Abstract
Biogenesis of the secondary cell wall in trees involves the massive biosynthesis of the phenylalanine-derived polymer lignin. Arogenate dehydratase (ADT) catalyzes the last, and rate-limiting, step of the main pathway for phenylalanine biosynthesis. In this study, we found that transcript levels for several members of the large ADT gene family, including ADT-A and ADT-D, were enhanced in compression wood of maritime pine, a xylem tissue enriched in lignin. Transcriptomic analysis of maritime pine silenced for PpMYB8 revealed that this gene plays a critical role in coordinating the deposition of lignin with the biosynthesis of phenylalanine. Specifically, it was found that ADT-A and ADT-D were strongly down-regulated in PpMYB8-silenced plants and that they were transcriptionally regulated through direct interaction of this transcription factor with regulatory elements present in their promoters. Another transcription factor, PpHY5, exhibited an expression profile opposite to that of PpMYB8 and also interacted with specific regulatory elements of ADT-A and ADT-D genes, suggesting that it is involved in transcriptional regulation of phenylalanine biosynthesis. Taken together, our results reveal that PpMYB8 and PpHY5 are involved in the control of phenylalanine formation and its metabolic channeling for lignin biosynthesis and deposition during wood formation in maritime pine.
Keywords: Amino acids, phenylalanine metabolism, phenylpropanoids, Pinus pinaster, transcription factors, transgenic trees
Functional genomics unravels the role of two arogenate dehydratases in maritime pine during wood formation, and provides new insights into how lignin biosynthesis and deposition are regulated in conifers.
Introduction
The biosynthesis of the amino acid phenylalanine (Phe) is a doubly essential process for land plants, as this amino acid serves both as a building block for proteins and as a main precursor for the biosynthesis of phenylpropanoids. Phenylpropanoids are a wide range of aromatic compounds that play key roles in plant growth, development, and the response to environmental cues. Land colonization by the first terrestrial plants would not have been possible without the emergence of specialized metabolic pathways for the biosynthesis of these secondary metabolites (Lowry et al., 1980). The metabolism of Phe is critical in carbon channeling from photosynthesis to the biosynthesis of phenylpropanoids in conifers, mainly lignin, which is an important constituent of wood (Pascual et al., 2016). A tight regulation of Phe metabolic flux would be expected depending on its alternative use for protein biosynthesis versus phenylpropanoid biosynthesis. In vascular plants, this second fate involves a massive carbon flux with as much as 30% of photosynthetically fixed carbon towards the synthesis of lignin and other compounds (Haslam, 1993).
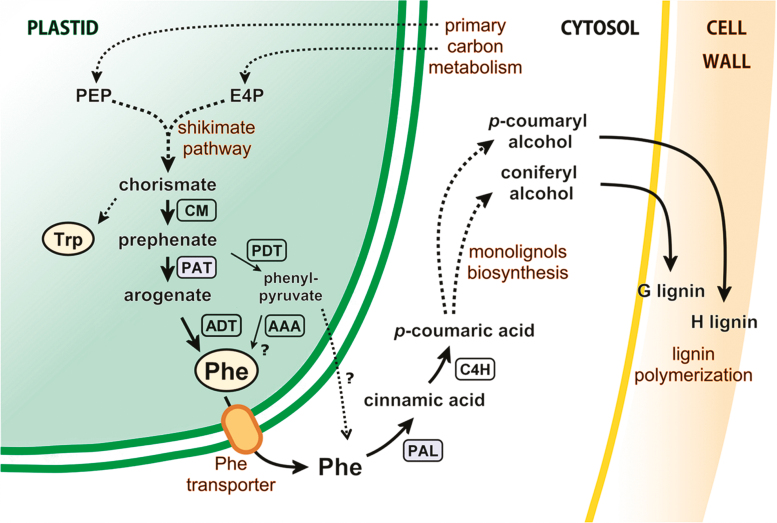
Within the plastids, primary carbon metabolism is connected to the biosynthesis of aromatic amino acids via the shikimate pathway (Fig. 1). This pathway starts with phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P), which are ultimately converted into chorismate, the direct precursor for the biosynthesis of aromatic amino acids and others compounds, such as vitamins K1 and B9 (Fig. 1; Tzin and Galili, 2010). Chorismate is alternatively used by anthranilate synthase in the initial reaction of the tryptophan biosynthetic pathway or by chorismate mutase (CM) to generate prephenate for the biosynthesis of Phe and tyrosine (Cotton and Gibson, 1965; Maeda and Dudareva, 2012). The biosynthesis of Phe occurs by transamination of prephenate to arogenate by the enzyme prephenate aminotransferase (PAT) and, in a second step, decarboxylation and dehydratation by the enzyme arogenate dehydratase (ADT) to produce Phe (Bonner and Jensen, 1987). Alternatively, Phe can also be synthesized through the decarboxylation and dehydration of prephenate into phenylpyruvate by the enzyme prephenate dehydratase (PDT), which is then followed by the conversion of phenylpyruvate into Phe by an aromatic amino acid aminotransferase (AAA) (Fischer and Jensen, 1987; Fig. 1). While in most microorganisms and fungi Phe biosynthesis occurs through the phenylpyruvate-dependent pathway, the arogenate pathway has been proposed as being predominant in plants (Maeda et al., 2010, 2011) (Fig. 1). Later, Phe is channeled into phenylpropanoid metabolism by the commitment reaction catalyzed by phenylalanine ammonia-lyase (PAL), producing cinnamic acid (Mottiar et al., 2016). Finally, the monolignols p-coumaroyl alcohol and coniferyl alcohol will constitute the building blocks for biosynthesis of H-lignins and G-lignins, respectively (Fig. 1). Sinapyl alcohol, the S-lignin monomer, is absent in conifers due to the lack of feruloyl-5-hydroxylase (F5H) activity in this group of plants (Wagner et al., 2015; Pascual et al., 2016).
Fig. 1.
Pathways for aromatic amino acid and lignin biosynthesis in conifers. Enzyme abbreviations are indicated within rectangles. Blue rectangles indicate enzymes encoded by genes previously demonstrated to be regulated by PpMYB8 (Craven-Bartle et al., 2013). PEP, phosphoenolpyruvate; E4P, erythrose 4-phosphate; CM, chorismate mutase; PAT, prephenate aminotransferase; PDT, prephenate dehydratase; ADT, arogenate dehydratase; ADH, arogenate dehydrogenase; AAA, aromatic amino acid aminotransferase; PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase. Phenylpyruvate transport outside the plastid needs to be confirmed.
The regulation of biosynthesis of phenylpropanoids and lignin in plants is a complex process that involves the coordinated expression of genes encoding enzymes located in different subcellular compartments and cellular types. Vascular plants, from herbaceous species to trees, share a relatively conserved ancestral xylem transcriptome (Li et al., 2010). Thus, our current knowledge highlights a general pyramid-shaped regulatory scenario in which a limited set of transcription factors (TFs), placed at the top of the regulatory network, act as master regulators of a larger set of downstream TFs with more specific regulatory functions over particular cell wall biosynthetic genes (Nakano et al., 2015). This hierarchical model is based on two extensively characterized TF families: NACs and MYBs (Zhong et al., 2011). In this model, NAC TFs act as master regulators, controlling the expression of MYB TFs, one of the largest TF families found in plants (Martin and Paz-Ares, 1997). In this regard, we have recently reported that the TF, PpNAC1, acts as a main regulator of Phe biosynthesis and utilization in maritime pine (Pascual et al., 2018).
Members of the MYB family regulate lignification through interactions with AC elements present in the promoter regions of phenylpropanoid and lignin biosynthetic genes (Zhong and Ye, 2009). In trees, several studies have demonstrated the roles of various TFs of this family in wood formation. For example, EgMYB88 and EgMYB1 have been reported to control lignin biosynthesis in eucalyptus (Soler et al., 2016, 2017). In conifers, PtMYB1, PtMYB4, and PtMYB8 have been shown to regulate phenylpropanoid metabolism and secondary cell wall biogenesis in Pinus taeda by controlling multiple genes encoding phenylpropanoid enzymes involved in lignin monomer synthesis (Patzlaff et al., 2003a, b; Bomal et al., 2008), similar to PgMYB1 and PgMYB8 in Picea glauca (Bomal et al., 2014). In addition, Craven-Bartle et al. (2013) reported the capacity of PpMYB8 from P. pinaster to coactivate the expression of PpPAT and PpPAL by specific binding to a conserved AC-II element in the promoter region of these genes. PpMYB8 transcripts were also abundant in lignifying stem tissues, suggesting the existence of a conserved transcriptional network controlling such processes in conifers.
Despite the fact that they can act as a bottleneck in Phe biosynthesis (Maeda and Dudareva, 2012; Corea et al., 2012), and therefore as a constraining factor in wood formation, little attention has been paid to the regulation of genes involved in Phe biosynthesis in trees. In the present work, we have characterized two ADT genes in maritime pine, PpADT-A and PpADT-D, which are highly expressed during formation of compression wood (CW), a specialized vascular tissue with an enhanced deposition of lignin (Villalobos et al., 2012). The analysis of transgenic pine silenced for PpMYB8, along with TF–promoter interaction experiments, revealed that PpMYB8 coordinates the expression of PpADT-A and PpADT-D with lignification, supporting the hypothesis that both ADT isoforms have a relevant role in the biosynthesis of Phe that is then channeled into lignin. Based on these studies, we have found that PpHY5, an ortholog of Arabidopsis AtHY5 (At5g11260) that belongs to the bZIP family of TFs, is also able to bind to the regulatory regions of the PpADT-A and PpADT-D genes. Interestingly, the gene expression pattern of PpHY5 is opposite to that of PpMYB8 in CW, suggesting an opposite role for these two genes in lignification.
