Abstract
Leaf angle (LA) is a critical agronomic trait in maize, with more upright leaves allowing higher planting density, leading to more efficient light capture and higher yields. A few genes responsible for variation in LA have been identified by map-based cloning. In this study, we cloned maize ZmIBH1-1, which encodes a bHLH transcription factor with both a basic binding region and a helix-loop-helix domain, and the results of qRT-PCR showed that it is a negative regulator of LA. Histological analysis indicated that changes in LA were mainly caused by differential cell wall lignification and cell elongation in the ligular region. To determine the regulatory framework of ZmIBH1-1, we conducted RNA-seq and DNA affinity purification (DAP)-seq analyses. The combined results revealed 59 ZmIBH1-1-modulated target genes with annotations, and they were mainly related to the cell wall, cell development, and hormones. Based on the data, we propose a regulatory model for the control of plant architecture by ZmIBH1-1 in maize.
Keywords: DAP-seq, leaf angle, maize, map-based cloning, plant architecture, regulatory network, RNA-seq, Zea mays
ZmIBH1-1 encodes a bHLH transcription factor that affects the leaf angle in maize primarily through differential cell wall lignification and cell elongation in the ligular region.
Introduction
The leaf angle (LA) in maize is formed by the leaf blade bending away from the main stem, and it has been used as an important agronomic trait for high-density planting in modern varietal breeding, as hybrids with upright leaves show a yield advantage in crop simulation models, field experiments, and breeding programs (Duncan, 1971; Pepper et al., 1977; Duvick, 2005; Duvick et al., 2010; Li et al., 2011; Wang et al., 2011). A better understanding of the molecular mechanisms underlying the determination of LA will therefore contribute to the breeding of new maize varieties with improved plant architectures.
Numerous studies have been conducted to investigate the natural variation in LA (Mickelson et al., 2002; Lu et al., 2007; Ku et al., 2010; Tian et al., 2011; Wang et al., 2011) and have demonstrated that it is a quantitative trait controlled by several quantitative trait loci (QTL)/genomic regions (Moreno et al., 1997; Yu et al., 2007; Ku et al., 2011; Dong et al., 2018). Most of the QTL studies of LA in maize have been based on primary mapping (Mickelson et al., 2002; Lu et al., 2007, 2018; Ku et al., 2010, 2012; Tian et al., 2011) and only a few genes have been characterized through map-based cloning (Ku et al., 2011; Zhang et al., 2014). Several genes regulating LA in maize have been identified by mutant screening, most of which are related to ligule development and hormone metabolism, and are members of the YABBY gene family. Genes for a liguleless phenotype in maize affect plant architecture through an auxin signaling pathway (Muehlbauer et al., 1999), and the absence of the ligular tissue results in erect leaves. Liguleless1 and Liguleless2 (Moreno et al., 1997; Walsh et al., 1998; Muehlbauer et al., 1999; Moon et al., 2013) are transcription factors that are necessary for the correct positioning of the blade–sheath boundary (Harper and Freeling, 1996). Liguleless narrow encodes a putative serine threonine kinase (Muehlbauer et al., 1999) involved in ligule formation in the maize B73 background. Maize mutants deficient in brassinosteroids (BRs) have shortened internodes and twisted, dark-green, erect leaves and feminized male flowers. Nana plant1 (na1) has a mutation in a gene homologous to DET2 that encodes a 5α-reductase enzyme in the BR biosynthesis pathway (Hartwig et al., 2011) and brd1 has a mutation in the gene encoding brC-6 oxidase (Makarevitch et al., 2012). Kir et al. (2015) used a transgenic RNAi approach to generate plants partially deficient in BRI1 which was the first report of BR signaling affecting plant development in maize. Zmbri1-RNAi plants have a dwarf phenotype with shortened internodes, erect leaves, and reduced auricle tissue formation, indicating a conserved function of BR signaling pathways in maize. DROOPING LEAF genes have also recently been reported in maize. Drooping leaf1 (drl1), encodes a putative transcriptional regulator with zinc-finger and YABBY domains. This is a pleiotropic mutant characterized by reduced midribs and distally extended auricles along the medial domain of the blade–sheath boundary, and it also affects leaf length and width, LA, and internode length and diameter (Strable et al., 2017). In contrast to rice, drls in maize play important roles not only in the leaf but also in the stem architecture.
Thus, although some studies have identified genes that alter LA, the regulatory mechanisms and networks that control this trait still remain unclear. In a previous study we demonstrated that the major QTL associated with LA in maize, qLA2-1, is mapped to the short arm of chromosome 2 in the bin 2.01 region between the markers umc1165 and bnlg1297, and that it could explain 18.43% of the LA variation in the F2 population (Ku et al., 2012). In this current study, we used a map-based approach to clone the candidate gene ZmIBH1-1 and characterized it as a typical basic helix-loop-helix (bHLH) transcription factor that negatively regulates LA in maize. DNA affinity purification (DAP)-seq analysis and transcriptomic analysis revealed that ZmIBH1-1 directly regulates genes involved in cell wall modification, cell development, and hormone responses. This results in alterations to cell wall lignification and cell size in the ligular region of the leaf. Our findings not only reveal a new molecular function for bHLH transcription factors in the regulation of maize plant architecture, but also provides a possible gene network that regulates LA. This provides the framework for a more comprehensive study of maize plant architecture and establishes a better understanding of the function of bHLH transcription factors.
Materials and methods
Plant materials and growth conditions
The fine-mapping populations of maize (Zea mays) were generated by using Yu82 as the donor and crossing to the recurrent parent Yu87-1. Since 2008, we have generated BC3F2 (6529 plants), BC4F2 (7477 plants), BC5F2 (1837 plants), and BC3F3 (1687 plants), BC4F3 (3465 plants), and BC5F3 (6682 plants). These populations were grown at two experimental locations, namely in Zhengzhou, Henan, in the spring of 2009 and 2010, and in Sanya, Hainan, in the winter of 2008, 2009, and 2010). Phenotyping of these populations has been described previously by Ku et al. (2010). DNA extraction for DNA affinity purification (DAP)-seq analysis was conducted using a whole leaf (leaf 7) from Yu82 at the V7 plant growth stage (when the height of the ligule in leaf 7 is the same as that of leaf 6).
Phenotypic analysis
Based on the results of fine-mapping, a Yu87-1 near-isogenic line (NIL) was obtained. The genetic background of this NIL was the same as Yu87-1 except for the region containing ZmIBH1-1, which was derived from Yu82 through foreground selection using flanking markers and background selection using 216 polymorphic SSR markers. The NIL was selected on the basis that it had a background recovery rate >95% with LA <15° (Ku et al., 2010), i.e. the same as Yu82 and considerably smaller than Yu87-1. The inbred lines Yu87-1, NIL, and Yu82 were planted in six-row plots at the same density. At 10 d after pollen shedding, 30 plants from each plot were chosen randomly for evaluation of LA (using all leaves above the uppermost ear).
Fine-mapping of qLA2-1
To develop molecular markers for fine-mapping, bacterial artificial chromosome (BAC) sequences of the B73 genome in the region flanked by um1165 and bnlg1297 on chromosome 2 were obtained from the Maize Genetics and Genomics Database (https://www.maizegdb.org/gbrowse). Simple sequence repeats (SSRs) were identified using the SSR Hunter software (https://en.bio-soft.net/dna/SSRHunter.html). Primers were designed using the Premier 5.0 software (Lalitha, 2000) with a PCR product size of <300 bp. Genomic DNA was extracted from fresh, immature maize leaves from all of the mapping populations. SSR markers were used initially to screen parental lines to identify polymorphic markers for linkage mapping and QTL analysis. PCR products were separated by electrophoresis on a 6% polyacrylamide gel followed by sliver staining for visualization.
ZmIBH1-1 cloning and phylogenetic analysis
To isolate full-length genomic DNA (gDNA) sequences for ZmIBH1-1, immature leaves were collected separately from Yu87-1, NIL, and Yu82. DNA extraction was performed according to the method described by Murray and Thompson (1980). The full-length gDNAs were amplified using the gIBH1-1 primers (Table S2 at Dryad Digital Repository; http://dx.doi.org/10.5061/dryad.18tk64p;Cao et al., 2020). The PCR products were purified and sequenced directly.
Immature leaves were collected from Yu87-1, NIL, and Yu82 and total RNA was prepared using TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from the total RNA and used as a template for reverse-transcriptase (RT)-PCR. The cIBH1-1 primer was designed to amplify the full-length ZmIBH1-1 cDNA (GenBank accession no. MN161779) using one-step RT-PCR (Table S2 at Dryad). The amplified products were purified and sequenced directly. ZmIBH1-1-like protein sequences from other species were obtained using BLAST searches at the NCBI database (http://www.ncbi.nlm.nih.gov). The identified sequences were aligned using the Mega6 software (https://www.megasoftware.net/).
