11,32,34,36,39,52]. These studies have relied on both conventional differential expression, as well as supervised machine learning algorithms to identify miRNA of interest from tumor tissue expression profiles [11,32,34,36,39,52]. However, studies that have incorporated miRNA expression profiling of melanoma cell lines have consistently identified a different subset of miRNAs associated with tumor- and invasion/metastasis-suppressive functions, including miR-211, and miR-204 [17,49,53]. This disconnect suggests that melanoma cell models may be poorly representative of tumor miRNA expression patterns, or that identification of candidate metastasis-suppressor miRNA from tissue-level data is influenced by cellular composition. After finding miRNA highly downregulated between primary and metastatic melanoma tumors to be undetectable in nearly all 131 melanoma cell lines examined here, we propose that stromal cell composition differences between primary and metastatic tumors dominate the tissue-level miRNA expression profiles, and is under reported.
When heterogeneous tissue samples are homogenized to extract RNA, critical spatial information is lost. Without prior knowledge of miRNA expression patterns at a cellular level, it becomes exceedingly difficult to determine the source of miRNA signals. For example, miR-143/145 are highly studied as tumor suppressors in colorectal cancer due to differential expression in tumor tissues, but were eventually shown to be highly expressed in tumor-adjacent fibroblasts and smooth muscle cells, and not in cancer cells [54]. In addition, tumors possess many properties that can further confound this issue. Due to altered vasculature, miRNAs that are highly expressed in endothelial cells and erythrocytes (miR-126, miR-144, miR-451a, and miR-486) are often identified as differentially expressed in solid tumors, and assigned tumor-intrinsic functions [[55], [56], [57], [58]]. Similarly, changes in tissue composition can occur with the recruitment of immune cells to tumors, leading to enrichment of immune cell miRNA signatures [58]. Keratinocytes make up approximately 90% of the cells in the epidermal layer [59]. Considering that all primary cutaneous melanoma tumors are situated in the skin, while metastatic tumors are derived from a diverse set of organs and tissues, it is logical that the most highly and consistently downregulated miRNA between primary and metastatic tumor tissue samples could arise from differences in stromal tissue composition.
The roles of miR-205, miR-203, and the miR-200 family have been well-documented in epithelial cells. The miR-200 family and miR-205 are known to suppress EMT by enhancing expression of E-cadherin through targeting of transcription factors ZEB1 and ZEB2, maintaining epithelial differentiation [47,60,61]. In addition, miR-203 and miR-200 members play a critical role in the terminal differentiation of keratinocytes [44,61]. Indeed, both miR-205 and miR-203 are prognostic markers in cutaneous squamous cell carcinomas [45], arising from keratinocytes [20]. In melanocytes, derived from the neural crest, the transcription factor MITF drives differentiation [50,51]. In the context of melanoma, low MITF expression is associated with invasion, loss of expression of epithelial genes, and increased phenotypic plasticity [13]. Melanoma cells are known to undergo an EMT-like process where epithelial markers and cell contacts are lost [76]. However, this process has been found to differ from the classical EMT paradigm established in epithelial cancers, whereby loss of MITF and miR-211 expression occurs with a switch from ZEB2 and SNAI2, to ZEB1 and TWIST1 [62]. Here, we found that miR-211 was downregulated in metastatic tumors, and was highly positively correlated with MITF, ZEB2, and SNAI2 in tumors and melanoma cell lines, providing further confirmation of its established role as a tumor and metastasis-suppressor [11,17,49,53]. However, given the lack of expression in cell lines, and limited association with markers of EMT and melanocyte differentiation, the role of epithelial-enriched miRNA like miR-205, miR-203, and the miR-200 family in melanoma requires further clarification.