Materials and methods
Generation of Pinus pinaster PpMYB8 transgenic lines
All P. pinaster transgenic plants were derived from the PN519 embryogenic line (Breton et al., 2006; Trontin et al., 2016). For the overexpression (OE) of PpMYB8, the corresponding cDNA was integrated into the pMBb7Fm21GW-UBIL vector (Karimi et al., 2002) and for the PpMYB8 RNAi-mediated silencing, a 243 bp fragment from the 3' end of P. taeda MYB8 was subcloned into the pB7GWIWG2(II) vector. Transformation was performed by co-cultivation of embryonal suspensor masses with Agrobacterium tumefaciens carrying the corresponding OE or RNAi vectors (Trontin et al., 2002). Transgenic lines were selected with phosphinothricin (PPT), confirmed by PCR assays (Trontin et al., 2007, 2013), and cryopreserved (Harvengt, 2005). Somatic embryo development from selected cryopreserved transgenic embryogenic lines was achieved on mLV-based maturation medium within 12 weeks following the method reported in Morel et al. (2014). A detailed description of methods used for maritime pine transformation and regeneration is provided in Supplementary Protocols S1 at JXB online.
Cloning of gene regulatory regions
PpADT-A and PpADT-D upstream regulatory sequences were obtained from SustainPineDB and cloned using 100 ng of genomic DNA of P. pinaster seedlings as template. A nested-PCR approach was used to amplify the ADT-A upstream region, using the primer pairs (see Supplementary Table S2): p1830.5Fwd/p1830.7Rvs (first PCR) and p1830.4Fwd/p1830.6Rvs (second PCR). The ADT-D upstream region was amplified in a single PCR by using the primer pair p3030.2Fwd/p3030Rvs. PCR products were purified from the gel, cloned into pJET1.2/blunt (Thermo Fisher Scientific), and confirmed by sequencing.
Reverse transcription–quantitative PCR (RT–qPCR) analysis
Total RNA was isolated from maritime pine tissues and cDNA was synthesized as described previously (de la Torre et al., 2014). Samples of compression (CW) and opposite (OW) wood were collected from 25-year-old pine trees growing in Sierra Bermeja (Estepona, Spain) and sampled in May 2008 (Villalobos et al., 2012). Bark and phloem were removed, and developing xylem was carefully scraped with a scalpel. Xylem scrapings were immediately frozen in liquid nitrogen after harvesting and stored at –80 °C. At least three biological replicates were used for transcript quantification. qPCR was performed on a CFX-384 Real Time System (Bio-Rad) with SsoFast EvaGreen Supermix (Bio-Rad) under the following conditions: 95 °C for 2 min (one cycle), followed by 95 °C for 1 s and 60 °C for 5 s (45 cycles). cDNAs corresponding to 10 ng of reverse-transcribed RNA were used as template. Raw fluorescence data from each well were fitted to the Mass Action Kinetic 2 model, which requires no assumptions about the amplification efficiency of a qPCR assay (Boggy and Woolf, 2010). The initial target concentration (D0 parameter) for each gene was deduced from the Mass Action Kinetic 2 model using the qPCR package for the R environment (Ritz and Spiess, 2008), and normalized to PpActin2 and PpEF1α. All primers used for RT–qPCR are listed in Supplementary Table S3.
Microarray hybridization and data analysis
The 60-mer oligonucleotides custom microarray PINARRAY3 (Cañas et al., 2015) based on the P. pinaster transcriptome (Canales et al., 2015) was used. Slides were made by Agilent Technologies and hybridization was performed as described by Cañas et al. (2015). Slides were scanned, and signal intensities were recorded using a GenePix 4100A microarray scanner (Molecular Devices, Sunnyvale, CA, USA). Differentially expressed genes (DEGs) were detected using the limma package for R (Smyth, 2005). Gene enrichment comparison was performed using the Mapman functional categories through the Mercator web tool (Lohse et al., 2014). The microarray data are accessible at NCBI’s Gene Expression Omnibus (Edgar et al., 2002) through the accession number GSE142093.
EMSA
The recombinant protein PpMYB8 (FN868598) was produced in Escherichia coli BL21-AI™ (Thermo Fisher Scientific) by overnight culture at 12 °C. The oligonucleotide probes described in Fig. 5 containing the AC elements, and those described in Fig. 7 containing ACGT elements were generated by annealing complementary biotinylated oligonucleotides designed to create 5′-biotinylated amplicons. At the end of the incubation period, the DNA–protein complexes were analysed by electrophoresis as previously described (Rueda-López et al., 2008). The binding specificity was evaluated using competition experiments with the corresponding non-biotinylated DNA probes.
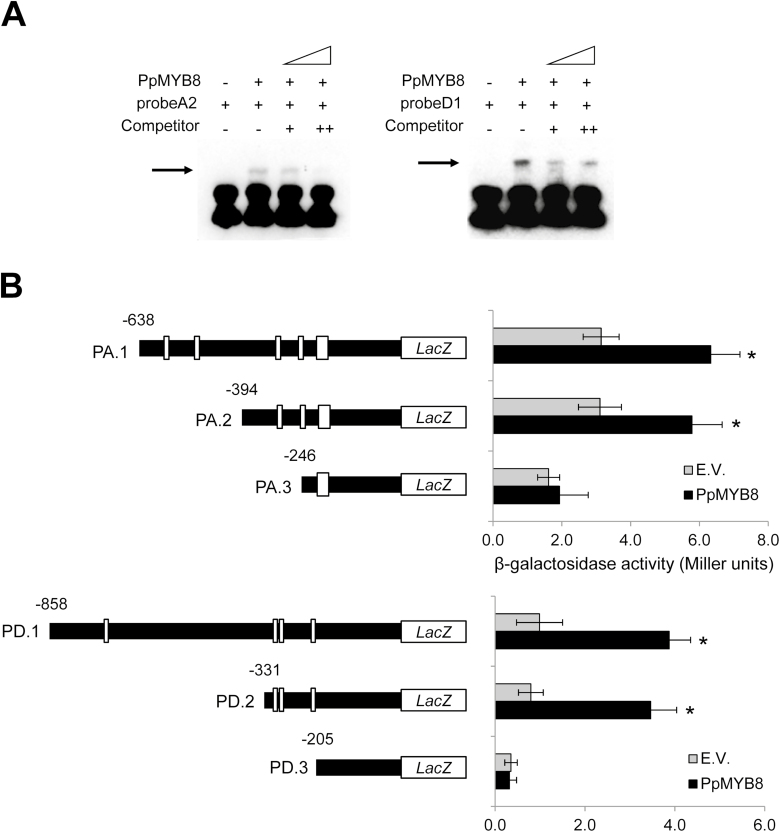
Fig. 5.
PpMYB8 interacts with putative regulatory regions of PpADT-A and PpADT-D. (A) EMSAs using recombinant PpMYB8 and the nucleotide probes A2 and D1 that contain potential AC-binding elements of the 5'-upstream regulatory region of PpADT-A, and D, respectively (see Supplementary Fig. S3). Band shifting after the formation of a probe–PpMYB8 complex is indicated with a black arrow. Lane 1, probe without PpMYB8; lane 2, probe and PpMYB8; lanes 3 and 4, probe, PpMYB8, and unlabeled probe as competitor. (B) β-Galactosidase reporter assay in yeast using subsequent deletions from regulatory regions of PpADT-A (PA.1, .2, and .3) and PpADT-D (PD.1, .2, and .3). White boxes indicate candidate AC-binding elements. Gray bars indicate β-galactosidase background activity (strains transformed with an empty construct, EV); black bars show β-galactosidase activity in strains transformed with the pDEST22-PpMYB8 yeast expression construct. Error bars represent the SD; asterisks indicate significant differences by t-test (α=0.01; n=3).
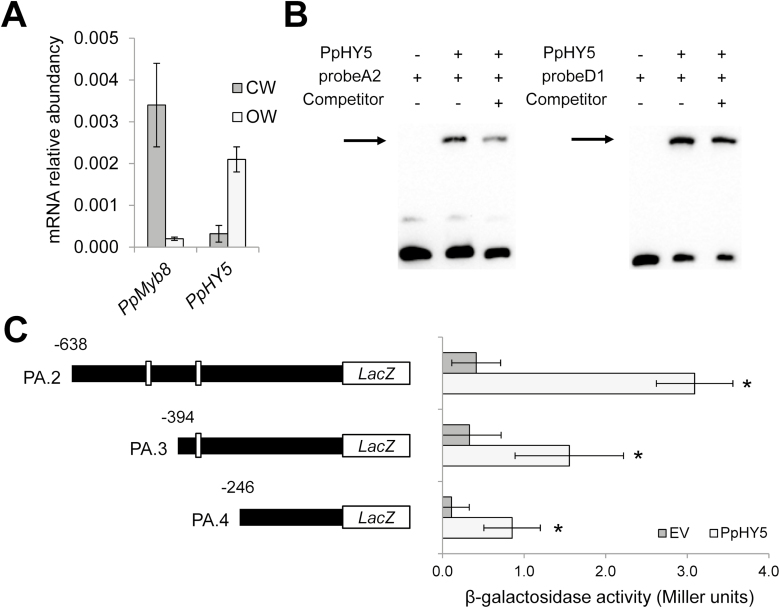
Fig. 7.
