Root architectural phenes have heritable and plastic responses, and genetic loci associated with stress and environmental plasticity are distinct from loci controlling phenotypic expression in water-stress and well-watered conditions.
Keywords: Architecture, association mapping, maize, plasticity, root, water deficit stress
Abstract
Root phenotypes regulate soil resource acquisition; however, their genetic control and phenotypic plasticity are poorly understood. We hypothesized that the responses of root architectural phenes to water deficit (stress plasticity) and different environments (environmental plasticity) are under genetic control and that these loci are distinct. Root architectural phenes were phenotyped in the field using a large maize association panel with and without water deficit stress for three seasons in Arizona and without water deficit stress for four seasons in South Africa. All root phenes were plastic and varied in their plastic response. We identified candidate genes associated with stress and environmental plasticity and candidate genes associated with phenes in well-watered conditions in South Africa and in well-watered and water-stress conditions in Arizona. Few candidate genes for plasticity overlapped with those for phenes expressed under each condition. Our results suggest that phenotypic plasticity is highly quantitative, and plasticity loci are distinct from loci that control phene expression in stress and non-stress, which poses a challenge for breeding programs. To make these loci more accessible to the wider research community, we developed a public online resource that will allow for further experimental validation towards understanding the genetic control underlying phenotypic plasticity.
Introduction
Crop varieties are generally developed for specific environmental and management scenarios. However, increasingly unpredictable growth environments due to climate change, decreasing freshwater availability, and rising costs of fuel and nitrogen fertilizer require the development of crop varieties that are resistant to abiotic stress and for increased production in marginal soils (Tebaldi and Lobell, 2008; Brisson et al., 2010; Woods et al., 2010; Sandhu et al., 2016). The occurrence of water deficit stress is likely to become increasingly frequent and unpredictable as a result of global climate change. Phenotypic plasticity is the ability of a plant to alter its phenotype in response to the environment and encompasses components of genotype by environment interaction (G×E). Plasticity may be morphological, anatomical, and developmental, involve changes in resource allocation (Sultan, 2000), and is under genetic control (e.g. Sandhu et al., 2016). Breeding programs have prioritized uniformity and yield stability in specific environments, and phenotypic plasticity has often been considered a challenge in this context (Basford and Cooper, 1998; Cooper et al., 1999). It has been proposed that crops that can adapt their growth in response to environmental signals may be a breeding target for increasing agricultural productivity (e.g. Nicotra et al., 2010; Topp, 2016), although the fitness impacts of phenotypic plasticity are poorly understood, and it has been proposed that plasticity may be maladaptive in some cases (Lynch, 2013, 2018).
Root architectural phenes (‘phene’ is to ‘phenotype’ as ‘gene’ is to ‘genotype’) (Lynch, 2011; Pieruschka and Poorter, 2012; York et al., 2013) have important roles in soil resource capture, particularly in environments with suboptimal water and nutrient availability. Root architectural phenes determine the temporal and spatial distribution of roots in specific soil domains and their ability to obtain mobile and immobile resources (Lynch, 1995, 2013, 2019; Hirel et al., 2007; Lynch and Brown, 2012). For example, root growth angle influences root distribution and depth, and therefore plant performance in nutrient and water-deficit stress conditions (Bonser et al., 1996; Uga et al., 2011; Trachsel et al., 2013; York et al., 2013; Dathe et al., 2016). Root phene states that enable exploration of deep soil domains enhance the capture of mobile soil resources, including water and nitrate (Lynch and Wojciechowski, 2015). Steep growth angles enable deeper rooting and the capture of mobile nutrients, such as nitrogen, in deep soil domains (Trachsel et al., 2013; Dathe et al., 2016), while shallow growth angles are more beneficial for the capture of immobile resources in the topsoil, such as phosphorus (Bonser et al., 1996; Lynch and Brown, 2001; Ho et al., 2005; Zhu et al., 2005a). Lateral root branching length and density have a significant effect on plant performance in water-stressed and low nitrogen environments, where longer, more dispersed lateral root branches are beneficial for the capture of mobile resources due to reduced inter- and intraplant competition for soil resources (Zhan and Lynch, 2015; Zhan et al., 2015), while greater density of lateral branching improves topsoil foraging and phosphorus capture (Postma et al., 2014; Jia et al., 2018). Reduced crown root number improves plant growth in low nitrogen (Saengwilai et al., 2014) and drought (Gao and Lynch, 2016) by reducing inter- and intraplant competition for internal and external resources, thereby increasing root depth and acquisition of deep soil resources. A reduced number of crown roots in modern maize lines increased plant growth in high nitrogen environments and was associated with increased nitrogen use efficiency compared with commercially successful lines a century ago (York et al., 2015). Greater crown root number improves plant growth in low phosphorus soil by reducing axial root elongation and improving topsoil foraging (Sun et al., 2018).
Root plasticity also varies spatially in response to soil conditions. In some genotypes and species, lateral root branches proliferate in response to localized patches of nitrogen and phosphorus availability (Drew, 1975; Zhu and Lynch, 2004) which has been proposed as a beneficial strategy for enhanced nitrogen acquisition (Mi et al., 2010). However, this response may be maladaptive if mobile resources, such as nitrogen or water, move faster through the soil profile than roots can proliferate. This is especially detrimental when proliferation in one soil domain diverts resources from other soil domains that will have greater resource availability later in the season, for example deeper soil domains in leaching precipitation regimes (Lynch, 2013, 2018). The plasticity of lateral root branching in response to local nutrient patches may be beneficial for nutrient resource capture in environments where the nutrient source is sustained or in conditions of interspecific competition (Robinson et al., 1999).
Phenotypic changes as a result of plasticity may be of short or long duration. For example, expression of nitrate transporters fluctuates rapidly in response to environmental signals including light and nitrogen availability (Feng et al., 2011). In contrast, initial root growth angles are established after root emergence, and possible changes to morphology are limited in mature tissue. Plasticity of root phenes that are established early in development, such as root growth angle, may be beneficial in conditions of sustained edaphic stress, such as low phosphorus availability, but may be maladaptive for stresses that fluctuate on shorter time scales, including drought, by generating sustained responses to ephemeral conditions (Lynch, 2013).
Although root architectural phenes can improve the capture of soil resources in specific environments, for example sustained nitrogen or phosphorus stress (Saengwilai et al., 2014; Dathe et al., 2016), some phene states can be functionally maladaptive in fluctuating environments (Ho et al., 2005; Poot and Lambers, 2008). In the field, the plant may be exposed to multiple, simultaneous, or successive stresses. For example, root phene states that improve topsoil foraging (e.g. shallow growth angle) are advantageous for phosphorus acquisition, but may be unfavorable for the capture of deep soil resources such as water (Ho et al., 2005). Trade-offs also exist for phene states for nitrogen and phosphorus acquisition. For example, in common bean (Phaseolus vulgaris), shallow growth angle and a greater number of basal root whorls and hypocotyl-borne roots increase total root length in the topsoil, resulting in greater phosphorus uptake (Rangarajan et al., 2018). However, as the number of axial roots and/or basal root whorls increase, the resulting carbon limitation leads to a reduced root depth and therefore trade-offs for nitrogen acquisition (Rangarajan et al., 2018). No single root phenotype is optimal across a range of environments (Tardieu, 2012; Dathe et al., 2016; Rangarajan et al., 2018). Understanding phenotypic plasticity and its genetic control will be useful in developing strategies to optimize soil resource capture under multiple, dynamic stresses.
Phenotypic plasticity may improve plant performance in variable environments; however, in high-input environments with intensive fertilization and greater nitrogen and phosphorus availability, root plasticity may be counterproductive. Crops and crop ancestors evolved in ecosystems with one or more edaphic stresses influencing growth and root function. Therefore, the ancestral strategies for soil resource capture may not be useful in high-input environments in which constraints to root function are mitigated (Lynch, 2018). Root phenotypes that explore deep soil domains, whether plastic or not, may be beneficial in most environments for the capture of water and nitrogen. In the majority of agricultural systems, deeper root phenotypes enhance water and nitrogen capture, despite the fact that water and nitrogen availability are sometimes greater in surface soils of high-input systems (Manschadi et al., 2006; Gowda et al., 2011; Henry et al., 2011).