In light of the poor of expression of miR-205, miR-203, and the miR-200 family in melanoma cell lines and normal melanocytes reported here, many of the models used to functionally characterize these miRNA in melanoma cell lines may represent systems of ectopic, rather than rescue of expression. In cases where there is a “down” phenotypic readout, for example, a decrease in cell invasion or reduced viability, results should be interpreted carefully. Such “down” readouts are vulnerable to off-target or non-specific effects on cellular fitness, as there are typically more ways to disrupt a complex phenotype than there are to enhance it [63]. Further to this, phenotypic and molecular changes documented with the use of miRNA inhibitors against miRNA not expressed in melanoma cell lines should be revisited by verifying endogenous expression levels and potential off target effects.
Although the miRNA expression patterns we report here for a large number of melanoma cell lines strongly implicates stromal composition as the major source of differential miRNA expression between primary and metastatic tumor tissues, a biological role for stromal-enriched miRNA in melanoma metastasis should not be immediately discounted. Melanocytes and keratinocytes are known to interact, and changes in these interactions have been linked to melanoma tumorigenesis and metastasis [64,65]. Furthermore, exosomes released from keratinocytes can modulate melanocyte function, and contain miRNA such as miR-200a and miR-203 [66]. It is also well established that cell lines do not always reflect the tumor biology [67], allowing the possibility that expression of miRNA such as miR-205, miR-203, and the miR-200 family are acquired in tumor progression through a mechanism that is not recapitulated in cultured cell models. However, the likelihood of this scenario must be weighed against one where expression of miRNA detected in homogenized tissue samples arises from other cell types that could be present in that tissue. Further investigation using miRNA fluorescent in situ hybridization (FISH) to assess the spatial expression patterns within fixed melanoma tumor sections can help define the source of miRNA expression [68]. Finally, recent advances in single-cell RNA-seq methods can allow population studies of cell types and miRNAs within melanoma tumors at different stages, and collected from different tissue sites with less confounding effects [69].
Our findings show that identification of metastasis-associated miRNA in melanoma tumors may be more challenging than currently appreciated. Although 80% of primary tumors were correctly classified before filtering of cutaneous stromal-enriched miRNA, this was reduced to 42% when these miRNAs were removed from the feature set, suggesting that changes in tumor-intrinsic miRNA expression upon metastasis are likely subtle and variable. This difficulty in separating classes was in part due to primary tumors being underrepresented in the cohort, making up only 21% of samples. Melanoma has often metastasized prior to diagnosis [70], and when primary tumors are resected, they are often too small to contribute materials to molecular analyses beyond diagnosis [71]. For these reasons, the primary samples included in the TCGA SKCM cohort may be advanced tumors with little differences to metastatic samples. As such, analyses focused on comparing primary and metastatic tumors may not provide the insights expected from a dataset of this size. In the future, survival-based comparisons could produce clinically relevant findings, and efforts to study longitudinal samples for individual patients should be prioritized to define metastasis-associated miRNAs.
Apart from stromal-enriched miRNA, dysregulation of large miRNA gene clusters may be a hallmark of the melanoma miRNA landscape. Expression of the miR-506/514 cluster has been reported in melanoma [31]. However, our identification of C19MC activation in melanoma is novel. Aberrant C19MC expression has been reported in several cancers, and these miRNA have immune-modulatory effects potentially relevant to melanoma [30,72]. However, little is known about how C19MC expression is acquired, or the functional consequence this has in tumors. Given the growing number of reports in other cancer types [[73], [74], [75]], further investigation into the role of C19MC in melanoma is highly warranted.
In conclusion, this work has demonstrated that many of the most highly studied metastasis-suppressor miRNAs in melanoma are derived from cutaneous stromal sources that differ between primary and metastatic tumor samples. These findings draw attention to the vulnerability of both differential expression analyses and supervised classification algorithms to spurious identification of metastasis-suppressor miRNA when consideration is not given to the cellular composition of tissue samples. We recommend that expression of candidate metastasis-suppressor miRNA be verified in appropriate cell lines to confirm tumor cell-intrinsic expression patterns, or that studies of relevant stromal mechanisms during tumor progression be included in experimental models. Currently, these expression validations can be labor-intensive, limited by data availability, and involve heuristic methods. Going forward, coordinated efforts to aggregate cell- and tissue-level miRNA sequencing, and the development of software tools to make this information widely accessible will be invaluable to improve the relevance and reproducibility of miRNA studies.