PpHY5 is able to bind to the PpADT-A and PpADT-D putative regulatory regions. (A) Expression pattern of PpMYB8 versus PpHY5 in CW and OW. (B) EMSAs using recombinant purified PpHY5 protein and nucleotide probes probeA2 and probeD1 (the same as present in Fig. 5A). (C) β-Galactosidase reporter assay in yeast using subsequent deletions of the putative promoter of PpADT-A (PA.1, .2, and .3). White boxes indicate the location of ACGT elements within the 5'-upstream sequence. Background β-galactosidase activity is represented in dark gray. Bars in pale gray represent β-galactosidase activity in yeast strains transformed with the pDEST22-PpHY5 yeast expression construct. Error bars represent the SD; asterisks indicate significant differences by t-test (α=0.01; n=3).
Plasmid constructions for transcriptional analysis in yeast
Deletions from the 5' end of PpADT-A were amplified from the pJET1.2 construct using the reverse primer PAAttB1R paired with three different forward primers: PA1AttB4 (638), PA2AttB4 (394), and PA3AttB4 (246). The corresponding PCR products were re-amplified using the primers AttB4 and AttB1R, and cloned into Gateway® pDONR-P4-P1R (Thermo Fisher Scientific) vector using BP Clonase® II, and subsequently recombined into pMW#3 (Deplancke et al., 2006) using the Gateway® LR Clonase® II mix. Deletions from the 5' end of PpADT-D were amplified using the primers: PD1AttB4, PD2AttB4, PD3AttB4 (forward), and PDAttB1R (reverse). PpADT-D promoter deletions were subcloned as described for PpADT-A. All primers used for promoter analysis in yeast are listed in Supplementary Table S2.
The PpMYB8 prey construct was done by amplifying the PpMYB8 ORF from a construct previously available in our laboratory (Craven-Bartle et al., 2013) using the primers MYB8AttB1 and MYB8AttB2. The PCR product was re-amplified using the primers AttB1 and AttB2, cloned into Gateway® pDONR207, and recombined into Gateway® pDEST22 as described.
Yeast manipulation and β-galactosidase assay
In vivo interaction between PpMYB8 and the regulatory regions of PADT-A and PADT-D was analyzed through a β-galactosidase reporter assay performed in Saccharomyces cerevisiae using o-nitrophenyl-galactopyranoside (ONPG) as substrate. More details can be found in Supplementary Protocols S1.
Microscopy
Stems previously preserved in liquid nitrogen were fixed overnight in a 4% paraformaldehyde solution in phosphate-buffered saline (PBS) and included in paraffin. Transversal sections (10 µm) were deparaffinized and rehydrated before visualization. For Wiesner (phloroglucinol-HCl) staining, a few drops of phloroglucinol-HCl solution (two parts of 1% phloroglucinol in 95% ethanol and one part of pure HCl) were deposited over the tissue sections and observed immediately under white light. For lignin autofluorescence visualization, an excitation range of 460–500 nm and an emission range of >510 nm were used. In both cases (Wiesner staining and lignin autofluorescence), a Leica TL3000 Ergo stereo-microscope was used to visualize and register the results.
Lignin determination
Acid-soluble and -insoluble lignin were determined in cell wall preparations from ~500 mg of fresh ground stems preserved at –80 °C. Acid-insoluble lignin was determined from ~100 mg of cell wall preparation using the Klason method (Dence, 1992) with the modifications described by Saleme et al. (2017). Acid-soluble lignin was determined spectrophotometrically as described (Dence, 1992).
Results
PpADT genes are up-regulated in compression wood
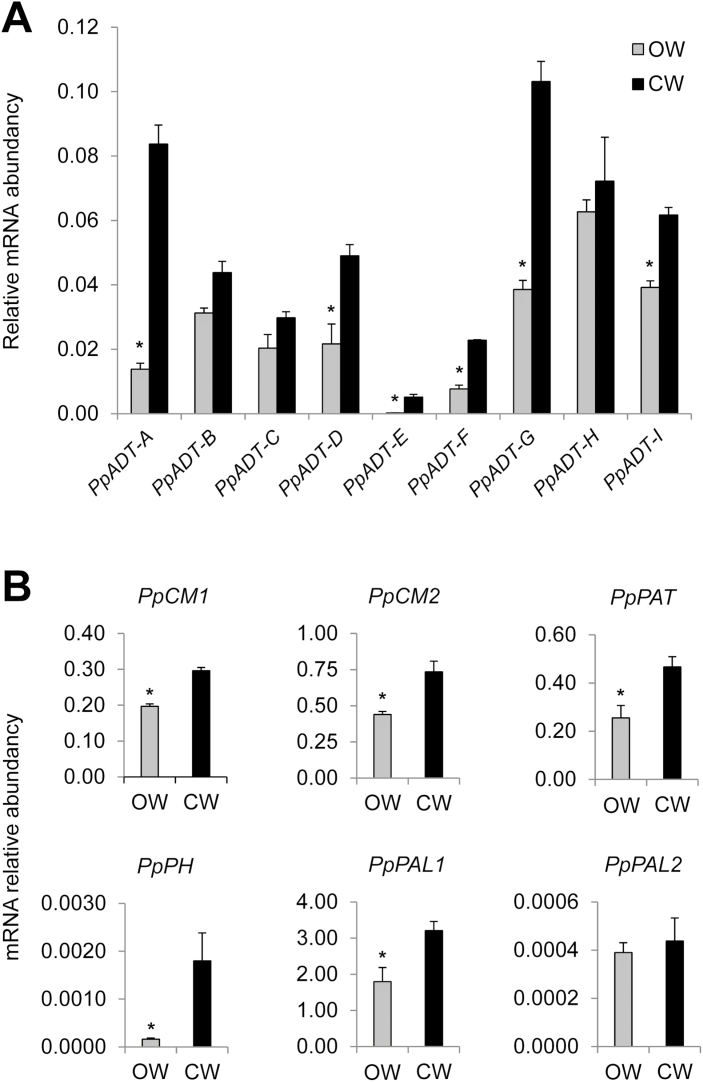
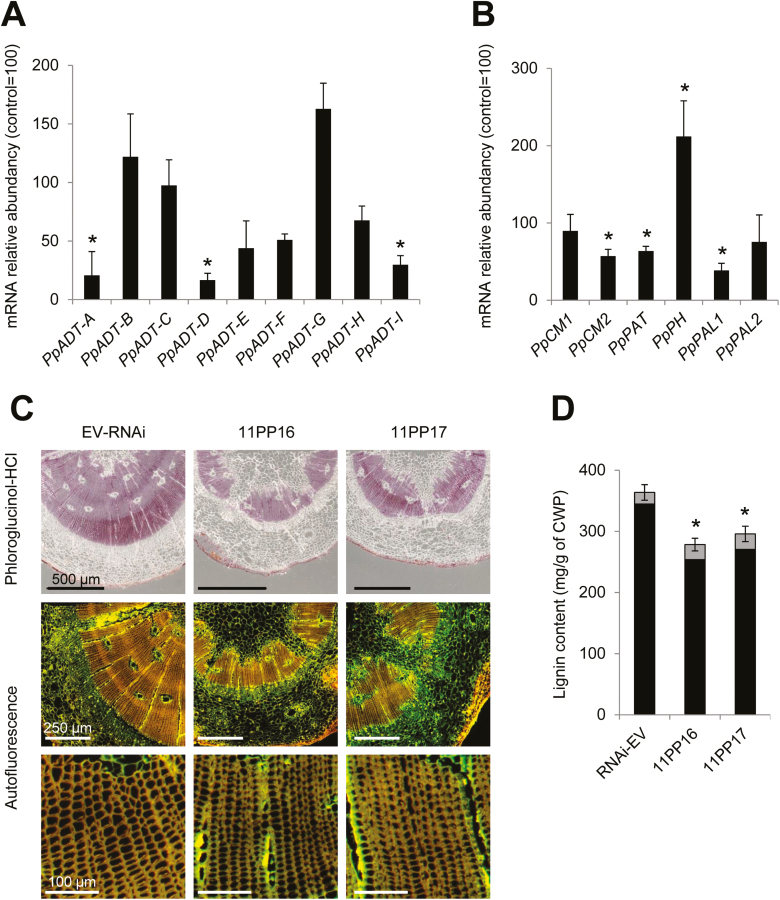
Conifers develop a specific woody tissue known as CW on the underside of branches and leaning stems. In parallel, facing the formation of the CW, these branches and stems develop OW (Groover, 2016). Compared with OW, in CW the lignin biosynthesis and deposition is enhanced and the cellulose content reduced, representing a good model to study the transcriptional regulation of the metabolic pathways that provide substrates for xylogenesis. In a previous study, we reported the characterization of the P. pinaster ADT gene family, consisting of nine identified isoforms (El-Azaz et al., 2016). To determine the PpADT isoforms that could be involved in xylogenesis, we have studied the expression profile of the ADT gene family in samples of CW and OW that were collected from 25-year-old trees mechanically stressed to produce such specialized vascular tissues (Villalobos et al., 2012). Six of the nine ADT genes showed significantly higher expression in CW when compared with OW: PpADT-A, PpADT-D, PpADT-E, PpADT-F, PpADT-G, and PpADT-I. Among them, PpADT-A, PpADT-D, and PpADT-G exhibited the greatest change (Fig. 2A). In CW, enhanced expression levels were also found for genes that encode other enzymes directly involved in the biosynthesis and utilization of Phe and monolignols such as chorismate mutases 1 and 2 (PpCM1 and PpCM2), prephenate aminotransferase (PpPAT), phenylalanine hydroxylase (PpPH), and phenylalanine ammonia lyase PpPAL1 (Fig. 2B).