Plasticity in root architecture may be advantageous for drought tolerance (e.g. Kano-Nakata et al., 2013; Prince et al., 2017). In drought conditions, plasticity in lateral root length and density (Kano et al., 2011; Kano-Nakata et al., 2013), root length density, and total root length (Kano-Nakata et al., 2011; Tran et al., 2014) correlated with greater shoot biomass, water uptake, and photosynthesis in rice. Plasticity in response to water deficit has also been observed for the number of nodal roots in rice (Suralta et al., 2010) and maize (Gao and Lynch, 2016), lateral branching density and length in maize (Zhan et al., 2015), and deep rooting in wheat (Ehdaie et al., 2012; Wasson et al., 2012), millet (Rostamza et al., 2013), rice (Hazman and Brown, 2018), and maize (Nakamoto, 1993). Plasticity in the positioning of lateral branches, root hairs, and aerenchyma towards available water has also been documented as a phenomenon called hydropatterning (Bao et al., 2014; Orosa-Puente et al., 2018). In addition, high yield stability correlated with high root plasticity in drought and low phosphorus environments in rice (Sandhu et al., 2016). Maize genotypes that increased root hair length in response to low phosphorus availability had better performance under low P than lines with constitutively long root hairs (Zhu et al., 2010). A few previous studies have demonstrated that genes associated with phene expression may be distinct from those associated with plasticity for that expression. Genetic regions controlling plasticity have been identified for root hair length (Zhu et al., 2005b) and lateral root branching and length (Zhu et al., 2005c) in maize under low phosphorus availability, and for lateral root growth (Niones et al., 2015), root anatomy (Kadam et al., 2017), and root length density and root dry weight (Sandhu et al., 2016) in rice in response to drought. The identification of genetic regions controlling plasticity could provide useful breeding targets for crop improvement and may aid in understanding the benefits and trade-offs of root plasticity (Collins et al., 2008).
The objectives of this research were to test the hypotheses that (i) the responses of root architectural phenes to water deficit (stress plasticity) and different environmental conditions (environmental plasticity) are under genetic control; and (ii) genetic loci associated with plasticity are distinct from loci controlling phenotypic expression in water-stress and well-watered conditions. Here we identify and characterize phenotypic plasticity in root architectural phenes in mature, field-grown maize and identify distinct genetic regions controlling these phenes in well-watered and water-stress conditions as well as genetic regions controlling the plastic stress and environmental response of these phenes.
Materials and methods
Field conditions, experimental design, and plant materials
Root architecture phenotypes were measured on the Wisconsin Diversity Panel (Hansey et al., 2011). The Wisconsin Diversity Panel is a large association panel composed of inbred maize lines that reach grain physiological maturity in the upper Midwest region of the USA and display uniformity and vigor. Experiments were conducted at the Apache Root Biology Center (ARBC) in Willcox, Arizona (32°153' 9.252''N, 109° 49' 56.928'' W) in well-watered (ARBC-WW) and water-stress (ARBC-WS) conditions (Supplementary Table S1 at JXB online, 383 genotypes planted) and at the Ukulima Root Biology Center (URBC) in Alma, Limpopo, South Africa (24°33'0012''S, 28°07'2584''E) under non-stress conditions (Supplementary Table S2, 641 genotypes planted). The Arizona experiments were conducted on a Grabe loam (coarse-loamy, mixed, thermic Typic Torrifluvent) from May to September 2014, 2015, and 2016. Genotypes were grown in two replications per treatment in a randomized complete block design each year. The experiments in South Africa were conducted on a Clovelly loamy sand (Typic Ustipsamment) from January to April in 2010, 2011, and 2012, and from November to February in 2013. Genotypes were grown in four replications in a randomized complete block design. For all experiments, each line was planted in a single row plot consisting of 20 plants per plot. Row width was 75 cm and distance between plants within a row was 23 cm. Soil nutrient levels were adjusted based on soil tests at the beginning of the season to meet the requirements for maize production. Pest control was carried out as needed. In South Africa, trials were irrigated using a center pivot system. In Arizona, trials were irrigated using drip irrigation in 2014 and a center pivot system in 2015 and 2016, and drought and well-watered treatments were grown in separate blocks. Water stress was confirmed by an ~20% vegetative biomass growth reduction and 40% yield reduction in water-stressed compared with well-watered conditions. Drought was induced ~4 weeks after planting. Drought was monitored throughout the growth season by PR2 multi-depth soil moisture probes (Dynamax, Houston, TX, USA).
Phenotypic analysis
Root architecture was phenotyped in all experiments. Evaluations of maize root crowns for architecture were performed based on the shovelomics method followed by manual phenotyping (Trachsel et al., 2011) in 2010–2012 and image analysis with Digital Imaging of Root Traits (DIRT) in 2013–2017 (Bucksch et al., 2014; Das et al., 2015). At anthesis, three representative plants were excavated from each plot for architectural analysis from 2010 to 2014, and one representative plant in 2015–2016. In brief, root crowns were excavated in a soil monolith using a standard shovel. Root crowns were soaked in water for 15 min to remove soil. The root crowns were then washed with low-pressure water to remove remaining soil. Four root architectural phenes were collected (Table 1) by imaging or manually phenotyping cleaned root crowns. Average lateral root length (LL), lateral branching frequency on the excised root (BF), and root angle (ANGLE) were measured in all experiments. Distance to the first lateral branch (DISTLAT) was only collected in 2013–2016. Excised root traits were measured on a representative third whorl crown root at the South Africa field site and on a representative fourth whorl crown root at the Arizona field site. Plant height at anthesis was measured in three plants per plot at anthesis in South Africa. Shoot dry biomass was collected for one plant per plot at anthesis in Arizona. Yield was collected at physiological maturity, and cobs from three plants per plot were bulked and weighed.
Table 1.
Description of architectural phenes measured at anthesis
| Trait | Description | Units |
|---|---|---|
| LL | Average lateral root length | mm |
| DISTLAT | Distance to the first lateral root from the root apex on the excised root | mm |
| BF | Lateral branching frequency on the excised root | Branches mm–1 |
| ANGLE | Angle of roots relative to the soil line | ° |
‘Excised root’ is a representative third whorl crown root in Arizona and a representative second whorl crown root in South Africa.
Data analysis
Plasticity in response to water deficit was calculated as a relative value compared with control growing conditions for each phene under no stress:
where water stress (WS) is the mean value of the phene in water-stress conditions for each replication and well-watered (WW) is the mean value of the phene in well-watered conditions for each replication.
In the case of environmental plasticity, plasticity was calculated as a relative phenotypic value of South Africa growing conditions compared with the Arizona growing conditions for each phene:
where SA is the mean value of the phene in the South Africa environment and AZ is the mean value of the phene in the Arizona environment.
Broad-sense heritability on an entry mean basis was calculated for each architectural phene according to Fehr (1993).
Spearman and Pearson correlations between replications and years suggested data could be combined by environment and treatment, therefore mean phenotypic values across all years were calculated and used for subsequent analysis. For phenotypes in water-stress and well-watered environments in Arizona, an average of two replications over 3 years within each treatment were combined. Plastic responses to water deficit were calculated by replication. For phenotypes in well-watered environments in South Africa, the averages of four replications over 4 years were combined. Plastic responses to the environment were calculated by year. Residuals were transformed according to boxcox analysis.
Architectural phenotypes in well-watered and water-stress conditions and their corresponding plasticity values were used in a Multiple Loci Linear Mixed Model for genome-wide association study (GWAS) analysis (Zhang et al., 2010) implemented in the FarmCPU R package (X. Liu et al., 2016). The model used 591 688 single nucleotide polymorphism (SNP) markers (Mazaheri et al., 2019). Allelic effects are estimated relative to the minor allele.
Significant SNPs were identified based on a genome-wide corrected Bonferroni threshold of –log(P)=7.07 based on the number of SNP markers used in the model. QQ-plots for each phene suggested a good model fit (Supplementary Fig. S1).
R Software (version 3.2.4) (R Core Team, 2018), Bioconductor (Bates et al., 2002), MapMan (Usadel et al., 2009), and MaizeGDB (Lawrence, 2005) were used to annotate genes and compare significant SNPs across treatments. Significant differences in MapMan ontologies between treatments were determined using a Student’s t-test. Candidate genes identified through significant GWAS hits were detected based on the physical position of genes in the version 4 B73 (AGPv4) reference sequence assembly (Jiao et al., 2017). To understand the functional relevance of associated candidate genes with root architectural traits, we examined the functional annotation and root expression of maize gene models and their respective orthologs in the genetically well-studied model plant Arabidopsis thaliana. In the WiDiv population, most LD mapping interval sizes are <2 kb (Hirsch et al., 2014); therefore, we only considered genes which had significant SNPs to be candidate genes and did not consider neighboring genes. We used Plaza 4.0 monocots (Van Bel et al., 2018) to determine one-to-one or one-to-many orthologs in A. thaliana, and TAIR (Rhee et al., 2003) and the Arabidopsis eFP Browser (Winter et al., 2007) to obtain ortholog function and its root tissue and root developmental expression patterns (Brady et al., 2007).