Funding
This research was funded by grants from Cancer Research Society (20142) and Canadian Institutes of Health Research (MOP 119562) to AWBC.
Acknowledgments
This research was funded by grants from Cancer Research Society (20142) and Canadian Institutes of Health Research (MOP 119562) to AWBC.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100802.
Appendix A. Supplementary data
Supplementary material
References
- 1.Statistics, C. C. S. s. A. C. O. C . Canadian Cancer Society; Toronto, ON: 2018. Canadian Cancer Statistics; A 2018 Special Report on cancer incidence by stage. [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Cancer, A. J. C. o. 7th ed. Springer; Chicago: 2010. AJCC Cancer Staging Handbook 7th Edition. [Google Scholar]
- 4.O'Donnell J.S., Long G.V., Scolyer R.A., Teng M.W., Smyth M.J. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat Rev. 2017;52:71–81. doi: 10.1016/j.ctrv.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Arozarena I., Wellbrock C. Overcoming resistance to BRAF inhibitors. Ann Transl Med. 2017;5:387. doi: 10.21037/atm.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pencheva N., Tavazoie S.F. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Lin C., Socci N.D., Hermida L., Fulci V., Chiaretti S., Foa R., Schliwka J., Fuchs U., Novosel A., Muller R.U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D.B., Choksi R., De Vita G., Frezzetti D., Trompeter H.I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C.E., Nagle J.W., Ju J., Papavasiliou F.N., Benzing T., Lichter P., Tam W., Brownstein M.J., Bosio A., Borkhardt A., Russo J.J., Sander C., Zavolan M., Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavazoie S.F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Brenn T., Brown E.R.S., Doherty V., Melton D.W. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentini V., Zelli V., Gaggiano E., Silvestri V., Rizzolo P., Bucalo A., Calvieri S., Grassi S., Frascione P., Donati P., Soda G., Ottini L., Richetta A.G. MiRNAs as potential prognostic biomarkers for metastasis in thin and thick primary cutaneous melanomas. Anticancer Res. 2019;39:4085–4093. doi: 10.21873/anticanres.13566. [DOI] [PubMed] [Google Scholar]
- 12.van Kempen L.C., van den Hurk K., Lazar V., Michiels S., Winnepenninckx V., Stas M., Spatz A., van den Oord J.J. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch. 2012;461:441–448. doi: 10.1007/s00428-012-1309-9. [DOI] [PubMed] [Google Scholar]
- 13.Akbani, R., Kadir, B, Albert, M., Ally, A., Samirkumar, Arachchi, H., Arora, A., J, Ayala, B., Baboud, J., Balasundaram, M., Balu, S., Barnabas, N., Bartlett, J., Bartlett, P., Boris, Stephen, Behera, M., Belyaev, D., Benz, C., Bernard, B., Beroukhim, R., Bir, N., Aaron, Bodenheimer, T., Boice, L., Genevieve, Bono, R., Moiz, Bosenberg, M., Bowen, J., Bowlby, R., Christopher, Brockway-Lunardi, L., Brooks, D., Brzezinski, J., Bshara, W., Buda, E., William, Yaron, Button, M., Calderone, T., Giancarlo, Carter, C., Scott, Cherney, L., Andrew, Chevalier, A., Chin, L., Cho, J., Raymond, Choi, Y.