We review modern light microscopy and computed projection tomography methods, their capabilities and limitations, and their current and potential applications to the study of flower development and fertilization.
Keywords: Bioimaging, confocal, flower development, light-sheet, microscopy, optical projection tomography, optical sectioning, super-resolution, tomography, two-photon, X-ray microscopy, X-ray tomography
Abstract
Developmental biology relies heavily on our ability to generate three-dimensional images of live biological specimens through time, and to map gene expression and hormone response in these specimens as they undergo development. The last two decades have seen an explosion of new bioimaging technologies that have pushed the limits of spatial and temporal resolution and provided biologists with invaluable new tools. However, plant tissues are difficult to image, and no single technology fits all purposes; choosing between many bioimaging techniques is not trivial. Here, we review modern light microscopy and computed projection tomography methods, their capabilities and limitations, and we discuss their current and potential applications to the study of flower development and fertilization.
Introduction
Angiosperms are one of the most successful groups on Earth. They have colonized six continents and thrive in a wide variety of environments and climates. This evolutionary success story is largely due to their reproductive structures: flowers. Most flowers are comprised of four types of organs: sepals, petals, stamens, and carpels; yet, flowers are extremely diverse in size, color, symmetry, scent, and number of organs, suggesting myriads of variations on a core developmental theme. Flowers also have a major agroeconomic importance: >80% of our food comes directly from plants, the vast majority of it fruits and seeds, which are parts and products of flowers. It is therefore critically important to understand the mechanisms underlying flower development and fertilization.
Richard Feynman, a theoretical physicist and Nobel laureate, famously said: ‘It is very easy to answer many of these fundamental biological questions; you just look at the thing!’ As biologists, we have daily reasons to scoff at that statement; however, ‘looking at the thing’ is undeniably a powerful way to try to understand biological phenomena. Developmental biology relies heavily on imaging to investigate how networks of genes and hormone signaling control organogenesis. This requires a precise four-dimensional (4D) knowledge of the model studied (i.e. the knowledge of the morphology of the model and its evolution through developmental time), and the ability to map gene activity and hormone response within these 4D structures. Early studies in flower development (e.g. Bowman et al., 1991, 1992; Jack et al., 1992; Tröbner et al., 1992) used mutant approaches combining SEM to study the morphology of wild-type and mutant flowers with in situ hybridization and immunostaining to localize the corresponding gene products. These techniques have significant limitations, which complicates access to precise 4D information: they lack cellular resolution and require fixation and, in the case of gene expression analysis, sectioning the specimen.
The development of optical sectioning techniques, and particularly laser scanning confocal microscopy (Amos et al., 1987; White et al., 1987; Amos and White, 2003), combined with the design of a wide variety of fluorescent proteins (FPs) (van Roessel and Brand, 2002), has made it possible to map gene activity and hormone signaling in 3D, with cellular resolution, in live specimens. Confocal microscopes have become the work horse of developmental biology laboratories, and have been extensively used to study flowers, and led to many advances in the field (e.g. Mayer et al., 1998; Heisler et al., 2005; Urbanus et al., 2009; Chandler et al., 2011; Milani et al., 2014; Sun et al., 2014; Prunet et al., 2017; Yamaguchi et al., 2017; Xu et al., 2018).
However, confocal microscopy is just one of many imaging techniques that can be used to study flowers. Biomedical imaging has seen an explosion of new techniques and provided us with a variety of tools for developmental biology. Here we review imaging tools available to flower developmental biologists, and cover both imaging modalities—from light microscopy to computed tomography—and reporters and sensors to detect gene expression and hormone gradients and response.
3D imaging with optical sectioning
Unlike single cells, flowers are thick specimens: a stage 2 Arabidopsis flower bud, for instance, is ~40 μm thick, and rapidly grows in size as it develops (stages as described in Smyth et al., 1990). Yet, Arabidopsis flowers are small compared with those of many other species. While widefield epifluorescence microscopy works well for thin specimens, out-of-focus light strongly reduces the contrast and resolution when imaging 3D, thick specimens (Webb, 1999). One option to circumvent this problem is to fix and section the specimen, image the serial sections, and computationally generate a 3D reconstruction. This approach is not compatible with live imaging and therefore not ideal to study development. The development of optical sectioning microscopy techniques in the 1990s has revolutionized developmental biology.
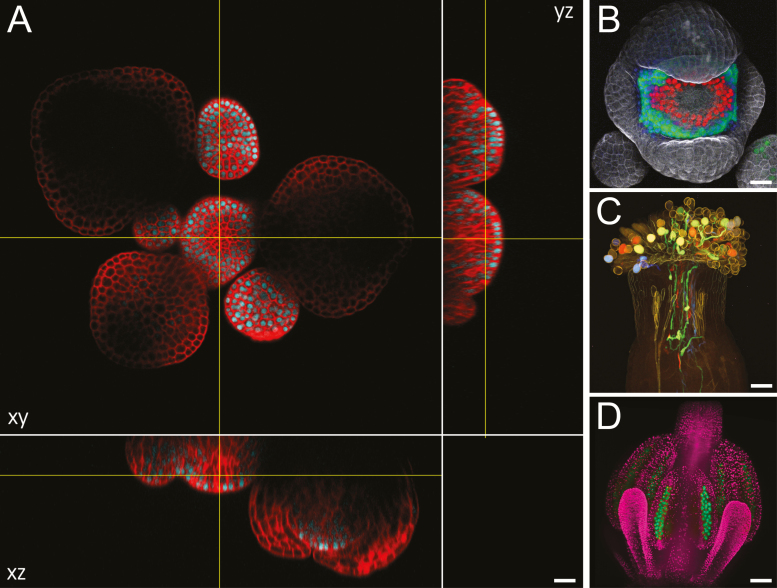
Confocal microscopy excites fluorophores within the specimen with a highly focused laser beam. Fluorescence emitted by the specimen is captured by the objective and filtered through a pinhole that only allows in-focus light to reach the detector (Webb, 1999; Murphy and Davidson, 2013). Successive optical sections are generated by scanning the laser beam over the specimen at different depths. These optical sections are then stacked together to generate a 3D reconstruction of the specimen, with a maximum optical resolution of ~250 nm laterally and ~500 nm axially (Table 1). Confocal microscopy has been extensively used to study flowers, and led to many advances in the field (e.g. Mayer et al., 1998; Heisler et al., 2005; Urbanus et al., 2009; Chandler et al., 2011; Milani et al., 2014; Sun et al., 2014; Prunet et al., 2017; Yamaguchi et al., 2017; Xu et al., 2018) (Fig. 1A, B). However, this technique has some caveats: it is a point-scanning technique (i.e. images are created pixel by pixel, in contrast to widefield microscopy in which the image is acquired all at once by a camera), and is therefore intrinsically slow; while the out-of-focus light is not captured, for each pixel generated, a whole z-column in the specimen is excited by the laser, which causes the progressive photobleaching of successive optical sections, and can be toxic for live samples; this photobleaching and phototoxicity issue is reinforced by the need for high laser power to compensate for the light lost through the confocal pinhole.
Table 1.