Results
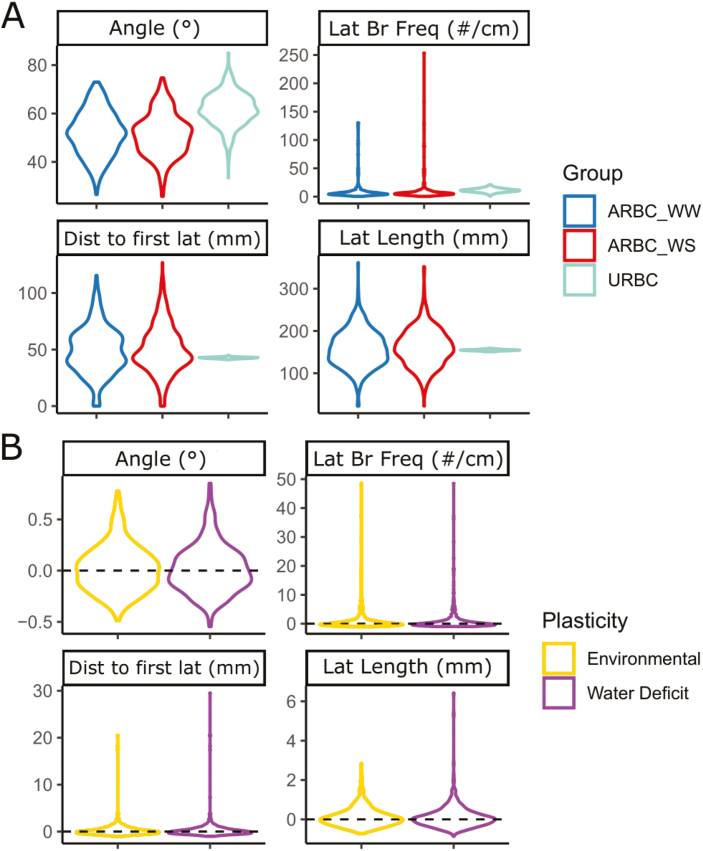
Plastic responses to drought and environment varied by phene (Fig. 1) and genotype (Fig. 2) for all four root phenes measured [angle (ANGLE), lateral branching frequency (BF), average lateral root length (LL), and distance to the first lateral branch (DISTLAT)] (Table 1). A wide range of natural variation was observed for architectural phenes, particularly in the Arizona environment (Supplementary Table S3). Water regime and environment had a significant effect on most root phenes (Supplementary Table S4) and water deficit on average reduced vegetative biomass by 21% and yield by 40% (Supplementary Table S3). LL was 4% greater in the Arizona environment compared with the South Africa environment, but no change in the phenotypic expression was observed between water-stress and well-watered conditions in the Arizona environment. In Arizona, phenotypic expression of nodal root angle (ANGLE), BF, and DISTLAT did not change between well-watered and water-stress conditions. BF and ANGLE were reduced by 46% and 38%, respectively in the Arizona environment compared with the South Africa environment. DISTLAT was 65% greater in the Arizona environment compared with the South Africa environment (Fig. 1). Expression of environmental and stress plasticity was not driven by a few genotypes, and most genotypes expressed plasticity to some degree. However, distinct genotypes expressed plasticity to different degrees for different phenes. Allometric relationships between root phenes and yield or vegetative biomass were not significant (Supplementary Table S5).
Fig. 1.
Distributions of genotypic means for each phene in (A) well-watered and water-stress conditions. (B) Distribution of the root phene stress and environmental plasticity. The y-axis represents the phene value in (A) and the relative difference in phene value between well-watered and water-stressed (stress plasticity) or relative difference between each environment (environmental plasticity) for each phene (B).
Fig. 2.
Genotypes vary in their plastic response to drought. Images of root crowns of a non-plastic genotype and a plastic genotype from well-watered and water-stress treatments. Architectural plasticity is shown for root angle and lateral branching length. Scale bar represents 1 cm (lateral branching length) and 2 cm (angle).
In both environments, LL and DISTLAT in well-watered and water-stress conditions were more heritable than their plastic responses (Table 2). The total phenotypic variation explained by the significant SNPs accounted for 36% of the variation of individual phenes. Most root architectural phenes were under highly quantitative genetic control, as demonstrated by the large number of significant SNPs identified with relatively small effect sizes (Table 2; Supplementary Tables S6–S10). Heritability for root architectural phenes was relatively low to moderate, ranging from 0.13 to 0.68 (Table 2).
Table 2.
Heritability of root architectural phenes in the Wisconsin Diversity panel
| LL | DISTLAT | BF | ANGLE | |||
|---|---|---|---|---|---|---|
| Arizona | Well-watered | Heritability | 0.19 | 0.21 | 0.13 | 0.16 |
| % Variation explained by SNPs | 25.09 | 19.31 | 163.84 | 23.18 | ||
| Water stress | Heritability | 0.15 | 0.17 | 0.16 | 0.22 | |
| % Variation explained by SNPs | 23.83 | 4.33 | 53.55 | 6.31 | ||
| Stress plasticity | Heritability | 0.14 | 0.13 | 0.17 | 0.18 | |
| South Africa | Well-watered | Heritability | 0.43 | 0.68 | 0.42 | 0.64 |
| % Variation explained by SNPs | 24.77 | 6.56 | 72.34 | 4.69 | ||
| Environmental plasticity | Heritability | 0.27 | 0.32 | 0.45 | 0.37 |
Broad-sense heritability on an entry mean basis and percentage variation explained by SNPs is the absolute sum of allelic effects of all significant SNPs for each phene. Explanations of abbreviations are given in Table 1.
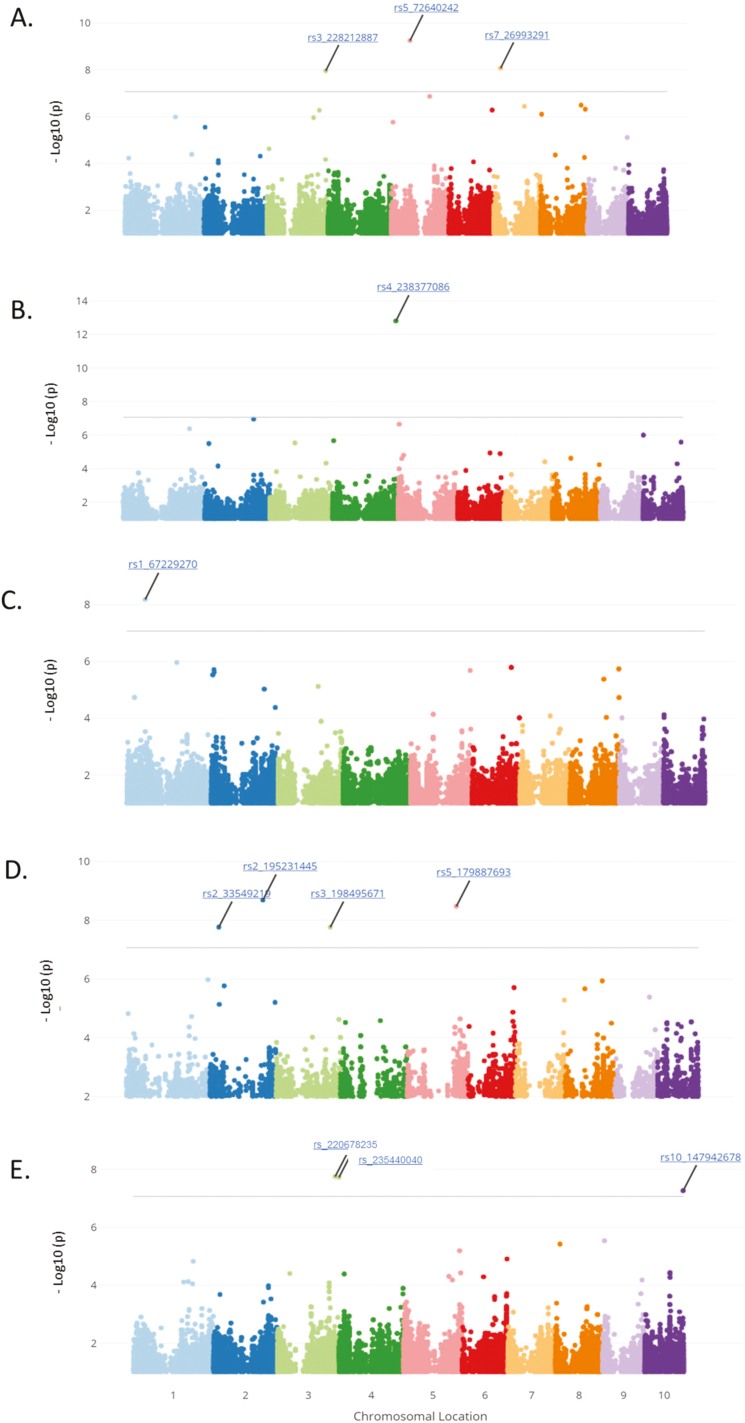
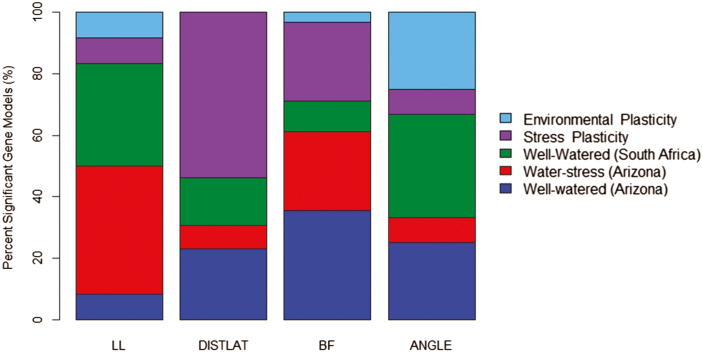
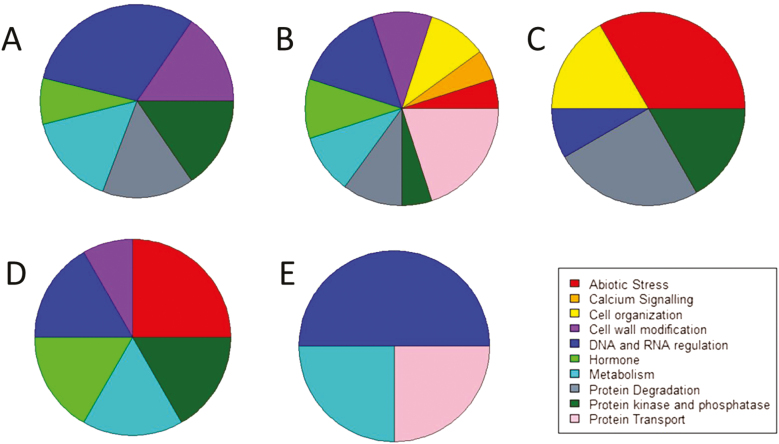
GWAS identified 69 significant SNPs associated with root architecture in well-watered and water-stressed plants and stress and environmental plastic responses, using a Bonferroni-corrected genome-wide threshold value of –log(P)=7.07 (Fig. 3; Supplementary Figs S2–S4). Gene models containing SNPs above the Bonferroni significance threshold were selected as candidate genes. Significant SNPs for root architecture phenes were located in 15, 19, and 17 unique gene models for water-stress, well-watered, and stress plasticity in Arizona, respectively (Fig. 4; Supplementary Tables S6–S8). Significant SNPs for root architecture phenes were located in 13 and five unique gene models for well-watered conditions in South Africa and environmental plasticity, respectively (Supplementary Tables S9, S10). Of the gene models identified as candidates controlling root phenes in well-watered and water-stress conditions and their plastic responses, ~64% were annotated for MapMan ontogenic categories (Supplementary Tables S6–S10). Environmental plasticity has gene models associated with mainly RNA regulation- and transport-related processes. Stress plasticity has gene models associated with RNA regulation, hormone metabolism, protein degradation, and protein translational modification. Water-stress environments have gene models associated with DNA and RNA regulation, and well-watered environments have gene models associated with abiotic stress and protein degredation in South Africa and protein transport in Arizona (Fig. 5).