-L., Chu, A., Chudamani, S., Cibulskis, K., Ciriello, G., Clarke, A., Coons, S., Cope, L., Crain, D., Curley, E., Danilova, L., D’Atri, S., Davidsen, T., Michael, Keith, John, Qixia, Yonathan, Dhalla, N., Dhir, R., Dicara, D., Dinikin, M., Dubina, M., J, Egea, S., Eley, G., Engel, J., Jennifer, Konstantin, Felau, I., Fennell, T., Martin, Fisher, S., Keith, Frazer, S., Frick, J., Fulidou, V., Stacey, Gao, J., Gardner, J., Levi, Julie, Gaudioso, C., Gehlenborg, N., Genovese, G., Gerken, M., Jeffrey, Getz, G., Gomez-Fernandez, C., Gribbin, T., Grimsby, J., Gross, B., Guin, R., Gutschner, T., Hadjipanayis, A., Halaban, R., Hanf, B., Haussler, D., Lauren, D, Nicholas, David, Herbert, L., James, Hersey, P., Katherine, Hodis, E., Robert, Dave, Hoppough, S., Alan, Franklin, Huang, M., Huang, S., Carolyn, Ibbs, M., Iype, L., Jacobsen, A., Jakrot, V., Janning, A., William, Stuart, Mark, Corbin, Steven, Ju, Z., Kakavand, H., Kang, H., Richard, Fadlo, Kim, J., John, Klode, J., Korkut, A., Korski, K., Krauthammer, M., Kucherlapati, R., Lawrence, Kycler, W., Ladanyi, M., Phillip, Peter, Lander, E., Michael, Alexander, Łaźniak, R., Lee, D., Jeffrey, Lee, J., Lee, K., Lee, S., Lee, W., Leporowska, E., Kristen, Haiyan, Tara, Lichtenstein, L., Lin, P., Ling, S., Liu, J., Liu, O., Liu, W., Georgina, Lu, Y., Ma, S., Ma, Y., Mackiewicz, A., Harshad, Malke, J., Mallery, D., Georgy, Graham, Marco, Matejka, B., Mayo, M., Mehrabi, S., Meng, S., Meyerson, M., Piotr, John, Martin, Gordon, Moiseenko, F., Richard, Morris, S., Morrison, C., Morton, D., Moschos, S., Lisle, Florian, Andrew, Murawa, D., Murawa, P., Bradley, Nezi, L., Ng, S., Nicholson, D., Michael, Osunkoya, A., Taofeek, Bradley, Pagani, E., Oxana, Pantazi, A., Parfenov, M., Parfitt, J., Peter, Park, W.-Y., Joel, Passarelli, F., Penny, R., Charles, Todd, Potapova, O., Victor, Protopopov, A., Michael, Radenbaugh, A., Rai, K., Suresh, Ayush, Nilsa, Ramirez, R., Rao, U., W, Ren, X., Sheila, Roach, J., A, Merrick, Roszik, J., Russo, G., Saksena, G., Saller, C., Samuels, Y., Sander, C., Sander, C., Sandusky, G., Santoso, N., Saul, M., Robyn, Schadendorf, D., Jacqueline, Schultz, N., Steven, Schwallier, C., Richard, Seidman, J., Pedamallu, Harmanjatinder, Senbabaoglu, Y., Seth, S., Kerwin, Sharpe, S., Norman, Kenna, Shelton, C., Shelton, T., Shen, R., Sheth, M., Shi, Y., Carolyn, Shmulevich, I., Gabriel, Janae, Sinha, R., Sipahimalani, P., Heidi, Matthew, Song, X., Sougnez, C., Andrew, Spychała, A., Jonathan, Stuart, J., Wiktoria, Sucker, A., S, Sun, Y., Synott, M., Tabak, B., Teresa, Tam, A., Tan, D., Tang, J., Tarnuzzer, R., Tarvin, K., Tatka, H., Barry, Teresiak, M., Thiessen, N., John, Thorne, L., Thorsson, V., Jeffrey, Timothy, Kenneth, Tsou, P., David, Eliezer, Veluvolu, U., Roeland, Voet, D., Voronina, O., Walter, V., Jessica, Wan, Y., Wang, Y., Wang, Z., Waring, S., Ian, Weinhold, N., John, Daniel, White, P., Matthew, James, Wise, L., Wiznerowicz, M., Scott, Wu, C.-J., Wu, C.-C., Wu, J., Wu, Y., Xi, R., Andrew, Yang, D., Yang, L., Yang, L., Travis, Jean, Zhang, H., Zhang, J., Zhang, W., Zhao, X., Zhu, J., Zhu, K., Zimmer, L., Zmuda, E., and Zou, L. (2015) Genomic classification of cutaneous melanoma. Cell 161, 1681-1696