Fig. 2.
Genes encoding enzymes from Phe metabolism and phenylpropanoid biosynthesis are induced in compression wood (CW) from Pinus pinaster. (A) PpADT gene relative mRNA levels in opposite wood (OW; gray bars) compared with CW (black bars). (B) Comparison of the expression level of some key Phe and phenylpropanoid biosynthetic genes between OW and CW. PpCM1, chorismate mutase 1; PpCM2, chorismate mutase 2; PpPAT, prephenate aminotransferase; PpPH, phenylalanine hydroxylase; PpPAL1, phenylalanine ammonia-lyase 1; PpPAL2, phenylalanine ammonia-lyase 2. Error bars represent the SD; asterisks indicate significant differences by t-test between OW and CW (α=0.01; n=3).
Production of transgenic lines with altered PpMYB8 expression in maritime pine
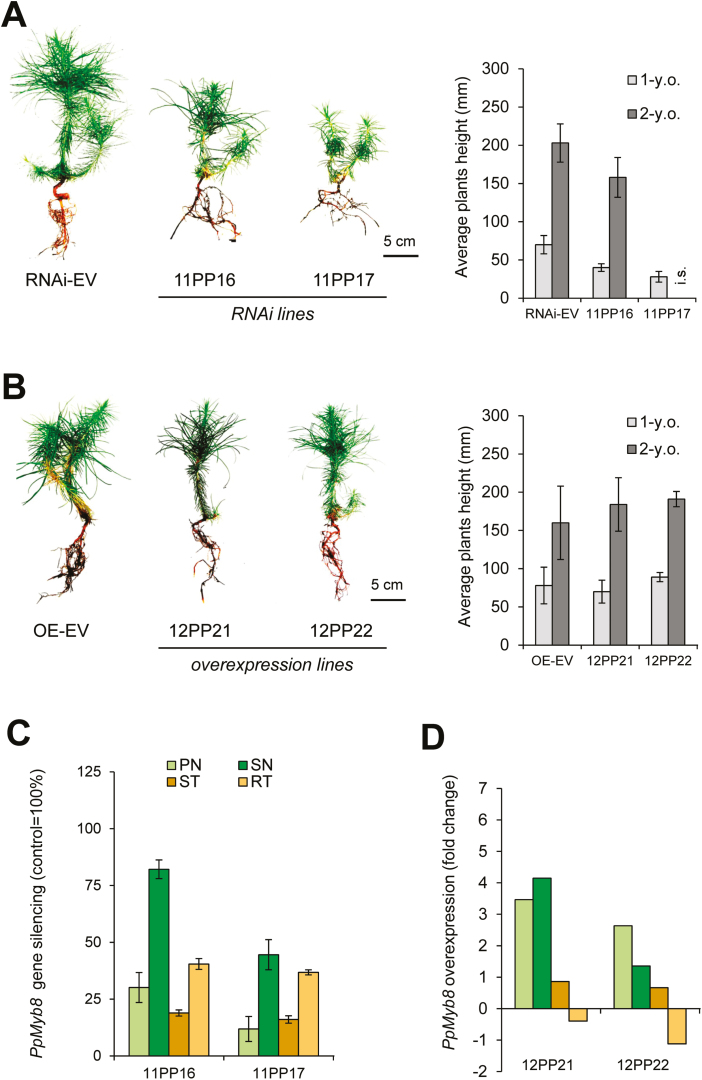
PpMYB8 has been suggested as a positive regulator of the expression of genes involved in Phe metabolism and nitrogen recycling during xylogenesis in CW (Craven-Bartle et al., 2013). To further explore processes controlled by this TF and identify members of the ADT gene family that could be involved in xylogenesis, transgenic plants of maritime pine overexpressing or silenced for PpMYB8 were obtained from cryopreserved transgenic lines via somatic embryogenesis (Fig. 3). Samples from primary and secondary needles, stems, and roots were collected twice after ~12 and 24 months of growth.
Fig. 3.
Characterization of transgenic Pinus pinaster PpMYB8 lines. (A) Morphology after 1 year in the greenhouse, and average height (left), of the transgenic lines (11PP16 and 11PP17) silenced for PpMYB8 (RNAi-PpMYB8). (B) Phenotype of PpMYB8 overexpression transgenic lines 12PP21 and 12PP22 (OE-PpMYB8). (C) Determination of the silencing level of PpMYB8 in different organs of 12-month-old RNAi-PpMYB8 plants, expressed as a percentage relative to the expression level of PpMYB8 in the transgenic control plants carrying the empty silencing vector (RNAi-EV). PN, primary needles; SN, secondary needles; ST, stems; RT, roots. (D) Expression level of PpMYB8 in the OE-PpMYB8 lines, as fold change relative to control plants transformed with the empty overexpression vector (OE-EV). Error bars represent the SD; asterisks indicate significant differences by t-test between the transgenic line and reference control line (α=0.01; n=3); i.s., inadequate too low sampling for 11PP17 (data excluded from the statistical analysis).
Compared with controls (RNAi-EV), RNAi-MYB8 plants exhibited a significant decrease in height after 1 year of growth, and differences were still detected after 2 years (Fig. 3A). In contrast, transgenic plants overexpressing OE-PpMYB8 did not exhibit differences in growth after either 1 or 2 years in the greenhouse (Fig. 3B). Endogenous PpMYB8 expression was analyzed by RT–qPCR in samples from RNAi-PpMYB8 plantlets to determine the extent of gene silencing compared with transgenic RNAi-EV control plants. As shown in Fig. 3C, significant differences in the relative abundance of the PpMYB8 transcripts were observed depending on the organ analyzed. PpMYB8 down-regulation was stronger in primary needles than in secondary needles. Interestingly, in stem and roots, PpMYB8 expression was decreased by ~80% and 60% compared with the reference level, respectively. The OE-PpMYB8 lines were found to significantly overexpress PpMYB8 in primary and secondary needles when compared with transgenic OE-EV controls [fold change (FC) from ~1.5 to 6; Fig. 3D]. However, much lower levels of overexpression were detected in the stem. No PpMYB8 overexpression was observed in roots where paradoxically a certain degree of silencing was perceptible, particularly in the line 12PP22.
Distinctive members of the PpADT gene family are down-regulated in RNAi-PpMYB8 plants
To investigate to what extent PpMYB8 regulates the expression of ADT genes during xylogenesis, the expression levels of the ADT genes were determined in the stems of the RNAi-PpMYB8 lines (Fig. 4A). The silencing of PpMYB8 resulted in a significant down-regulation of three ADT genes: PpADT-A, PpADT-D, and PpADT-I. Interestingly, PpADT-A and PpADT-D were among the most up-regulated ADT genes in CW (Fig. 2A), suggesting that PpMYB8 could directly, or indirectly, regulate the transcription of these particular ADT genes during xylogenesis. On the other hand, when the ADT expression profiles were also analyzed in the OE-PpMYB8 plants, there were no significant differences between the transgenic plants and controls (Supplementary Fig. S1). The impact of PpMYB8 silencing on the expression levels of other genes coding for enzymes directly involved in Phe biosynthesis was also determined. The results indicate a significant down-regulation of PpPAT, PpCM2, and PpPAL1 in RNAi-PpMYB8 plants, whereas the gene encoding phenylalanine hydroxylase (PpPH) was found to be overexpressed (Fig. 4B). The down-regulation of PpPAT, PpCM2, PpPAL1, and ADT genes is in good agreement with a potential role for PpMYB8 as a regulator of multiple genes involved in Phe biosynthesis and channeling of this amino acid into phenylpropanoids.
Fig. 4.
Effect of PpMYB8 RNAi-mediated silencing in the regulation of Phe and phenylpropanoid biosynthesis and lignin deposition. (A) Average expression level of the nine ADT genes from P. pinaster in the RNAi-PpMYB8 lines, expressed as a percentage relative to the RNAi-EV control. (B) Equivalent analysis to (A) for some key genes involved in Phe biosynthesis and the first steps of the biosynthesis of phenylpropanoids. (C) Vascular anatomy in cross-sections of stems from RNAi-PpMYB8 transgenic plants (lines 11PP16 and 11PP17) and control plants (RNAi-EV). The first row shows phloroglucinol-HCl staining. The second and third rows show blue light autofluorescence. (D) Klason (black) and acid-soluble (gray) lignin determination in the stems from RNAi-EV control and RNAi-PpMYB8 plants (11PP16, 11PP17). Gene abbreviations in (A) and (B) are as described in the legend of Fig. 2. Error bars represent the SD; asterisks indicate significant differences by t-test (α=0.01; n=3) between transgenic and control RNAi-EV lines.
We hypothesized that down-regulation of genes related to the metabolism of Phe in the RNAi-PpMYB8 transgenic lines could alter lignin content and/or structure, potentially affecting the formation of secondary cell walls, and therefore the formation of the tracheary elements of the xylem. Wiesner staining (phloroglucinol-HCl), which reveals the presence of lignin in a quantitative manner, and lignin autofluorescence after excitation under blue light showed that the RNAi-PpMYB8 plants were strongly affected in their xylematic tissues (Fig. 4C). A severely thinned and irregular, discontinuous xylem cylinder was observed in the stems from these plants. In addition, individual tracheids in the stems of RNAi-PpMYB8 plants were also observed to be narrower than in control plants (Fig. 4C, lower panels).