CRISPR-Cas9-mediated genome editing in Setaria
CRISPR/Cas9-mediated genome editing was applied to Setaria viridis to induce targeted mutagenesis of orthologous SvIBH1-1 to create loss-of-function alleles. gRNAs were designed using the CRISPOR online tool http://crispor.tefor.net/. Agrobacterium-mediated transformation of mature seed-derived callus was applied to transform the destination vectors to the ME34 background (Van Eck et al., 2017).
Protoplast transient assays
Protoplasts of tobacco (Nicotiana tabacum) were prepared from BY-2 suspension cells by following the method of Miao and Jiang (2007), with modifications. Tobacco plants were grown at 24 °C and subcultured every 7 d. Protoplasts were prepared from the cells 5 d after subculture. Cell walls were digested at room temperature for 2 h in a solution containing 1% (w/v) cellulase Onozuka R-10 (Serva), 0.1% (w/v) pectinase (Sigma), 0.5% (w/v) Macerozyme RS (Serva), and 0.25 M mannitol. The isolated protoplasts were transformed with 20 µg each of the reporter and effector constructs or mock DNAs using the polyethylene glycol (PEG) method.
35S-ZmIBH1-1 contains the Cauliflower Mosaic Virus (CaMV) 35S promoter driving ZmIBH1-1 expression, and 35S-LUC contains the firefly luciferase driven by the constitutive CaMV-35S promoter. The reporter gene construct (UAS-GUS) and effector constructs (VP16, Gal4, and IAA17) have been described previously by Tiwari et al. (2001). The ZmIBH1-1-GAL4 effector construct contains the full-length ZmIBH1-1 coding sequence fused to the N-terminus of the Gal4 DNA-binding domain under the control of the CaMV-35S promoter. The 35S-LUC construct was co-transformed as an internal control to normalize the GUS reporter gene expression. GUS and LUC enzymatic assays were performed according to Gampala et al. (2001).
Subcellular localization
Full-length cDNA sequences of ZmIBH1-1 were cloned into the vectors pSAT1-cCFP-C and pSAT1-nVenus-C (Biovector NTCC Inc., Beijing, China) to create ZmIBH1-1-YFP. The ZmIBH1-1-YFP plasmids were co-transformed into tobacco protoplasts following standard protocols. After incubation overnight in the dark at 24 °C, nuclei were stained with DAPI. eYFP and DAPI fluorescence in the protoplasts were monitored sequentially with a confocal microscope (Zeiss) using the following respective wavelengths: excitation, 514 nm and 405 nm; and detection, 527 nm and 488 nm.
Safranin staining
Safranin staining for lignin was performed on fully expanded leaf 7 at the V7 stage. The ligular tissue was sectioned by hand to provide both cross- and longitudinal sections. Fresh sections were placed in 0.01% safranin solution for 12 min, then rinsed with 70% ETOH and ddH2O for 1 min each. The samples were mounted in paraffin oil and imaged under a light dissecting microscope.
RNA-seq sampling
Samples of tissue were taken from the leaf base (1 cm length), the middle of the leaf blade (3 cm), and the leaf tip (10 cm) of NIL and Yu87-1 plants. In each case, the samples from 10 plants were pooled together to form one biological replicate. Three biological replicates were used for the RNA extraction. The RNA-seq library was prepared using a QuantSeq 3´ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, Austria). Library DNA was checked for concentration and size distribution in an Agilent 2100 Bioanalyzer before sequencing with an Illumina HiSeq4000 system according to the manufacturer’s instructions.
RNA-seq data analysis
Raw reads were adapter and quality trimmed (Q≥20) using Trim Galore (www.bioinformatics.babraham.ac.uk/projects/trim_galore/). One of the replicates each from the NIL samples for the base and mid-region of leaf 7 yielded very low read counts and hence only two replicates were used for these samples. Expression values (TPM) were generated using Salmon (Patro et al., 2017) in quasi-mapping mode with the following parameters: --noLengthCorrection, --incompatPrio 0.0, and --libType SF. Maize genome annotation v3.31 was used. Gene level read counts were obtained from Salmon and used for analysis of differential gene expression by the R-based EdgeR package v3.16.5 (Robinson et al., 2010). Genes with more than 5 counts per million (cpm) reads in at least three samples were kept. Estimation dispersion was performed using the glmQLFit function with robust =TRUE parameter in EdgeR (Robinson et al., 2010). Significance tests were performed using glmTreat with lfc=log2(1.5). Significant genes with more than two-fold change and FDR≥0.05 were selected for further analysis.
Real-time PCR analysis
Quantitative real-time PCR was performed using an iCycler IQ real-time PCR detection system (Bio-Rad) with SYBR Green Real-Time PCR Master Mix (ABI) according to the standard protocol. Specific primers were designed (Table 2 at Dryad), and the experiments were performed using two sets of independent RNA samples with tubulin as the internal control gene. All the RNA samples used were the same samples as for the RNA-seq assay. Quantifiable differences in gene expression were analysed using the 2–∆∆CT method (Livak and Schmittgen, 2001).
DAP-seq sampling
DAP-seq was performed by following the method described by O’Malley et al. (2016). First, a DAP-seq genomic DNA (gDNA) library was prepared by attaching a short DNA sequencing adaptor on to purified and fragmented gDNA. The adapter sequences were truncated Illumina TruSeq adapters; the TruSeq Universal and Index adapters corresponded to the DAP-seq Adapter A: CACGACGCTCTTCCGATCT; and Adapter B: GATCGGAAGAGCACACGTCTG. The DAP gDNA library was prepared using a kit from NEBNext® DNA Library Prep Master Mix Set for Illumina® (NEB, #E6040S/L). ZmIBH1-1 was fused to the HaloTag using a kit from pFN19K HaloTag T7 SP6 Flexi Vecto (Promega #G184A). ZmIBH1-1 fused to HaloTag was expressed using a TnT SP6 High-Yield Wheat Germ Protein Expression System (L3260, Promega), and then was purified using Magne HaloTag Beads (G7281, Promega). The Magne HaloTag Beads and ZmIBH1-1-HaloTag mixture were incubated with 500 ng DNA library in 40 µl PBS buffer with slow rotation in a cold room for 1.5 h. The beads were washed five times with 200 µl PBS + NP40 (0.005%), resuspended in PBS buffer, the supernatant was removed, and 25 µl EB buffer was added and incubated for 10 min at 98 °C to elute the bound DNA from the beads. The correct DAP-seq library concentration to achieve a specific read count was calculated based on library fragment size. Negative control mock DAP-seq libraries were prepared as described above without the addition of protein to the beads.
DAP-seq data analysis
We defined target genes as those that contained DAP-seq peaks located within the transcribed regions of genes, in introns, or 5 kb upstream of the transcription start site (TSS), or 5 kb downstream of the transcription termination site (TTS). DAP-seq reads were aligned to the maize genome using Bowtie 2 (Langmead and Salzberg, 2012), which supports gapped and paired-end alignment modes. We ran Bowtie version 2.2.3 with default parameters and reported only unique alignments. DAP-seq peaks were detected by MACS2 (Zhang et al., 2008). We used MACS version 2.0.10 with default parameters, as duplicates were allowed, and the q-value <0.05. Core motifs were identified by MEME-ChIP (Machanick and Bailey, 2011).
3´-UTR transcription level test assay
To verify the association between the extracted single-nucleotide polymorphisms (SNPs)/indels and LA, the 3´-untranslated regions (UTRs) of ZmIBH1-1 were amplified from Yu82 and Yu87-1 and cloned into the pGreenII0800-LUC vector. Transient dual-luciferase assays were performed in N. benthamiana leaves. After infiltration, plants were kept at room temperature with a 14/10-h light/dark regime. Leaf protein was extracted 48 h later using a passive lysis buffer (E1910, Promega). The LUC activity was measured using a GloMax®20/20 Luminometer (Promega). Then, 100 µl of Stop and Glow Buffer was added to the reaction and Renilla luciferase (REN) activity was measured. The ratios between LUC and REN activities were measured three times.
Electrophoretic mobility shift assays
32P-labeled DNA fragments containing specific DNA sites were incubated with the ZmIBH1-1 protein, and electrophoretic mobility shift assays (EMSAs) were performed according to Carey et al. (2013).