- 14.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V., Akbani R., Bowlby R., Wong C.K., Wiznerowicz M., Sanchez-Vega F., Robertson A.G., Schneider B.G., Lawrence M.S., Noushmehr H., Malta T.M., Cancer Genome Atlas N., Stuart J.M., Benz C.C., Laird P.W. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304. doi: 10.1016/j.cell.2018.03.022. e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews M.C., Cursons J., Hurley D.G., Anaka M., Cebon J.S., Behren A., Crampin E.J. Systems analysis identifies miR-29b regulation of invasiveness in melanoma. Mol Cancer. 2016;15:72. doi: 10.1186/s12943-016-0554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behren A., Anaka M., Lo P.-H., Vella L.J., Davis I.D., Catimel J., Cardwell T., Gedye C., Hudson C., Stan R., Cebon J. The Ludwig Institute for Cancer Research Melbourne Melanoma Cell Line Panel. Pigment Cell Melanoma Res. 2013;26:597–600. doi: 10.1111/pcmr.12097. [DOI] [PubMed] [Google Scholar]
- 17.Boyle G.M., Woods S.L., Bonazzi V.F., Stark M.S., Hacker E., Aoude L.G., Dutton-Regester K., Cook A.L., Sturm R.A., Hayward N.K. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24:525–537. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 18.Caramuta S., Egyhazi S., Rodolfo M., Witten D., Hansson J., Larsson C., Lui W.O. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 19.McCall M.N., Kim M.S., Adil M., Patil A.H., Lu Y., Mitchell C.J., Leal-Rojas P., Xu J., Kumar M., Dawson V.L., Dawson T.M., Baras A.S., Rosenberg A.Z., Arking D.E., Burns K.H., Pandey A., Halushka M.K. Toward the human cellular microRNAome. Genome Res. 2017;27:1769–1781. doi: 10.1101/gr.222067.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizrahi A., Barzilai A., Gur-Wahnon D., Ben-Dov I.Z., Glassberg S., Meningher T., Elharar E., Masalha M., Jacob-Hirsch J., Tabibian-Keissar H., Barshack I., Roszik J., Leibowitz-Amit R., Sidi Y., Avni D. Alterations of microRNAs throughout the malignant evolution of cutaneous squamous cell carcinoma: the role of miR-497 in epithelial to mesenchymal transition of keratinocytes. Oncogene. 2018;37:218–230. doi: 10.1038/onc.2017.315. [DOI] [PubMed] [Google Scholar]
- 21.Panarelli N., Tyryshkin K., Wong J.J.M., Majewski A., Yang X., Scognamiglio T., Kim M.K., Bogardus K., Tuschl T., Chen Y.T., Renwick N. Evaluating gastroenteropancreatic neuroendocrine tumors through microRNA sequencing. Endocr Relat Cancer. 2019;26:47–57. doi: 10.1530/ERC-18-0244. [DOI] [PubMed] [Google Scholar]
- 22.López-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan G.A., T. F. Routledge; New York: 2013. IBM SPSS for Introductory Statistics: Use and Interpretation. [Google Scholar]
- 25.Mathworks. Statistics and Machine Learning Toolbox Documentation (R2018a).