Characteristics of bioimaging techniques
| Technique | Lateral resolution | Axial resolution | Imaging depth | Live imaging | Fluorescent reporters |
|---|---|---|---|---|---|
| Point-scanning confocal | 250 nm | 500 nm | 100 µm | +++ | Yes |
| Spinning disc confocal | 250 nm | 500 nm | 100 µm | +++++ | Yes |
| Two-photon | 250 nm | 500 nm | 500 µm | ++++ | Yes, but not ideal for multiple colors |
| Light-sheet | 300 nm | Depends on thickness of sheet | 60 µm | ++++++ | Yes |
| SIM | 100 nm | 200 nm | 15 µm | ++ | Yes |
| STED | 20 nm | Variable but <500 nm | 20 µm | +++ | Yes |
| SMLM | 20 nm | 20 nm | 5 µm | – | PALM, yes; dSTORM, no |
| OPT | 1 µm | 1 µm | 15 mm | +++ | Yes |
| Macro-OPT | 6.5 µm | 6.5 µm | 45 mm | +++ | Yes |
| XRM | 500 nm | 500 nm | 1 cma | – | No |
| XRT | 20 µm | 20 µm | 1 ma | ++ | No |
As far as possible the resolution and imaging depth values shown in this table are based on published data specific to flowers, or plant aerial tissues, and do not necessarily reflect the true resolution and imaging depth limit of the techniques. XRT and XRM resolution values for instance are estimates relating to imaging low-contrast biological samples; true instrument resolution for high-contrast materials, such as metals, ceramics, and geological samples, is higher.
a Imaging depth is not relevant to X-ray imaging in the same way as with optical imaging methods. Rather, sample size varies inversely with achievable resolution and is dependent upon source–sample–detector geometry: the larger the sample or region of interest, the lower the voxel resolution.
Fig. 1.
Arabidopsis flowers imaged with optical sectioning techniques. (A) Optical xy section and reconstructed xz and yz sections of a live Arabidopsis inflorescence expressing a transcriptional SHOOT MERISTEMLESS reporter (cyan) imaged with a point-scanning confocal microscope; cell walls were stained with propidium iodide (red); these images show the limitation of imaging depth with confocal microscopy. (B) Maximum intensity projection of a live, stage 5 Arabidopsis flower expressing a transcriptional reporter for APETALA3 (AP3; green) and translational reporters for AP3 (green) and SUPERMAN (red); cell walls were stained with propidium iodide (gray); note the differences in expression of the transcriptional and translational AP3 reporters. (C) Maximum intensity projection of an Arabidopsis pistil pollinated with pollen expressing different transcriptional reporters (mTFP1, sGFP, Venus, and mApple) for LAT52, treated with ClearSee for 5 months, and imaged with two-photon excitation microscopy; this image, courtesy of Drs Yoko Mizuta and Daisuke Kurihara, was originally published in Kurihara et al. (2015). (D) Maximum intensity projection of a live Arabidopsis floral bud expressing reporters for the ASY1 (green) and H2B (pink) genes; sepals were removed; image courtesy of Sona Valuchova and Pavlina Mikulkova. Scale bars=50 µm in (A–C), 100 µm in (D).
Spinning disc microscopy is a faster, gentler approach to confocal microscopy. Instead of scanning the specimen with a laser beam, hundreds of discrete points are simultaneously excited through hundreds of pinholes spirally arranged on a rapidly rotating disc, and the light emitted by these points is collected through the same pinholes and captured by a fast camera (Oreopoulos et al., 2014). As the disc rotates, the whole specimen is covered much faster than with point-scanning confocal microscopy to generate an optical section; moreover, cameras are more sensitive than point-scanning detectors, and less excitation light is needed. One disadvantage of spinning disc confocal microscopy compared with point-scanning confocal microscopy is the fixed sized of the pinholes, which cannot be adjusted to alter optical sectioning and resolution (Oreopoulos et al., 2014).
Another way to generate optical sections of a thick specimen is to selectively excite in-focus parts of the specimen, which considerably reduces photobleaching and phototoxicity: each optical section is only excited once. This can be done with either two-photon excitation microscopy (also referred to as multiphoton excitation microscopy) or light-sheet microscopy (Huisken et al., 2004; Helmchen and Denk, 2005; Murphy and Davidson, 2013; Weber et al., 2014). Conventional fluorescence uses single-photon excitation: a fluorophore is excited with a photon of a specific wavelength, and rapidly emits a photon of longer wavelength and lower energy. In two-photon excitation, a fluorophore is excited by the simultaneous absorption of two photons of longer wavelength and lower energy than the photon it would absorb with single-photon excitation (Helmchen and Denk, 2005). Two-photon excitation is obtained by focusing a powerful femto-second pulse laser to a diffraction-limited spot within the specimen. Only in that spot are photons concentrated enough to generate two-photon excitation. Because the wavelength of the laser is too short to generate single-photon excitation of the fluorophore, excitation only occurs at this in-focus spot: there is no out-of-focus light. Two-photon excitation microscopy offers a similar resolution and speed to confocal microscopy (it is also a point-scanning technique) (Table 1).
Confocal and two-photon microscopy use the reflected light path (i.e. excitation and emission light are, respectively, shone onto the specimen through, and collected by, the objective lens). Conversely, light-sheet fluorescence microscopy separates the excitation and emission light paths: the specimen is illuminated by a sheet of light, generated either with a cylindrical lens, which focuses the light along one axis, or by rapidly scanning a laser beam along one axis within the exposure time of the camera (digitally scanned, or virtual light sheet); fluorescence emitted by the specimen is collected by an objective lens that is orthogonal to the illuminated plane and captured by a fast camera, generating an optical section (Huisken et al., 2004; Weber et al., 2014). 3D reconstructions can be obtained from stacking successive optical sections acquired from a single angle or through computational reconstruction of the specimen imaged at different angles—the latter option compensates uneven illumination of the focal plane as the light sheet is absorbed as it goes deeper into the tissues. Light-sheet microscopy offers an optimal lateral resolution of ~300 nm; axial resolution depends on the thickness of the light sheet, and can potentially be higher than that of confocal microscopy (Table 1). Light-sheet microscopy is much faster than point-scanning techniques. It is also gentler: cameras are much more sensitive than detectors used for point-scanning systems, and all the emitted light is collected (there is no pinhole), so less laser power is needed for illumination. These two characteristics make light-sheet microscopy an ideal optical sectioning technique for live specimens (Weber and Huisken, 2011; Weber et al., 2014). It has emerged as a major tool for imaging live specimen in developmental biology in animals (Weber and Huisken, 2011), and has been very useful in plants to study roots, which are transparent (Ovečka et al., 2018). Applying light-sheet microscopy to aerial tissues, which are opaque and highly autofluorescent, has proven more difficult (Ovečka et al., 2015). However, light-sheet microscopy was recently successfully used to study germ cell development in Arabidopsis flowers over a period of days (Fig. 1D) (Valuchova et al., 2020). We should expect a more widespread use of light-sheet microscopy to study flowers in the near future.
3D imaging with computed tomography
The word tomography comes from ancient Greek tomos, which means section. In a broad sense, any imaging technique that generates digital sections of a 3D object—including the optical sectioning techniques described above—could be considered tomography. Computed tomography, however, uses a reverse approach compared with optical sectioning: a 3D image of the specimen is computationally reconstructed from 2D projections that do not contain information about precisely where they come from within the specimen (Sharpe, 2004). The term tomography has typically been associated with X-ray tomography (XRT; also referred to as computed tomography, or CT scan), but many different tomography techniques use different part of the electromagnetic spectrum, including visible light in the case of optical projection tomography (OPT), but also sound waves or electric or magnetic fields (Sharpe, 2004). Here, we will focus on the use of XRT and OPT to study flowers.
XRT imaging has an X-ray source and detector enclosed in a lead cabinet to contain X-ray energy, and the patient or sample is placed between source and detector for imaging. The source generates X-rays that are directed through the sample toward the detector. Digital 2D images—radiographs—are projected onto the detector as X-rays pass through the sample and are differentially absorbed due to variation in sample density. Hundreds or thousands of radiographs are captured as the system or the sample rotates over (typically) 360°. All the 2D radiographs are then computationally reconstructed into a single 3D volume that can be manipulated and analyzed using advanced image analysis software. Human XRT imaging places the patient motionless in the center of the instrument while the source and detector rotate around the subject, whereas industrial XRT systems use a turntable upon which the sample rotates while positioned between source and detector.