Results
Fine-mapping of qLA2-1
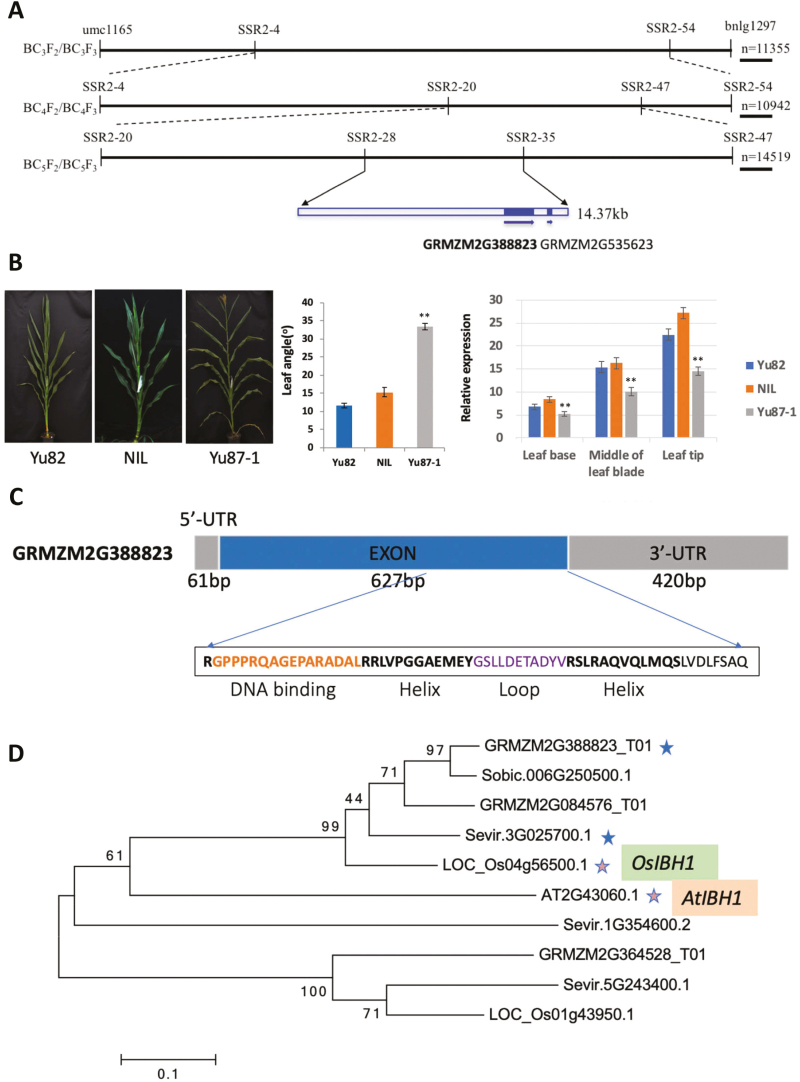
qLA2-1 in the 2.02 region has previously been identified as a major QTL that contributes 18.43% of phenotypic variation in leaf angle (LA) (Ku et al., 2012; Table S1 at Dryad). For fine-mapping, the Yu82 (donor) and Yu87-1 (receptor) parental inbred lines were used to create NILs of Yu87-1 (Yu87-1_NIL). The LA of Yu87-1 was 35° whilst that of Yu87-1_NIL was 18.24° greater, confirming that qLA2-1 contains an important locus controlling LA in maize (Fig. 1B).
Fig. 1.
Fine-mapping of maize ZmIBH1-1. (A) Summary of the fine-mapping. Recombinant populations are shown on the left, the molecular markers are labeled, and the shaded boxes represent exons in the genes. The arrows indicate the direction of the genes. (B) The images show the plant architecture and leaf angles of the parental lines Yu82 and Yu87-1, and the near-isogenic line (NIL), which has the same genetic background as Yu87-1 except for the region containing ZmIBH1-1, which is derived from Yu82. The graphs show mean values (±SD) of leaf angle for all the leaves above the upper ear measured in 30 plants at 10 d after pollen shedding, and the relative expression levels of ZmIBH1-1 in three regions of leaf 7, sampled at growth stage V7. Data are means (±SD) of n=10 replicates and are relative to expression of tubulin. Significant differences between the NIL and Yu87-1 were determined using Student’s t-test: **P<0.01. (C) Structure and sequence and of the ZmIBH1-1 protein. The sequence of the bHLH domain is shown. (D) Phylogenic analysis of IBH1 in rice (Oryza sativa, Os), sorghum (Sorghum bicolor, Sobic), Arabidopsis (AT), Setaria virdis (Sevir), and in maize.
In order to fine-map the qLA2-1 locus, multiple backcross and self-pollinated mapping populations were developed (see Methods). A total of 60 newly developed polymorphic SSR markers (Table S2 at Dryad) located in the qLA2-1 interval were used to identify recombinant genotypes for development of near-isogenic lines (NILs). The qLA2-1 locus was located to a 14.37-kb region between the markers SSR2-33 and SSR2-34 (Fig. 1A).
Scans of this 14.37-kb interval of the maize B73_V3 reference genome identified two candidate genes in this region, GRMZM2G388823 and GRMZM2G535623. Characterization of gene expression was performed by examining published RNA-seq datasets (Wang et al., 2014b) and by qRT-PCR analysis. The results suggested that GRMZM2G535623 was not expressed in developing leaf tissues, while GRMZM2G388823 was expressed in leaf tissues at relatively high levels, with the highest expression near the leaf tip (Fig. 1B).
GRMZM2G388823 encodes a 208-amino bHLH transcription factor that has both a basic binding region and a helix-loop-helix domain (Fig. 1C). Protein BLAST searches and phylogenic analysis showed that GRMZM2G388823 is one of the two homologous genes of rice OsIBH1 and Arabidopsis AtIBH1 (Fig. 1D), and therefore we named it as ZmIBH1-1.
Functional verification using CRISPR-Cas9-mediated genome editing in Setaria
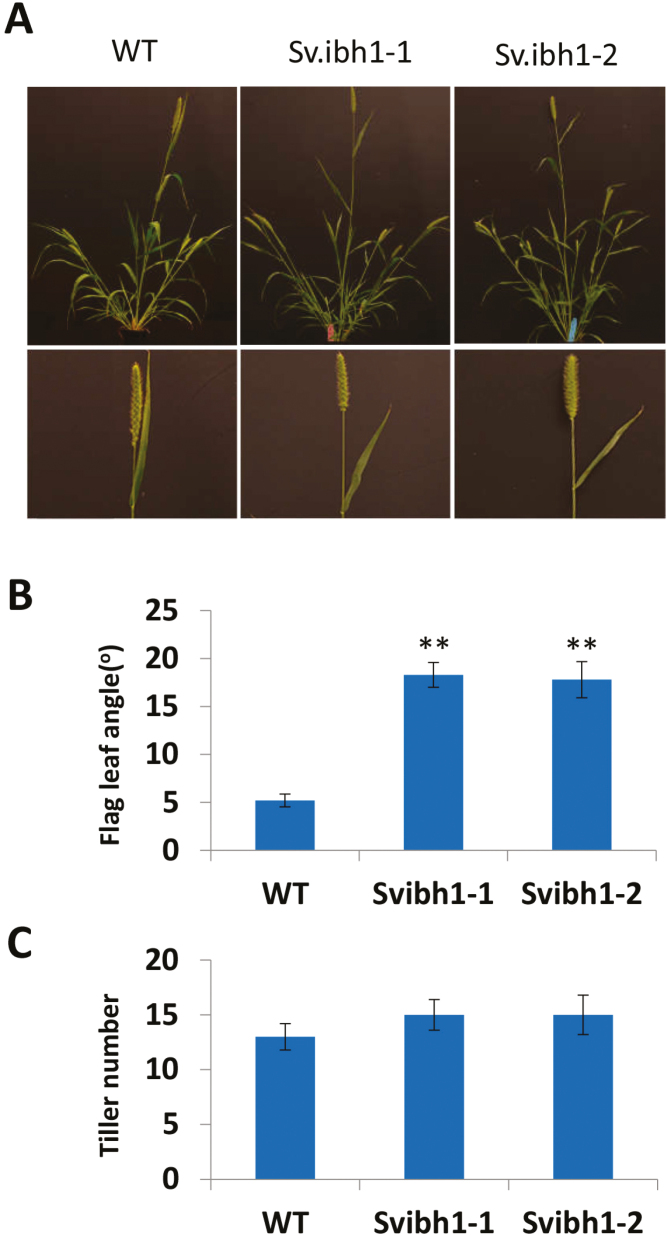
Setaria viridis is a panicoid grass that is evolutionarily related to major crops such as maize, rice, and sorghum (Huang et al., 2016). It has a small and simple genome, short stature, short life cycle, produces seeds prolifically, and has simple growing requirements. With the development of genetic and genomic resources over recent years, Setaria has been promoted as a model system to address fundamental questions in crop plants. More importantly, Setaria has the same leaf architecture as maize, as both are composed of a proximal sheath and a distal blade that are joined together by a hinge-like auricle and membranous ligule. CRISPR-Cas9-mediated genome editing was therefore applied to induce targeted mutagenesis to SvIBH1-1 to create loss-of-function alleles. Phylogenetic analysis indicated that Sevir3g025700 (SvIBH1-1) was the most closely related orthologue to ZmIBH1-1(Fig. 1D), and protein sequence alignment showed that it had the same conserved bHLH domain (Fig. S2 at Dryad). Setaria viridis ME34 was chosen as the landrace background because it has an erect leaf architecture that makes it easier to screen for altered leaf angle. The knockout mutant had a flag leaf that was bent away from the main panicle rather than wrapping around it (Fig. 2A), the mean LA of which was ~15° (range 10–25°) compared with 5.2° (0–8°) in the wild-type (WT) (Fig. 2B). The mean LA of the next leaf down the stem (second leaf) in Svibh1-1 was 27.5° (18–36°), compared with 22° in the WT (data not shown). These results indicated that SvIBH1-1 negatively regulates LA in Setaria. No significant differences were found for tiller number (Fig. 2C).