- 26.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., Beroukhim R., Pellman D., Levine D.A., Lander E.S., Meyerson M., Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A language and environment for statistical computing. [Google Scholar]
- 29.Li M., Lee K.F., Lu Y., Clarke I., Shih D., Eberhart C., Collins V.P., Van Meter T., Picard D., Zhou L., Boutros P.C., Modena P., Liang M.L., Scherer S.W., Bouffet E., Rutka J.T., Pomeroy S.L., Lau C.C., Taylor M.D., Gajjar A., Dirks P.B., Hawkins C.E., Huang A. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bullerdiek J., Flor I. Exosome-delivered microRNAs of "chromosome 19 microRNA cluster" as immunomodulators in pregnancy and tumorigenesis. Mol Cytogenet. 2012;5:27. doi: 10.1186/1755-8166-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streicher K.L., Zhu W., Lehmann K.P., Georgantas R.W., Morehouse C.A., Brohawn P., Carrasco R.A., Xiao Z., Tice D.A., Higgs B.W., Richman L., Jallal B., Ranade K., Yao Y. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2011;31:1558. doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Sendra B., Martinez-Ciarpaglini C., Gonzalez-Munoz J.F., Murgui A., Terradez L., Monteagudo C. Downregulation of intratumoral expression of miR-205, miR-200c and miR-125b in primary human cutaneous melanomas predicts shorter survival. Sci Rep. 2018;8:17076. doi: 10.1038/s41598-018-35317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzaei H., Gholamin S., Shahidsales S., Sahebkar A., Jaafari M.R., Mirzaei H.R., Hassanian S.M., Avan A. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer. 2016;53:25–32. doi: 10.1016/j.ejca.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu S., Tetzlaff M.T., Liu A., Liegl-Atzwanger B., Guo J., Xu X. Loss of microRNA-205 expression is associated with melanoma progression. Lab Invest. 2012;92:1084–1096. doi: 10.1038/labinvest.2012.62. [DOI] [PubMed] [Google Scholar]
- 35.Liu S., Tetzlaff M.T., Cui R., Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012;181:1823–1835. doi: 10.1016/j.ajpath.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanna J.A., Hahn L., Agarwal S., Rimm D.L. In situ measurement of miR-205 in malignant melanoma tissue supports its role as a tumor suppressor microRNA. Lab Invest. 2012;92:1390–1397. doi: 10.1038/labinvest.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dar A.A., Majid S., de Semir D., Nosrati M., Bezrookove V., Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bu P., Yang P. MicroRNA-203 inhibits malignant melanoma cell migration by targeting versican. Exp Ther Med. 2014;8:309–315. doi: 10.3892/etm.2014.1708. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Bhalla S., Kaur H., Dhall A., Raghava G.P.S. Prediction and analysis of skin cancer progression using genomics profiles of patients. Sci Rep. 2019;9:15790. doi: 10.1038/s41598-019-52134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caramel J., Papadogeorgakis E., Hill L., Browne G.J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J.H., Ansieau S., Tulchinsky E. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 41.Seiffert C., Khoshgoftaar T.M., Hulse J.V., Napolitano A. RUSBoost: a hybrid approach to alleviating class imbalance. IEEE Transactions on Systems, Man, and Cybernetics - Part A: Systems and Humans. 2010;40:185–197. [Google Scholar]
- 42.Cheng J.B., Sedgewick A.J., Finnegan A.I., Harirchian P., Lee J., Kwon S., Fassett M.S., Golovato J., Gray M., Ghadially R., Liao W., Perez White B.E., Mauro T.M., Mully T., Kim E.A., Sbitany H., Neuhaus I.M., Grekin R.C., Yu S.S., Gray J.W., Purdom E., Paus R., Vaske C.J., Benz S.C., Song J.S., Cho R.J. Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. 2018;25:871–883. doi: 10.1016/j.celrep.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haltaufderhyde K.D., Oancea E. Genome-wide transcriptome analysis of human epidermal melanocytes. Genomics. 2014;104:482–489. doi: 10.1016/j.ygeno.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hildebrand J., Rutze M., Walz N., Gallinat S., Wenck H., Deppert W., Grundhoff A., Knott A. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol. 2011;131:20–29. doi: 10.1038/jid.2010.268. [DOI] [PubMed] [Google Scholar]