To further determine the consequences of PpMYB8 down-regulation on lignin deposition, acid-soluble and insoluble (Klason) lignin contents were estimated in the crude cell wall residue from stems of RNAi-PpMYB8 plantlets. The silenced lines exhibited an ~30% decrease in the total lignin content (acid-soluble+acid-insoluble lignin) when compared with the control plants (Fig. 4D), confirming previous anatomical observations (Fig. 4C). In addition, the ratio of the H- and G-derived monomers of lignin was evaluated by thioacidolysis, demonstrating a significant decrease in the H:G ratio in the RNAi-PpMYB8 plants (Supplementary Table S1).
The 5'-flanking regions of the PpADT genes contain putative R2R3-MYB-binding sites
Considering the current lack of a whole-genome assembly for P. pinaster, the reference genome of the closely related conifer P. taeda (V1.01 Genomic Scaffolds: https://pinerefseq.faculty.ucdavis.edu/; Wegrzym et al., 2014) was searched to identify the 5'-flanking regions of the ADT genes and used as a template to identify and assemble their P. pinaster relatives (SustainPineDB: http://www.scbi.uma.es/sustainpine/) (Supplementary Fig. S2). The length of the sequences assembled ranged from a minimum of 517 bp for PpADT-B to a maximum of 4804 bp for PpADT-E. In silico analysis of the PpADT 5'-flanking regions showed the presence of putative R2R3-MYB-binding sites in all of these sequences, with the single exception of PpADT-B and PpADT-H. Interestingly, this analysis also revealed the presence of several ACGT-based motifs in most of these 5'-flanking regions, which are the consensus binding sites for bZIP TFs (Deppmann et al., 2006).
A close-up analysis of these 5'-flanking sequences showed a particular enrichment of putative R2R3-MYB-binding sites in the proximal region of PpADT-A and PpADT-D (Fig. 5; Supplementary Fig. S2). The 5'-flanking region of PpADT-A (PADT-A) analyzed contains 1907 bp upstream of the predicted translation start codon, presenting three AC elements in this region at positions 301 (AC-III element), 247 (AC-II element), and 209 (tandem repeated AC-II element) (see Gómez-Maldonado et al., 2004 for a description of these AC elements). The respective 5'-flanking region of PpADT-D (PADT-D) comprises 1226 bp from the predicted translation start codon, with three AC-II class elements in its proximal region at positions 298, 283, and 206.
PpMYB8 is able to bind PpADT-A and PpADT-D regulatory regions
To investigate whether the putative cis-AC elements found in the 5'-upstream regions are important for PpMYB8 recognition, we used an EMSA to establish a physical link between PpMYB8 and synthetic DNA probes from PADT-A and PADT-D. The following probes were designed as follows: for PADT-A, probe A1 from position 225 to 329 (105 bp) and probe A2 from 281 to 370 (90 bp); for PADT-D, probe D1 from 253 to 331 (66 bp) and probe D2 from 192 to 318 (127 bp) (Supplementary Fig. S3). Band shifting was observed for probe A2 and probe D1 in the presence of recombinant PpMYB8 protein (Fig. 5A). The band shift was reduced (probes A2 and D1) when unlabeled competitor DNA was added. In contrast, no shift was detected for probe A1 and probe D2 fragments. These results point to the existence of a physical interaction between PpMYB-8 and the putative AC-binding elements included in these probes.
To confirm an in vivo direct interaction between PpMYB8 and the putative regulatory regions PADT-A and PADT-D, a β-galactosidase reporter assay was performed in the yeast S. cerevisiae. For PADT-A the following 5'-sequential deletions were assayed: 638 (PA.1), 394 (PA.2), and 246 (PA.3) (Fig. 5B). A significant increase in β-galactosidase activity in the presence of the prey construct was detected for the baits PA.1 (638) and PA.2 (394) compared with controls. In contrast, β-galactosidase activity was strongly reduced in yeast cells carrying the PA.3 (246) deletion. These results are consistent with previous EMSAs, indicating that AC elements located at 301 and 247 play a critical role in the transcriptional regulation of PpADT-A by PpMYB8. The following bait constructs were designed to analyze the regulatory properties of PADT-D: PD.1 (858), PD.2 (331), and PD.3 (205). Similarly, as found for PADT-A, the AC-II-binding elements located at positions 298, 283, and 206 of the 5'-flanking region of PADT-D were found to be critical for the recognition by PpMYB8, as demonstrated by the drastic reduction of β-galactosidase activity in PD.3 (Fig. 5B).
Transcriptional reprogramming induced by silencing of PpMYB8
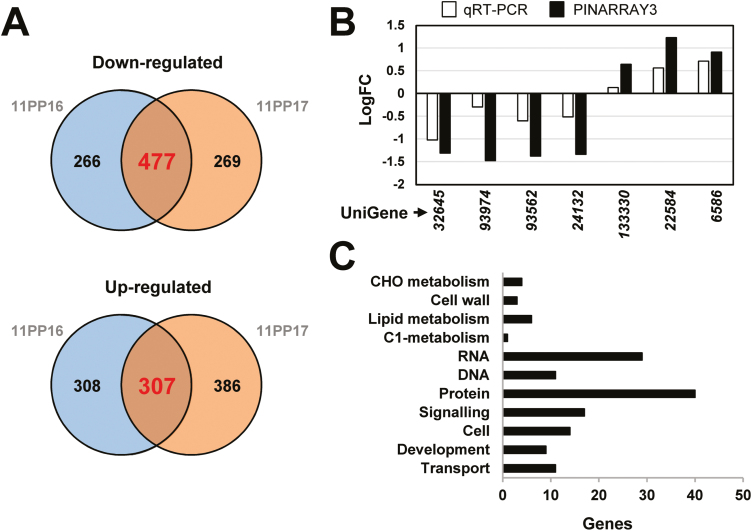
To identify the transcriptional network under the control of PpMYB8 in more depth, we performed transcriptomic analysis of RNAi-PpMYB8 plants. A 60-mer oligonucleotide microarray (PINARRAY3) developed in our laboratory and manufactured by Agilent (Santa Clara, CA, USA; Cañas et al., 2015) was used. Twelve microarray fields were hybridized with four biological replicates corresponding to stems from RNAi-EV and four replicates of each of the RNAi-PpMYB8 transgenic lines 11PP16 and 11PP17. Taken together, our analysis indicated 784 spots considered to represent DEGs (adjusted P-value <0.05; logFC>0.5). Among these genes, 307 were up-regulated in plants silenced for PpMYB8 as compared with controls, whereas 477 were down-regulated (Fig. 6). Among the down-regulated genes, we identified a broad set of TFs including two MYBs, (sp_v3.0_unigene19258 and sp_v3.0_unigene3263), a bZIP (sp_v3.0_unigene8197), a RAV/NGATHA (sp_v3.0_unigene34155), a MADS box (sp_v3.0_unigene32645), a C3H zinc finger (sp_v3.0_unigene17894), a GARP (sp_v3.0_unigene19789), and a bHLH (sp_v3.0_unigene29985). Consistent with the strong effect on xylem tissue in plants silenced for PpMYB8, we detected down-regulation of genes directly involved in cell wall modification such as xyloglucan endotransglucosylase/hydrolase (sp_v3.0_unigene35083), a wall polysaccharide-specific O-acetyltransferase (TBL-TRICHOME BIREFRINGENCE-LIKE, sp_v3.0_unigene7027), a pectinesterase (sp_v3.0_unigene9201), an oxidoreductase (sp_v3.0_unigene2867), a chitinase (sp_v3.0_unigene29478), and up to three S-adenosyl-l-methionine-dependent methyltransferases putatively involved in cell wall biogenesis (sp_v3.0_unigene1437, sp_v3.0_unigene96035, and sp_v3.0_unigene93562). Also consistent with the observed phenotype, multiple genes with a role in plant development were down-regulated, including NO VEIN (sp_v3.0_unigene63788), the actin cytoskeleton organizer PIROGI (sp_v3.0_unigene13374), and the mRNA decapping DCP2 (sp_v3.0_unigene183375) whose silencing in Arabidopsis results in disorganized veins. Additionally, an important group of genes involved in cell organization and division were also down-regulated including PROHIBITIN 3 (sp_v3.0_unigene24132), CRUMPLED LEAF (sp_v3.0_unigene1324), and multiple cytoskeleton-related genes (sp_v3.0_unigene5018, sp_v3.0_unigene93974, sp_v3.0_unigene13374, sp_v3.0_unigene2327, and sp_v3.0_unigene6084) (Supplementary Table S4).
Fig. 6.
Transcriptome analysis of RNAi-PpMYB8 plant stems. (A) Venn diagram showing both unique and overlapping (bold) expressed genes of significantly up-regulated and down-regulated genes between transgenic lines 11PP16/11PP17 and control plants (empty vector). (B) Validation of microarray results by RT–qPCR. Fold changes (logFC) of gene expression in control and RNAi-PpMYB8 lines (mean of 11PP16 and 11PP17) analyzed using PINARRAY3 (black bars) and qPCR (white bars) are shown. (C) Enrichment analysis of functional categories (MapMan/Mercator web tool). The horizontal bars represent the number of genes included in each functional category.