Fig. 2.
Phenotypic analysis of Setaria viridis Svibh1-1 mutants. (A) Plant architecture of two mutant lines compared with the wild-type (WT). (B) Flag leaf angle. Data are means (±SD), n=100. Significant differences compared with the WT were determined using Student’s t-test: **P<0.01. (C) Tiller numbers.
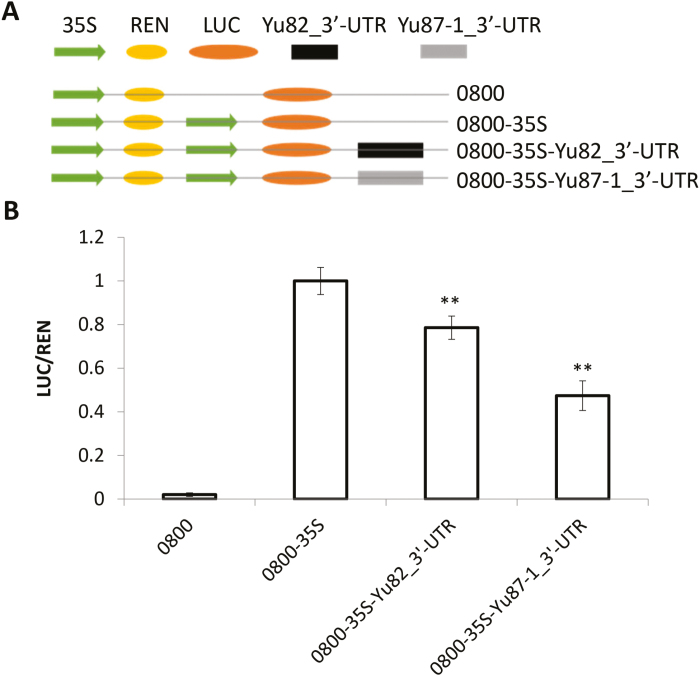
Comparison of the genomic sequences of ZmIBH1-1 in the two parent lines, Yu82 and Yu87-1, showed that there were no variations that would cause amino acid changes in the coding region and no nucleotide changes in the promoter and 5′-UTR. This suggested that variations in the non-coding sequences from the 3´-UTR may underlie the variation in the expression level and/or functionality of ZmIBH1-1. Indeed, the main variations between Yu82 and Yu87-1 were found in the region from 1.8 kb to 338 bp downstream of the translational end of ZmIBH1-1 (Fig. S1 at Dryad). The 3´-UTR is involved in post‐transcriptional regulation, such as mRNA decay rates and the level of mRNA transcripts (Olivas and Parker, 2000; Yu et al., 2007). To test a possible regulatory role of this region in the control of expression, we conducted dual-luciferase transient assays in N. benthamiana leaves. The results showed that the LUC reporter gene driven by the 3´-UTR of Yu82 had significantly higher expression than when it was driven by the 3´-UTR of Yu87-1 (Fig. 3), indicating that variations in the non-coding sequences from 3´-UTR could underlie the variations in the expression level and/or functionality of ZmIBH1-1.
Fig. 3.
Expression difference test of the 3´-UTRs of ZmIBH1-1 from the maize Yu82 and Yu87-1 lines in leaves of Nicotiana benthamiana. (A) Diagram of the constructs. REN, Renilla luciferase; LUC, firefly luciferase; 0800 is the vector. (B) Results of dual-luciferase assays showing the relative expression of LUC/REN. Data are means (±SD). Significant differences compared with the 0800-35S control were determined using Student’s t-test: **P<0.01.
Elongation, number, and lignification of cells contribute to the variation in leaf angle
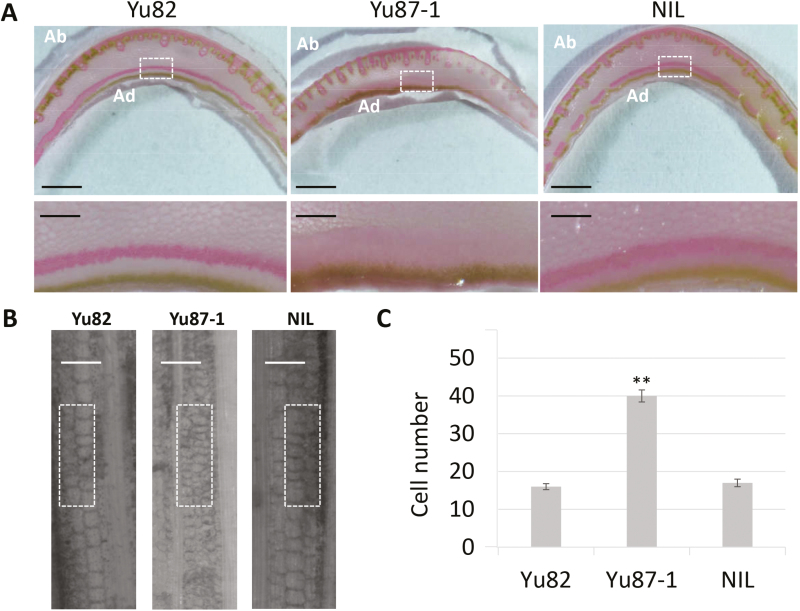
To investigate cytological changes associated with the decreased LAs in the NILs, safranin staining was performed on fully expanded leaf 7 at the V7 plant growth stage. The NIL and Yu82 had more layers of hypodermal sclerenchyma cells on the adaxial side of the ligular region than Yu87-1 (Fig. 4A), and these cells in the NIL and Yu82 were more lignified than those of Yu87-1. Longitudinal sections of the abaxial surface of the leaf base indicated that cell lengths were smaller and cells were significantly more numerous in Yu87-1 than in the NIL and Yu82 (Fig. 4B, C). These results indicated that the decreased LAs in NILs and Yu82 were caused by more lignified hypodermal sclerenchyma cells in the ligular region, and by increased cell length at the leaf base region.
Fig. 4.
Cytological examination of the parental maize varieties Yu82 and Yu87-1 and the near-isogenic line (NIL) using safranin staining for lignin. The NIL has the same genetic background as Yu87-1 except for the region containing ZmIBH1-1, which is derived from Yu82. Sections were taken from fully expanded leaf 7 at the V7 growth stage. (A) Cross-sections of the ligular tissue. The enlarged areas are denoted by the white rectangles. Ab, abaxial; Ad, adaxial. Scale bars are 5 mm in the images above and 0.5 mm in the images below. (B) Longitudinal sections of the ligular tissue. Scale bars are 150 µm. (C) Analysis of cell numbers in the areas denoted by the white rectangles in (B). Data are means (±SD) of n=10 replicates. The significant difference between Yu87-1 and the NIL was determined using Student’s t-test: **P<0.01.
RNA-seq identification of genes affected by the differential expression of ZmIBH1-1
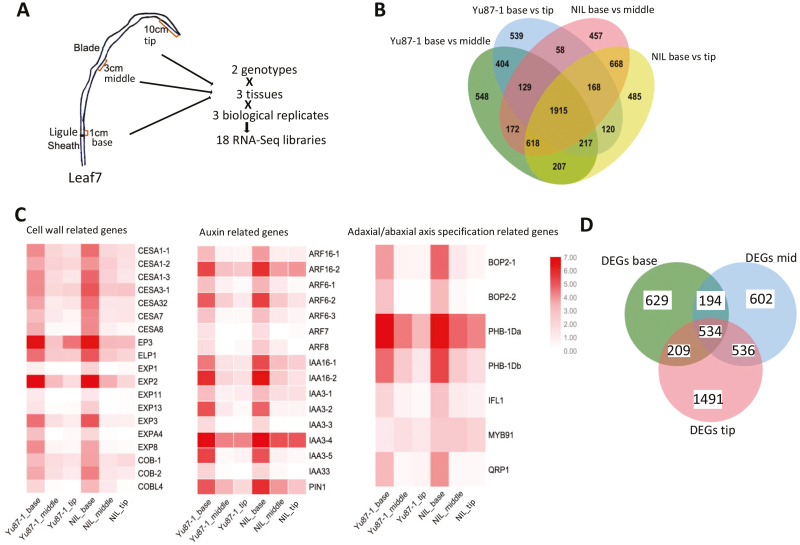
To understand the regulatory network of ZmIBH1-1 during the establishment of LA, RNA-seq analysis was conducted using total RNA extracted from different regions of leaf 7 of the Yu87-1 line and the NIL at the V7 growth stage (Fig. 5A). We generated ~8.06 million single-end 100-nt reads for 18 cDNA libraries in total, and 82.51% of the reads uniquely mapped to the B73 reference genome V3 (Table S3 at Dryad).