- 45.Canueto J., Cardenoso-Alvarez E., Garcia-Hernandez J.L., Galindo-Villardon P., Vicente-Galindo P., Vicente-Villardon J.L., Alonso-Lopez D., De Las Rivas J., Valero J., Moyano-Sanz E., Fernandez-Lopez E., Mao J.H., Castellanos-Martin A., Roman-Curto C., Perez-Losada J. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br J Dermatol. 2017;177:168–178. doi: 10.1111/bjd.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn S.M., Cha J.Y., Kim J., Kim D., Trang H.T., Kim Y.M., Cho Y.H., Park D., Hong S. Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene. 2012;31:3051–3059. doi: 10.1038/onc.2011.484. [DOI] [PubMed] [Google Scholar]
- 47.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 48.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazar J., DeYoung K., Khaitan D., Meister E., Almodovar A., Goydos J., Ray A., Perera R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehrotra A., Mehta G., Aras S., Trivedi A., de la Serna I.L. SWI/SNF chromatin remodeling enzymes in melanocyte differentiation and melanoma. Crit Rev Eukaryot Gene Expr. 2014;24:151–161. doi: 10.1615/critreveukaryotgeneexpr.2014007882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steingrimsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 52.Babapoor S., Wu R., Kozubek J., Auidi D., Grant-Kels J.M., Dadras S.S. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab Invest. 2017;97:636–648. doi: 10.1038/labinvest.2017.5. [DOI] [PubMed] [Google Scholar]
- 53.Galasso M., Morrison C., Minotti L., Corrà F., Zerbinati C., Agnoletto C., Baldassari F., Fassan M., Bartolazzi A., Vecchione A., Nuovo G.J., Di Leva G., D’Atri S., Alvino E., Previati M., Nickoloff B.J., Croce C.M., Volinia S. Loss of miR-204 expression is a key event in melanoma. Molecular Cancer. 2018;17:71. doi: 10.1186/s12943-018-0819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent O.A., McCall M.N., Cornish T.C., Halushka M.K. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42:7528–7538. doi: 10.1093/nar/gku461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCall M.N., Kent O.A., Yu J., Fox-Talbot K., Zaiman A.L., Halushka M.K. MicroRNA profiling of diverse endothelial cell types. BMC Med Genomics. 2011;4:78. doi: 10.1186/1755-8794-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng R., Chen X., Yu Y., Su L., Yu B., Li J., Cai Q., Yan M., Liu B., Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Witwer K.W., Halushka M.K. Toward the promise of microRNAs - Enhancing reproducibility and rigor in microRNA research. RNA Biol. 2016;13:1103–1116. doi: 10.1080/15476286.2016.1236172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGrath J.A., Eady R.A.J., Pope F.M. 2008. Anatomy and Organization of Human Skin. Rook's Textbook of Dermatology. [Google Scholar]
- 60.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi R., Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 2010;17:229–235. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li F.Z., Dhillon A.S., Anderson R.L., McArthur G., Ferrao P.T. Phenotype switching in melanoma: implications for progression and therapy. Front Oncol. 2015;5:31. doi: 10.3389/fonc.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaelin W.G., Jr. Common pitfalls in preclinical cancer target validation. Nat Rev Cancer. 