The routine use of XRT in plant biology, particularly for imaging and analysis of complicated floral structures, is relatively recent. XRT imaging relies on differential density within a sample to generate an image. However, plant material typically is homogeneous low-density tissue, which makes high-resolution imaging difficult. Plants have been successfully imaged using large format industrial CT instruments simply by placing samples in appropriate devices that keep them stable during the course of the scan (see, for example, Bray and Topp, 2018; Roberston et al., 2017; Li et al., 2019). Nevertheless, there is a practical limit to sample size and resolution in large format instruments, at best approaching 20 µm voxel resolution for large plant samples (Table 1).
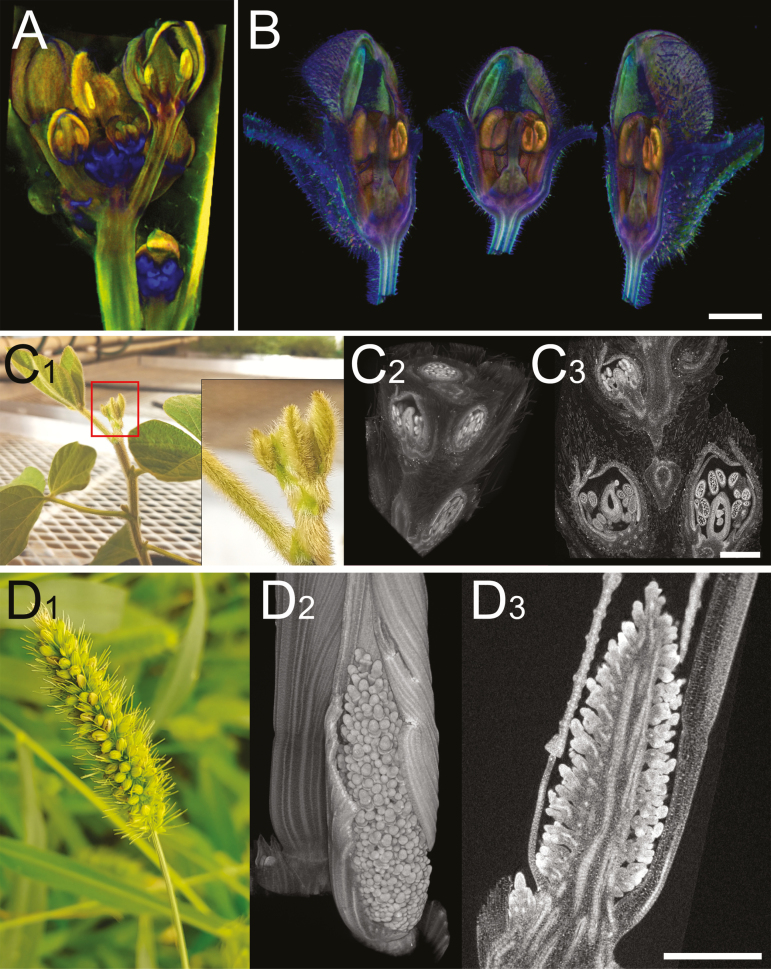
For high-resolution imaging (i.e. microCT or nanoCT), floral structures are typically fixed and contrast enhanced (Staedler et al., 2013). Recent examples have demonstrated the utility of fixation and contrast enhancement to image and analyze complicated floral and other plant biological tissues (Fig. 2C1–D3) (van der Niet et al., 2010; Staedler et al., 2013; Ijiri et al., 2014; Wang et al., 2015; Tracy et al., 2017; Jeiter et al., 2018; Mathers et al., 2018; Staedler et al., 2018; Duncan et al., 2019). In particular, X-ray microscopy (XRM) has proven a valuable tool for imaging the complex biology of floral structures, providing high-resolution, data-rich biological information in 3D that is not practical or possible with other imaging technologies. The XRM adds a series of sophisticated microscope lenses to the traditional X-ray beam path (X-ray source–sample–detector); each lens has a coating that converts the X-ray signal into light, which is then magnified by the lens, and a high-resolution CCD camera functions as the detector to capture the final images. Again, hundreds or thousands of 2D digital radiographs are collected and computationally reconstructed into a detailed 3D volume with potentially submicrometer resolution for well-contrasted fixed samples. This provides excellent cellular detail in a full 3D volume with samples much larger and more complex than is typically possible with fluorescence microscopy. For example, entire large floral structures—1 cm3—can be imaged with XRM at 10 µm resolution, and specific regions of interest within that volume can be imaged at 1 µm resolution, all without physically cutting the sample into slices (Fig. 2C1–D3). This 3D imaging capability provides visualization and analysis of complicated floral morphology across scales, allowing unprecedented insight into floral developmental biology.
Fig. 2.
Computed tomography images of flowers. (A) Transmission OPT image of an Arabidopsis inflorescence expressing a GUS reporter for LEAFY (blue); image courtesy of Karen Lee. (B) Three views of an Antirrhinum flower imaged with emission OPT and virtual dissecting with a clipping plane to reveal internal structures; image courtesy of Karen Lee. C1–C3. Photograph (C1) and XRM images (C2 and C3) of a young soybean axillary bud containing numerous young florets that will eventually develop into soybean pods; C2 shows a virtual dissection with three clipping planes; C3 shows a computationally reconstructed section. (D1–D3) Photograph (D1), 3D computed reconstruction (D2), and computationally reconstructed section (D3) of young inflorescences from foxtail millet (Setaria viridis). Scale bars=500 µm.
OPT uses light instead of X-rays. Like light microscopy, OPT can use either transmitted light (transmission OPT) or fluorescence (emission OPT). In transmission OPT, the specimen is placed between a widefield, diffuse light source and an optical lens system that focuses the transmitted light onto a CCD camera (Sharpe, 2004). In contrast, emission OPT uses UV light, and the emitted fluorescence is only collected from a certain angle, usually on the same side as for excitation. As in XRT, a 3D image of the specimen is computationally reconstructed from hundreds of projections acquired as the sample rotates over 360°, with a near-cellular maximum resolution (~1 µm voxel; Table 1) (Sharpe, 2004; Lee et al., 2006). Lee et al. applied both transmission and emission OPT to plant tissues, including flowers (Fig. 2A, B), and developed a version designed to image larger specimens, called macro-OPT (Lee et al., 2006, 2017).
From subcellular structures to large flowers—a matter of scale
While in most cases, imaging at cellular resolution is sufficient, some developmental studies require the ability to resolve much smaller, subcellular structures. However, the resolution of classic microscopy techniques is limited by the diffraction of light, which spreads the light from each point within the specimen into a diffraction pattern: the image of even an infinitely narrow point is thus not captured as a single point, but a point-spread function (PSF) (Murphy and Davidson, 2013). When two separate points of the specimen are too close, their PSFs overlap in the image, and the two points cannot be resolved as separate. The resolution limit depends both on the numerical aperture of the objective and the wavelength of light; typically, the resolution limit is ~300 nm for widefield microscopy techniques, and ~250 nm for point-scanning techniques.