Fig. 5.
Transcriptomic study of ZmIBH1-1 of the parental maize variety and Yu87-1 and a near-isogenic line (NIL) having the same genetic background except for the region containing ZmIBH1-1, which is derived from Yu82. (A) Summary of the tissues collected and RNA-seq library preparation. (B) Venn diagram showing a comparison of the differentially expressed genes (DEGs) identified in the ligular tissues in Yu87-1 and the NIL. (C) Heat-map show the differences in expression of genes related to the cell wall, auxin, and adaxial/abaxial axis specification. (D) Venn diagram showing a comparison of DEGs in all the tissues between Yu87-1 and NIL.
For quality control, we chose a set of genes related to ligule development to verify tissue specificity. LG1 is specifically expressed in the ligule region (Johnston et al., 2014), and our results for Yu87-1 and NIL corresponded with this. We found that genes that are known to be preferentially expressed in the ligular region such as ZmPIN1a, BOP-Like, BEL14, and GA2Ox (Johnston et al., 2014) also showed significantly high expression levels in the leaf-base tissues of our samples. A total of 214 differentially expressed genes (DEGs) in the ligular tissue (57.3%, out of 373) were detected in our study, which matched the results of Johnston et al. (2014) (leaf base; Table S4 at Dryad). Taken together, the results indicated that our data were suitable for further analysis.
The anatomical structures of the ligular region at the base of the leaf blade on both the adaxial and abaxial surface play key roles in leaf development. In order to understand how changes in gene expression regulate development in the ligule region, we first examined the number of genes that were differentially expressed in Yu87-1 and the NIL (Fig. 5B). Genes that showed significant changes in expression (>2-fold, log2 fold-change >1) were selected for further analysis, and this resulted in 1915 genes being identified. The genes were functionally annotated using GO enrichment analysis with the online tool agriGO (http://bioinfo.cau.edu.cn/agriGO/). A total of 1347 out of the 1915 genes were functionally annotated, and the most significant subcategory was ‘cell wall organization or biogenesis’ (GO:0071554), which included cellulose synthase genes, such as ESAs and COBs, and the cell-wall modification genes EXPs (Li et al., 2003; Kam et al., 2005) (Fig. 5C). Two other significant subcategories were ‘response to stimulus’ (GO:0050896) and ‘anatomical structure development’ (GO:0048856). Under ‘response to stimulus’, auxin transport and auxin-mediated signaling genes, such as ARFs and IAAs, were significantly enriched, whilst under ‘anatomical structure development’, genes related to adaxial/abaxial axis specification and to cell growth were significantly enriched (Table S5 at Dryad). A total of 1566 DEGs were identified between Yu87-1 and NIL, 629 of which were specific to the ligular region (leaf base tissue) (Fig. 5D), and 434 (68.9%) of which could be functionally annotated. We conducted GO analysis for these 434 DEGs and determined which ones were related to cellular components. These corresponded to three GO terms, namely ‘membrane’ (GO:0016020, P-value=3.9×10–5), ‘plant-type cell wall’ (GO:0009505, P-value=1.5×10–5), and ‘cytoplasmic part’ (GO:0005737, P-value=3.5×10–4). A total of 132 genes were involved with cellular membranes, 66 of which were specifically associated with the plasma membrane, including ZmPIP1;6 (plasma-membrane intrinsic protein1; also named as Aquaporin PIP1-6) and Bm3 (brown midrib3, AC196475.3_FG004). A total of 15 DEGs were related to ‘plant-type cell wall’, including Expa2 (alpha expansin2, GRMZM2G105844), Expansin A6 (GRMZM2G445169), and Pectinesterase (GRMZM2G175499; Arora et al., 2017). A total of 142 genes were related to the cytoplasm, 31 of which were involved in vacuole development (Table S6 at Dryad).
DAP-seq identification of genes directly targeted by ZmIBH1-1
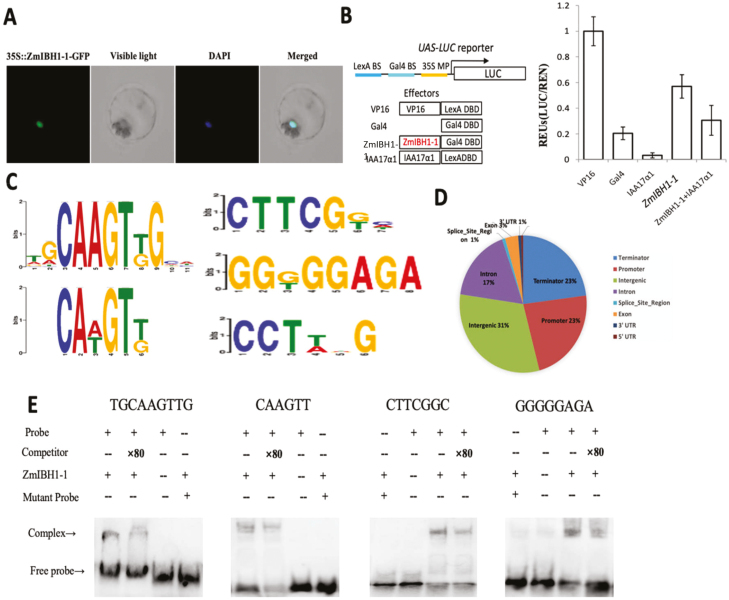
Sequence analysis suggested that ZmIBH1-1 encodes a bHLH transcription factor, so we first applied the tobacco protoplast system to verify that ZmIBH1-1 is localized to the nucleus (Merkle and Nagy, 1997). The merged images of the GFP and the control DAPI signals indeed suggested that ZmIBH1-1 was localized in the nucleus (Fig. 6A). To confirm that ZmIBH1-1 had the ability to activate or inhibit gene expression, we conducted transcriptional reporter gene assays using a promoter-reporter gene that contained binding sequences for the LexA and Gal4 DNA-binding proteins, The reporter gene was expressed at high levels when co-transformed into protoplasts with the LexA-VP16 fusion construct (Fig. 6B), which contained the coding sequences for the LexA DNA-binding domain (DBD) fused in-frame with the coding sequence of the VP16 transcriptional activation domain. Similar to results reported previously (Tiwari et al., 2001), the fusion LexA DBD with the transcriptional repressor domain of IAA17 (IAA17a1) strongly reduced the activation of the reporter gene. Similarly, co-transformation of the reporter gene with a construct for the ZmIBH1-1-Gal4 fusion protein (ZmIBH1-1-Gal4) significantly increased the expression of the reporter gene (Fig. 6B). These results demonstrated that ZmIBH1-1 specifically induced the expression of LUC under the IAA17 repressor domain.
Fig. 6.
DAP-seq analysis of maize ZmIBH1-1. (A) Nuclear localization of ZmIBH1-1 in tobacco protoplasts, as visualized with green fluorescent protein (GFP), with DAPI as the control. (B) Transient assays of transcriptional activity of ZmIBH1-1. Protoplasts from a tobacco BY-2 cell line were transformed with the reporter (UAS-LUC) and effector constructs (left), and the reporter gene expression was determined (right). Data are means (±SD), n=3. UAS-LUC, reporter construct containing Gal4 and LexA binding sites and a 35S minimal promoter upstream of the coding sequence of LUC; VP16, VP16 fused to the LexA DNA binding domain (DBD); Gal4, Gal4 DBD; IAA17a1, the transcription repression domain of IAA17 fused to the Gal4 DBD; ZmIBH1-1, full-length ZmIBH1-1 fused to Gal4 DBD. The LUC reporter gene expression was normalized to the Renilla luciferase activity and presented as values relative to the VP16 control, the value of which was set as 1. (C) ZmIBH1-1 binding to TGCAAGTTGCA, CAAGTT, CTTCGNN, and GGNGGAGA core motifs as identified by MEME-ChIP. (D) Distribution of ZmIBH1-1 binding sites. (E) Results of EMSAs confirming ZmIBH1-1 binding to TGCAAGTTGCA, CAAGTT, CTTCGNN, and GGNGGAGA.