Fig. 3.
GWAS results for root angle (ANGLE) for plants grown in (A) well-watered conditions, (B) water-stressed conditions, (C) water-stress plasticity in Arizona, (D) well-watered conditions in South Africa, and (E) environmental plasticity. See Supplementary Figs S2–S4 for plots of other architectural phenes.
Fig. 4.
Relative proportion of unique gene models associated with well-watered, water-stress, stress plasticity, and environmental plasticity.
Fig. 5.
Mapman ontogenic categories for annotated gene models associated with significant SNPs in (A) water stress in Arizona (57% annotated), (B) well-watered in Arizona (48% annotated), (C) well-watered in South Africa (70% annotated), (D) stress plasticity in Arizona (29% annotated), and (E) environmental plasticity (80% annotated).
A few significant SNPs co-localized between well-watered, water-stress, and/or plastic response groups. Two SNPs co-localized between stress plasticity and water-stress groups. Both of these SNPs are associated with BF and are located in a gene (Zm00001d026191) that is associated with RNA regulation of an ethylene-responsive element-binding protein family (Supplementary Tables S6, S8).
Significant SNPs for well-watered, water-stress, and stress and environmental plasticity groups were detected for all architectural phenes measured, with the exception of DISTLAT. Approximately 46, 22, 24, and 7% of unique gene models were associated with well-watered, water-stress, stress plasticity, and environmental plasticity, respectively (Fig. 4).
A publicly available online resource was developed for users to easily view and explore GWAS results. The website, which can be accessed at https://rootplasticitygwas.nottingham.ac.uk/, provides information about the phenes measured, interactive Manhattan plots, and information about candidate gene annotations and organ and tissue expression patterns in maize and rice, and in Arabidopsis orthologs. Such collective information could help users to prioritize candidate genes for further experimental validation towards understanding the genetic control underlying phenotypic plasticity.
Discussion
Root architectural phenes are plastic in response to drought and environment. We observed heritable responses of root phenes (Table 2) and large variation in the extent and direction of phene plasticity in response to drought and environment (Fig. 1). Most genetic loci associated with stress or environmental plasticity were distinct from loci controlling phenotypic expression in water-stressed or well-watered conditions (Figs 3, 4; Supplementary Figs S2–S4; Supplementary Tables S6–S10). The genetic architecture and phenotypic characterization of the plastic response of root phenes have important implications in understanding plant adaptation to edaphic stress.
Although significant plasticity was observed in both directions (Fig. 1B), there was no significant overall trend in the direction of plasticity for root angle (ANGLE) between well-watered and water-stress conditions. However, significant ranges of stress and environmental plasticity were observed for root angle (Fig. 1B). In maize, root angle can be plastic in response to nitrogen availability, and in the majority of genotypes studied angles became steeper in nitrogen stress (Trachsel et al., 2013). Steeper root angles enable greater root biomass in deep soil domains and thus greater N capture in leaching environments (Trachsel et al., 2013; Dathe et al., 2016) and increased water capture in drought environments (Mace et al., 2012; Uga et al., 2013). In addition, genetic loci associated with nodal root angle in sorghum co-localized with genes associated with the stay-green drought tolerance mechanism (Mace et al., 2012). DRO1, a gene associated with steep root angle in rice, contributes to avoiding drought stress by increasing deep rooting and thus increasing yield in drought environments (Uga et al., 2013). In wheat, steeper seminal roots were associated with plants with increased drought tolerance (Manschadi et al., 2008; Maccaferri et al., 2016).
Lateral root length (LL) became longer under water stress, and no trend was observed for lateral branching frequency (BF) under stress. Few, long lateral roots compared with many, short lateral roots are beneficial for maize under water and nitrogen stress (Zhan and Lynch, 2015; Zhan et al., 2015). For mobile soil resources, such as nitrogen and water, resource depletion zones surrounding roots are relatively large. Short, dense lateral roots create overlapping resource depletion zones around roots of the same plant, decreasing resource capture efficiency (Ge et al., 2000). Long, dispersed lateral phenotypes along the axial roots optimize the capture of mobile resources as they reduce inter- and intraplant competition (Zhu et al., 2005c; Postma et al., 2014; Zhan and Lynch, 2015; Zhan et al., 2015). No significant differences in the mean and range in expression of BF or distance to the first lateral root (DISTLAT) were observed between well-watered and water-stress conditions. While there was no significant overall trend in the direction of plasticity of BF and DISTLAT, a wide range of phenotypic plasticity was observed, and a few individuals had extreme responses to water deficit. Presumably, reduced BF and greater DISTLAT would be beneficial for the capture of mobile resources as they would decrease root length in shallow soil domains, thereby enabling exploration of deeper soil strata (York and Lynch, 2015).