Among the up-regulated genes, we detected several genes related to protein post-translational modification, protein degradation, development, and cell organization (Supplementary Table S4). Additionally, the set of up-regulated genes included further TFs putatively involved in development and differentiation processes such as an MYB (sp_v3.0_unigene133569) orthologous to the Arabidopsis AtMYB35, DEFECTIVE IN MERISTEM DEVELOPMENT AND FUNCTION 1, a TCP putatively orthologous to the Arabidopsis PLASTID TRANSCRIPTION FACTOR 1 (PTF1) involved in chloroplast regulation of leaf differentiation (sp_v3.0_unigene6586), and a HIGH MOBILITY GROUP (HMG) box protein (sp_v3.0_unigene24902). Interestingly, we also detected up-regulation of tryptophan synthase (sp_v3.0_unigene133330) and enoyl-CoA hydratase/isomerase D (sp_v3.0_unigene22584), key enzymes involved, respectively, in the biosynthesis of Trp and vitamin K1, through two alternative pathways that, as with the Phe/Tyr pathway, start from chorismate. Also related to amino acids, two additional up-regulated genes were detected, an ACT domain repeat protein (sp_v3.0_unigene8593), involved in feedback regulation of amino acid metabolism, and ornithine delta-aminotransferase (sp_v3.0_unigene5775), involved in arginine catabolism. The microarray results were validated by RT–qPCR analysis of a set of DEGs (Fig. 6B). A functional enrichment analysis, using the Mapman categories through the Mercator web tool, was performed, revealing a set of categories with significant differences between RNAi-PpMYB8 plants and controls, including, among others, Cell wall, Development, Signaling, or Cell organization (Fig. 6C). We also analyzed the transcriptome of stems from OE-PpMYB8 plants to characterize the putative impact of PpMYB8 overexpression. In contrast to what was observed in RNAi-PpMYB8 plants, little impact was observed as a consequence of PpMYB8 overexpression (Supplementary Table S5).
PpHY5 interacts with the regulatory regions of PpADT-A and PpADT-D
Microarray analysis of PpMYB8-silenced plants showed that in the 11PP16 line, PpHY5 (sp_v3.0_unigene6404), a pine ortholog of Arabidopsis HY5 (ELONGATED HYPOCOTYL 5), was significantly up-regulated (adjusted P-value=0.04) while, in the 11PP17 line, the adjusted P-value was 0.09. HY5 is a central TF that promotes, among other processes, the accumulation of Phe-derived compounds and the repression of critical genes for cell wall biogenesis. Moreover, there is evidence demonstrating the coordination of HY5 and MYB TFs in different processes related to the synthesis of Phe-derived compounds (Shin et al., 2013; Nguyen et al., 2015; Czemmel et al., 2017); on the other hand, our previous observation revealed the existence of multiple potential binding sites for HY5 (ACGT elements) in the 5'-flanking regions of most PpADTs (Supplementary Fig. S3). Thus, we decided to investigate whether this gene might be associated with lignin accumulation in pine by analyzing its expression in CW versus OW. In this regard, RT–qPCR analysis demonstrated that, unlike what was observed for PpMYB8, the expression of PpHY5 in CW is much lower than it is in OW (Fig. 7A).
The upstream regions of PpADT-A and PpADT-D are particularly enriched in ACGT elements, so we performed EMSAs using recombinant PpHY5 protein to test a putative interaction. Our results revealed that recombinant PpHY5 can bind to PpADT-A and PpADT-D gene promoters in a manner similar to PpMYB8 (Fig. 7B). In addition, a β-galactosidase assay in yeast confirmed that ACGT boxes located in ADT-A at positions 347 and 464 contribute to PpHY5 recognition in an accumulative manner (Fig. 7C).
Discussion
The biosynthesis of lignin in trees requires a massive supply of its essential precursor Phe. Therefore, the biosynthesis of this amino acid requires fine regulation presumably at the transcriptional and post-transcriptional levels. In the present work, we have investigated the transcriptional regulation of ADT enzymes related to lignification in P. pinaster. The use of a conifer tree to perform these studies is particularly relevant, due to the quantitative importance of lignin biosynthesis in these plants. The role of ADTs could be especially relevant, as these enzymes have been proposed to be a rate-limiting step of Phe biosynthesis (Maeda and Dudareva, 2012). Moreover, the existence of a single PAT gene in most plants and multiple ADT isogenes strongly suggests that different ADT isoforms could be involved in the biosynthesis of Phe for different and specific metabolic fates. Consistently, Corea et al. (2012) used different combinations of ADT knockouts in Arabidopsis thaliana and found that the six Arabidopsis ADT isoforms differ profoundly in their contribution to lignin accumulation, with AtADT5 being the most important contributor.
The comparative study of CW and OW in conifers provides an excellent model for dissecting the regulatory cues that coordinate metabolic pathways providing the substrates for lignification and xylogenesis. CW is characterized by the deposition of a thicker secondary cell wall, with more lignin content, which is structurally enriched in p-hydroxyphenylpropane units (H-lignin) and reduced levels of cellulose (Groover, 2016). Previous studies have also shown that the development of CW is correlated with the up-regulation of genes coding for enzymes involved in the construction of the secondary cell wall (Villalobos et al., 2012). To identify PpADT genes putatively involved in the biosynthesis of Phe for the later biosynthesis of lignin, we compared the expression of these genes in CW and OW. Our results showed an increased expression in CW of six out of the nine ADT genes in maritime pine (Fig. 2A). However, PpADT-A and, to a lesser extent, PpADT-G and PpADT-D, were the most highly expressed genes. At the same time, a set of genes directly linked to Phe metabolism, those encoding the chorismate mutase isoforms 1 and 2 (PpCM1 and PpCM2), prephenate aminotransferase (PpPAT), phenylalanine hydroxylase (PpPH), and phenylalanine ammonia lyase (PpPAL1), were up-regulated in CW compared with OW (Fig. 2B).
Previous studies have supported a role for MYB8 as a key transcriptional regulator of phenylpropanoid metabolism and secondary cell wall biogenesis in conifers (Bomal et al., 2008; Craven-Bartle et al., 2013). Accordingly, the heterologous overexpression of PtMYB8 from P. taeda in young plantlets of P. glauca resulted in the up-regulation of a set of genes involved in the biosynthesis of monolignols and the parallel down-regulation of other genes putatively involved in the same pathway (Bomal et al., 2008). These authors further reported the reduced root growth phenotype of in vitro transgenic plantlets that did not survive to subsequent transfer to soil. However, in the present work, it has been found that the homologous overexpression of PpMYB8 in maritime pine did not affect the phenotype of the 16-month-old transgenic plants, and it consistently exhibited only small changes in the transcriptome (Fig. 3B; Supplementary Table S5). These results indicate that an increase in the endogenous levels of this TF does not have a relevant effect on the regulation of its target genes, including ADT genes. In addition, we gained little evidence in maritime pine for adverse effects of MYB8 overexpression on somatic embryo production and conversion into rooted somatic seedlings or growth of trangenic plants after 12 and 24 months (Fig. 3B). In contrast, P. pinaster plantlets silenced for PpMYB8 exhibited concomitant down-regulation of three ADTs: PpADT-A, PpADT-D, and PpADT-I (Fig. 4A). Interestingly, these isoforms were transcriptionally activated in CW compared with OW, an effect that was particularly strong for PpADT-A and PpADT-D (Fig. 2A). Additional genes linked to Phe metabolism were also down-regulated, with the exception of PpPH, whose expression was enhanced in the silenced plants (Fig. 4B). This finding suggests that Phe hydroxylation is not associated with the metabolic activity of the ADT-A and ADT-D isoforms.
The RNAi-PpMYB8 lines exhibited a significant decrease in growth, reduced levels of lignin, and an altered H-:G-lignin ratio (Fig. 3A; Supplementary Table S1). Consistently, the silencing of PpMYB8 was observed in different organs, with particularly reduced levels of transcripts in the stems of the transgenic lines (Fig. 3C). Microscopic anatomy of the stems and histochemical detection of lignin support the hypothesis that PpMYB8 down-regulation hampers the proper development of the xylem, significantly reducing lignin deposition and presumably affecting plant growth (Fig. 4C, D). It is still unclear whether a fine-tuning in PpMYB8 down-regulation could result in a reduction in lignin content without severely affecting the overall plant growth rate.
The marked developmental alterations observed in RNAi-PpMYB8 plants strongly point to a significant underlying modification of the plant transcriptome. In this regard, we investigated whether the observed down-regulation of certain PpADT genes in these plants occurs through the direct activity of PpMYB8 on the corresponding promoters, or whether the reduction observed in their expression was a consequence of a pleiotropic effect. In this regard, in silico analysis of the 5'-upstream regions of the pine ADT genes revealed the occurrence of putative R2R3-MYB-binding sites in all of them, with the exception of those corresponding to PpADT-B and PpADT-H (Supplementary Fig. S2). This result is consistent with the broadly characterized regulation of phenylpropanoid biosynthesis in plants through MYB TFs (Zhong and Ye, 2007; Nakano et al., 2015; Ye and Zhong, 2015; Lamara et al., 2016). The particular enrichment of R2R3-MYB-binding sites in the 5'-flanking regions of PpADT-A and PpADT-D mimics the previously described promoter architecture and PpMYB8 regulation of PpPAT and PpPAL1 genes coding, respectively, for enzymes catalyzing the preceding and subsequent reactions of ADT (Craven-Bartle et al., 2013). These observations strongly suggest that these ADT isoforms are involved in the lignin-associated Phe biosynthetic pathway in maritime pine. To determine whether the putative regulatory regions of PpADT-A and PpADT-D physically interact with PpMYB8, a complementary strategy using EMSAs and gene reporter assays in yeast was carried out. Both approximations have provided concurrent evidence about the direct regulation of PpADT-A and PpADT-D expression by PpMYB8 through the recognition of a region enriched in AC elements (Fig. 5A, B).