Several microscopy methods, commonly referred to as super-resolution microscopy, push or bend the resolution limit. Structured illumination microscopy (SIM) uses the principle of Moiré fringes: the specimen is illuminated through a grid, which is rotated and translated over the specimen; the superimposition of the grid over the specimen causes the formation of coarse, resolvable details in the image: Moiré fringes. Because the spatial frequency of the grid is known, these resolvable patterns can be used to computationally deduce fine details in the specimen that are smaller than the resolution limit (Fiolka, 2014). SIM pushes the resolution limit down to 100 nm laterally and 200 nm axially (Table 1) (Komis et al., 2015b). Other techniques such as stimulated emission depletion microscopy (STED) and single molecule localization microscopy (SMLM) completely circumvent diffraction to break the resolution limit. STED is a point-scanning technique that uses PSF engineering: the specimen is scanned with two laser beams; one excites the fluorophore, while the other, shaped into a donut surrounding the excitation beam, depletes fluorescence around the excitation point and restricts it to a very small spot (Murphy and Davidson, 2013). STED pushes the resolution limit down to 20 nm laterally (Table 1) (Komis et al., 2015b). SMLM stochastically separates the emission of fluorophores in time, so that only a limited number of them emit light at any given time; in that way, the PSFs of neighboring fluorophores can be acquired separately instead of overlapping, and each fluorophore can be localized precisely to the center of its PSF (Murphy and Davidson, 2013). The final super-resolution image is generated from hundreds to thousands of successive images; SMLM is therefore a slow technique. Different SMLM modalities use different types of fluorophores. Photoactivation localization microscopy (PALM) relies on photoactivatable, photoswitchable, or photoreversible, genetically encoded FPs, and is therefore compatible with the imaging of live specimens. Conversely, direct stochastic optical reconstruction microscopy (dSTORM) uses immunostaining with organic fluorophores, which are induced to blink by high-level excitation in a redox buffer; dSTORM is thus not compatible with live imaging. SMLM pushes the resolution limit down to 20 nm (Table 1) (Murphy and Davidson, 2013).
Only a few studies have used super-resolution on plant tissues, and it has yet to be applied to flower development. However, super-resolution methods have the potential to resolve subcellular structures that are critical for flower development and their dynamics much better than traditional microscopy techniques. The cytoskeleton is one example: microtubules regulate plant development through their association with cellulose synthase and cell wall biosynthesis, as well as biomechanical constraints (Sampathkumar et al., 2013, 2014a, b; Hervieux et al., 2016), and precisely resolving the organization of the microtubule network requires super-resolution (Komis et al., 2014, 2018). Another example is the flow of auxin through the PIN-FORMED (PIN) transporters: subcellular localization of the PIN proteins is highly polar, yet confocal microscopy does not have the power to resolve the plasma membranes and the shared cell wall of two neighboring cells; the localization of PIN proteins is usually inferred from the shape of the fluorescence from PIN fluorescent reporters, with no certitude regarding on which side of the cell wall the PIN proteins actually are. STED was used to precisely analyze the dynamic, polar distribution of PIN proteins in root tissues (Kleine-Vehn et al., 2011), and similar approaches would shed a new light on auxin flows in developing flowers. SIM, STED, and PALM were also used for visualization of diverse subcellular compartments including the actin cytoskeleton, endoplasmic reticulum, endosomes, nuclei, plasma membrane subdomains, and nuclear nanodomains in living plant cells (Komis et al., 2018).
On the opposite side of the scale, many flowers are too big to be imaged with a microscope. Arabidopsis thaliana has been by far the most studied plant model over the last 30 years. The small size of the Arabidopsis flower makes it possible to image it integrally within the field of view of a microscope objective, with cellular resolution, for a large portion of its development. This is not the case for bigger ornamental species such as Antirrhinum majus (snapdragon) and Petunia hybrida, which have long been used for flower development studies, or for crop species such as Solanum lycopersicon (tomato) or Zea mays (maize). Light microscopy is limited to relatively small specimens: the typical field of view of a 10× objective lens is ~2 mm. While it is possible to acquire and tile multiple, overlapping images to generate a 3D reconstruction, with cellular resolution, of samples that do not fit within this field of view, significantly larger specimens require the use of CT techniques.
OPT typically allows for the imaging of live specimens ranging from 0.5 mm to 16 mm in size, and therefore slightly too big for light microscopy, with near-cellular resolution (Fig. 2A, B), while macro-OPT can be used for specimens up to 60 mm, with a spatial resolution ranging from ~6.5 µm to 62.5 µm (Lee et al., 2006, 2017). XRM can be used to image specimens up to 1 cm3 in size with cellular resolution; a low-magnification scan (~20 µm voxel) of the entire sample can be combined with a high-resolution scan (500 nm voxel) (Table 1; Fig. 2C1–D3) to visualize features of interest. The primary drawback is the fixation and contrast enhancement required for XRM imaging; living tissue is difficult to immobilize sufficiently for the long scan lengths required for such high-resolution imaging. Finally, commercial XRT can accommodate much larger specimens, up to several meters in size, but without cellular resolution (20 µm voxel at best; Table 1).
Imaging deeper in the tissues
Plants, and particularly aerial tissues, are difficult specimens in terms of light microscopy. Their epidermis is covered in a cuticle and, unlike animal cells, plant cells are surrounded by a cell wall of variable thickness, and contain a vacuole as well as plastids (plant cells range in size from ~3 µm to 100 µm, with cell walls ~0.1–10 µm thick). All these compartments and organelles have different refractive indices, making plant tissues strongly scattering (light rays are deviated at the interface between media of different refractive indices). Most floral organs also contain pigments that absorb light. Moreover, photosynthetic organs—which include sepals and carpels—are also strongly autofluorescent due to the presence of chlorophyll. This combination of scattering, absorption, and autofluorescence significantly hinders our ability to image flowers using optical microscopy: both quality and intensity of the signal degrade rapidly with imaging depth within plant tissues. Confocal microscopy typically allows for imaging at depths up to ~80 μm (Haseloff et al., 1997), but imaging through sepals and carpels is further limited by chlorophyll absorption and autofluorescence (Fig. 1A). Similarly, light-sheet microscopy works well to image outer structures in flowers, but does not provide sufficient penetration to resolve inner tissues and organs through the sepals or carpels (Ovečka et al., 2015; Valuchova et al., 2020).
One possible approach to circumvent this issue is to remove sepals or carpel valves to image the underlying tissues and organs. This can be achieved through manual dissection or laser ablation, and was successfully used in Arabidopsis to study the establishment of the boundary between stamens and carpels (Prunet et al., 2017; Xu et al., 2018), pollen tube growth, and male gamete release (Rotman et al., 2003) with confocal microscopy, and germline differentiation with light-sheet microscopy (Fig. 1D) (Valuchova et al., 2020), for instance. However, organ dissection is stressful for the specimen and results in less physiological imaging conditions. Another approach to get a better 3D reconstruction of deep tissues is to image the specimen from several angles, and combine the images computationally (Fernandez et al., 2010; Ovečka et al., 2018; Valuchova et al., 2020).
Two-photon microscopy is a better alternative to confocal and light-sheet microscopy for deep tissue imaging of intact, non-optically cleared specimens. It uses near-IR light, which penetrates deeper in tissues and scatters less than visible light, for excitation (Gilroy, 1997; Benninger and Piston, 2013). Moreover, two-photon excitation is restricted to the focal plane, thus each optical section is only excited once; this considerably reduces photobleaching compared with confocal microscopy, in which optical sections are excited repeatedly, causing the progressive bleaching of successive sections (Blancaflor and Gilroy, 2000). Two-photon microscopy allows for imaging several hundreds of micrometers deep within scattering specimens (Centonze and White, 1998; Helmchen and Denk, 2005). Two-photon microscopy enabled imaging in plant tissues at twice the depth obtained with confocal microscopy (Fig. 1C) (Mizuta et al., 2015); it made it possible to image development processes that occur underneath several cell layers, such as pollen tube growth and double fertilization, in vivo (Cheung et al., 2010; Mizuta et al., 2015). Two-photon imaging at higher wavelength (>1000 nm) also strongly reduces autofluorescence in plant tissues (Mizuta et al., 2015).