Our results indicate that root architectural phenes and their plastic response to water stress and environment are genetically controlled and highly quantitative. A total of 69 unique gene models were identified as being associated with root architecture within well-watered and water-stressed environments and for stress and environmental plasticity. Many genes with relatively small effect sizes were associated with these phenes, but additively these genes accounted for ~36% of total phenotypic variation. Heritability for phenotypic plasticity was lower than heritability for root phenes in water-stressed and well-watered conditions. This could result from short-term variation in phene plasticity to track fluctuating environmental signals. Heritable plasticity responses indicate that root plasticity is genetically controlled. Heritable plastic responses have also been reported for other species and phenes including flower formation (Bradshaw, 2006; Lande, 2009). Root phenes are highly quantitative, and plasticity in response to edaphic stress and different environments may enable breeding efforts for plastic or non-plastic lines in specific phenes. Understanding root phenotypic plasticity and its genetic control may permit the selection of lines with optimal plasticity to improve plant growth in specific environments. For example, plants with greater phenotypic plasticity for BF may be more useful in environments with fluctuating drought, but reduced phenotypic plasticity for ANGLE may be more beneficial in environments with sustained stress including low nitrogen.
Heritability for root architectural phenes ranged from 0.13 to 0.68 (Table 2). The relatively low percentage of variation explained by significant SNPs (Table 2) can partially be explained by the relatively low heritability of root phenes. However, heritability of field-grown root phenes of mature plants is generally low (Bucksch et al., 2014; Burridge et al., 2016). Low heritabilities may reduce the power of SNP detection, inflate gene effect sizes, and increase the chance of detecting false positives. In the current study, the relatively low heritability values of root phenes can be attributed to the highly heterogenous environment of field-grown maize and the highly quantitative nature of these phenes.
While the overall trend was a decrease in vegetative biomass and yield in drought, significant plasticity was observed in both directions (Supplementary Fig. S5). Plasticity is not simply a growth reduction due to edaphic stress. Plants may have different strategies to achieve drought tolerance, and the plastic responses of root phenes or phene aggregates to drought or different environments could be adaptive or maladaptive. Unresponsiveness of lateral root branching to localized availability of resources, such as water, would be advantageous under drought. Localized proliferation in response to mobile resources, such as water or nitrogen, may be counterproductive as these resources are subject to leaching, movement, and depletion, while root growth is relatively slow and involves significant construction and maintenance costs (Lynch, 2018). In addition, root production in shallow soil domains in response to ephemeral resources may divert resources from root construction in deeper soil domains with greater water availability (Lynch, 2013, 2018).
Specific root phenes are important in plant stress tolerance; however, root phenes do not function in isolation (York et al., 2013; Miguel et al., 2015). Synergisms exist between phene states with a large metabolic cost, for example lateral root branching density, with phenes that reduce the metabolic cost of the root, including fewer basal root whorls in bean. A decreased number of basal root whorls is more beneficial in plants with more dense lateral branching density (Rangarajan et al., 2018). Synergisms exist with phenes that affect the placement of roots in the soil domains. For example, basal root growth angle interacts with root hair density and length to determine the placement of root hairs in the soil profile and increase plant growth up to twice the expected additive effects (York et al., 2013; Miguel et al., 2015). Understanding phene synergisms and their plastic interactions may be an important consideration for breeders.
Auxin has been well studied for its role in regulating root gravitropism (Su et al., 2017) and plays a role in the establishment of root angle (Toal et al., 2018). We identified two candidate genes annotated to auxin-related processes, associated with the phenotypic expression of ANGLE under well-watered conditions (Zm00001d019311; IAA-amino acid hydrolase ILR1-like 7) and stress plasticity (Zm00001d029356; O-methyltransferase ZRP4) in Arizona. These genes are orthologs of Arabidopsis ILR1 (IAA-LEUCINE RESISTANT 1) and ASMT (N-ACETYLSEROTONIN O-METHYLTRANSFERASE), respectively. ILR1-like family hydrolases are known to modulate auxin response by regulating auxin homeostasis (Sanchez Carranza et al., 2016). ASMT is involved in the melatonin biosynthetic process, and has been recently implicated in regulating root architecture and gravitropism by modulating auxin response in rice (Liang et al., 2017). Melatonin stimulates several physiological responses to environmental conditions including water deficit (Ding et al., 2018; Debnath et al., 2019).
Another candidate gene associated with the phenotypic expression of DISTLAT under stress plasticity (Zm00001d024644) is a MYB-related transcription factor family protein. Due to its role in auxin biosynthesis, the overexpression of this MYB gene showed a significantly increased number of lateral roots and elongated hypocotyl (Kwon et al., 2013). Auxin has known roles in the establishment of root angle and development of lateral roots, and presumably is an important regulator of the development of other root phenes.
A candidate gene associated with lateral root branching frequency (BF) under well-watered conditions in South Africa (Zm00008a029231) is a GRAS transcription factor family protein. Its Arabidopsis ortholog encodes SHR (Short Root), is involved in asymmetric cell division and radial pattern formation in root, and is required for the initiation and patterning of lateral root primordia (Lucas et al., 2011). Additionally, our study associated Zm00001d043612 (orthologous to Arabidopsis FLZ10) with BF under well-watered conditions in Arizona. This gene is expressed in young primordia during lateral root development and its mutant showed reduced lateral roots (Jamsheer et al., 2018). The expression of FLZ genes is highly regulated by energy status and abiotic stress (Jamsheer and Laxmi, 2015).
Cytokinin metabolism and signaling genes are known to form a redundant network to modulate lateral root initiation and outgrowth of young primordia. A candidate gene associated with the stress plasticity response of LL is involved in cytokinin metabolism and signaling. Cytokinin is also known to act antagonistically to other hormones (e.g. brassinosteroids) and affect lateral root elongation in cytokinin receptor mutants (Chang et al., 2015). Multilevel redundancy of cytokinin modulating lateral root phenes is believed to reflect the role of cytokinin in mediating environmental signals.
A candidate gene associated with the phenotypic expression of DISTLAT under stress plasticity (Zm00001d038366) is involved in lipid metabolism. The Arabidopsis ortholog of this gene encodes GPAT5, a glycerol-3-phosphate SN-2-acyltransferase that is involved in suberin biosynthesis. GPAT5 was found to be specifically expressed in the lateral root formation zone in the root and may be required for lateral root formation (Beisson et al., 2007). Suberin functions as an apoplastic diffusion barrier at lateral root emergence sites (Li et al., 2017), and developmental variations of apoplastic barriers within lateral root system could be an important trait in response to abiotic stress factors (Tylová et al., 2017).
A cytochrome P450 gene differentially expressed in nitrogen stress conditions in maize leaves and ears (Zm00001d048702) (Arp, 2017) has implications in auxin formation (Irmisch et al., 2015) and was associated with DISTLAT in well-watered conditions. Lateral branching phenes including LL and BF have roles in plant performance under nitrogen stress (Zhan and Lynch, 2015). Presumably, DISTLAT also influences plant performance in edaphic stress by affecting the metabolic cost of soil exploration (York et al., 2015). A gene controlling aquaporin expression in maize (Zm00008a000537) (Yue et al., 2012) was associated with the plasticity of LL between different environments. LL has been demonstrated to have a large role in drought tolerance (Zhan et al., 2015) and presumably is associated with genes controlling drought tolerance including aquaporin genes. A gene associated with environmental plasticity of root angle (Zm00008a014805) was up-regulated in leaves in water deficit and cold stress (F. Liu et al., 2016). Root angle influences rooting depth and thus the capture of deep soil water, increasing drought tolerance (Mace et al., 2012; Uga et al., 2013).
A gene associated with the plasticity of root angle in different environments (Zm00008a038792) was up-regulated during phosphate starvation in maize (Xu et al., 2018). A gene associated with stress plasticity of DISTLAT (Zm00001d038366) is up-regulated during phosphorus stress and is involved in phosphorus metabolism and utilization (Yu et al., 2019), and may also be involved in suberin synthesis (Zhu et al., 2019). A number of root phenes influence phosphorus capture under suboptimal phosphorus availability, including the density of lateral branching (Zhu and Lynch, 2004; Postma et al., 2014), root cortical aerenchyma (RCA) (Galindo-Castañeda et al., 2018), root angle (Zhu et al., 2005a), the number of crown roots (Sun et al., 2018), and root hair length (Zhu et al., 2010). Presumably, genes that are differentially expressed in low phosphorus availability could contribute to controlling root phenes under many edaphic stresses through common signaling pathways such as auxin and ethylene (Ma et al., 2003; Giri et al., 2018).