To further explore whether the DEGs identified through RNA-seq analysis were direct targets of ZmIBH1-1, we used DNA affinity purification sequencing (DAP-seq) assays. Using the Illumina platform (150-bp pair-end reads), this produced ~10 million reads in total, 60% of which uniquely mapped to the maize genome V3. ZmIBH1-1 binding sites were examined using MACS2 (Zhang et al., 2008; q-value <0.005, based on a Poisson distribution comparing the ZmIBH1-1 sample and the control). Peaks located 5 kb upstream from the transcription start site (TSS) were defined as the peaks located in the promoter region. Peaks located 5kb downstream of the transcription termination site (TTS) were defined as those located in the terminator region. A total of 3189 genes were identified from 4927 peaks identified across the whole genome. Two typical ZmIBH1-1 binding motifs (NNCAAGTNG and CANGTN; Jones, 2004) and two novel binding sites (CTTCGNN and GGNGGAGA) were identified (Fig. 6C). We next analysed the distribution of the peaks within these genes and found that 23% were located within the promoter region (–5 kb to the TSS), 1% were located in the 5´-UTRs, 3% were located in the exon regions, 17% were located in the intron regions, 1% were located in the 3´-UTRs, and 23% were located in the terminator regions (Fig. 6D). To confirm the typical ZmIBH1-1-binding motifs NNCAAGTN and CANGTN, EMSAs were performed using purified ZmIBH1-1 protein and a labelled DNA probe containing the ZmIIBH1-1-binding sites (TGCAAGTTGCA and CAAGTT). As shown in Fig. 6E, ZmIBH1-1 bound to TGCAAGTTGCA and CAAGTT, the addition of 80× unlabeled competitors reduced the detected binding, and it did not bind to mutant probes (TGTGGACTGCA and TAGATAT). Without the ZmIBH1-1 protein only the band for the free probe was observed. The results confirmed the specific binding of ZmIBH1-1 to TGCAAGTTGCA and CAAGTT. In addition, we also found two new ZmIBH1-1-binding motifs, CTTCGNN and GGNGGAGA. An EMSA was performed using a purified ZmIBH1-1 protein and a labelled DNA probe containing the new ZmIIBH1-1-binding sites, CTTCGGC and GGGGAGA. As shown in Fig. 6E, ZmIBH1-1 bound to the two new motifs. The addition of 80× unlabeled competitors reduced the detected binding of ZmIBH1-1, and it did not bind to the mutant probes (ACCTAGC and TTGTTCGA). Without the ZmIBH1-1 protein only the band for the free probe was observed. The results confirmed the specific binding of ZmIBH1-1 to CTTCGGC and GGGGAGA.
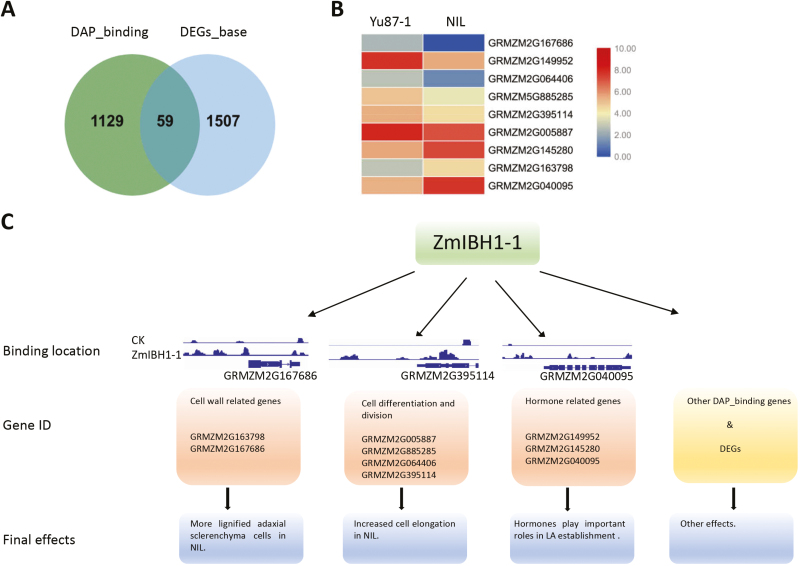
Of the 3189 target genes bound by ZmIBH1-1, only 1188 bound to the upstream regions. RNA-seq analysis revealed 1566 DEGs between the leaf base of Yu87-1 and the NIL, and DAP-seq identified 59 of these as putative target genes where ZmIBH1-1 bound to upstream regions (Fig. 7A). Of these 59 genes, 29 were up-regulated in Yu87-1 and 30 were down-regulated, and genes related to the cell wall, , cell development, and hormone metabolism were identified as being the major components (Fig. 7B; Table S7 at Dryad).
Fig. 7.
Differentially expressed genes (DEGs) that are bound by ZmIBH1-1, and a proposed ZmIBH1-1 regulatory network. (A) Venn diagram showing a comparison of ZmIBH1-1 binding targets and the DEGs in the ligular region between the parental line Yu87-1 and a near-isogenic line (NIL) having the same genetic background except for the region containing ZmIBH1-1, which is derived from Yu82. DAP_binding, ZmIBH1-1 binding targets in the promoter region; DEGs_base, DEGs identified in the ligular region between Yu87-1 and NIL. (B) Heat-map showing the differences in expression of genes related to the cell wall, cell differentiation, and hormones. (C) A hierarchical genome-wide transcriptional regulatory framework for ZmIBH1-1. CK, negative control; LA, leaf angle.
ZmIBH1-1 directly regulates genes involved in cell wall metabolism
By aligning the target genes of ZmIBH1-1 to the DEGs, two genes were identified as related to cell wall biosynthesis or modification (Fig. 7C). Walls Are Thin1 (WAT1, GRMZM2G163798) was significantly up-regulated in the NIL and corresponded to more lignified adaxial sclerenchyma cells and more elongated cells in the NIL (Fig. 4). AtWAT1 is a gene required for secondary wall formation in Arabidopsis, and the wat1 mutant shows defects in stem-fiber cell wall thickness and cell elongation, and has reduced lignin and stem strength (Ranocha et al., 2011). In addition to WAT1, a gene related cell wall modification, GRMZM2G167686, was expressed in Yu87-1 but not in the NIL. To determine whether ZmIBH1-1 directly regulated these genes we performed dual-luciferase transient transcriptional activity assays in N. benthamiana leaves with ZmIBH1-1driven by the CaMV 35S promoter as the effecter and LUC as the reporter gene (Fig. S3A at Dryad). The results showed that ZmIBH1-1 specifically increased the expression of LUC with the WAT1 promoter and it repressed the promoter of GRMZM2G167686, indicating these genes are indeed targets of ZmIBH1-1. The results were consistent with the corresponding gene expression results.
ZmIBH1-1 directly regulates genes related to cell proliferation
Four genes related to cell development were targeted directly by ZmIBH1-1, and all of them were up-regulated in Yu87-1 compared to the NIL (Table S7 at Dryad). GRMZM2G005887 and GRMZM5G885285 were annotated as a proteins related to cell differentiation, and GRMZM2G064406 and GRMZM2G395114 were annotated as proteins related to cell division. We performed dual-luciferase transient transcriptional activity assays with the ZmIBH1-1 protein and LUC driven by the promoter sequences of the four genes as reporters in N. benthamiana (Fig. S3A at Dryad). The results showed that the promoters of all the genes were significantly induced by ZmIBH1-1, indicating the genes are targets of ZmIBH1-1 (Fig. S3B at Dryad). These data confirmed the important roles of genes related to cell differentiation and proliferation in the regulation of leaf architecture.
ZmIBH1-1 is involved in hormone regulation
Plant hormones play pivotal roles in the formation of LA (Luo et al., 2016). Previous studies have shown that cytokinins and jasmonates affect the inclination of the lamina by interacting with other phytohormones, such as auxins and brassinosteroids (Eizo, 1965; Wada et al., 1984; Gan et al., 2015). In our analysis of the transcriptome in the leaf ligular region, genes involved in cytokinin, jasmonate, and ethylene synthesis and signal transduction pathways were detected (Table S7 at Dryad). The genes GRMZM2G145280 and GRMZM2G149952 were both responsive to cytokinin, but GRMZM2G149952 was down-regulated in the mutant NIL whilst GRMZM2G145280 was up-regulated. A jasmonate biosynthesis gene, GRMZM2G040095, was detected and had 16-fold higher abundance of transcript accumulation in the NIL than in Yu87-1.
Discussion
Map-based cloning of ZmIBH1-1
Using a nested-association mapping population, Tian et al. (2011) detected three major QTLs related to leaf angle (LA) on chromosome 2, and qLA2-1 was located in one of these. Cloning the candidate gene within this region may therefore contribute to the understanding of the molecular mechanisms controlling LA in maize.