Even with two-photon imaging, scattering caused by the variety of refractive indices in plant cells limits deep-tissue imaging. Chemical treatments can be used to clear fixed tissues by reducing refractive mismatch and removing pigments. Chloral hydrate has long been used to clear plant tissues, but it is not compatible with the use of FPs (Kurihara et al., 2015), which have become a major tool in developmental biology. Kurihara and colleagues used chemical screening to design ClearSee, a clearing solution for plant tissues that maintains the stability of FPs (Kurihara et al., 2015). ClearSee has a high refractive index, and limits scattering; it is also highly efficient at removing chlorophyll, thus strongly reducing absorption and autofluorescence. ClearSee significantly increases confocal imaging depth in plant tissues, but the best results were obtained with two-photon microscopy of ClearSee-treated specimens (Fig. 1C): it is possible to image through an entire Arabidopsis pistil (~500 µm in diameter) (Kurihara et al., 2015). To our knowledge, ClearSee has not yet been used in combination with light-sheet microscopy, but would undoubtedly increase the penetration depth of the light-sheet.
While it lacks cellular resolution, OPT allows for imaging much deeper than classic optical sectioning microscopy techniques (Sharpe, 2004). Regular OPT has been used to image plant specimens >10 mm thick, and macro-OPT plant specimens up to 45 mm thick (Lee et al., 2006, 2017). OPT and macro-OPT can be used to image live specimens; however, larger specimens typically need to be optically cleared to properly resolve deeper tissues (Lee et al., 2006, 2017). X-rays can penetrate much deeper into biological tissues than visible light, and XRT can therefore be used for intact 3D imaging for very thick specimens. XRM, however, is limited by the size of the specimen that can fit in the instrument, which can only accommodate samples up to 1 cm3 when sub-micron resolution is required. The most efficient system for X-ray tomography of plant biology across scales would combine a large format XRT—which can still image down to ~20–30 µm voxel (Table 1)—and the high resolution XRM.
Adding the fourth dimension: live imaging
All the optical sectioning techniques described here, as well as OPT, are compatible with the imaging of live samples. However, they differ greatly in speed and photodamage of the specimen (Table 1). Point-scanning confocal and two-photon microscopy are slow, and confocal imaging can cause significant photodamage to the specimen, as successive sections are excited multiple times. Both methods can still be used for time-lapse imaging of flowers [for examples of time-lapse confocal studies of flower development, see Heisler et al., 2005; Meyer et al., 2017; Tsugawa et al., 2017; detailed protocols for live confocal imaging of Arabidopsis (Fernadez et al., 2010; Barbier de Reuille et al., 2015; Prunet et al., 2016; Prunet, 2017) and Brachypodium flowers (O’Connor, 2018) are available, as are protocols for live confocal imaging of the shoot apical meristem of soybean and tomato (Geng and Zhou, 2019), which could easily be applied to imaging flowers]. However, samples can only be imaged every few hours at best; prolonged, repeated imaging of the specimen results in both phototoxicity and bleaching. Light-sheet fluorescence microscopy and, to a lesser extent, spinning disc microscopy, are much faster, making these methods better for live imaging. Indeed, light-sheet microscopy was used to image live Arabidopsis flowers nearly continuously over a period of 5 d without causing any significant bleaching, which would not be possible with point-scanning techniques (Valuchova et al., 2020; protocols for live light-sheet imaging of plant tissues and flowers can be found in Ovečka et al., 2015, 2018; Valuchova et al., 2020).
Computed tomography techniques require the acquisition of hundreds to thousands of projections to generate a 3D reconstruction of the sample, which makes data acquisition slow. Yet, OPT is compatible with time-lapse imaging (Table 1) (Lee et al., 2006, 2017). XRT can also be performed on live specimens. Various X-ray instrument manufacturers are exploring high-speed tomographic acquisition systems to allow some level of 4D XRT, but detector technology is still limiting this work to relatively low-resolution imaging (e.g. 100 µm voxel). XRM of plant samples, however, requires fixation and contrasting, and cannot be used for live imaging (Table 1) (Staedler et al., 2013).
Super-resolution techniques are not all compatible with live imaging (Table 1). SMLM requires the acquisition of hundreds to thousands of images to reconstruct the final, super-resolution image and is therefore extremely slow and not geared towards live imaging (Murphy and Davidson, 2013). Still, PALM is technically compatible with the use of live specimen; dSTORM, however, uses fixed, immunostained samples. SIM also requires the acquisition of multiple images, but not nearly as many as SMLM (up to 25 images per channel per optical section depending on the number of phases used), and can be used for the live imaging of phenomena changing at moderate rates (e.g. microtubule growth) (Komis et al., 2015a, b). Finally, STED is a point-scanning technique and, as such, has the same restrictions as confocal microscopy; it is compatible with time-lapse imaging.
Mapping gene activity and hormone signaling
The ability to transform plants (for a review of plant transformation methods, see Keshavareddy et al., 2013) made it possible to generate transgenic reporter lines to analyze the patterns of gene expression. β-Glucuronidase (GUS) (Jefferson et al., 1987), an enzyme that catalyzes the formation of a colored product from a colorless substrate, has been extensively used to study flower development (e.g. Jack et al., 1994; Jenik and Irish, 2000; Ito et al., 2003), but this approach suffers from the same limitations as in situ hybridization and immunostaining: it does not provide cellular resolution, and is not compatible with the study of live specimens.
The development of an extensive array of FPs of different colors, brightness, folding requirements, stability, and environment sensitivity from jellyfish Aequora victoria’s green fluorescent protein (GFP) and coral Discosoma sp.’s DsRed provided us with a wide variety of tools to design genetically encoded reporters that are compatible with biological imaging of live specimens (for the story behind the engineering of FP variants, see https://www.ibiology.org/talks/fluorescent-proteins/; for a guide of how to choose your FP, see Shaner et al., 2005; for a database of FPs, see www.fpbase.org [Lambert, 2019]).
FPs have been used to generate reporters to monitor the expression of many genes that regulate various aspects of flower development, including the initiation of flower buds (e.g.Heisler et al., 2005; Goldshmidt et al., 2008; Besnard et al., 2014), floral organ positioning (e.g. Chandler et al., 2011), growth (e.g. Meyer et al., 2017), identity (e.g. Urbanus et al., 2009; Yamaguchi et al., 2017), and polarity (e.g. Yamaguchi et al., 2017), boundary formation (e.g. Takeda et al., 2004; Prunet et al., 2017; Xu et al., 2018), and floral stem cell termination (e.g. Sun et al., 2014; Yamaguchi et al., 2017) using live confocal imaging. Transcriptional reporters (in which an FP gene is fused to the promoter of a gene of interest) and translational reporters (in which an FP gene is fused to both promoter and coding region of a gene of interest) can be used to map the domains where the corresponding mRNA and protein accumulate, respectively. Several genes involved in flower development show differences between these domains, due either to the ability of the protein to move between cells through plasmodesmata (e.g. ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6) (Besnard et al., 2014) or to protein instability in the absence of a partner (e.g. APETALA3 and PISTILLATA; Fig. 1B) (Jack et al., 1994; Krizek and Meyerowitz, 1996). Intercellular protein movement is also to be considered when designing transcriptional reporters, as the molecular size of single FPs allows them to move freely through plasmodesmata, generating a fluorescent domain that does not necessarily reflect the expression pattern of the gene of interest. This problem can be circumvented by adding a nuclear localization signal or an endoplasmic reticulum signal peptide to the FP, or using tandem FP fusions.