A gene associated with the plastic response of BF and the phenotypic expression of BF in water stress (Zm00001d026191) co-localized with a gene up-regulated in maize cortical cells upon ethylene exposure which encoded an ethylene response factor class of transcription factor (Takahashi et al., 2015). A gene associated with BF in well-watered conditions is annotated to APETALA2/Ethylene-responsive element binding protein (AP2/EREBP). A differentially expressed gene in root cortical cells during ethylene-induced RCA formation (Zm00008a029231) (Takahashi et al., 2015) was associated with BF in well-watered conditions in the South Africa field site. Hypoxia induces RCA formation (Evans, 2003), and presumably common signaling pathways (e.g. ethylene) induce RCA formation and control expression of other root phenes under a range of edaphic stresses. For example, ethylene inhibits root branching at the earliest stages of lateral root initiation (Negi et al., 2008; Lewis et al., 2011).
Of the gene models identified, 45% were annotated. The stress plasticity group had significantly more genes associated with hormones and abiotic stress compared with the well-watered and water-stressed groups (Fig. 5). Hormone signaling, particularly ABA and ethylene signaling, has important implications in drought tolerance (Sharp and LeNoble, 2002; Shi et al., 2015). In addition, a gene associated with cytokinin signal transduction (Zm00001d037694) was associated with the water stress response of lateral root length. Cytokinin signal transduction is associated with adaptation to stress and interacts with ABA signaling (Ha et al., 2012). The co-localization of a few SNPs between the well-watered, water-stressed, and stress and environmental plasticity groups indicates that plasticity is controlled by many different genes in distinct pathways (Figs 4, 5).
The fact that there was no co-localization of significant SNPs between the Arizona and South Africa field sites for the same root phenes indicates that the expression of these phenes, and subsequent identification of associated genetic loci, is highly dependent on the environment. Factors including differences in soil texture, photoperiod, and irrigation regimes may account for some of these differences. In the South Africa environment, root angles were steeper and lateral root phenes had considerably less variation compared with the Arizona environment. Highly quantitative traits with small effects, such as root architectural phenes, may not be ideal for GWAS models that have historically been successful with qualitative traits or highly heritable quantitative traits (e.g. flowering time). Consideration of multiple phenes, gene networks, and dynamic responses may result in stronger associations of phenes with genetic loci or regulatory pathways.
Environmental plasticity has widespread implications in interpretation and extrapolation of data from different growing systems and environments. With a few notable exceptions (e.g. Schneider et al., 2020), the majority of root quantitative trait loci (QTLs) and GWAS studies use artificial growth systems (Zhu et al., 2005c, 2006; Trachsel et al., 2009) that do not realistically represent field conditions and therefore cannot adequately address questions related to root architecture and its relationship to nutrient uptake. In addition, many studies examine embryonic root systems (Hund et al., 2004; Zhu et al., 2005c; Trachsel et al., 2009), which are poor predictors of mature root system architecture (Zhu et al., 2011) and may be under genetic control distinct from that of post-embryonic root systems (Hochholdinger and Feix, 1998; Hochholdinger et al., 2001). Root growth in artificial systems may be constrained by the size of the growth media or container, and is buffered from the atmospheric environment in a completely different way when compared with field-grown conditions. In addition, elongation and trajectory of growing roots are affected by changes in soil bulk density as a result of sieving and compacting soil, relative to undisturbed soil. The spatiotemporal dynamics of nutrient and water regimes in soil are difficult to mimic in artificial media. Therefore, root growth in artificial media may be artifactual relative to field conditions (Walter et al., 2009).
In this study, root architectural phenotypes were profiled in diverse maize lines with and without water stress in the field in multiple environments. Significant and substantial variation was observed for all phenes in well-watered and water-stressed environments, and for plasticity, namely phenotypic responses to water availability or phenotypic responses to different environmental conditions. Root architectural phenes and their responses to drought and environment are heritable and genetically controlled. Phenotypic plasticity and interactions between root phenes may be synergistic or antagonistic. Phenotypic plasticity and interactions between phenes will require further research to understand their implications for edaphic stress tolerance. Identifying genes that control root phenes and their plastic expression under edaphic stress will enable the selection of lines with greater or reduced plasticity in breeding programs to increase plant productivity. Identification of genes underlying root plasticity can provide breeders with novel opportunities to develop crop varieties better suited to a wide range of environments and agroecosystems.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Genotypes in the Wisconsin Diversity Panel grown in Arizona.
Table S2. Genotypes in the Wisconsin Diversity Panel grown in South Africa.
Table S3. Phenotypic variation of root architectural traits.
Table S4. Summary of the analysis of variance for root architectural phenes.
Table S5. Allometric analysis of root phenes.
Table S6. Gene models identified for root phenes in water-stressed conditions in Arizona.
Table S7. Gene models identified for root phenes in well-watered conditions in Arizona.
Table S8. Gene models identified to control plasticity in drought for root phenes.
Table S9. Gene models identified for root phenes in well-watered conditions in South Africa.
Table S10. Gene models identified to control environmental plasticity for root phenes.
Fig. S1. Q-Q plots assessing the fitness of K model for GWAS of root phenes.
Fig. S2. GWAS results for DISTLAT for plants grown in well-watered and water-stress conditions, and their plasticity.
Fig. S3. GWAS results for BF for plants grown in well-watered and water-stress conditions, and their plasticity.
Fig. S4. GWAS results for LL for plants grown in well-watered and water-stress conditions, and their plasticity.
Fig. S5. Violin plots showing the distribution of vegetative biomass and yield.
Acknowledgements
This research was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, award #2014-67013-21572, USDOE ARPA-E ROOTS Award Number DE-AR0000821, Hatch project #4582, University of Nottingham Future Food Beacon of Excellence Nottingham Research, and a BBSRC discovery fellowship. This research was also supported by the Howard G. Buffet Foundation. We thank Robert Snyder, Curtis Fredrick, Johan Prinsloo, and Patricio Cid for technical support, and Malcolm Bennett for commenting on the manuscript.
Glossary
Abbreviations
- ANGLE
angle of roots relative to the soil line
- BF
lateral branching frequency on the excised root
- DISTLAT
distance to the first lateral root from the root apex on the excised root
- GWAS
genome-wide association study
- LL
average lateral root length
- QTL
quantitative trait locus
- WS
water stress
- WW
well-watered
References
- Arp J. 2017. Discovery of novel regulators and genes in nitrogen utilization pathways in maize. Dissertation, University of Illinois at Urbana-Champaign. [Google Scholar]
- Bao Y, Aggarwal P, Robbins NE 2nd, et al. . 2014. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proceedings of the National Academy of Sciences, USA 111, 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basford KE, Cooper M. 1998. Genotype × environment interactions and some considerations of their implications for wheat breeding in Australia. Australian Journal of Agricultural Research 49, 153–174. [Google Scholar]
- Bates D, Carey V, Dettling M, et al. . 2002. Bioconductor www.bioconductor.org.