2017;17:425–440. doi: 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 64.Golan T., Messer A.R., Amitai-Lange A., Melamed Z., Ohana R., Bell R.E., Kapitansky O., Lerman G., Greenberger S., Khaled M., Amar N., Albrengues J., Gaggioli C., Gonen P., Tabach Y., Sprinzak D., Shalom-Feuerstein R., Levy C. Interactions of melanoma cells with distal keratinocytes trigger metastasis via notch signaling inhibition of MITF. Mol Cell. 2015;59:664–676. doi: 10.1016/j.molcel.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 65.Mescher M., Jeong P., Knapp S.K., Rubsam M., Saynisch M., Kranen M., Landsberg J., Schlaak M., Mauch C., Tuting T., Niessen C.M., Iden S. The epidermal polarity protein Par3 is a non-cell autonomous suppressor of malignant melanoma. J Exp Med. 2017;214:339–358. doi: 10.1084/jem.20160596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cicero A.L., Delevoye C., Gilles-Marsens F., Loew D., Dingli F., Guéré C., André N., Vié K., van Niel G., Raposo G. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nature Communications. 2015;6:7506. doi: 10.1038/ncomms8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renwick N., Cekan P., Masry P.A., McGeary S.E., Miller J.B., Hafner M., Li Z., Mihailovic A., Morozov P., Brown M., Gogakos T., Mobin M.B., Snorrason E.L., Feilotter H.E., Zhang X., Perlis C.S., Wu H., Suarez-Farinas M., Feng H., Shuda M., Moore P.S., Tron V.A., Chang Y., Tuschl T. Multicolor microRNA FISH effectively differentiates tumor types. J Clin Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., Trapnell C., Shendure J. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braeuer R.R., Watson I.R., Wu C.J., Mobley A.K., Kamiya T., Shoshan E., Bar-Eli M. Why is melanoma so metastatic? Pigment Cell Melanoma Res. 2014;27:19–36. doi: 10.1111/pcmr.12172. [DOI] [PubMed] [Google Scholar]
- 71.Cancer Genome Atlas N. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flor I., Bullerdiek J. The dark side of a success story: microRNAs of the C19MC cluster in human tumours. J Pathol. 2012;227:270–274. doi: 10.1002/path.4014. [DOI] [PubMed] [Google Scholar]
- 73.Augello C., Colombo F., Terrasi A., Trombetta E., Maggioni M., Porretti L., Rossi G., Guerneri S., Silipigni R., Bosari S., Vaira V. Expression of C19MC miRNAs in HCC associates with stem-cell features and the cancer-testis genes signature. Dig Liver Dis. 2018;50:583–593. doi: 10.1016/j.dld.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 74.Jinesh G.G., Flores E.R., Brohl A.S. Chromosome 19 miRNA cluster and CEBPB expression specifically mark and potentially drive triple negative breast cancers. PLoS One. 2018;13 doi: 10.1371/journal.pone.0206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sin-Chan P., Mumal I., Suwal T., Ho B., Fan X., Singh I., Du Y., Lu M., Patel N., Torchia J., Popovski D., Fouladi M., Guilhamon P., Hansford J.R., Leary S., Hoffman L.M., Mulcahy Levy J.M., Lassaletta A., Solano-Paez P., Rivas E., Reddy A., Gillespie G.Y., Gupta N., Van Meter T.E., Nakamura H., Wong T.T., Ra Y.S., Kim S.K., Massimi L., Grundy R.G., Fangusaro J., Johnston D., Chan J., Lafay-Cousin L., Hwang E.I., Wang Y., Catchpoole D., Michaud J., Ellezam B., Ramanujachar R., Lindsay H., Taylor M.D., Hawkins C.E., Bouffet E., Jabado N., Singh S.K., Kleinman C.L., Barsyte-Lovejoy D., Li X.N., Dirks P.B., Lin C.Y., Mack S.C., Rich J.N., Huang A. A C19MC-LIN28A-MYCN oncogenic circuit driven by hijacked super-enhancers is a distinct therapeutic vulnerability in ETMRs: a lethal brain tumor. Cancer Cell. 2019;36:51–67. doi: 10.1016/j.ccell.2019.06.002. e57. [DOI] [PubMed] [Google Scholar]
- 76.Jayachandran A., Anaka M., Prithviraj P., Hudson C., McKeown S.J., Lo P.H., Vella L.J., Goding C.R., Cebon J., Behren A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5:5782–5797. doi: 10.18632/oncotarget.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material