As we have shown, silencing of PpMYB8 resulted in a large alteration of plant growth and development. These findings suggest that the RNAi silencing of PpMYB8 could be triggering a complex combination of direct and indirect alterations of the stem transcriptome. To further identify other genes putatively co-regulated with PpADT-A and PpADT-D and also with PpPAT and PpCM2, in the lignin-associated Phe biosynthesis pathway, the transcriptomes of the RNAi-PpMYB8 plantlets were analyzed. Consistent with our hypothesis, we identified a large number (784) of DEGs compared with the controls that were shared by both the 11PP16 and 11PP17 RNAi-PpMYB8 lines, of which 307 genes were up-regulated and 477 were down-regulated (Fig. 6). Markedly, we detected that the silencing of PpMYB8 results in the parallel down-regulation and up-regulation of a complex network of TFs belonging to different families: R2R3-MYBs, bZIP, RAV/NGATHA, MADS box, C3H zinc finger, GARP, HLH TCP, or HMG-Box, suggesting that a significant proportion of the DEGs are regulated in this process through TFs other than PpMYB8. Interestingly, the down-regulated GARP TF belongs to a family of TFs related to the MYB superfamily that have been suggested to be central coordinators of plant nutrition and development (Safi et al., 2017), so its putative involvement in the channeling of nitrogen resources towards the biosynthesis of Phe and later synthesis of lignin is of particular interest.
Interestingly, among the down-regulated DEGs, we have detected several genes coding for enzymatic activities directly related to cell wall modification including xyloglucan endotransglucosylase/hydrolase, pectinesterase, chitinase, wall polysaccharide O-acetyltransferase, and oxidoreductases, and up to three S-adenosyl-l-methionine-dependent methyltransferases putatively involved in monolignol biosynthesis. Down-regulation of these activities could be associated with an altered biogenesis of the secondary cell wall in stem cells that, in turn, results in disorganization of the xylematic tissues (Fig. 4C). Consistently, we also found down-regulation of multiple genes directly associated with the architecture of the cytoskeleton, which could be directly related to the same process of xylem disorganization. In this context, it should be mentioned that there was down-regulation of a gene orthologous to the Arabidopsis NO VEIN, encoding a plant-specific nuclear factor required for leaf vascular development (Tsugeki et al., 2009), and we hypothesize that its deregulation may be directly involved in the altered phenotype of vascular development in pine stems.
Among the up-regulated genes, we have detected two genes that encode key enzymes participating in the biosynthetic pathways of Trp and vitamin K1: tryptophan synthase and enoyl-CoA hydratase/isomerase D. Both pathways use chorismate as a precursor and therefore compete with the Phe/Tyr pathway for its use. We have shown that the silencing of PpMYB8 involves down-regulation of important genes involved in the Phe pathway so presumably, as a result, the availability of chorismate increases and could be channeled towards the synthesis of Trp and vitamin K1. Additionally, Trp is a precursor for the biosynthesis of auxin, a hormone related to cell elongation, and in this regard we have identified up to seven auxin-related genes putatively involved in functions such as hormone transport, vascular pattern, gravitropism, and dormancy (Supplementary Table S4). These results suggest that a set of auxin-related genes are transcriptionally activated in response to molecular cues associated with cell elongation defects triggered by the silencing of PpMYB8.
The observed up-regulation of an ACT domain repeat (ACR) protein is particularly interesting since these proteins are involved in feedback regulation of amino acid metabolism and function in modulating the activity of amino acid biosynthetic enzymes (Singh et al., 2018). For example, in Arabidopsis, AtACR11 is coordinately expressed with the gene encoding chloroplastic glutamine synthetase (GS2), and the AtACR11 protein activates GS2 activity (Singh et al., 2018). In this regard, sp_v3.0_unigene8593 encodes the ortholog of Arabidopsis AtACR4, a gene strongly co-expressed with that coding for tryptophan aminotransferase (http://atted.jp/), which is essential for auxin biosynthesis, and thus this supports the possible involvement of auxin in the RNAi-PpMYB8-mediated phenotype.
As previously mentioned, the biosynthesis of Phe in plants is associated with multiple processes of enormous physiological importance that therefore requires fine and complex regulation at different transcriptional, post-translational, and metabolic levels. Thus, we have demonstrated here that during xylogenesis, PpMYB8 plays a fundamental role in the transcriptional regulation of several important genes involved in this process. However, the participation of these enzymes in the synthesis of Phe associated with other metabolic processes suggests that the corresponding genes may have alternative mechanisms of transcriptional regulation through the action of other TFs or combinations of TFs. In this work, we provide experimental data suggesting that the expression levels of PpADT-A and PpADT-D are also regulated by PpHY5. We have shown that this TF is able to bind the regulatory regions of PpADT-A and PpADT-D, and also that the expression profile of this gene is opposite to that of PpMYB8 in CW, OW, and in RNAi-PpMYB8 plants, suggesting an antagonistic role. Functional studies in transgenic plants are required to address whether PpHY5 could be repressing the expression of PpADT-A and PpADT-D in competition with activator TFs such as PpMYB8.
In summary, our results support the hypothesis that distinctive members of the ADT gene family are transcriptionally regulated by PpMYB8 for the provision of precursors for the synthesis of lignin that is necessary for the proper development of xylematic tissues in maritime pine as previously proposed (Craven-Bartle et al., 2013). Recently, we have shown that PpNAC1 is a central regulator governing secondary cell wall formation, and thus Phe biosynthesis and utilization, in P. pinaster (Pascual et al., 2018). In that work, we also demonstrated that PpNAC1 partially mediates its activity through direct activation of PpMYB8. Future research will allow for deepening our knowledge of the complementary or alternative regulatory mechanisms that govern Phe biosynthesis dependently and independently of PpMYB8 and PpNAC1 in this conifer species.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Estimated H- and G-lignin content in RNAi-PpMYB8 silenced lines compared with the control (RNAi-EV).
Table S2. Primer sequences used for cloning.
Table S3. Primer pairs used for RT-qPCR.
Table S4. Transcriptome analysis of transgenic plants silenced for PpMYB8 (RNAi- PpMYB8).
Table S5. Transcriptome analysis of transgenic plants overexpressing PpMYB8 (OE-PpMYB8).
Fig. S1. Average expression level of the nine ADT genes from P. pinaster in the OE-PpMYB8 lines, expressed as a percentage of controls
Fig. S2. In silico analysis of candidate AC- and bZIP-binding elements within the 5'-flanking region of the ADT genes in Pinus pinaster.
Fig. S3. Probes designed for the EMSA in the PADT-A and PADT-D sequences.
Protocols S1.
Acknowledgements
CW and OW samples were kindly provided by Dr Francisco J. Ruiz Cantón (Universidad de Málaga). We are indebted to Dr Javier Canales Carrasco for help with microarray analysis, Dr Philippe Label (INRA, UMR PIAF, France) for helpful support in selecting MYB8 target sequences for RNAi, and Ana Alvarez for collaboration in the PpHY5 work. We would also like to thank Dr Alberto de Marcos Serrano (Universidad de Castilla-La Mancha) for the pMW#3 vector and Dr Christophe Plomion (INRA, UMR BIOGECO) for access to a P. taeda MYB8 EST clone. This work was supported by grants from the European Commission Seventh Framework PROCOGEN (FP7-KBBE-2011-5), Minsterio de Ciencia e Innovación (MICINN) (BIO2015-69285-R and RTI2018-094041-B-I00), and Junta Andalucía (BIO2012-0474). The RNAi-PpMYB8 vector was obtained during the GENOQB project funded by ANR, the French National Research Agency (ANR GNP05013C). Genetic transformation of maritime pine with OE-PpMYB8 and RNAi-PpMYB8 vectors was funded by the SUSTAINPINE project (EU, Plant KBBE multinational programme, PLE2009-0016) through a specific grant in France from ANR (ANR-09-KBBE-007-001). OE-PpMYB8 and RNAi-PpMYB8 plant regeneration and study at FCBA benefited from the support of the XYLOBIOTECH technical facility (ANR-10-EQPX-16 XYLOFOREST).