Our study was based on the development of 300 new SSR markers and 60 polymorphic primers were identified from the screening of the parental lines (Table S2 at Dryad). The large fine-mapping populations that were used in this study increased the recombination rate of different chromosome segments, and thus improved the accuracy of the mapping results. In addition, we combined the phenotype with the genotype in the process of constructing the NIL population, and a single plant with a background recovery rate >95% with LA <15° was selected (Ku et al., 2010). As well as increasing the size of the fine-mapping population, we also increased the mapping efficiency and accuracy. Map-based cloning has not only been widely used in the isolation of small-genome crop species such as rice, it has also become an important method for gene isolation of large-genome crops, such as maize, and this will benefit crop breeding projects.
bHLH transcription factors play important roles in regulating LA in maize
In plants, bHLH family proteins such as MYCs, BEEs, PRE (PACLOBUTRAZOL RESISTANCE), BIM (BES1-INTERACTING MYC-LIKE), and PIFs (PHYTOCHROME INTERACTING FACTORS) have been shown to function in multiple cellular processes, including plant morphogenesis, epidermal cell differentiation, shade-avoidance responses, and stress responses (Ledent and Vervoort, 2001; Heim, 2003; Duek and Fankhauser, 2005). The maize genome encodes a total of 262 bHLH proteins grouped into 21 subfamilies (Feller et al., 2011). bHLH transcription factors are a group of proteins containing 60 amino acids with two distinctive regions, the basic region and the HLH region. The former consists of 13–17 highly conserved residues (His, Glu, Arg) and is arranged at the domain N-terminal end, and mediates the recognition process between the bHLH monomer and the DNA–protein binding site. The HLH region carries two amphipathic alpha-helices linked by a variable loop, which allows them to bind as dimers (Atchley and Fitch, 1997). Previous studies have indicated that bHLH proteins are involved in a wide range of plant developmental and physiological activities, including plant morphogenesis, stomatal differentiation, and cell elongation (Nadeau, 2009; Zhang et al., 2009; Ikeda et al., 2013). There are no reports that bHLH functions in the regulation of maize leaf architecture. It has been reported that all six members of the bHLH subfamily in rice have a conserved function in regulating the LA of the flag leaf (Dong et al., 2018). In maize, the first bHLH family member identified was the R gene, which is essential for anthocyanin synthesis (Ludwig and Wessler, 1990); however, only a few bHLH proteins have been functionally characterized so far in maize. Here, we cloned ZmIBH1-1, encoding a bHLH transcription factor through map-based cloning. Cytological examination revealed an increased number of sclerenchymatous cells in the leaf base area of the NIL (Fig. 4). The abnormal mechanical tissues in the NIL leaf-base area were also more lignified, which would lead to stronger mechanical support for the leaves. Thus, ZmIBH1-1 regulated LA through affecting the number of sclerenchymatous cells and producing abnormal mechanical tissues. In addition, a mutant of Setaria generated using CRISPR-Cas9 technology to knockdown SevirIBH1 showed increased LA (Fig. 2), confirming a negative role of IBH1 in leaf angle and thus implying that ZmIBH1-1 negatively regulates LA in maize. In rice, OsIBH1 also acts as a negative regulator and plays an important role in the regulation of LA (Zhang et al., 2009).
ZmIBH1-1 directly regulates specific genes
Histological analysis combined with RNA-seq and DAP-seq results indicated that ZmIBH1-1 regulated cell wall lignification, proliferation, and elongation within the ligular region of the maize leaf to affect the LA (Figs 4–6). ZmIBH1-1 directly bound to WAT1 (Fig. 7), which encodes a plant-specific protein that dictates secondary cell wall thickness of fibers in Arabidopsis (Ranocha et al., 2011). The wat1 mutant leads to decreased fiber cell wall thickness in Arabidopsis stems. We found that the expression level WAT1 was significantly higher in the NIL than in Yu87-1 and that cell wall lignification was also higher in the NIL. Taken together, this suggests that fiber cell wall thickness is a main factor determining LA. Jasmonate acts in plant growth and development (Huang et al., 2017), and bHLH transcription factors in Arabidopsis including MYC2 and its homologs MYC3–5 all interact with JAZ proteins (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Qi et al., 2015). The jasmonate synthesis/degradation gene GRMZM2G040095 had higher abundance of transcripts in the NIL than in Yu87-1 (Fig. 7). In addition, cytokinin has been reported to regulate leaf axil establishment in maize (Wang et al., 2014a), and GRMZM2G145280 (BBC1) and GRMZM2G149952 were both annotated as ‘response to cytokinin’ (Table S7 at Dryad). BBC1 activity is positively correlated with active cell division in Arabidopsis (Bertauche et al., 1994). The expression level of BBC1 was higher in NIL than in Yu87-1, which implies that it also affected the formation of LA through cell division.
Conclusions
We have used map-based cloning to identify ZmIBH1-1 as a negative regulator of leaf angle (LA) in maize. Histological analysis showed that differences in LA between two parental lines and a NIL were mainly the result of differential cell wall lignification and cell elongation in the ligular region. RNA-seq and DAP-seq analyses identified 59 target genes of ZmIBH1-1, with annotation that were related to either the cell wall, cell development, or hormones. Our results from GO enrichment analysis and cytological studies indicated that ZmIBH1-1 affected LA mainly through regulation of cell wall lignification and cell elongation. We present a model of gene regulation by ZmIBH1-1 to explain the control of maize LA (Fig. 7C).
Our results demonstrate that ZmIBH1-1 encodes a transcription activator of multiple metabolic pathways related to cell metabolism during the establishment of LA in maize, and it acts in both direct and indirect ways to affect the patterns of gene transcription. Our data suggest that ZmIBH1-1 not only directly activates genes associated with a variety of genes related to cell development, but also controls transcription factors that, in turn, regulate various other aspects of plant metabolism. A better understanding of the regulatory pathways of ZmIBH1-1 will provide insights into the pleiotropic effects of the bHLH transcription factors on the establishment of plant architecture in maize, and will help to support future efforts to breed high-yielding varieties.
Data availability
The following data are available at Dryad Data Repository: http://dx.doi.org/10.5061/dryad.18tk64p.
Fig. S1. Sequence alignment of Yu82 and Yu87-1.
Fig. S2. Protein sequence alignment of ZmIBH1-1 and SvIBH1-1.
Fig. S3. Results of dual-luciferase transient transcriptional activity assays
Table S1. Summary of the primary mapping of qla2-1.
Table S2. Primers used in this study.
Table S3. Summary of RNA-seq mapping.
Table S4. Verification of RNA-seq results.
Table S5. AgriGo analysis of genes differentially expressed in the ligular region of both NIL and Yu87-1.
Table S6. AgriGo analysis of genes differentially expressed in the ligular region in either NIL and Yu87-1.
Table S7. Differentially expressed genes detected in the DAP-seq analysis.
Acknowledgements
This research was supported by grants from the National Key Research and Development Program of China (2016YFD0101001), Central Plains Science and Technology Innovation Leading Talents (194200510021), and National Natural Science Foundation of China (31571678, 31871639). The authors declare that they have no conflicts of interest.
Author contributions:
LK, HZ, and YCao performed the fine-mapping of ZmIBH1-1, and generated and characterized the transgenic plants; ZR, YH, HS, DD, HL, YD, FZ, TY, and QZ conducted the molecular biology experiments; YChen, LK, YCao, and HZ designed the experiments and wrote the manuscript.