Phytohormones are also major regulators of flower development (Aloni et al., 2006; Sundberg and ØOstergaard, 2009; Wybouw and De Rybel, 2019). Transgenic approaches have also been used to monitor hormone response and accumulation. Reporters for auxin (DR5rev) and cytokinin (TCSn) response were generated using synthetic promoters with multiple tandem repeats of response elements found in the promoters of genes responsive to hormones (Ulmasov et al., 1997; Müller and Sheen, 2008; Zürcher et al., 2013), and fluorescent versions of these reporters are available (Benková et al., 2003; Brunoud et al., 2012; Zürcher et al., 2013; Liao et al., 2015). DR5rev and TCSn share common limitations: both promoters were designed from response elements identified in a single gene (Ulmasov et al., 1997; Müller and Sheen, 2008; Zürcher et al., 2013), raising the question of whether these response elements are high- or low-affinity binding sites. While this question remains unsolved in the case of TCSn, it has been shown that DR5 is a low-affinity binding site (Boer et al., 2014). A higher affinity auxin response element has been identified (Boer et al., 2014), and used to design a more sensitive reporter for auxin response: DR5v2, which reveals active auxin response in domains where auxin was predicted to accumulate but are not marked by the DR5 reporter (Liao et al., 2015).
Reporters based on hormone response elements are unlikely to reflect the complexity of hormone responses and do not directly reflect hormone accumulation patterns. Several hormone sensors have been designed to address this issue. An auxin sensor, DII, was built by fusing three Venus FPs to the auxin-interacting domain II (DII) of the IAA28 protein under the control of a constitutive promoter (Brunoud et al., 2012). This domain is ubiquitylated in the presence of auxin and triggers the degradation of the protein. Absence of fluorescence in specific domains in plants expressing the DII reporter therefore indicates the presence of auxin (Brunoud et al., 2012). An enhanced auxin sensor, R2D2, combines DII-Venus with mDII-tdTomato, in which a mutated, degradation-proof version of the DII domain is fused to a tdTomato FP (Liao et al., 2015; for a comprehensive review on monitoring auxin concentration, transport, and response, see Parizkova et al., 2017). The ratio between red and yellow fluorescence in different domains of plants expressing the R2D2 reporter allows for the semi-quantitative measurement of auxin levels. Genetically encoded, ratiometric Förster resonance energy transfer (FRET) sensors can also be used to quantify the concentration of hormones (for a review on FRET sensors, see Okumoto et al., 2012). FRET sensors combine two FPs, a donor and an acceptor, with a sensory domain. Binding of this sensory domain to its ligand induces a change of conformation that brings the donor and acceptor closer together, allowing the excitation of the acceptor FP by the light emitted by the donor FP. Changes in the ratio between donor and acceptor fluorescence intensity upon excitation of the donor reflect the concentration of the ligand. Such FRET sensors were recently designed for high-resolution quantification of abscisic acid, gibberellins, and now auxin (Jones et al., 2014; Waadt et al., 2014; Rizza et al., 2017; Herud-Sikimic, 2020, Preprint). FRET sensors can also be used to monitor calcium concentration (Miyawaki et al., 1997; Nagai et al., 2004), and were used to detect rapid calcium fluctuations in ovules during double fertilization (Hamamura et al., 2014).
Any light microscopy technique that produces optical sectioning can potentially be used to map gene expression and hormone signaling using multiple reporters. It is worth noting, however, that multicolor imaging is not trivial with two-photon microscopy, which uses a single, high-energy laser. To date, the majority of flower development studies have used confocal microscopy (e.g. Mayer et al., 1998; Urbanus et al., 2009; Chandler et al., 2011; Sun et al., 2014; Monniaux et al., 2017; Prunet et al., 2017; Yamaguchi et al., 2017; Xu et al., 2018) or two-photon microscopy (e.g. Mizuta et al., 2015; Kimata et al., 2016). While light-sheet microscopy was extensively used on underground tissues, it has been more difficult to apply to aerial tissues. The first study to use light-sheet microscopy to image flowers produced good surface rendering of the external morphology of the flower, but not of inner tissues (Ovečka et al., 2018); however, light-sheet microscopy was recently successfully used to study germ cell development in Arabidopsis flowers (Fig. 1C) (Valuchova et al., 2020), and can be expected to be widely used in the future. OPT and macro-OPT can also be used to map gene expression and hormone response in flowers in 3D (Lee et al., 2006, 2017); it works both with reporters that catalyze the formation of a colored product from a colorless substrate, like GUS (with transmitted OPT; Fig. 2A), and with fluorescent reporters (with fluorescence OPT), and both types of reporters can be imaged in the same specimen. Conversely, X-ray-based imaging techniques do not currently allow for the imaging of either colored or fluorescent reporters. However, it may be possible in the future to design reporters based on enzymes catalyzing a reaction that produces contrasting agents and could therefore be visualized with CT scan and XRM. Further, the use of nanogold particles as contrast agents is being explored for contrasting plant and fungal structures (KD, personal communication) as has been done recently for studying soil properties with XRT (Scotson et al., 2019). In the meanwhile, the recent development of correlative microscopy, and in particular fluorescence XRM (FXM), allows for combining fluorescence and XRM of the same specimen.
Qualitative and quantitative image analysis
Modern bioimaging techniques rapidly generate large, complex 3D and 4D data sets, which are not trivial to analyze. A variety of open-access (e.g. MorphographX, FiJi, and MARS-ALT) and commercial (e.g. Aivia and Imaris) software makes it possible to segment cells, detect organelles, trace cytoskeleton filaments, and track cell lineages in developing specimens (Fernandez et al., 2010; Barbier de Reuille et al., 2014, 2015). There is much more to microscopy and tomography images than meets the eye: providing that imaging is done correctly, image analysis software can also be used to extract a wealth of quantitative data from these bioimaging data, such as levels of gene expression, hormone accumulation and response, ratio between donor and acceptor fluorescence in FRET sensors, area and volume of different compartments, and cell growth (for examples of growth quantification in sepals and leaves, see Hervieux et al., 2016; Kierzkowski et al., 2019). Access to quantitative information over developmental time is critical to further our understanding of flower development.
Concluding remarks
The explosion of new bioimaging technologies that we have seen over the last two decades is still ongoing: scientists are developing ‘mesoscopes’ that use larger objectives or mirrors instead of lenses to resolve cellular and subcellular details in specimens more than 1 cm in size (Perkel, 2019); expansion microscopy lets biologists resolve finer details by expanding their specimens (Wassie et al., 2019); adaptative optics correct for light scattering in deep tissues (Marx, 2017). While some of these new techniques might prove difficult to adapt to plants (e.g. expansion microscopy), others will make their way into the plant field. In the meanwhile, this explosion of new imaging techniques has already provided biologists with invaluable tools to ‘look at the thing’. We now have the ability to study the development of live flowers, from a few dividing cells at the tip of a stem to mature flowers undergoing pollination and fertilization, with access to gene expression and hormone signaling, over a wide range of physical and temporal scales. When picking an imaging modality, we are also facing a complex choice. Ideally, we would like to generate sharp and well-contrasted 4D reconstructions of developing flowers at high spatial and temporal resolution, but no size fits all, and bioimaging requires compromises, between signal, speed, and resolution. We hope this review helps flower developmental biologists pick the imaging technique(s) that best fit their purpose.
Acknowledgements
The authors wish to thank Yoko Mizuta, Daisuke Kurihara, Tetsuya Higashiyama, Karen Lee, Enrico Coen, Sona Valuchova, Pavlina Mikulkova, and Karel Riha for providing images for the figures, Dan Chitwood and Michelle Quigley for their insights about XRT, Pavak Shah for his comments on the manuscript, and Frank Wellmer for patiently waiting for this manuscript for more than 3 years. NP’s work at UCLA is supported by the Department of Molecular, Cell and Development Biology and the Broad Stem Cell Research Center. KD’s work in Christopher Topp’s lab at the Donald Danforth Plant Science Center, and the two X-ray instruments are funded by NSF grants and by collaborative agreements with Sumitomo Chemical Company and Valent BioSciences.