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. The Plant Cell 19, 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonser AM, Lynch J, Snapp S. 1996. Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytologist 132, 281–288. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD. 2006. Unravelling phenotypic plasticity—why should we bother? New Phytologist 170, 644–648. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Brisson N, Gate P, Gouache D, Charmet G, Oury FX, Huard F. 2010. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Research 119, 201–212. [Google Scholar]
- Bucksch A, Burridge J, York LM, Das A, Nord E, Weitz JS, Lynch JP. 2014. Image-based high-throughput field phenotyping of crop roots. Plant Physiology 166, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge J, Jochua CN, Bucksch A, Lynch JP. 2016. Legume shovelomics: high-throughput phenotyping of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp. unguiculata) root architecture in the field. Field Crops Research 192, 21–32. [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2015. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. Journal of Experimental Botany 66, 4759–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Tardieu F, Tuberosa R. 2008. Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiology 147, 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M, Rajatasereekul S, Immark S, Fukai S, Basnayake J. 1999. Rainfed lowland rice breeding strategies for Northeast Thailand. I. Genotypic variation and genotype × environment interactions for grain yield. Field Crops Research 64, 131–151. [Google Scholar]
- Das A, Schneider H, Burridge J, Ascanio AK, Wojciechowski T, Topp CN, Lynch JP, Weitz JS, Bucksch A. 2015. Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe A, Postma JA, Postma-Blaauw MB, Lynch JP. 2016. Impact of axial root growth angles on nitrogen acquisition in maize depends on environmental conditions. Annals of Botany 118, 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath B, Islam W, Li M, Sun Y, Lu X, Mitra S, Hussain M, Liu S, Qiu D. 2019. Melatonin mediates enhancement of stress tolerance in plants. International Journal of Molecular Sciences 20, 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Zhao Y, Xu JW, Zhao P, Li T, Ma H, Reiter RJ, Yu X. 2018. Melatonin: a multifunctional molecule that triggers defense responses against high light and nitrogen starvation stress in Haematococcus pluvialis. Journal of Agricultural and Food Chemistry 66, 7701–7711. [DOI] [PubMed] [Google Scholar]
- Drew M. 1975. Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytologist 75, 479–490. [Google Scholar]
- Ehdaie B, Layne AP, Waines JG. 2012. Root system plasticity to drought influences grain yield in bread wheat. Euphytica 186, 219–232. [Google Scholar]
- Evans DE. 2003. Aerenchyma formation. New Phytologist 161, 35–49. [Google Scholar]
- Fehr W. 1993. Principles of cultivar development. New York: Macmillan Publishing Company. [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G. 2011. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. Journal of Experimental Botany 62, 2319–2332. [DOI] [PubMed] [Google Scholar]
- Galindo-Castañeda T, Brown KM, Lynch JP. 2018. Reduced root cortical burden improves growth and grain yield under low phosphorus availability in maize. Plant, Cell & Environment 41, 1579–1592. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lynch JP. 2016. Reduced crown root number improves water acquisition under water deficit stress in maize (Zea mays L.). Journal of Experimental Botany 67, 4545–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Rubio G, Lynch JP. 2000. The importance of root gravitropism for inter-root competition and phosphorus acquisition efficiency: results from a geometric simulation model. Plant and Soil 218, 159–171. [DOI] [PubMed] [Google Scholar]
- Giri J, Bhosale R, Huang G, et al. . 2018. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nature Communications 9, 1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R. 2011. Root biology and genetic improvement for drought avoidance in rice. Field Crops Research 122, 1–13. [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. 2012. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends in Plant Science 17, 172–179. [DOI] [PubMed] [Google Scholar]
- Hansey CN, Johnson JM, Sekhon RS, Kaeppler SM de Leon N. 2011. Genetic diversity of a maize association population with restricted phenology. Crop Science 51, 704–715. [Google Scholar]
- Hazman M, Brown KM. 2018. Progressive drought alters architectural and anatomical traits of rice roots. Rice 11, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Gowda VRP, Torres RO, Mcnally KL, Serraj R. 2011. Variation in root system architecture and drought response in rice (Oryza sativa): phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crops Research 120, 205–214. [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. 2007. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany 58, 2369–2387. [DOI] [PubMed] [Google Scholar]
- Hirsch CN, Foerster JM, Johnson JM, et al. . 2014. Insights into the maize pan-genome and pan-transcriptome. The Plant Cell 26, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MD, Rosas JC, Brown KM, Lynch JP. 2005. Root architectural tradeoffs for water and phosphorus acquisition. Functional Plant Biology 32, 737–748. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Feix G. 1998. Early post-embryonic root formation is specifically affected in the maize mutant lrt1. The Plant Journal 16, 247–255. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Feix H. 2001. Cooperative action of SLR1 and SLR2 is required for lateral root-specific cell elongation in maize 1. Plant Physiology 125, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund A, Fracheboud Y, Soldati A, Frascaroli E, Salvi S, Stamp P. 2004. QTL controlling root and shoot traits of maize seedlings under cold stress. Theoretical and Applied Genetics 109, 618–629. [DOI] [PubMed] [Google Scholar]
- Irmisch S, Müller AT, Schmidt L, Günther J, Gershenzon J, Köllner TG. 2015. One amino acid makes the difference: the formation of ent-kaurene and 16α-hydroxy-ent-kaurane by diterpene synthases in poplar. BMC Plant Biology 15, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer MK, Laxmi A. 2015. Expression of Arabidopsis FCS-like Zinc finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Frontiers in Plant Science 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer MK, Shukla BN, Jindal S, Gopan N, Mannully CT, Laxmi A. 2018. The FCS-like zinc finger scaffold of the kinase SnRK1 is formed by the coordinated actions of the FLZ domain and intrinsically disordered regions. Journal of Biological Chemistry 293, 13134–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Liu P, Lynch JP. 2018. Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. Journal of Experimental Botany 69, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, et al. . 2017. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam NN, Tamilselvan A, Lawas LMF, et al. . 2017. Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiology 174, 2302–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano M, Inukai Y, Kitano H, Yamauchi A. 2011. Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant and Soil 342, 117–128. [Google Scholar]
- Kano-Nakata M, Gowda VRP, Henry A, Serraj R, Inukai Y, Fujita D, Kobayashi N, Suralta RR, Yamauchi A. 2013. Functional roles of the plasticity of root system development in biomass production and water uptake under rainfed lowland conditions. Field Crops Research 144, 288–296. [Google Scholar]
- Kano-Nakata M, Inukai Y, Wade LJ, Siopongco JD, Yamauchi A. 2011. Root development, water uptake, and shoot dry matter production under water deficit conditions in two CSSLs of rice: functional roles of root plasticity. Plant Production Science 14, 307–317. [Google Scholar]
- Kwon Y, Kim JH, Nguyen HN, Jikumaru Y, Kamiya Y, Hong SW, Lee H. 2013. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. Journal of Experimental Botany 64, 3911–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology 22, 1435–1446. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Seigfried TE, Brendel V. 2005. The maize genetics and genomics database. The community resource for access to diverse maize data. Plant Physiology 138, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. 2011. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138, 3485–3495. [DOI] [PubMed] [Google Scholar]
- Li B, Kamiya T, Kalmbach L, et al. . 2017. Role of LOTR1 in nutrient transport through organization of spatial distribution of root endodermal barriers. Current Biology 27, 758–765. [DOI] [PubMed] [Google Scholar]
- Liang C, Li A, Yu H, Li W, Liang C, Guo S, Zhang R, Chu C. 2017. Melatonin regulates root architecture by modulating auxin response in rice. Frontiers in Plant Science 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu Y, Han G, Zhou L, Ali A, Zhu S, Li X. 2016. Molecular evolution and genetic variation of G2-like transcription factor genes in maize. PLoS One 11, e01617630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z. 2016. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genetics 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, et al. . 2011. Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiology 155, 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. 1995. Root architecture and plant productivity. Plant Physiology 109, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2011. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiology 156, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2018. Rightsizing root phenotypes for drought resistance. Journal of Experimental Botany 69, 3279–3292. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2019. Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytologist 223, 548–564. [DOI] [PubMed] [Google Scholar]
- Lynch J, Brown K. 2001. Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant and Soil 237, 225–237. [Google Scholar]
- Lynch JP, Brown KM. 2012. New roots for agriculture: exploiting the root phenome. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 1598–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Wojciechowski T. 2015. Opportunities and challenges in the subsoil: pathways to deeper rooted crops. Journal of Experimental Botany 66, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. 2003. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiology 131, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, El-Feki W, Nazemi G, Salvi S, Canè MA, Colalongo MC, Stefanelli S, Tuberosa R. 2016. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. Journal of Experimental Botany 67, 1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace ES, Singh V, Van Oosterom EJ, Hammer GL, Hunt CH, Jordan DR. 2012. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theoretical and Applied Genetics 124, 97–109. [DOI] [PubMed] [Google Scholar]
- Manschadi AM, Christopher J, deVoil P, Hammer GL. 2006. The role of root architectural traits in adaptation of wheat to water-limited environments. Functional Plant Biology 33, 823. [DOI] [PubMed] [Google Scholar]