References
- Arora K, Panda KK, Mittal S, Mallikarjuna MG, Thirunavukkarasu N. 2017. In silico characterization and functional validation of cell wall modification genes imparting waterlogging tolerance in maize. Bioinformatics and Biology Insights 11, doi: 10.1177/1177932217747277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proceedings of the National Academy of Sciences, USA 94, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertauche N, Leung J, Giraudat J. 1994. Conservation of the human breast basic conserved 1 gene in the plant kingdom: characterization of a cDNA clone from Arabidopsis thaliana. Gene 141, 211–214. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zeng H, Ku L, et al. 2020. Data from: ZmIBH1-1 regulates plant architecture in maize. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.18tk64p. [Google Scholar]
- Carey MF, Peterson CL, Smale ST. 2013. Electrophoretic mobility-shift assays. Carey MF, Peterson CL, Smale ST. 2013. Electrophoretic mobility-shift assays. Cold Spring Harbor Protocols 2013, doi: 10.1101/pdb.prot075861. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. 2011. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Molecular Plant 4, 279–288. [DOI] [PubMed] [Google Scholar]
- Dong H, Zhao H, Li S, Han Z, Hu G, Liu C, Yang G, Wang G, Xie W, Xing Y. 2018. Genome-wide association studies reveal that members of bHLH subfamily 16 share a conserved function in regulating flag leaf angle in rice (Oryza sativa). PLoS Genetics 14, e1007323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. 2005. bHLH class transcription factors take centre stage in phytochrome signalling. Trends in Plant Science 10, 51–54. [DOI] [PubMed] [Google Scholar]
- Duncan WG. 1971. Leaf angles, leaf area, and canopy photosynthesis1. Crop Science 11, 482. [Google Scholar]
- Duvick DN. 2005. Genetic progress in yield of United States maize (Zea mays L.). Maydica 50, 193–202. [Google Scholar]
- Duvick DN, Smith JSC, Cooper M. 2010. Long-term selection in a commercial hybrid maize breeding program. In: Janick J. ed. Long-term selection: crops, animals, and bacteria. Plant Breeding Reviews, vol. 24, part II . Hoboken, NJ: John Wiley & Sons, Inc., 109–151. [Google Scholar]
- Eizo M. 1965. Inhibition of lamina inclination by cytokinin in excised rice leaves. Plant & Cell Physiology 6, 653–660. [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SSL, Hagenbeek D, Rock CD. 2001. Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. The Journal of Biological Chemistry 276, 9855–9860. [DOI] [PubMed] [Google Scholar]
- Gan L, Wu H, Wu D, et al. 2015. Methyl jasmonate inhibits lamina joint inclination by repressing brassinosteroid biosynthesis and signaling in rice. Plant Science 241, 238–245. [DOI] [PubMed] [Google Scholar]
- Harper L, Freeling M. 1996. Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 144, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B. 2011. Brassinosteroid control of sex determination in maize. Proceedings of the National Academy of Sciences, USA 108, 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA. 2003. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Huang H, Liu B, Liu L, Song S. 2017. Jasmonate action in plant growth and development. Journal of Experimental Botany 68, 1349–1359. [DOI] [PubMed] [Google Scholar]
- Huang P, Shyu C, Coelho CP, Cao Y, Brutnell TP. 2016. Setaria viridis as a model system to advance millet genetics and genomics. Frontiers in Plant Science 7, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. 2013. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signaling & Behavior 8, e23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ. 2014. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. The Plant Cell 26, 4718–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biology 5, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam MJ, Yun HS, Kaufman PB, Chang SC, Kim SG-K. 2005. Two expansins, EXP1 and EXPB2, are correlated with the growth and development of maize roots. Journal of Plant Biology 48, 304–310. [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW. 2015. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiology 169, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku L, Wei X, Zhang S, Zhang J, Guo S, Chen Y. 2011. Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PloS ONE 6, e20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku LX, Zhang J, Guo SL, Liu HY, Zhao RF, Chen YH. 2012. Integrated multiple population analysis of leaf architecture traits in maize (Zea mays L.). Journal of Experimental Botany 63, 261–274. [DOI] [PubMed] [Google Scholar]
- Ku LX, Zhao WM, Zhang J, Wu LC, Wang CL, Wang PA, Zhang WQ, Chen YH. 2010. Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays L.). Theoretical and Applied Genetics 121, 951–959. [DOI] [PubMed] [Google Scholar]
- Lalitha S. 2000. Primer Premier 5. Biotech Software & Internet Report 1, 270–272. [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. 2001. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Research 11, 754–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. 2003. Expansins and cell growth. Current Opinion in Plant Biology 6, 603–610. [DOI] [PubMed] [Google Scholar]
- Li Y, Ma X, Wang T, et al. 2011. Increasing maize productivity in China by planting hybrids with germplasm that responds favorably to higher planting densities. Crop Science 51, 2391. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu M, Zhou F, Xie CX, Li MS, Xu YB, Marilyn W, Zhang SH. 2007. Construction of a SSR linkage map and mapping of quantitative trait loci (QTL) for leaf angle and leaf orientation with an elite maize hybrid. Hereditas (Beijing) 29, 1131–1138. [In Chinese.] [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang M, Zhang Z, Wang Z, Wu N, Song Y, Wang P. 2018. Screening and verification of genes associated with leaf angle and leaf orientation value in inbred maize lines. PloS ONE 13, e0208386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Wessler SR. 1990. Maize R gene family: tissue-specific helix-loop-helix proteins. Cell 62, 849–851. [DOI] [PubMed] [Google Scholar]
- Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X. 2016. Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Reports 35, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM. 2012. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PloS ONE 7, e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T, Nagy F. 1997. Nuclear import of proteins: putative import factors and development of in vitro import systems in higher plants. Trends in Plant Science 2, 458–464. [Google Scholar]
- Miao Y, Jiang L. 2007. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols 2, 2348–2353. [DOI] [PubMed] [Google Scholar]
- Mickelson SM, Stuber CS, Senior L, Kaeppler SM. 2002. Quantitative trait loci controlling leaf and tassel traits in a B73×Mo17 population of maize. Crop Science 42, 1902. [Google Scholar]
- Moon J, Candela H, Hake S. 2013. The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development 140, 405–412. [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M. 1997. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes & Development 11, 616–628. [DOI] [PubMed] [Google Scholar]
- Muehlbauer GJ, Fowler JE, Girard L, Tyers R, Harper L, Freeling M. 1999. Ectopic expression of the maize homeobox gene liguleless3 alters cell fates in the leaf. Plant Physiology 119, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA. 2009. Stomatal development: new signals and fate determinants. Current Opinion in Plant Biology 12, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. 2011. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. Journal of Experimental Botany 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. The EMBO Journal 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SS, Song L, et al. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper GE, Pearce RB, Mock JJ. 1977. Leaf orientation and yield of maize. Crop Science 17, 883. [Google Scholar]
- Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D. 2015. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. The Plant Cell 27, 1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, Dima O, Felten J, et al. 2011. WAT1 (WALLS ARE THIN1) defines a novel auxin transporter in plants and integrates auxin signaling in secondary wall formation in Arabidopsis fibers. BMC Proceedings 5, O24. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable J, Wallace JG, Unger-Wallace E, Briggs S, Bradbury PJ, Buckler ES, Vollbrecht E. 2017. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture. The Plant Cell 29, 1622–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genetics 43, 159–162. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. 2001. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. The Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck J, Swartwood K, Pidgeon K, Maxson-Stein K. 2017. Agrobacterium tumefaciens-mediated transformation of Setaria viridis. In: Doust A., Diao X eds. Genetics and genomics of Setaria. Plant genetics and genomics: crops and models, vol 19. Cham, Switzerland: Springer, 343–356. [Google Scholar]
- Wada K, Marumo S, Abe H, Morishita T, Nakamura K, Uchiyama M, Mori M. 1984. A rice lamina inclination test—a micro-quantitative bioassay for brassinosteroids. Agricultural and Biological Chemistry 48, 719–726. [Google Scholar]
- Walsh J, Waters CA, Freeling M. 1998. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade–sheath boundary. Genes & Development 12, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chen Y, Guo B, Kabir MR, Yao Y, Peng H, Xie C, Zhang Y, Sun Q, Ni Z. 2014a Expression and functional analysis of genes encoding cytokinin receptor-like histidine kinase in maize (Zea mays L.). Molecular Genetics and Genomics 289, 501–512. [DOI] [PubMed] [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, et al. 2014. b Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nature Biotechnology 32, 1158–1165. [DOI] [PubMed] [Google Scholar]
- Wang T, Ma X, Li Y, et al. 2011. Changes in yield and yield components of single-cross maize hybrids released in China between 1964 and 2001. Crop Science 51, 512. [Google Scholar]
- Yu B, Lin Z, Li H, et al. 2007. TAC1, a major quantitative trait locus controlling tiller angle in rice. The Plant Journal 52, 891–898. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ku LX, Han ZP, Guo SL, Liu HJ, Zhang ZZ, Cao LR, Cui XJ, Chen YH. 2014. The ZmCLA4 gene in the qLA4-1 QTL controls leaf angle in maize (Zea mays L.). Journal of Experimental Botany 65, 5063–5076. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, et al. 2009. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. The Plant Cell 21, 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following data are available at Dryad Data Repository: http://dx.doi.org/10.5061/dryad.18tk64p.
Fig. S1. Sequence alignment of Yu82 and Yu87-1.
Fig. S2. Protein sequence alignment of ZmIBH1-1 and SvIBH1-1.
Fig. S3. Results of dual-luciferase transient transcriptional activity assays
Table S1. Summary of the primary mapping of qla2-1.
Table S2. Primers used in this study.
Table S3. Summary of RNA-seq mapping.
Table S4. Verification of RNA-seq results.
Table S5. AgriGo analysis of genes differentially expressed in the ligular region of both NIL and Yu87-1.
Table S6. AgriGo analysis of genes differentially expressed in the ligular region in either NIL and Yu87-1.
Table S7. Differentially expressed genes detected in the DAP-seq analysis.