Author contributions
NP contributed the sections about light microscopy and OPT, as well as the confocal microscopy images in Fig. 1. KD contributed the sections about XRT and XRM, and the XRM images in Fig. 2.
References
- Aloni R, Aloni E, Langhans M, Ullrich CI. 2006. Role of auxin in regulating Arabidopsis flower development. Planta 223, 315–328. [DOI] [PubMed] [Google Scholar]
- Amos WB, White JG, Fordham M. 1987. Use of confocal imaging in the study of biological structures. Applied Optics 26, 3239–3243. [DOI] [PubMed] [Google Scholar]
- Amos WB, White JG. 2003. How the confocal laser scanning microscope entered biological research. Biology of the Cell 95, 335–342. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Robinson S, Smith RS. 2014. Quantifying cell shape and gene expression in the shoot apical meristem using MorphoGraphX. Methods in Molecular Biology 1080, 121–134. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, et al. 2015. MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife 4, 05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Benninger RK, Piston DW. 2013. Two-photon excitation microscopy for the study of living cells and tissues. Current Protocols in Cell Biology Chapter 4, Unit 4.11.1–Unit 4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, et al. 2014. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Gilroy S. 2000. Plant cell biology in the new millennium: new tools and new insights. American Journal of Botany 87, 1547–1560. [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. 1991. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. The Plant Cell 3, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. 1992. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615. [DOI] [PubMed] [Google Scholar]
- Bray AL, Topp CN. 2018. The quantitative genetic control of root architecture in maize. Plant & Cell Physiology 59, 1919–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, et al. 2012. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. [DOI] [PubMed] [Google Scholar]
- Centonze VE, White JG. 1998. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophysical Journal 75, 2015–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Jacobs B, Cole M, Comelli P, Werr W. 2011. DORNRÖSCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Molecular Biology 76, 171–185. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Boavida LC, Aggarwal M, Wu HM, Feijó JA. 2010. The pollen tube journey in the pistil and imaging the in vivo process by two-photon microscopy. Journal of Experimental Botany 61, 1907–1915. [DOI] [PubMed] [Google Scholar]
- Duncan KE, Bray AL, Dowd TG, Topp CN. 2019. Using 3D X-ray microscopy to study crown root development and primary root tip growth in diverse maize (Zea mays L.) lines. Microscopy and Microanalysis 25, 1032–1033.31134876 [Google Scholar]
- Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C. 2010. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nature Methods 7, 547–553. [DOI] [PubMed] [Google Scholar]
- Fiolka R. 2014. Seeing more with structured illumination microscopy. Methods in Cell Biology 123, 295–313. [DOI] [PubMed] [Google Scholar]
- Geng Y, Zhou Y. 2019. Confocal live imaging of shoot apical meristems from different plant species. Journal of Visualized Experiments 145, e5936. [DOI] [PubMed] [Google Scholar]
- Gilroy S. 1997. Fluorescence microscopy of living plant cells. Annual Review of Plant Physiology and Plant Molecular Biology 48, 165–190. [DOI] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. 2008. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. The Plant Cell 20, 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura Y, Nishimaki M, Takeuchi H, Geitmann A, Kurihara D, Higashiyama T. 2014. Live imaging of calcium spikes during double fertilization in Arabidopsis. Nature Communications 5, 4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W. 2005. Deep tissue two-photon microscopy. Nature Methods 2, 932–940. [DOI] [PubMed] [Google Scholar]
- Herud-Sikimic O, Stiel AC, Ortega-Perez M, Shanmugaratnam S, Höcker B, Jürgens G. 2020. Design of a biosensor for direct visualisation of auxin. bioRxiv doi.org/10.1101/2020.01.19.911735. [Preprint]. [Google Scholar]
- Hervieux N, Dumond M, Sapala A, Routier-Kierzkowska AL, Kierzkowski D, Roeder AH, Smith RS, Boudaoud A, Hamant O. 2016. A mechanical feedback restricts sepal growth and shape in Arabidopsis. Current Biology 26, 1019–1022. [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. 2004. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. [DOI] [PubMed] [Google Scholar]
- Ijiri T, Yoshizawa S, Yokota H, Igarashi T. 2014. Flower modeling via X-ray computed tomography. ACM Transcations on Graphics 33, 48. [Google Scholar]
- Ito T, Sakai H, Meyerowitz EM. 2003. Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Current Biology 13, 1524–1530. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM. 1994. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76, 703–716. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeiter J, Staedler YM, Schönenberger J, Weigend M, Luebert F. 2018. Gynoecium and fruit development in Heliotropium sect. Heliothamnus (Heliotropiaceae). International Journal of Plant Sciences 179, 275–286. [Google Scholar]
- Jenik PD, Irish VF. 2000. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB. 2014. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3, e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavareddy G, Rohini S, Ramu SV, Sundaresha S, Kumar AR, Kumar PA, Udayakumar M. 2013. Transgenics in groundnut (Arachis hypogaea L.) expressing cry1AcF gene for resistance to Spodoptera litura (F.). Physiology and Molecular Biology of Plants 19, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, et al. 2019. A growth-based framework for leaf shape development and diversity. Cell 177, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Higaki T, Kawashima T, Kurihara D, Sato Y, Yamada T, Hasezawa S, Berger F, Higashiyama T, Ueda M. 2016. Cytoskeleton dynamics control the first asymmetric cell division in Arabidopsis zygote. Proceedings of the National Academy of Sciences, USA 113, 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, et al. 2011. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Molecular Systems Biology 7, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Mistrik M, Samajová O, Doskočilová A, Ovečka M, Illés P, Bartek J, Samaj J. 2014. Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant Physiology 165, 129–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Mistrik M, Šamajová O, Ovečka M, Bartek J, Šamaj J. 2015a Superresolution live imaging of plant cells using structured illumination microscopy. Nature Protocols 10, 1248–1263. [DOI] [PubMed] [Google Scholar]
- Komis G, Novák D, Ovečka M, Šamajová O, Šamaj J. 2018. Advances in imaging plant cell dynamics. Plant Physiology 176, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Šamajová O, Ovečka M, Šamaj J. 2015b Super-resolution microscopy in plant cell imaging. Trends in Plant Science 20, 834–843. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T. 2015. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert TJ. 2019. FPbase: a community-editable fluorescent protein database. Nature Methods 16, 277–278. [DOI] [PubMed] [Google Scholar]
- Lee K, Avondo J, Morrison H, Blot L, Stark M, Sharpe J, Bangham A, Coen E. 2006. Visualizing plant development and gene expression in three dimensions using optical projection tomography. The Plant Cell 18, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJI, Calder GM, Hindle CR, Newman JL, Robinson SN, Avondo JJHY, Coen ES. 2017. Macro optical projection tomography for large scale 3D imaging of plant structures and gene activity. Journal of Experimental Botany 68, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Klein LL, Duncan KE, Jiang N, Chitwood DH, Londo JP, Miller AJ, Topp CN. 2019. Characterizing 3D inflorescence architecture in grapevine using X-ray imaging and advanced morphometrics: implications for understanding cluster density. Journal of Experimental Botany 70, 6261–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nature Methods 12, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. 2017. Microscopy: hello, adaptive optics. Nature Methods 14, 1133–1136. [DOI] [PubMed] [Google Scholar]