- Manschadi AM, Hammer GL, Christopher JT, DeVoil P. 2008. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant and Soil 303, 115–129. [Google Scholar]
- Mazaheri M, Heckwolf M, Vaillancourt B, et al. . 2019. Genome-wide association analysis of stalk biomass and anatomical traits in maize. BMC Plant Biology 19, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi G, Chen F, Wu Q, Lai N, Yuan L, Zhang F. 2010. Ideotype root architecture for efficient nitrogen acquisition by maize in intensive cropping systems. Science China Life Sciences 53, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Miguel MA, Postma JA, Lynch JP. 2015. Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant Physiology 167, 1430–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto T. 1993. Effect of soil water content on the gravitropic behavior of nodal roots in maize. Plant and Soil 152, 261–267. [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. 2008. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. The Plant Journal 55, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, et al. . 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15, 684–692. [DOI] [PubMed] [Google Scholar]
- Niones JM, Inukai Y, Suralta RR, Yamauchi A. 2015. QTL associated with lateral root plasticity in response to soil moisture fluctuation stress in rice. Plant and Soil 391, 63–75. [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, et al. . 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362, 1407–1410. [DOI] [PubMed] [Google Scholar]
- Pieruschka R, Poorter H. 2012. Phenotyping plants: genes, phenes and machines. Functional Plant Biology 39, 813–820. [DOI] [PubMed] [Google Scholar]
- Poot P, Lambers H. 2008. Shallow-soil endemics: adaptive advantages and constraints of a specialized root-system morphology. New Phytologist 178, 371–381. [DOI] [PubMed] [Google Scholar]
- Postma JA, Dathe A, Lynch JP. 2014. The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiology 166, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince SJ, Murphy M, Mutava RN, Durnell LA, Valliyodan B, Shannon JG, Nguyen HT. 2017. Root xylem plasticity to improve water use and yield in water-stressed soybean. Journal of Experimental Botany 68, 2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan H, Postma JA, Lynch JP. 2018. Co-optimization of axial root phenotypes for nitrogen and phosphorus acquisition in common bean. Annals of Botany 122, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Version 3.3.1. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, et al. . 2003. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Hodge A, Gri BS, Fitter AH. 1999. Plant root proliferation in nitrogen-rich patches confers competitive advantage. Proceedings of the Royal Society B: Biological Sciences 266, 431–435. [Google Scholar]
- Rostamza M, Richards RA, Watt M. 2013. Response of millet and sorghum to a varying water supply around the primary and nodal roots. Annals of Botany 112, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengwilai P, Tian X, Lynch JP. 2014. Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiology 166, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Carranza AP, Singh A, Steinberger K, Panigrahi K, Palme K, Dovzhenko A, Dal Bosco C. 2016. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Scientific Reports 6, 24212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu N, Raman KA, Torres RO, Audebert A, Dardou A, Kumar A, Henry A. 2016. Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiology 171, 2562–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider HS, Klein SP, Hanlon MT, Brown KM, Kaeppler S, Lynch JP. 2020. Genetic control of root anatomical plasticity in maize. Plant Genome (in press). [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME. 2002. ABA, ethylene and the control of shoot and root growth under water stress. Journal of Experimental Botany 53, 33–37. [PubMed] [Google Scholar]
- Shi J, Habben JE, Archibald RL, Drummond BJ, Chamberlin MA, Williams RW, Lafitte HR, Weers BP. 2015. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiology 169, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Gibbs NM, Jancewicz AL, Masson PH. 2017. Molecular mechanisms of root gravitropism. Current Biology 27, R964–R972. [DOI] [PubMed] [Google Scholar]
- Sultan SE. 2000. Phenotypic plasticity for plant development, function and life history. Trends in Plant Science 5, 537–542. [DOI] [PubMed] [Google Scholar]
- Sun B, Gao Y, Lynch JP. 2018. Large crown root number improves topsoil foraging and phosphorus acquisition. Plant Physiology 177, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suralta RR, Inukai Y, Yamauchi A. 2010. Dry matter production in relation to root plastic development, oxygen transport, and water uptake of rice under transient soil moisture stresses. Plant and Soil 332, 87–104. [Google Scholar]
- Takahashi H, Yamauchi T, Rajhi I, Nishizawa NK, Nakazono M. 2015. Transcript profiles in cortical cells of maize primary root during ethylene-induced lysigenous aerenchyma formation under aerobic conditions. Annals of Botany 115, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F. 2012. Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. Journal of Experimental Botany 63, 25–31. [DOI] [PubMed] [Google Scholar]
- Tebaldi C, Lobell DB. 2008. Towards probabilistic projections of climate change impacts on global crop yields. Geophysical Research Letters 41, 1–14. [Google Scholar]
- Toal TW, Ron M, Gibson D, et al. . 2018. Regulation of root angle and gravitropism. G3 8, 3841–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN. 2016. Hope in change: the role of root plasticity in crop yield stability. Plant Physiology 172, 5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. 2011. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant and Soil 341, 75–87. [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. 2013. Maize root growth angles become steeper under low N conditions. Field Crops Research 140, 18–31. [Google Scholar]
- Trachsel S, Messmer R, Stamp P, Hund A. 2009. Mapping of QTLs for lateral and axile root growth of tropical maize. Theoretical and Applied Genetics 119, 1413–1424. [DOI] [PubMed] [Google Scholar]
- Tran TT, Kano-Nakata M, Suralta RR, Menge D, Mitsuya S, Inukai Y, Yamauchi A. 2014. Root plasticity and its functional roles were triggered by water deficit but not by the resulting changes in the forms of soil N in rice. Plant and Soil 386, 65–76. [Google Scholar]
- Tylová E, Pecková E, Blascheová Z, Soukup A. 2017. Casparian bands and suberin lamellae in exodermis of lateral roots: an important trait of roots system response to abiotic stress factors. Annals of Botany 120, 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Okuno K, Yano M. 2011. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. Journal of Experimental Botany 62, 2485–2494. [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, et al. . 2013. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. 2009. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, maize. Plant, Cell & Environment 32, 1211–1229. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. 2018. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46, D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Silk WK, Schurr U. 2009. Environmental effects on spatial and temporal patterns of leaf and root growth. Annual Review of Plant Biology 60, 279–304. [DOI] [PubMed] [Google Scholar]
- Wasson AP, Richards RA, Chatrath R, Misra SC, Prasad SV, Rebetzke GJ, Kirkegaard JA, Christopher J, Watt M. 2012. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. Journal of Experimental Botany 63, 3485–3498. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J, Williams A, Hughes JK, Black M, Murphy R. 2010. Energy and the food system. Philosophical Transactions of the Royal Society B: Biological Sciences 365, 2991–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu F, Han G, Cheng B. 2018. Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Reports 37, 711–726. [DOI] [PubMed] [Google Scholar]
- York LM, Galindo-Castañeda T, Schussler JR, Lynch JP. 2015. Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. Journal of Experimental Botany 66, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York LM, Lynch JP. 2015. Intensive field phenotyping of maize (Zea mays L.) root crowns identifies phenes and phene integration associated with plant growth and nitrogen acquisition. Journal of Experimental Botany 66, 5493–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York LM, Nord EA, Lynch JP. 2013. Integration of root phenes for soil resource acquisition. Frontiers in Plant Science 4, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Liu C, Lu X, Bai Y, Zhou L, Cai Y. 2019. ZmAPRG, an uncharacterized gene, enhances acid phosphatase activity and Pi concentration in maize leaf during phosphate starvation. Theoretical and Applied Genetics 132, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Yue X, Zhao X, Fei Y, Zhang X. 2012. Correlation of aquaporins and transmembrane solute transporters revealed by genome-wide analysis in developing maize leaf. Comparative and Functional Genomics 2012, 546930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Lynch JP. 2015. Reduced frequency of lateral root branching improves N capture from low-N soils in maize. Journal of Experimental Botany 66, 2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Schneider H, Lynch JP. 2015. Reduced lateral root branching density improves drought tolerance in maize. Plant Physiology 168, 1603–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, et al. . 2010. Mixed linear model approach adapted for genome-wide association studies. Nature Genetics 42, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Ingram PA, Benfey PN, Elich T. 2011. From lab to field, new approaches to phenotyping root system architecture. Current Opinion in Plant Biology 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005a Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Functional Plant Biology 32, 749–762. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005b Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant and Soil 270, 299–310. [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005c Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theoretical and Applied Genetics 111, 688–695. [DOI] [PubMed] [Google Scholar]
- Zhu J, Lynch JP. 2004. The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Functional Plant Biology 31, 949–958. [DOI] [PubMed] [Google Scholar]
- Zhu J, Mickelson SM, Kaeppler SM, Lynch JP. 2006. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theoretical and Applied Genetics 113, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang C, Lynch JP. 2010. The utility of phenotypic plasticity of root hair length for phosphorus acquisition. Functional Plant Biology 37, 313–322. [Google Scholar]
- Zhu T, Wu S, Zhang D, Li Z, Xie K, An X, Ma B, Hou Q. 2019. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theoretical and Applied Genetics 132, 2137–2154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.