References
- Boggy GJ, Woolf PJ. 2010. A mechanistic model of PCR for accurate quantification of quantitative PCR data. PLoS One 5, e12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, et al. 2008. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. Journal of Experimental Botany 59, 3925–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Duval I, Giguère I, Fortin É, Caron S, Stewart D, Boyle B, Séguin A, MacKay JJ. 2014. Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. Journal of Experimental Botany 65, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C, Jensen R. 1987. Prephenate aminotransferase. Methods in Enzymology 142, 479–487. [DOI] [PubMed] [Google Scholar]
- Breton D, Harvengt L, Trontin J-F, Bouvet, Favre J-M. 2006. Long-term subculture randomly affects morphology and subsequent maturation of early somatic embryos in maritime pine. Plant Cell, Tissue and Organ Culture 87, 95–108. [Google Scholar]
- Cañas RA, Feito I, Fuente-Maqueda JF, Ávila C, Majada J, Cánovas FM. 2015. Transcriptome-wide analysis supports environmental adaptations of two Pinus pinaster populations from contrasting habitats. BMC Genomics 16, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corea OR, Ki C, Cardenas CL, Kim SJ, Brewer SE, Patten AM, Davin LB, Lewis NG. 2012. Arogenate dehydratase isoenzymes profoundly and differentially modulate carbon flux into lignins. Journal of Biological Chemistry 287, 11446–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton RG, Gibson F. 1965. The biosynthesis of phenylalanine and tyrosine; enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochimica et Biophysica Acta 100, 76–88. [DOI] [PubMed] [Google Scholar]
- Craven-Bartle B, Pascual MB, Cánovas FM, Avila C. 2013. A Myb transcription factor regulates genes of the phenylalanine pathway in maritime pine. The Plant Journal 74, 755–766. [DOI] [PubMed] [Google Scholar]
- Czemmel S, Höll J, Loyola R, Arce-Johnson P, Alcalde JA, Matus JT, Bogs J. 2017. Transcriptome-wide identification of novel UV-B- and light modulated flavonol pathway genes controlled by VviMYBF1. Frontiers in Plant Science 8, 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F, El-Azaz J, Avila C, Cánovas FM. 2014. Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiology 164, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dence CW. 1992. The determination of lignin. In: Lin SY, Dence CW, eds. Methods in lignin chemistry. Berlin: Springer-Verlag, 33–61. [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, et al. 2006. A gene-centered C. elegans protein–DNA interaction network. Cell 125, 1193–1205. [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Alvania RS, Taparowsky EJ. 2006. Cross-species annotation of basic leucine zipper factor interactions: insight into the evolution of closed interaction networks. Molecular Biology and Evolution 23, 1480–1492. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Azaz J, de la Torre F, Ávila C, Cánovas FM. 2016. Identification of a small protein domain present in all plant lineages that confers high prephenate dehydratase activity. The Plant Journal 87, 215–229. [DOI] [PubMed] [Google Scholar]
- Fischer R, Jensen R. 1987. Prephenate dehydratase (monofunctional). Methods in Enzymology 142, 507–512. [DOI] [PubMed] [Google Scholar]
- Gómez-Maldonado J, Cánovas FM, Avila C. 2004. Molecular analysis of the 5'-upstream region of a gibberellin-inducible cytosolic glutamine synthetase gene (GS1b) expressed in pine vascular tissue. Planta 218, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Groover A. 2016. Gravitropisms and reaction woods of forest trees—evolution, functions and mechanisms. New Phytologist 211, 790–802. [DOI] [PubMed] [Google Scholar]
- Harvengt L. 2005. Somatic embryogenesis in maritime pine (Pinus pinaster Ait.). In: Jain SM, Gupta PK, eds. Protocols of somatic embryogenesis in woody plants. Berlin: Springer Verlag, 107–120. [Google Scholar]
- Haslam E. 1993. Shikimic acid. Metabolism and metabolites. Chichester: John Wiley and Sons. [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Lamara M, Raherison E, Lenz P, Beaulieu J, Bousquet J, MacKay J. 2016. Genetic architecture of wood properties based on association analysis and co-expression networks in white spruce. New Phytologist 210, 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu HX, Southerton SG. 2010. Comparative genomics reveals conservative evolution of the xylem transcriptome in vascular plants. BMC Evolutionary Biology 10, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B. 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell & Environment 37, 1250–1258. [DOI] [PubMed] [Google Scholar]
- Lowry B, Lee D, Hébant C. 1980. The origin of land plants: a new look at an old problem. Taxon 29, 183–197. [Google Scholar]
- Maeda H, Dudareva N. 2012. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology 63, 73–105. [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N. 2010. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. The Plant Cell 22, 832–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N. 2011. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nature Chemical Biology 7, 19–21. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. 1997. MYB transcription factors in plants. Trends in Genetics 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Morel A, Trontin JF, Corbineau F, et al. 2014. Cotyledonary somatic embryos of Pinus pinaster Ait. most closely resemble fresh, maturing cotyledonary zygotic embryos: biological, carbohydrate and proteomic analyses. Planta 240, 1075–1095. [DOI] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD. 2016. Designer lignins: harnessing the plasticity of lignification. Current Opinion in Biotechnology 37, 190–200. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M. 2015. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Frontiers in Plant Science 6, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong SW, Lee H. 2015. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. The Plant Journal 84, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Pascual MB, El-Azaz J, de la Torre FN, Cañas RA, Avila C, Cánovas FM. 2016. Biosynthesis and metabolic fate of phenylalanine in conifers. Frontiers in Plant Science 7, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual MB, Llebrés MT, Craven-Bartle B, Cañas RA, Cánovas FM, Ávila C. 2018. PpNAC1, a main regulator of phenylalanine biosynthesis and utilization in maritime pine. Plant Biotechnology Journal 16, 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, et al. 2003. a Characterisation of a pine MYB that regulates lignification. The Plant Journal 36, 743–754. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM. 2003b Characterisation of Pt MYB1, an R2R3-MYB from pine xylem. Plant Molecular Biology 53, 597–608. [DOI] [PubMed] [Google Scholar]
- Ritz C, Spiess AN. 2008. qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 24, 1549–1551. [DOI] [PubMed] [Google Scholar]
- Rueda-López M, Crespillo R, Cánovas FM, Avila C. 2008. Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. The Plant Journal 56, 73–85. [DOI] [PubMed] [Google Scholar]
- Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B, Krouk G. 2017. The world according to GARP transcription factors. Current Opinion in Plant Biology 39, 159–167. [DOI] [PubMed] [Google Scholar]
- Saleme MLS, Cesarino I, Vargas L, et al. 2017. Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar. Plant Physiology 175, 1040–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI. 2013. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters 587, 1543–1547. [DOI] [PubMed] [Google Scholar]
- Singh SK, Sung TY, Chung TY, Lin SY, Lin SC, Liao JC, Hsieh WY, Hsieh MH. 2018. ACR11 modulates levels of reactive oxygen species and salicylic acid-associated defense response in Arabidopsis. Scientific Reports 8, 11851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S eds. Bioinformatics and computational biology solutions using R and Bioconductor. statistics for biology and health. New York: Springer, 397–420. [Google Scholar]
- Soler M, Plasencia A, Larbat R, Pouzet C, Jauneau A, Rivas S, Pesquet E, Lapierre C, Truchet I, Grima-Pettenati J. 2017. The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation. New Phytologist 213, 287–299. [DOI] [PubMed] [Google Scholar]
- Soler M, Plasencia A, Lepikson-Neto J, Camargo EL, Dupas A, Ladouce N, Pesquet E, Mounet F, Larbat R, Grima-Pettenati J. 2016. The woody-preferential gene EgMYB88 regulates the biosynthesis of phenylpropanoid-derived compounds in wood. Frontiers in Plant Science 7, 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin J-F, Debille S, Canlet F, et al. 2013. Somatic embryogenesis as an effective regeneration support for reverse genetics in maritime pine: the Sustainpine collaborative project as a case study. In: Park Y-S, Bonga JM, eds. Proceeding of the IUFRO Working Party 2.09.02 conference on Integrating vegetative propagation, biotechnology and genetic improvement for tree production and sustainable forest management, 25–28/06/2012. Brno, Czech Republic, 184–187.#
- Trontin JF, Harvengt L, Garin E, Vernaza ML, Arancio L, Hoebeke J, Canlet F, Paques M. 2002. Towards genetic engineering of maritime pine (Pinus pinaster Ait.). Annals of Forest Science 59, 687–697. [Google Scholar]
- Trontin J-F, Teyssier C, Morel A, Harvengt L, Lelu-Walter M-A. 2016. Prospects for new variety deployment through somatic embryogenesis in maritime pine. In: Park Y-S, Bonga JM, Moon H-K, eds. Vegetative propagation of forest trees. Seoul, Korea: Korea Forest Research Institute, 572–606. [Google Scholar]
- Trontin J-F, Walter C, Klimaszewska K, Park YS, Lelu-Walter MA. 2007. Recent progress on genetic transformation of four Pinus spp. Transgenic Plant Journal 1, 314–329. [Google Scholar]
- Tsugeki R, Ditengou FA, Sumi Y, Teale W, Palme K, Okada K. 2009. NO VEIN mediates auxin-dependent specification and patterning in the Arabidopsis embryo, shoot, and root. The Plant Cell 21, 3133–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Galili G. 2010. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Molecular Plant 3, 956–972. [DOI] [PubMed] [Google Scholar]
- Villalobos DP, Díaz-Moreno SM, Said el-SS, Cañas RA, Osuna D, Van Kerckhoven SH, Bautista R, Claros MG, Cánovas FM, Cantón FR. 2012. Reprogramming of gene expression during compression wood formation in pine: coordinated modulation of S-adenosylmethionine, lignin and lignan related genes. BMC Plant Biology 12, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Geddes B, Lu F, Ralph J. 2015. Syringyl lignin production in conifers: proof of concept in a pine tracheary element system. Proceedings of the National Academy of Sciences, USA 112, 6218–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Liechty JD, Stevens KA, et al. 2014. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196, 891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Zhong R. 2015. Molecular control of wood formation in trees. Journal of Experimental Botany 66, 4119–4131. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. 2011. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant & Cell Physiology 52, 1856–1871. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2007. Regulation of cell wall biosynthesis. Current Opinion in Plant Biology 10, 564–572. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2009. Transcriptional regulation of lignin biosynthesis. Plant Signaling & Behavior 4, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.