- Mathers AW, Hepworth C, Baillie AL, Sloan J, Jones H, Lundgren M, Fleming AJ, Mooney SJ, Sturrock CJ. 2018. Investigating the microstructure of plant leaves in 3D with lab-based X-ray computed tomography. Plant Methods 14, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Meyer HM, Teles J, Formosa-Jordan P, Refahi Y, San-Bento R, Ingram G, Jonsson H, Locke JC, Roeder AH. 2017. Fluctuations of the transcription factor ATML1 generate the pattern of giant cells in the Arabidopsis sepal. eLife 6, e19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani P, Mirabet V, Cellier C, Rozier F, Hamant O, Das P, Boudaoud A. 2014. Matching patterns of gene expression to mechanical stiffness at cell resolution through quantitative tandem epifluorescence and nanoindentation. Plant Physiology 165, 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Mizuta Y, Kurihara D, Higashiyama T. 2015. Two-photon imaging with longer wavelength excitation in intact Arabidopsis tissues. Protoplasma 252, 1231–1240. [DOI] [PubMed] [Google Scholar]
- Monniaux M, McKim SM, Cartolano M, Thévenon E, Parcy F, Tsiantis M, Hay A. 2017. Conservation vs divergence in LEAFY and APETALA1 functions between Arabidopsis thaliana and Cardamine hirsuta. New Phytologist 216, 549–561. [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DB, Davidson MW. 2013. Fundamentals of light microscopy and electronic imaging. Hoboken, NJ: Wiley-Blackwell. [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. 2004. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proceedings of the National Academy of Sciences, USA 101, 10554–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor D. 2018. Live confocal imaging of brachypodium spikelet meristems. Bio-Protocol 8, e3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Jones A, Frommer WB. 2012. Quantitative imaging with fluorescent biosensors. Annual Review of Plant Biology 63, 663–706. [DOI] [PubMed] [Google Scholar]
- Oreopoulos J, Berman R, Browne M. 2014. Spinning-disk confocal microscopy: present technology and future trends. Methods in Cell Biology 123, 153–175. [DOI] [PubMed] [Google Scholar]
- Ovečka M, Vaškebová L, Komis G, Luptovčiak I, Smertenko A, Šamaj J. 2015. Preparation of plants for developmental and cellular imaging by light-sheet microscopy. Nature Protocols 10, 1234–1247. [DOI] [PubMed] [Google Scholar]
- Ovečka M, von Wangenheim D, Tomančák P, Šamajová O, Komis G, Šamaj J. 2018. Multiscale imaging of plant development by light-sheet fluorescence microscopy. Nature Plants 4, 639–650. [DOI] [PubMed] [Google Scholar]
- Parizkova B, Pernisova M, Novak O. 2017. What has been seen cannot be unseen—detecting auxin in vivo. International Journal of Molecular Sciences 18, 2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM. 2019. The microscope makers putting ever-larger biological samples under the spotlight. Nature 575, 715–717. [DOI] [PubMed] [Google Scholar]
- Prunet N. 2017. Live confocal imaging of developing Arabidopsis flowers. Journal of Visualized Experiments 122, e55156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Jack TP, Meyerowitz EM. 2016. Live confocal imaging of Arabidopsis flower buds. Developmental Biology 419, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. 2017. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proceedings of the National Academy of Sciences, USA 114, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM. 2017. In vivo gibberellin gradients visualized in rapidly elongating tissues. Nature Plants 3, 803–813. [DOI] [PubMed] [Google Scholar]
- Roberston D, Julias M, Yang Lee S, Cook D. 2017. Maize stalk lodging: morphological determinants of stalk strength. Crop Science 57, 927–934. [Google Scholar]
- Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure JE. 2003. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Current Biology 13, 432–436. [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Gutierrez R, McFarlane HE, Bringmann M, Lindeboom J, Emons AM, Samuels L, Ketelaar T, Ehrhardt DW, Persson S. 2013. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiology 162, 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jönsson H, Meyerowitz EM. 2014. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3, e01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Yan A, Krupinski P, Meyerowitz EM. 2014. Physical forces regulate plant development and morphogenesis. Current Biology 24, R475–R483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotson CP, Munoz-Hernando M, Duncan SJ, Ruiz SA, Keyes SD, van Veelen A, Dunlop IE, Roose T. 2019. Stabilizing gold nanoparticles for use in X-ray computed tomography imaging of soil systems. Royal Society Open Science 6, 190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nature Methods 2, 905–909. [DOI] [PubMed] [Google Scholar]
- Sharpe J. 2004. Optical projection tomography. Annual Review of Biomedical Engineering 6, 209–228. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. 1990. Early flower development in Arabidopsis. The Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedler YM, Kreisberger T, Manafzadeh S, Chartier M, Handschuh S, Pamperl S, Sontag S, Paun O, Schönenberger J. 2018. Novel computed tomography-based tools reliably quantify plant reproductive investment. Journal of Experimental Botany 69, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedler YM, Masson D, Schönenberger J. 2013. Plant tissues in 3D via X-ray tomography: simple contrasting methods allow high resolution imaging. PLoS One 8, e75295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Looi LS, Guo S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T. 2014. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343, 1248559. [DOI] [PubMed] [Google Scholar]
- Sundberg E, Østergaard L. 2009. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harbor Perspectives in Biology 1, a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K. 2004. RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425–434. [DOI] [PubMed] [Google Scholar]
- Tracy SR, Gómez JF, Sturrock CJ, Wilson ZA, Ferguson AC. 2017. Non-destructive determination of floral staging in cereals using X-ray micro computed tomography (µCT). Plant Methods 13, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. 1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. The EMBO Journal 11, 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa S, Hervieux N, Kierzkowski D, Routier-Kierzkowska AL, Sapala A, Hamant O, Smith RS, Roeder AHK, Boudaoud A, Li CB. 2017. Clones of cells switch from reduction to enhancement of size variability in Arabidopsis sepals. Development 144, 4398–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus SL, de Folter S, Shchennikova AV, Kaufmann K, Immink RG, Angenent GC. 2009. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biology 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valuchova S, Mikulkova P, Pecinkova J, Klimova J, Krumnikl M, Bainar P, Heckmann S, Tomancak P, Riha K. 2020. Imaging plant germline differentiation within Arabidopsis flower by light sheet microscopy. eLife 9, e52546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Niet T, Zollikofer CP, León MS, Johnson SD, Linder HP. 2010. Three-dimensional geometric morphometrics for studying floral shape variation. Trends in Plant Science 15, 423–426. [DOI] [PubMed] [Google Scholar]
- van Roessel P, Brand AH. 2002. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nature Cell Biology 4, E15–E20. [DOI] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI. 2014. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3, e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CN, Hsu HC, Wang CC, Lee TK, Kuo YF. 2015. Quantifying floral shape variation in 3D using microcomputed tomography: a case study of a hybrid line between actinomorphic and zygomorphic flowers. Frontiers in Plant Science 6, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassie AT, Zhao Y, Boyden ES. 2019. Expansion microscopy: principles and uses in biological research. Nature Methods 16, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb RH. 1999. Theoretical basis of confocal microscopy. Methods in Enzymology 307, 3–20. [DOI] [PubMed] [Google Scholar]
- Weber M, Huisken J. 2011. Light sheet microscopy for real-time developmental biology. Current Opinion in Genetics & Development 21, 566–572. [DOI] [PubMed] [Google Scholar]
- Weber M, Mickoleit M, Huisken J. 2014. Light sheet microscopy. Methods in Cell Biology 123, 193–215. [DOI] [PubMed] [Google Scholar]
- White JG, Amos WB, Fordham M. 1987. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. Journal of Cell Biology 105, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw B, De Rybel B. 2019. Cytokinin—a developing story. Trends in Plant Science 24, 177–185. [DOI] [PubMed] [Google Scholar]
- Xu Y, Prunet N, Gan ES, et al. 2018. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. The EMBO Journal 37, e97499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Huang J, Xu Y, Tanoi K, Ito T. 2017. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nature Communications 8, 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B. 2013. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiology 161, 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]